Abstract
Proteins are crucial for medicine, pharmaceuticals, food, and environmental applications since they are used in various fields such as synthesis of drugs, industrial enzyme production, biodegradable plastics, bioremediation processes, etc. Xylanase is an important and versatile enzyme with applications across various industries, including pulp and paper, biofuel production, food processing, textiles, laundry detergents, and animal feed. Key parameters in biotechnological protein production include temperature, pH, and working volumes and especially medium compositions where optimization is crucial for large-scale applications due to cost considerations. Machine learning methods have emerged as effective alternatives to traditional statistical approaches in optimization. This study focuses on optimizing xylanase production via bioprocesses by employing regression analysis on datasets from various studies. The extreme gradient boosting (XGBoost) regression model was applied to predict xylanase activity under different experimental conditions, accurately predicting xylanase activity and identifying the significance of each variable. This study utilized experimentally derived datasets from peer-reviewed publications, in which the corresponding laboratory experiments had already been conducted and validated. The results demonstrate that machine learning methods can effectively optimize protein production processes, offering a strong alternative to traditional statistical approaches.
1. Introduction
Protein production by bioprocesses has become essential in modern medicine, pharmaceuticals, food production, and environmental activities. These proteins can be utilized in several applications, including the synthesis of essential pharmaceuticals like insulin and monoclonal antibodies, as well as the production of industrial enzymes, biodegradable polymers, and bioremediation agents [1,2]. The effective and sustainable production of these proteins is essential for addressing global issues such as food security, illness prevention, and environmental sustainability [3,4].
Xylanase has emerged as a versatile and essential enzyme among biotechnologically produced proteins, with applications in various industries, including pulp and paper, biofuel production, food processing, textiles, and animal feed [5,6]. Due to its extensive industrial utility, xylanase is regarded as a highly significant protein in both biotechnological and industrial applications [7]. Its typical applications span the food industry, animal feed, bioconversion, textiles, and the paper and pulp sectors [8]. Additionally, xylanase is used to improve the clarity of fruit juices and wine; assist in the extraction of vegetable oils, coffee, and starch; facilitate oligosaccharide synthesis; and enhance the nutritional value of animal feed [9]. This enzyme is synthesized by various microorganisms, including fