In Silico Prediction of Secondary Metabolites and Biosynthetic Gene Clusters Analysis of Streptomyces thinghirensis HM3 Isolated from Arid Soil
Abstract
1. Introduction
2. Materials and Methods
2.1. Soil Properties, Cultural Conditions, and Antimicrobial Assay
- pH was measured in the soil aqueous extract (soil:water) at a ratio of (1:2.5) using a pH meter type (Jenway, model 3310), according to the method of Page et al. [36].
- EC (electrical conductivity, dS m−1) was measured in the saturated soil paste extract and estimated using an EC-meter type (ELE, model 470), according to the method recommended in the study of Jackson [37].
- Total N was determined by the Kjeldahl method; P was extracted using the Olsen method; K by 1 N NH4OAc at pH 7, Page et al. [36].
- Organic matter (OM, %) was determined in soil by the modified Walkley–Black method, according to Nelson and Sommers [38].
- Particle size analysis and soil texture were measured by Gee [39].
2.2. Genomic DNA Extraction and Whole Genome Sequencing
2.3. Genome Assembly, Annotation, and Data availability
2.4. Phylogenetic Analysis Based on 16S rRNA Gene and Whole Genome Sequencing
2.5. Metabolites Analysis and Identification of Putative Biosynthetic Gene Clusters (BGCs)
3. Results
3.1. Antimicrobial Activity of S. thinghirensis Strain HM3
3.2. Whole Genome Sequencing and General Genome Features
3.3. Phylogenetic Analysis and Genome Comparison with Other Streptomyces
3.4. Overview of the Putative Secondary Metabolites’ Biosynthetic Gene Clusters
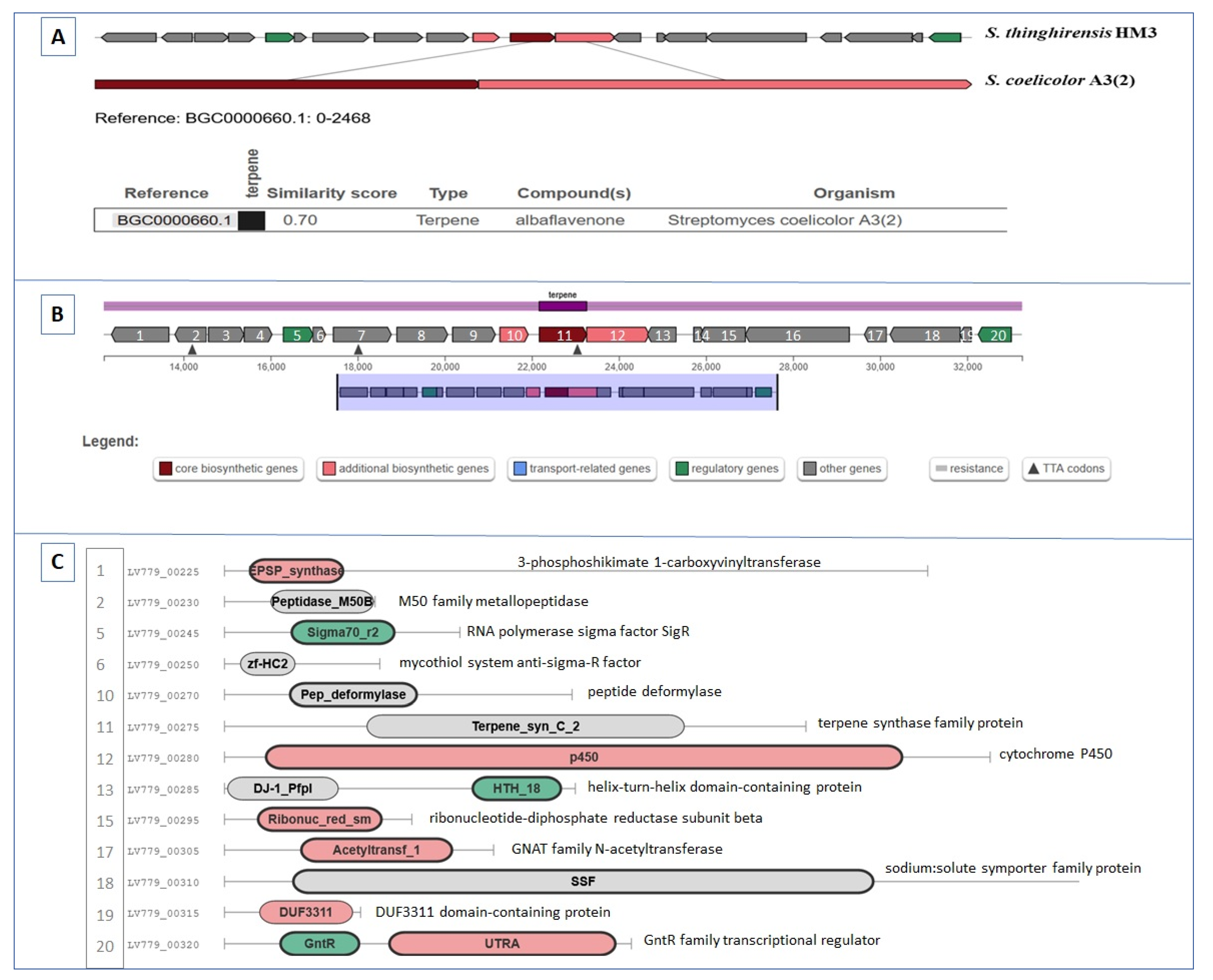
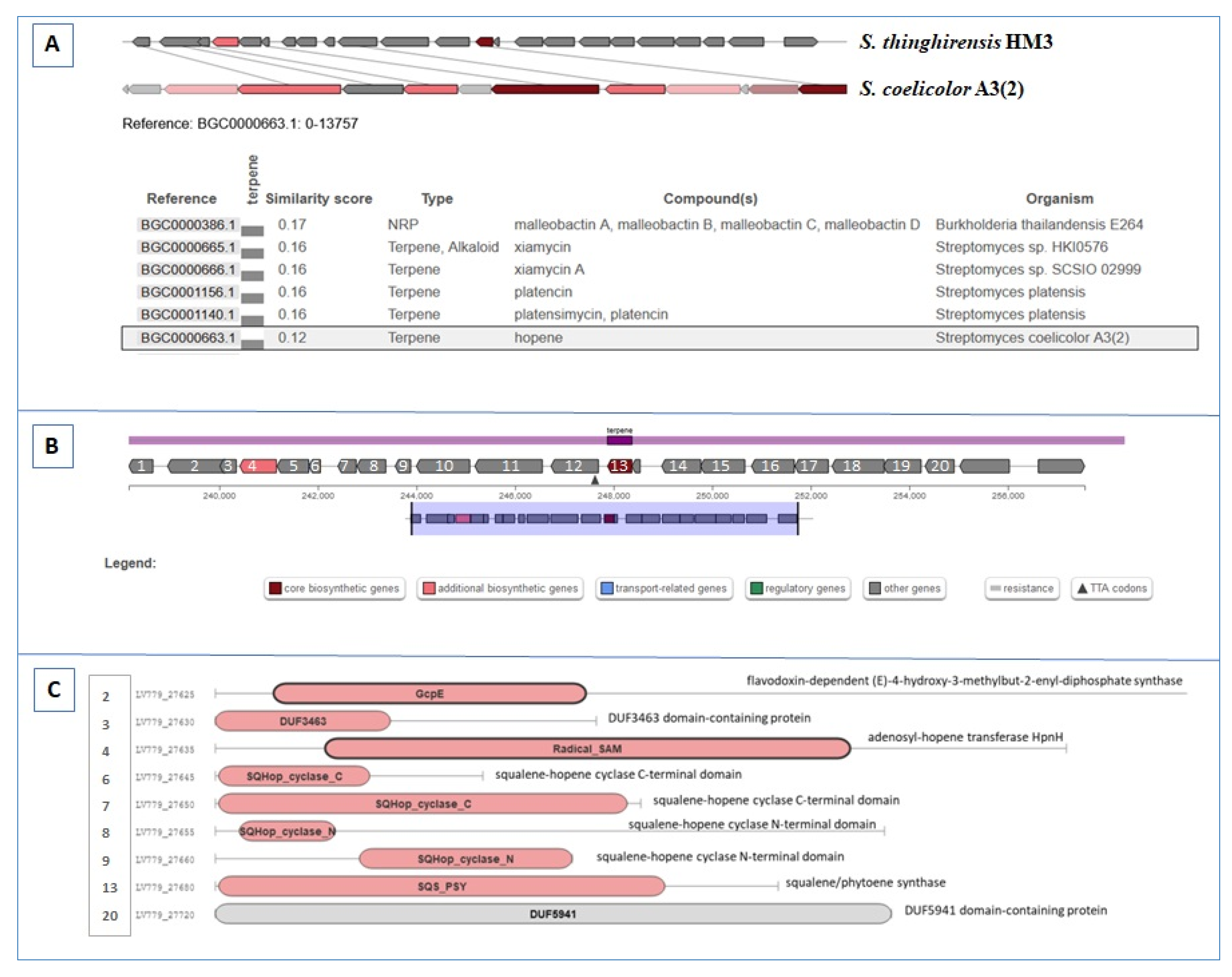
3.5. BGCs Involving Terpene Biosynthesis
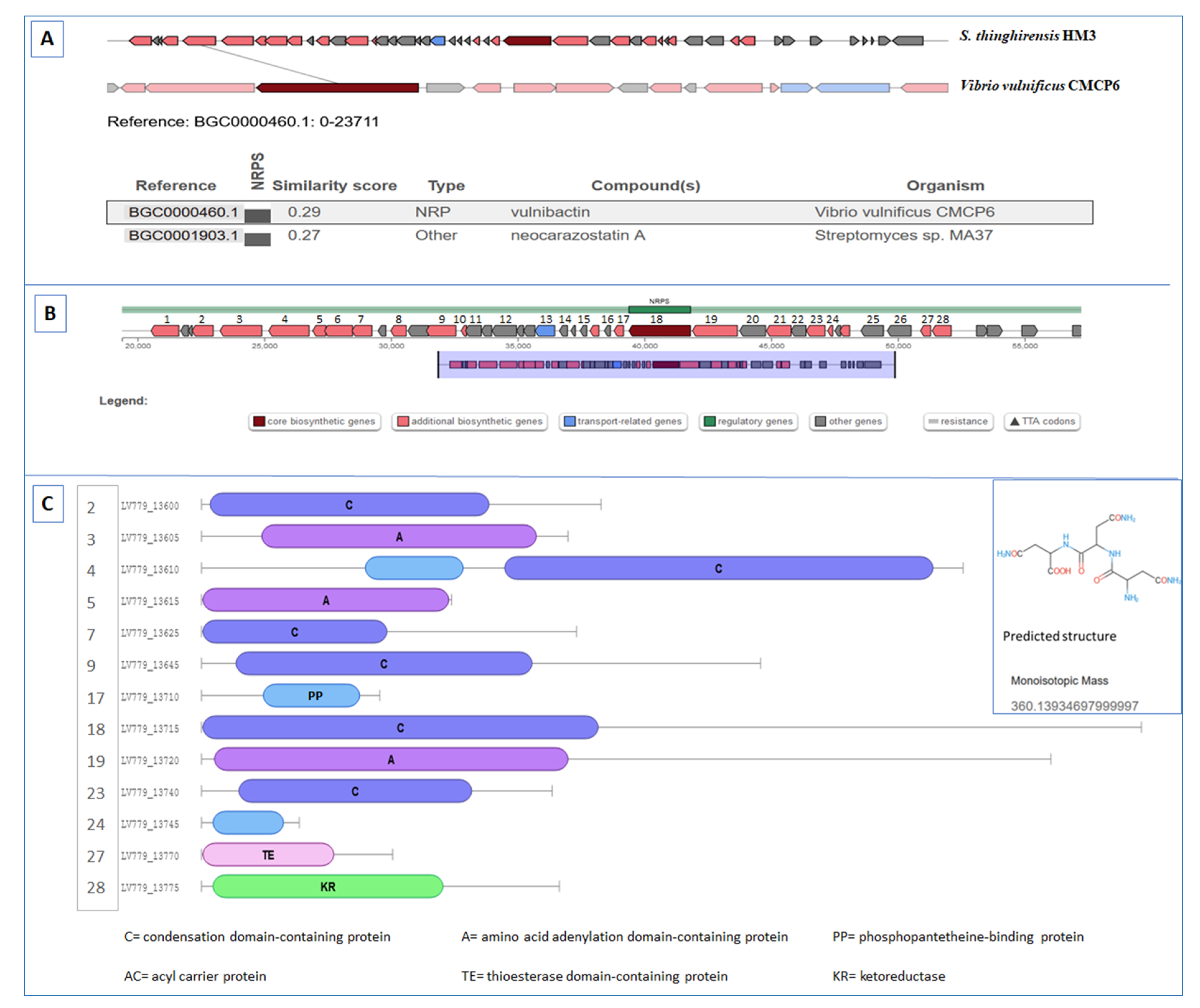
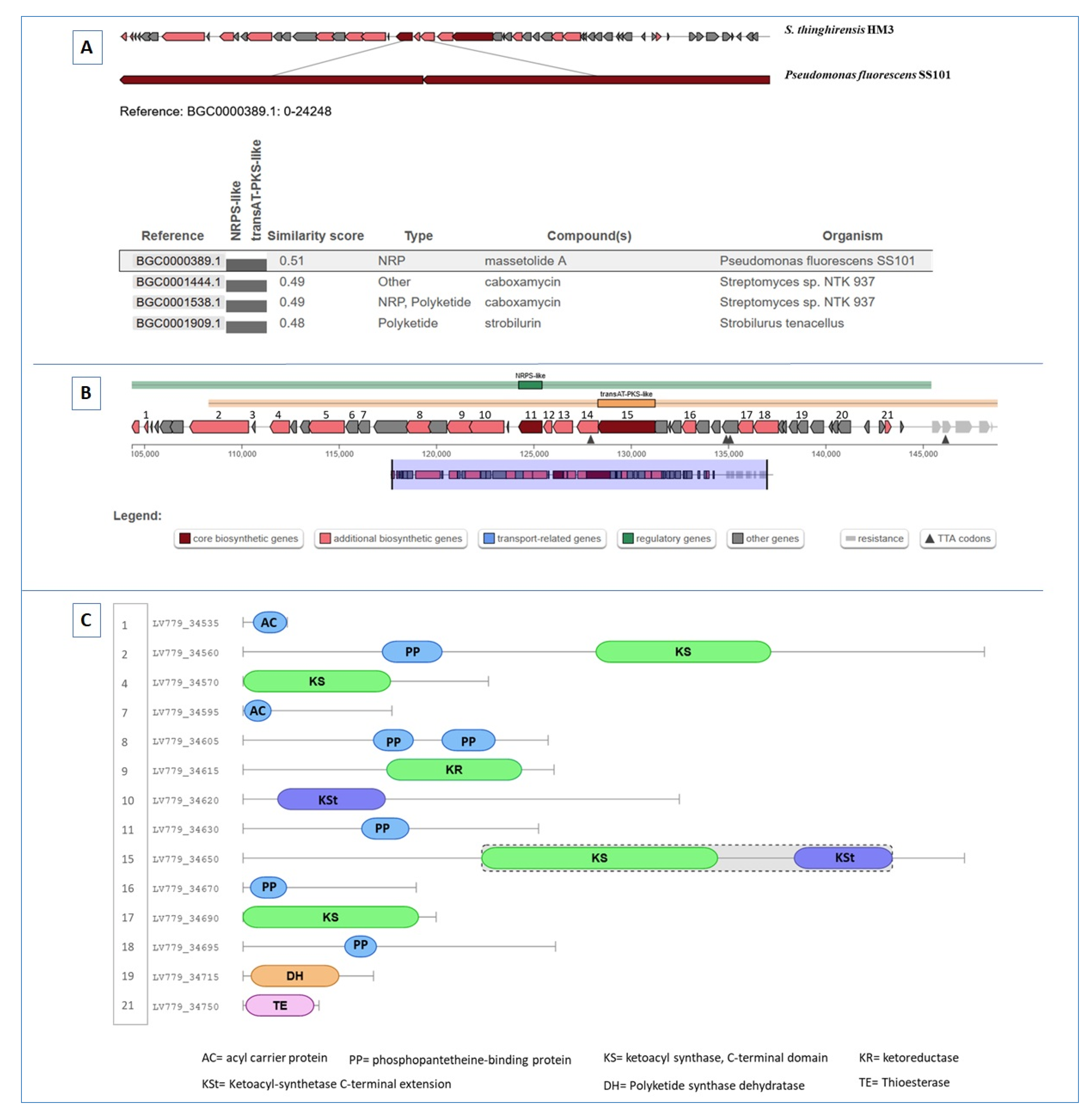
3.6. BGCs Involving NRPS, PKS, and NRPS/PKS Hybrid
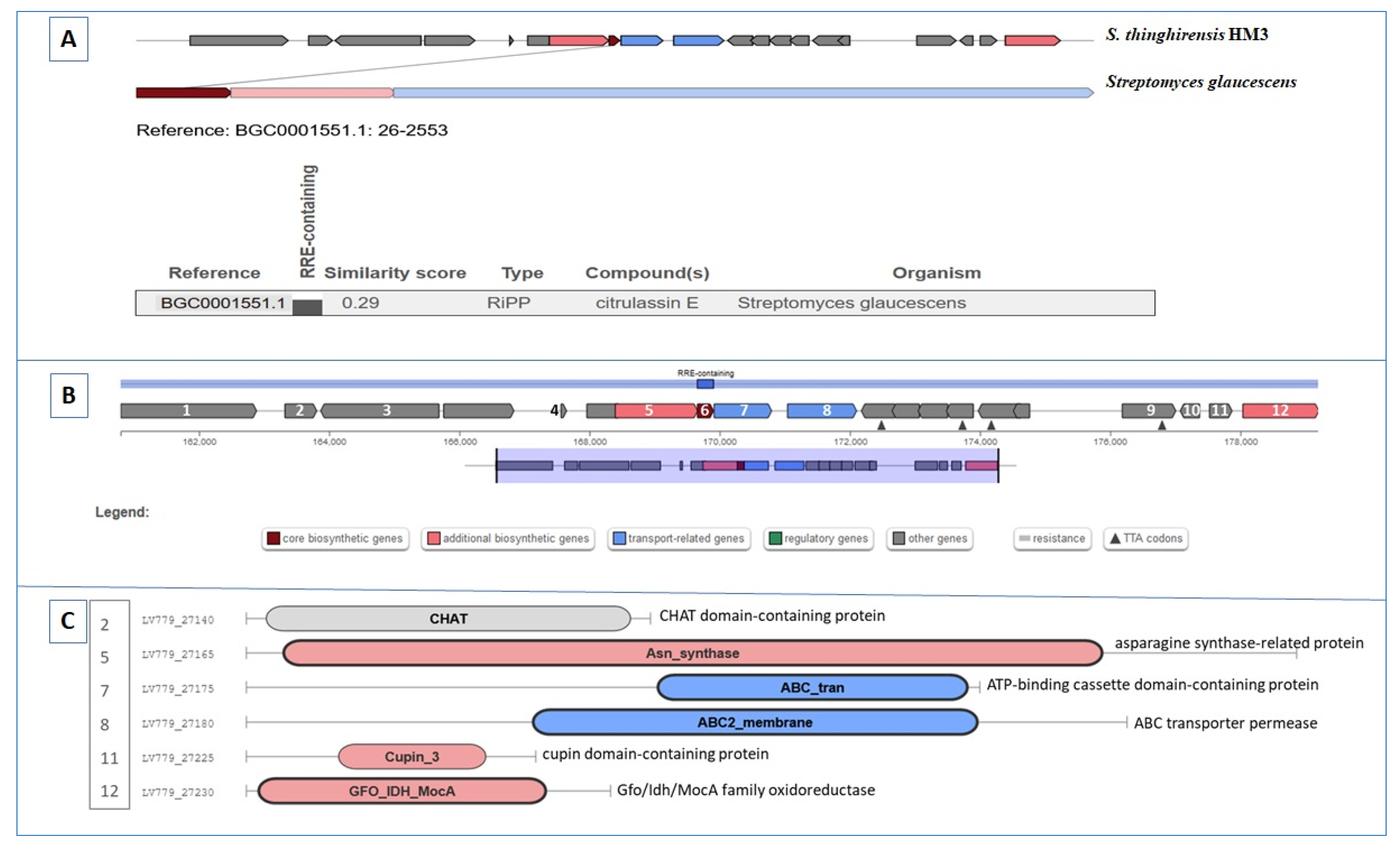
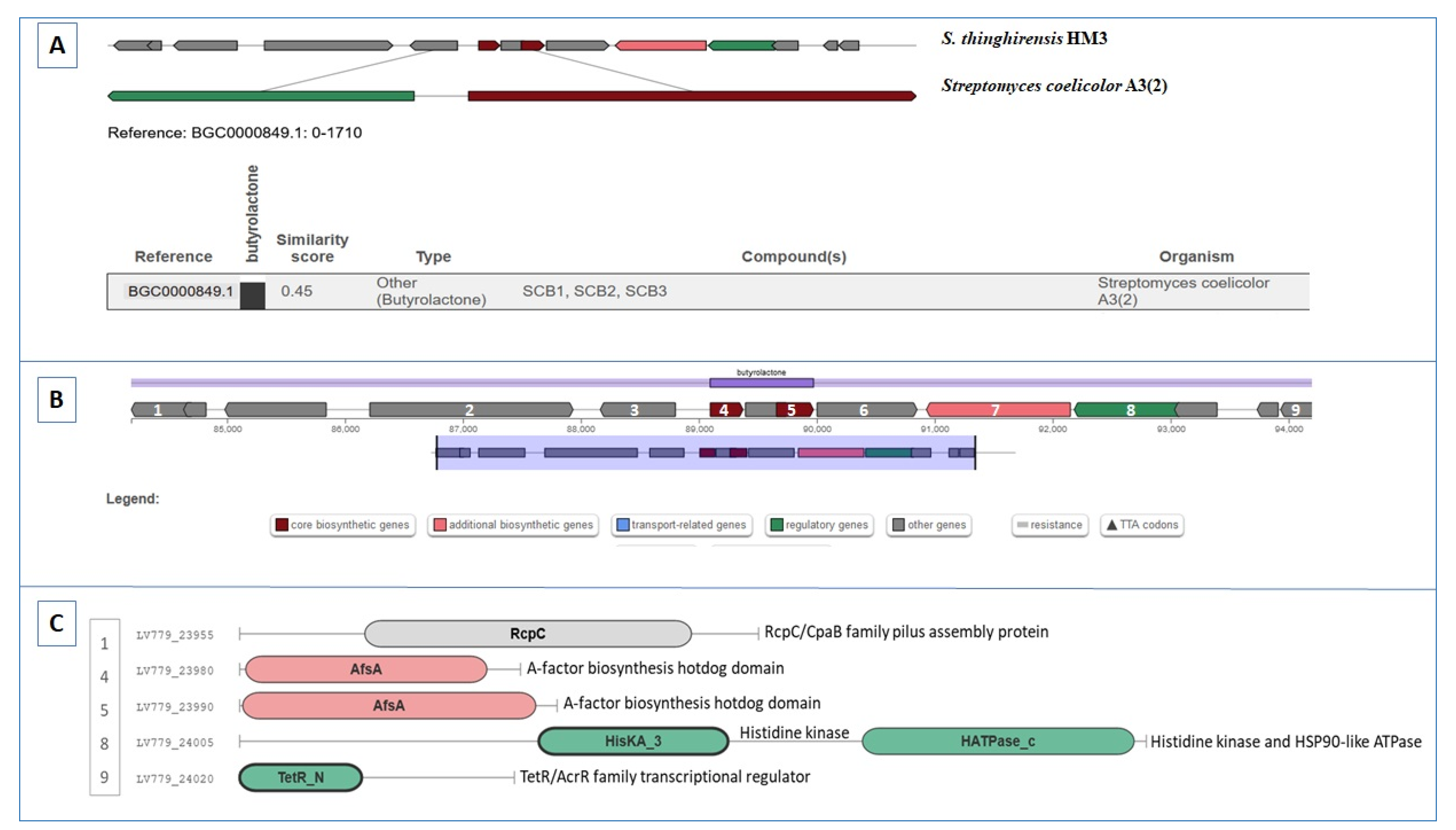
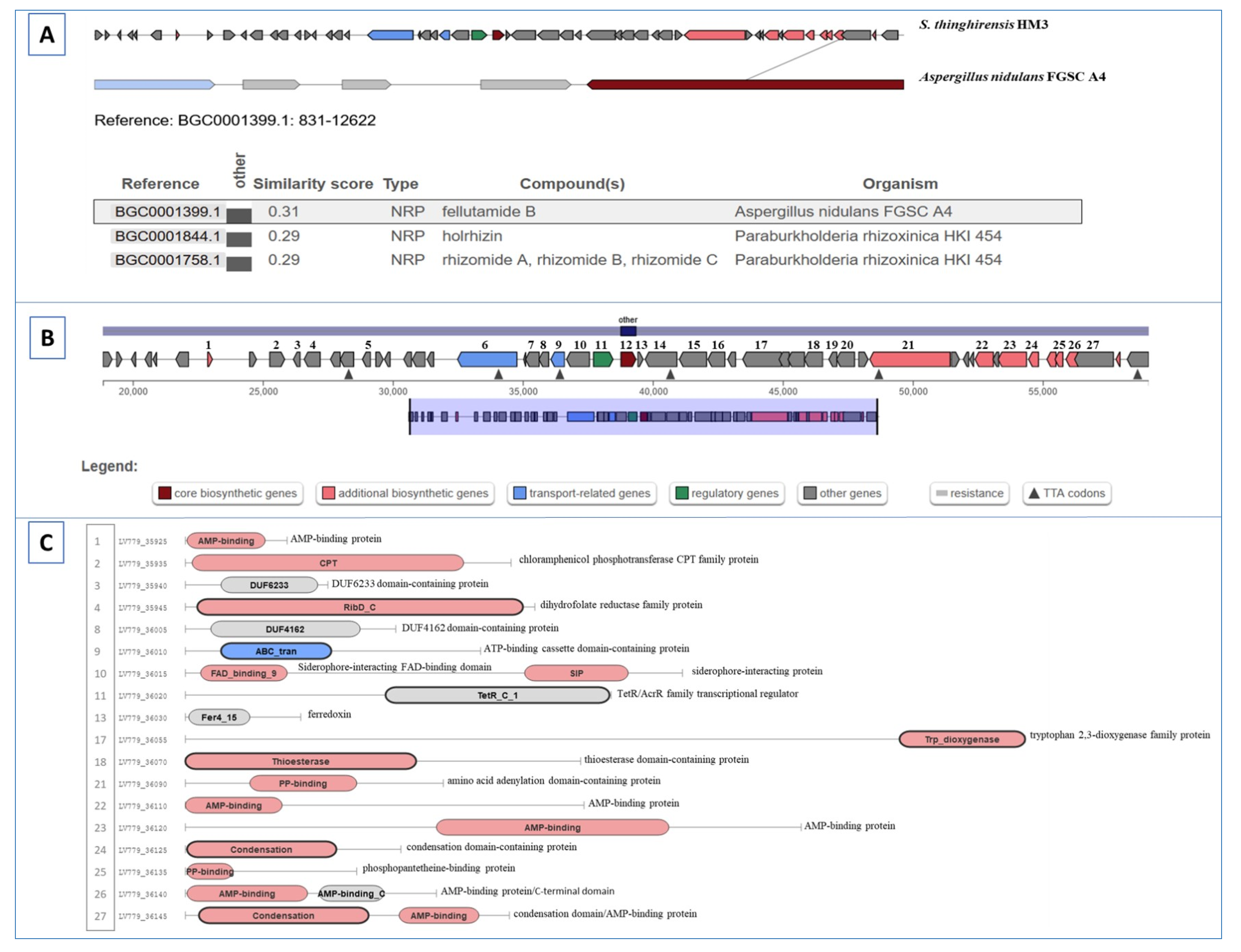
3.7. Gene Clusters Involving RiPP, Butyrolactone, and Other Biosynthesis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Waksman, S.A.; Henrici, H.T. The Nomenclature and Classification of the Actinomycetes. J. Bacteriol. 1943, 46, 337–341. [Google Scholar] [CrossRef] [PubMed]
- Dewi, T.K.; Agustiani, D.; Antonius, S. Secondary Metabolites Production by Actinomycetes and Their Antifungal Activity. KnE Life Sci. 2017, 3, 256–264. [Google Scholar] [CrossRef]
- Lacey, H.J.; Rutledge, P.J. Recently Discovered Secondary Metabolites from Streptomyces Species. Molecules 2022, 27, 887. [Google Scholar] [CrossRef] [PubMed]
- Salwan, R.; Sharma, V. Chapter 15—Bioactive Compounds of Streptomyces: Biosynthesis to Applications. In Studies in Natural Products Chemistry; Atta-Ur-Rahman, M., Ed.; Elsevier: Amsterdam, The Netherlands, 2020; Volume 64, pp. 467–491. ISBN 1572-5995. [Google Scholar]
- Selim, M.S.M.; Abdelhamid, S.A.; Mohamed, S.S. Secondary Metabolites and Biodiversity of Actinomycetes. J. Genet. Eng. Biotechnol. 2021, 19, 72. [Google Scholar] [CrossRef]
- Ahsan, T.; Chen, J.; Zhao, X.; Irfan, M.; Wu, Y. Extraction and Identification of Bioactive Compounds (Eicosane and Dibutyl Phthalate) Produced by Streptomyces Strain KX852460 for the Biological Control of Rhizoctonia Solani AG-3 Strain KX852461 to Control Target Spot Disease in Tobacco Leaf. AMB Express 2017, 7, 54. [Google Scholar] [CrossRef] [PubMed]
- Stackebrandt, E.; Rainey, F.A.; Ward-Rainey, N.L. Proposal for a New Hierarchic Classification System, Actinobacteria Classis Nov. Int. J. Syst. Evol. Microbiol. 1997, 47, 479–491. [Google Scholar] [CrossRef]
- Kämpfer, P.; Glaeser, S.P.; Parkes, L.; van Keulen, G.; Dyson, P. The Family Streptomycetaceae BT—The Prokaryotes: Actinobacteria; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 889–1010. ISBN 978-3-642-30138-4. [Google Scholar]
- Seipke, R.F.; Kaltenpoth, M.; Hutchings, M.I. Streptomyces as Symbionts: An Emerging and Widespread Theme? FEMS Microbiol. Rev. 2012, 36, 862–876. [Google Scholar] [CrossRef]
- Bérdy, J. Thoughts and Facts about Antibiotics: Where We Are Now and Where We Are Heading. J. Antibiot. 2012, 65, 385–395. [Google Scholar] [CrossRef]
- Watve, M.G.; Tickoo, R.; Jog, M.M.; Bhole, B.D. How Many Antibiotics Are Produced by the Genus Streptomyces? Arch. Microbiol. 2001, 176, 386–390. [Google Scholar] [CrossRef]
- Antoraz, S.; Santamaria, R.; Díaz, M.; Sanz, D.; Rodríguez, H. Towards a New Focus in Antibiotic and Drug Discovery from Streptomyces Arsenal. Front. Microbiol. 2015, 6, 461. [Google Scholar] [CrossRef]
- Takahashi, Y.; Nakashima, T. Actinomycetes, an Inexhaustible Source of Naturally Occurring Antibiotics. Antibiot. 2018, 7, 45. [Google Scholar] [CrossRef]
- Yao, L.; Liu, Q.; Wu, Z.M.; Li, K.T. Discovery and Evaluation on the Antibacterial and Cytotoxic Activities of a Novel Antifungalmycin N2 Produced from Streptomyces Sp. Strain N2. Nat. Prod. Res. 2019, 35, 2090–2094. [Google Scholar] [CrossRef] [PubMed]
- Gohil, N.; Bhattacharjee, G.; Singh, V. Chapter 1—An Introduction to Microbial Cell Factories for Production of Biomolecules; Gohil, N., Bhattacharjee, G., Singh, V., Eds.; Academic Press: Cambridge, MA, USA, 2021; pp. 1–19. ISBN 978-0-12-821477-0. [Google Scholar]
- Margalith, P.; Beretta, G. Rifomycin. XI. Taxonomic Study on Streptomyces Mediterranei Nov. Sp. Mycopathol. Mycol. Appl. 1960, 13, 321–330. [Google Scholar] [CrossRef]
- Shapiro, S.; Vining, L.C. Nitrogen Metabolism and Chloramphenicol Production in Streptomyces Venezuelae. Can. J. Microbiol. 1983, 29, 1706–1714. [Google Scholar] [CrossRef] [PubMed]
- Shih, H.D.; Liu, Y.C.; Hsu, F.L.; Mulabagal, V.; Dodda, R.; Huang, J.W. Fungichromin: A Substance from Streptomyces Padanus with Inhibitory Effects on Rhizoctonia Solani. J. Agric. Food Chem. 2003, 51, 95–99. [Google Scholar] [CrossRef] [PubMed]
- Brautaset, T.; Sletta, H.; Degnes, K.F.; Sekurova, O.N.; Bakke, I.; Volokhan, O.; Andreassen, T.; Ellingsen, T.E.; Zotchev, S.B. New Nystatin-Related Antifungal Polyene Macrolides with Altered Polyol Region Generated via Biosynthetic Engineering of Streptomyces Noursei. Appl. Environ. Microbiol. 2011, 77, 6636–6643. [Google Scholar] [CrossRef] [PubMed]
- Park, S.R.; Park, J.W.; Ban, Y.H.; Sohng, J.K.; Yoon, Y.J. 2-Deoxystreptamine-Containing Aminoglycoside Antibiotics: Recent Advances in the Characterization and Manipulation of Their Biosynthetic Pathways. Nat. Prod. Rep. 2013, 30, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Barreales, E.G.; Payero, T.D.; de Pedro, A.; Aparicio, J.F. Phosphate Effect on Filipin Production and Morphological Differentiation in Streptomyces Filipinensis and the Role of the PhoP Transcription Factor. PLoS ONE 2018, 13, e0208278. [Google Scholar] [CrossRef]
- Õmura, S.; Crump, A. The Life and Times of Ivermectin—A Success Story. Nat. Rev. Microbiol. 2004, 2, 984–989. [Google Scholar] [CrossRef]
- Sadeghi, A.; Karimi, E.; Dahaji, P.A.; Javid, M.G.; Dalvand, Y.; Askari, H. Plant Growth Promoting Activity of an Auxin and Siderophore Producing Isolate of Streptomyces under Saline Soil Conditions. World J. Microbiol. Biotechnol. 2012, 28, 1503–1509. [Google Scholar] [CrossRef]
- Armin, R.; Zühlke, S.; Grunewaldt-Stöcker, G.; Mahnkopp-Dirks, F.; Kusari, S. Production of Siderophores by an Apple Root-Associated Streptomyces Ciscaucasicus Strain GS2 Using Chemical and Biological OSMAC Approaches. Molecules 2021, 26, 3517. [Google Scholar] [CrossRef] [PubMed]
- Nozari, R.M.; Ortolan, F.; Astarita, L.V.; Santarém, E.R. Streptomyces Spp. Enhance Vegetative Growth of Maize Plants under Saline Stress. Braz. J. Microbiol. 2021, 52, 1371–1383. [Google Scholar] [CrossRef] [PubMed]
- El-Naggar, N.E.-A. Chapter 11—Streptomyces-Based Cell Factories for Production of Biomolecules and Bioactive Metabolites. In Microbial Cell Factories Engineering for Production of Biomolecules; Singh, V., Ed.; Academic Press: Cambridge, MA, USA, 2021; pp. 183–234. ISBN 978-0-12-821477-0. [Google Scholar]
- Sanjivkumar, M.; Silambarasan, T.; Ananthi, S.; ThangaTharani, K. Biosynthesis and Characterization of Zinc Oxide Nanoparticles from an Estuarine-Associated Actinobacterium Streptomyces Spp. and Its Biotherapeutic Applications. Arch. Microbiol. 2021, 204, 17. [Google Scholar] [CrossRef]
- Mechouche, M.S.; Merouane, F.; Messaad, C.E.H.; Golzadeh, N.; Vasseghian, Y.; Berkani, M. Biosynthesis, Characterization, and Evaluation of Antibacterial and Photocatalytic Methylene Blue Dye Degradation Activities of Silver Nanoparticles from Streptomyces Tuirus Strain. Environ. Res. 2022, 204, 112360. [Google Scholar] [CrossRef]
- Khattab, A.; Babiker, E.; Saeed, H. Streptomyces: Isolation, Optimization of Culture Conditions and Extraction of Secondary Metabolites. Int. Curr. Pharm. J. 2016, 5, 27. [Google Scholar] [CrossRef]
- Ding, W.; Tu, J.; Zhang, H.; Wei, X.; Ju, J.; Li, Q. Genome Mining and Metabolic Profiling Uncover Polycyclic Tetramate Macrolactams from Streptomyces Koyangensis SCSIO 5802. Mar. Drugs 2021, 19, 440. [Google Scholar] [CrossRef]
- Duhsaki, L.; Mukherjee, S.; Rani, T.S.; Madhuprakash, J. Genome Analysis of Streptomyces Sp. UH6 Revealed the Presence of Potential Chitinolytic Machinery Crucial for Chitosan Production. Environ. Microbiol. Rep. 2022, 14, 431–442. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Chen, Y.; Li, Y.; Song, Y.; Ma, J.; Ju, J. Secondary Metabolites and Biosynthetic Gene Clusters Analysis of Deep-Sea Hydrothermal Vent-Derived Streptomyces Sp. SCSIO ZS0520. Mar. Drugs 2022, 20, 393. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Ren, Z.; Chunyu, W.X.; Li, G.D.; Chen, X.; Zhang, Z.T.L.; Sun, H.B.; Wang, M.; Xie, T.P.; Wang, M.; et al. Exploration of Diverse Secondary Metabolites From Streptomyces Sp. YINM00001, Using Genome Mining and One Strain Many Compounds Approach. Front. Microbiol. 2022, 13, 831174. [Google Scholar] [CrossRef] [PubMed]
- Rehan, M.; ALSohim, A.S.; Abidou, H.; Rasheed, Z.; Abdulmonem, W.A.L. Isolation, Identification, Biocontrol Activity, and Plant Growth Promoting Capability of a Superior Streptomyces Tricolor Strain HM10. Pol. J. Microbiol. 2021, 70, 245–256. [Google Scholar] [CrossRef]
- Omar, A.F.; Abdelmageed, A.H.A.; Al-Turki, A.; Abdelhameid, N.M.; Sayyed, R.Z.; Rehan, M. Exploring the Plant Growth-Promotion of Four Streptomyces Strains from Rhizosphere Soil to Enhance Cucumber Growth and Yield. Plants 2022, 11, 3316. [Google Scholar] [CrossRef] [PubMed]
- Page, A.L. Methods of Soil Analysis. In Part 2—Chemical and Microbiological Properties; The American Society of Agronomy, Inc.: Madison, WI, USA; Soil Science Society of America: Madison, WI, USA, 1982. [Google Scholar]
- Rathje Jackson, M.L. Soil Chemical Analysis. Verlag: Prentice Hall, Inc., Englewood Cliffs, NJ. 1958, 498 S. DM 39.40. Zeitschrift für Pflanzenernährung Düngung Bodenkd 1959, 85, 251–252. [Google Scholar] [CrossRef]
- Nelson, D.W.; Sommers, L.E. Total Carbon, Organic Carbon, and Organic Matter. In Methods of Soil Analysis; SSSA Book Series; American Society of Agronomy, Inc.: Madison, WI, USA; Soil Science Society of America, Inc.: Madison, WI, USA, 1996; pp. 961–1010. ISBN 9780891188667. [Google Scholar]
- Gee, G.W.; Or, D. 2.4 Particle-Size Analysis. In Methods of Soil Analysis; SSSA Book Series; Soil Science Society of America, Inc.: Madison, WI, USA, 2002; pp. 255–293. ISBN 9780891188933. [Google Scholar]
- Qureshi, K.A.; Bholay, A.D.; Rai, P.K.; Mohammed, H.A.; Khan, R.A.; Azam, F.; Jaremko, M.; Emwas, A.-H.; Stefanowicz, P.; Waliczek, M.; et al. Isolation, Characterization, Anti-MRSA Evaluation, and in-Silico Multi-Target Anti-Microbial Validations of Actinomycin X2 and Actinomycin D Produced by Novel Streptomyces Smyrnaeus UKAQ_23. Sci. Rep. 2021, 11, 14539. [Google Scholar] [CrossRef] [PubMed]
- William, S.; Feil, H.; Copeland, A. Bacterial Genomic DNA Isolation Using CTAB. Sigma 2012, 50, 6876. [Google Scholar]
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of Long, Error-Prone Reads Using Repeat Graphs. Nat. Biotechnol. 2019, 37, 540–546. [Google Scholar] [CrossRef] [PubMed]
- Vaser, R.; Sović, I.; Nagarajan, N.; Šikić, M. Fast and Accurate de Novo Genome Assembly from Long Uncorrected Reads. Genome Res. 2017, 27, 737–746. [Google Scholar] [CrossRef]
- Oosterbroek, S.; Doorenspleet, K.; Nijland, R.; Jansen, L. Decona: From Demultiplexing to Consensus for Nanopore Amplicon Data. ARPHA Conf. Abstr. 2021, 4, e65029. [Google Scholar] [CrossRef]
- Mikheenko, A.; Prjibelski, A.; Saveliev, V.; Antipov, D.; Gurevich, A. Versatile Genome Assembly Evaluation with QUAST-LG. Bioinformatics 2018, 34, i142–i150. [Google Scholar] [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the Quality of Microbial Genomes Recovered from Isolates, Single Cells, and Metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI Prokaryotic Genome Annotation Pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Göker, M. TYGS Is an Automated High-Throughput Platform for State-of-the-Art Genome-Based Taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef] [PubMed]
- Ondov, B.D.; Treangen, T.J.; Melsted, P.; Mallonee, A.B.; Bergman, N.H.; Koren, S.; Phillippy, A.M. Mash: Fast Genome and Metagenome Distance Estimation Using MinHash. Genome Biol. 2016, 17, 132. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.H.; Ha, S.; Lim, J.; Kwon, S.; Chun, J. A Large-Scale Evaluation of Algorithms to Calculate Average Nucleotide Identity. Antonie Van Leeuwenhoek 2017, 110, 1281–1286. [Google Scholar] [CrossRef] [PubMed]
- Lagesen, K.; Hallin, P.; Rødland, E.A.; Staerfeldt, H.-H.; Rognes, T.; Ussery, D.W. RNAmmer: Consistent and Rapid Annotation of Ribosomal RNA Genes. Nucleic Acids Res. 2007, 35, 3100–3108. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and Applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; van Wezel, G.P.; Medema, M.H.; Weber, T. AntiSMASH 6.0: Improving Cluster Detection and Comparison Capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef]
- Skinnider, M.A.; Johnston, C.W.; Gunabalasingam, M.; Merwin, N.J.; Kieliszek, A.M.; MacLellan, R.J.; Li, H.; Ranieri, M.R.M.; Webster, A.L.H.; Cao, M.P.T.; et al. Comprehensive Prediction of Secondary Metabolite Structure and Biological Activity from Microbial Genome Sequences. Nat. Commun. 2020, 11, 6058. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The Protein Families Database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Kautsar, S.A.; Blin, K.; Shaw, S.; Navarro-Muñoz, J.C.; Terlouw, B.R.; van der Hooft, J.J.J.; van Santen, J.A.; Tracanna, V.; Suarez Duran, H.G.; Pascal Andreu, V.; et al. MIBiG 2.0: A Repository for Biosynthetic Gene Clusters of Known Function. Nucleic Acids Res. 2020, 48, D454–D458. [Google Scholar] [CrossRef]
- Lefort, V.; Desper, R.; Gascuel, O. FastME 2.0: A Comprehensive, Accurate, and Fast Distance-Based Phylogeny Inference Program. Mol. Biol. Evol. 2015, 32, 2798–2800. [Google Scholar] [CrossRef]
- Farris, J.S. Estimating Phylogenetic Trees from Distance Matrices. Am. Nat. 1972, 106, 645–668. [Google Scholar] [CrossRef]
- Zhao, B.; Lin, X.; Lei, L.; Lamb, D.C.; Kelly, S.L.; Waterman, M.R.; Cane, D.E. Biosynthesis of the Sesquiterpene Antibiotic Albaflavenone in Streptomyces Coelicolor A3(2). J. Biol. Chem. 2008, 283, 8183–8189. [Google Scholar] [CrossRef] [PubMed]
- Bentley, S.D.; Chater, K.F.; Cerdeño-Tárraga, A.M.; Challis, G.L.; Thomson, N.R.; James, K.D.; Harris, D.E.; Quail, M.A.; Kieser, H.; Harper, D.; et al. Complete Genome Sequence of the Model Actinomycete Streptomyces Coelicolor A3(2). Nature 2002, 417, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Holmes, T.C.; May, A.E.; Zaleta-Rivera, K.; Ruby, J.G.; Skewes-Cox, P.; Fischbach, M.A.; DeRisi, J.L.; Iwatsuki, M.; Ōmura, S.; Khosla, C. Molecular Insights into the Biosynthesis of Guadinomine: A Type III Secretion System Inhibitor. J. Am. Chem. Soc. 2012, 134, 17797–17806. [Google Scholar] [CrossRef]
- Zhang, W.; Wang, L.; Kong, L.; Wang, T.; Chu, Y.; Deng, Z.; You, D. Unveiling the Post-PKS Redox Tailoring Steps in Biosynthesis of the Type II Polyketide Antitumor Antibiotic Xantholipin. Chem. Biol. 2012, 19, 422–432. [Google Scholar] [CrossRef]
- Xu, F.; Nazari, B.; Moon, K.; Bushin, L.B.; Seyedsayamdost, M.R. Discovery of a Cryptic Antifungal Compound from Streptomyces Albus J1074 Using High-Throughput Elicitor Screens. J. Am. Chem. Soc. 2017, 139, 9203–9212. [Google Scholar] [CrossRef]
- Mohimani, H.; Kersten, R.D.; Liu, W.T.; Wang, M.; Purvine, S.O.; Wu, S.; Brewer, H.M.; Pasa-Tolic, L.; Bandeira, N.; Moore, B.S.; et al. Automated Genome Mining of Ribosomal Peptide Natural Products. ACS Chem. Biol. 2014, 9, 1545–1551. [Google Scholar] [CrossRef]
- Pawlik, K.; Kotowska, M.; Chater, K.F.; Kuczek, K.; Takano, E. A Cryptic Type I Polyketide Synthase (Cpk) Gene Cluster in Streptomyces Coelicolor A3(2). Arch. Microbiol. 2007, 187, 87–99. [Google Scholar] [CrossRef]
- O’Rourke, S.; Wietzorrek, A.; Fowler, K.; Corre, C.; Challis, G.L.; Chater, K.F. Extracellular Signalling, Translational Control, Two Repressors and an Activator All Contribute to the Regulation of Methylenomycin Production in Streptomyces Coelicolor. Mol. Microbiol. 2009, 71, 763–778. [Google Scholar] [CrossRef]
- Metsä-Ketelä, M.; Palmu, K.; Kunnari, T.; Ylihonko, K.; Mäntsälä, P. Engineering Anthracycline Biosynthesis toward Angucyclines. Antimicrob. Agents Chemother. 2003, 47, 1291–1296. [Google Scholar] [CrossRef]
- Palmu, K.; Ishida, K.; Mäntsälä, P.; Hertweck, C.; Metsä-Ketelä, M. Artificial Reconstruction of Two Cryptic Angucycline Antibiotic Biosynthetic Pathways. ChemBioChem 2007, 8, 1577–1584. [Google Scholar] [CrossRef] [PubMed]
- Kersten, R.D.; Yang, Y.-L.; Xu, Y.; Cimermancic, P.; Nam, S.-J.; Fenical, W.; Fischbach, M.A.; Moore, B.S.; Dorrestein, P.C. A Mass Spectrometry-Guided Genome Mining Approach for Natural Product Peptidogenomics. Nat. Chem. Biol. 2011, 7, 794–802. [Google Scholar] [CrossRef]
- Fagerholm, A.E.; Habrant, D.; Koskinen, A.M.P. Calyculins and Related Marine Natural Products as Serine-Threonine Protein Phosphatase PP1 and PP2A Inhibitors and Total Syntheses of Calyculin A, B, and C. Mar. Drugs 2010, 8, 122–172. [Google Scholar] [CrossRef] [PubMed]
- Wakimoto, T.; Egami, Y.; Nakashima, Y.; Wakimoto, Y.; Mori, T.; Awakawa, T.; Ito, T.; Kenmoku, H.; Asakawa, Y.; Piel, J.; et al. Calyculin Biogenesis from a Pyrophosphate Protoxin Produced by a Sponge Symbiont. Nat. Chem. Biol. 2014, 10, 648–655. [Google Scholar] [CrossRef] [PubMed]
- Pfennig, F.; Schauwecker, F.; Keller, U. Molecular Characterization of the Genes of Actinomycin Synthetase I and of a 4-Methyl-3-Hydroxyanthranilic Acid Carrier Protein Involved in the Assembly of the Acylpeptide Chain of Actinomycin in Streptomyces. J. Biol. Chem. 1999, 274, 12508–12516. [Google Scholar] [CrossRef]
- Keller, U.; Lang, M.; Crnovcic, I.; Pfennig, F.; Schauwecker, F. The Actinomycin Biosynthetic Gene Cluster of Streptomyces Chrysomallus: A Genetic Hall of Mirrors for Synthesis of a Molecule with Mirror Symmetry. J. Bacteriol. 2010, 192, 2583–2595. [Google Scholar] [CrossRef]
- Schauwecker, F.; Pfennig, F.; Grammel, N.; Keller, U. Construction and in Vitro Analysis of a New Bi-Modular Polypeptide Synthetase for Synthesis of N-Methylated Acyl Peptides. Chem. Biol. 2000, 7, 287–297. [Google Scholar] [CrossRef]
- Xiao, C.; Xu, C.; Zhang, J.; Jiang, W.; Zhang, X.; Yang, C.; Xu, J.; Zhang, Y.; Zhou, T. Soil Microbial Communities Affect the Growth and Secondary Metabolite Accumulation in Bletilla Striata (Thunb.) Rchb. F. Front. Microbiol. 2022, 13, 916418. [Google Scholar] [CrossRef]
- Zhao, Q.; Wang, L.; Luo, Y. Recent Advances in Natural Products Exploitation in Streptomyces via Synthetic Biology. Eng. Life Sci. 2019, 19, 452–462. [Google Scholar] [CrossRef]
- Liu, R.; Deng, Z.; Liu, T. Streptomyces Species: Ideal Chassis for Natural Product Discovery and Overproduction. Metab. Eng. 2018, 50, 74–84. [Google Scholar] [CrossRef]
- Sarmiento-Vizcaíno, A.; Martín, J.; Reyes, F.; García, L.A.; Blanco, G. Bioactive Natural Products in Actinobacteria Isolated in Rainwater From Storm Clouds Transported by Western Winds in Spain. Front. Microbiol. 2021, 12, 773095. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; He, J.; Wei, X.; Ju, J.; Ma, J. Exploration and Genome Mining of Natural Products from Marine Streptomyces. Appl. Microbiol. Biotechnol. 2020, 104, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Paulus, C.; Rebets, Y.; Tokovenko, B.; Nadmid, S.; Terekhova, L.P.; Myronovskyi, M.; Zotchev, S.B.; Rückert, C.; Braig, S.; Zahler, S.; et al. New Natural Products Identified by Combined Genomics-Metabolomics Profiling of Marine Streptomyces Sp. MP131-18. Sci. Rep. 2017, 7, 42382. [Google Scholar] [CrossRef]
- Lee, N.; Kim, W.; Hwang, S.; Lee, Y.; Cho, S.; Palsson, B.; Cho, B.-K. Thirty Complete Streptomyces Genome Sequences for Mining Novel Secondary Metabolite Biosynthetic Gene Clusters. Sci. Data 2020, 7, 55. [Google Scholar] [CrossRef]
- Jaroszewicz, W.; Bielańska, P.; Lubomska, D.; Kosznik-Kwaśnicka, K.; Golec, P.; Grabowski, Ł.; Wieczerzak, E.; Dróżdż, W.; Gaffke, L.; Pierzynowska, K.; et al. Antibacterial, Antifungal and Anticancer Activities of Compounds Produced by Newly Isolated Streptomyces Strains from the Szczelina Chochołowska Cave (Tatra Mountains, Poland). Antibiotics 2021, 10, 1212. [Google Scholar] [CrossRef]
- Santos-Beneit, F.; Ceniceros, A.; Nikolaou, A.; Salas, J.A.; Gutierrez-Merino, J. Identification of Antimicrobial Compounds in Two Streptomyces Sp. Strains Isolated from Beehives. Front. Microbiol. 2022, 13, 742168. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.J.; Kim, J.; Rho, J.-Y. Antifungal Activities of Streptomyces Blastmyceticus Strain 12-6 Against Plant Pathogenic Fungi. Mycobiology 2019, 47, 329–334. [Google Scholar] [CrossRef]
- Le, K.D.; Yu, N.H.; Park, A.R.; Park, D.-J.; Kim, C.-J.; Kim, J.-C. Streptomyces sp. AN090126 as a Biocontrol Agent against Bacterial and Fungal Plant Diseases. Microorganisms 2022, 10, 791. [Google Scholar] [CrossRef]
- Tomaseto, A.A.; Alpiste, M.C.; de Castro Nassar, A.F.; Destéfano, S.A.L. Antibacterial activity of phytopathogenic Streptomyces strains against bacteria associated to clinical diseases. Arq. Inst. Biol. 2020, 87, e0142020. [Google Scholar] [CrossRef]
- Shah, M.; Gul, S.; Amjad, A.; Javed, M.S.; Fatima, B.; Nawaz, H.; Ishaq, J. Genome Mining of Streptomyces Formicae KY5 for Potential Drug like Natural Products Characterizations. J. Proteomics Bioinform. 2019, 12, 122–127. [Google Scholar] [CrossRef]
- Komaki, H.; Ishikawa, A.; Ichikawa, N.; Hosoyama, A.; Hamada, M.; Harunari, E.; Nihira, T.; Panbangred, W.; Igarashi, Y. Draft Genome Sequence of Streptomyces Sp. MWW064 for Elucidating the Rakicidin Biosynthetic Pathway. Stand. Genom. Sci. 2016, 11, 83. [Google Scholar] [CrossRef] [PubMed]
- Moser, S.; Pichler, H. Identifying and Engineering the Ideal Microbial Terpenoid Production Host. Appl. Microbiol. Biotechnol. 2019, 103, 5501–5516. [Google Scholar] [CrossRef] [PubMed]
- Peralta-Yahya, P.P.; Ouellet, M.; Chan, R.; Mukhopadhyay, A.; Keasling, J.D.; Lee, T.S. Identification and Microbial Production of a Terpene-Based Advanced Biofuel. Nat. Commun. 2011, 2, 483. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Liu, Q.; Cao, Y.; Feng, X.; Zheng, Y.; Zou, H.; Liu, H.; Yang, J.; Xian, M. Microbial Production of Sabinene—A New Terpene-Based Precursor of Advanced Biofuel. Microb. Cell Fact. 2014, 13, 20. [Google Scholar] [CrossRef] [PubMed]
- Phelan, R.M.; Sekurova, O.N.; Keasling, J.D.; Zotchev, S.B. Engineering Terpene Biosynthesis in Streptomyces for Production of the Advanced Biofuel Precursor Bisabolene. ACS Synth. Biol. 2015, 4, 393–399. [Google Scholar] [CrossRef]
- Oldfield, E.; Lin, F.Y. Terpene Biosynthesis: Modularity Rules. Angew. Chem. Int. Ed. Engl. 2012, 51, 1124–1137. [Google Scholar] [CrossRef]
- Yamada, Y.; Kuzuyama, T.; Komatsu, M.; Shin-ya, K.; Omura, S.; Cane, D.E.; Ikeda, H. Terpene Synthases Are Widely Distributed in Bacteria. Proc. Natl. Acad. Sci. USA 2015, 112, 857–862. [Google Scholar] [CrossRef]
- Wenski, S.L.; Thiengmag, S.; Helfrich, E.J.N. Complex Peptide Natural Products: Biosynthetic Principles, Challenges and Opportunities for Pathway Engineering. Synth. Syst. Biotechnol. 2022, 7, 631–647. [Google Scholar] [CrossRef]
- Minowa, Y.; Araki, M.; Kanehisa, M. Comprehensive Analysis of Distinctive Polyketide and Nonribosomal Peptide Structural Motifs Encoded in Microbial Genomes. J. Mol. Biol. 2007, 368, 1500–1517. [Google Scholar] [CrossRef]
- Silakowski, B.; Kunze, B.; Müller, R. Multiple Hybrid Polyketide Synthase/Non-Ribosomal Peptide Synthetase Gene Clusters in the Myxobacterium Stigmatella Aurantiaca. Gene 2001, 275, 233–240. [Google Scholar] [CrossRef]
- Minami, A.; Ugai, T.; Ozaki, T.; Oikawa, H. Predicting the Chemical Space of Fungal Polyketides by Phylogeny-Based Bioinformatics Analysis of Polyketide Synthase-Nonribosomal Peptide Synthetase and Its Modification Enzymes. Sci. Rep. 2020, 10, 13556. [Google Scholar] [CrossRef] [PubMed]
- Hwang, S.; Lee, N.; Cho, S.; Palsson, B.; Cho, B.-K. Repurposing Modular Polyketide Synthases and Non-Ribosomal Peptide Synthetases for Novel Chemical Biosynthesis. Front. Mol. Biosci. 2020, 7, 87. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Zhang, L.; Zhou, Z.; Yan, X. Diversity of Gene Clusters for Polyketide and Nonribosomal Peptide Biosynthesis Revealed by Metagenomic Analysis of the Yellow Sea Sediment. Front. Microbiol. 2018, 9, 295. [Google Scholar] [CrossRef] [PubMed]
- Letzel, A.-C.; Pidot, S.J.; Hertweck, C. Genome Mining for Ribosomally Synthesized and Post-Translationally Modified Peptides (RiPPs) in Anaerobic Bacteria. BMC Genom. 2014, 15, 983. [Google Scholar] [CrossRef]
- Komaki, H.; Tamura, T. Polyketide Synthase and Nonribosomal Peptide Synthetase Gene Clusters in Type Strains of the Genus Phytohabitans. Life 2020, 10, 257. [Google Scholar] [CrossRef]
- Zheng, Y.; Saitou, A.; Wang, C.-M.; Toyoda, A.; Minakuchi, Y.; Sekiguchi, Y.; Ueda, K.; Takano, H.; Sakai, Y.; Abe, K.; et al. Genome Features and Secondary Metabolites Biosynthetic Potential of the Class Ktedonobacteria. Front. Microbiol. 2019, 10, 893. [Google Scholar] [CrossRef]
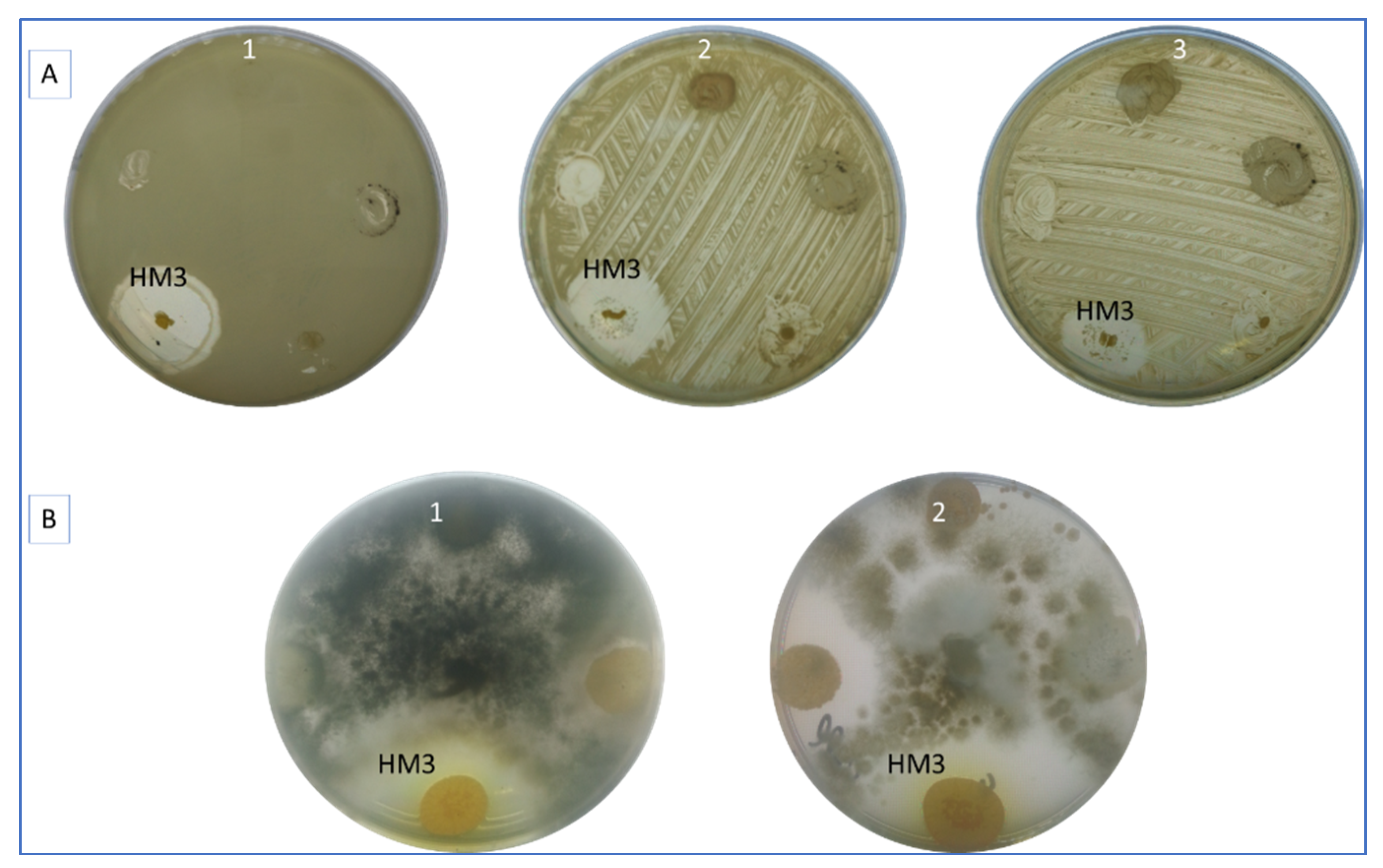


| pH | EC | OM | C/N | N | P | K | Particle Size Analysis | Texture | ||
|---|---|---|---|---|---|---|---|---|---|---|
| (dS m−1) | (%) | Ratio | mg kg−1 | Sand (%) | Silt (%) | Clay (%) | ||||
| 7.90 | 3.2 | 0.25 | 35 | 42 | 4.6 | 35 | 98.0 | 1.2 | 0.8 | Sand |
| Statistics | S. thinghirensis Strain HM3 |
|---|---|
| contigs number | 69 |
| Largest contig (bp) | 478,557 |
| Total length (bp) | 7,139,324 |
| N50 | 174,705 |
| N75 | 88,698 |
| L50 | 13 |
| L75 | 27 |
| GC (%) | 71.49 |
| Genes (total) | 7949 |
| Coding genes | 3362 |
| Genes (RNA) | 75 |
| rRNAs | 4, 4, 4 (5S, 16S, 23S) |
| tRNA | 60 |
| Pseudo genes (total) | 4512 |
| Region (Contigs) | Type | Nucleotide Sequences (From–To) | Most Similar Known Cluster | Microorganism | Similarity Percentage | Reference |
|---|---|---|---|---|---|---|
| Region 2.1 | Terpene | 12,171–33,262 | Albaflavenone | Streptomyces coelicolor A3(2) | 100 | [59] |
| Region 6.1 | Terpene | 8731–27,414 | - | - | - | - |
| Region 17.1 | Terpene | 39,937–60,815 | - | - | - | - |
| Region 50.2 | Terpene | 237,876–258,370 | Hopene | Streptomyces coelicolor A3(2) | 38 | [60] |
| Region 8.1 | Lantipeptide class V | 75,517–117,727 | Guadinomine (NRP + polyketide) | Streptomyces sp. K01-0509 | 7 | [61] |
| Region 9.1 | Siderophore | 31,741–44,344 | - | - | - | - |
| Region 15.1 | Type II PKS (Polyketide synthase) | 42,349–113,381 | Xantholipin (Polyketide) | Streptomyces flavogriseus | 8 | [62] |
| Region 24.1 | NRPS | 19,390–61,821 | Surugamide A/D (NRP) | Streptomyces albidoflavus | 23 | [63] |
| Region 35.1 | RiPP-like | 4066–14,248 | Informatipeptin (RiPP: Lantipeptide) | Streptomyces viridochromogenes DSM 40736 | 28 | [64] |
| Region 41.1 | Butyrolactone | 84,097–94,972 | SCB1 | Streptomyces coelicolor A3(2) | 100 | [65] |
| Region 54.1 | Butyrolactone | 33,375–44,304 | Methylenomycin A | Streptomyces coelicolor A3(2) | 9 | [66] |
| Region 44.1 | Type II PKS | 44,648–107,146 | Prejadomycin ( Polyketide/ Saccharide) | Streptomyces sp. PGA64 | 41 | [67,68] |
| Region 50.1 | RiPP | 159,658–179,906 | SSV-2083 (RiPP:Lantipeptide) | Streptomyces sviceus ATCC 29083 | 27 | [69] |
| Region 59.1 | CDPS | 1–13,469 | - | - | - | - |
| Region 59.2 | NRPS-like | 104,234–151,246 | Calyculin A (NRP + polyketide: Trans-AT type I) | Uncultured Candidatus Entotheonella sp. | 28 | [70,71] |
| Region 62.1 | Other | 18,773–59,354 | Actinomycin D (NRP) | Streptomyces anulatus | 57 | [72,73,74] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rehan, M.; Gueddou, A.; Alharbi, A.; Ben Abdelmalek, I. In Silico Prediction of Secondary Metabolites and Biosynthetic Gene Clusters Analysis of Streptomyces thinghirensis HM3 Isolated from Arid Soil. Fermentation 2023, 9, 65. https://doi.org/10.3390/fermentation9010065
Rehan M, Gueddou A, Alharbi A, Ben Abdelmalek I. In Silico Prediction of Secondary Metabolites and Biosynthetic Gene Clusters Analysis of Streptomyces thinghirensis HM3 Isolated from Arid Soil. Fermentation. 2023; 9(1):65. https://doi.org/10.3390/fermentation9010065
Chicago/Turabian StyleRehan, Medhat, Abdellatif Gueddou, Abdulaziz Alharbi, and Imen Ben Abdelmalek. 2023. "In Silico Prediction of Secondary Metabolites and Biosynthetic Gene Clusters Analysis of Streptomyces thinghirensis HM3 Isolated from Arid Soil" Fermentation 9, no. 1: 65. https://doi.org/10.3390/fermentation9010065
APA StyleRehan, M., Gueddou, A., Alharbi, A., & Ben Abdelmalek, I. (2023). In Silico Prediction of Secondary Metabolites and Biosynthetic Gene Clusters Analysis of Streptomyces thinghirensis HM3 Isolated from Arid Soil. Fermentation, 9(1), 65. https://doi.org/10.3390/fermentation9010065









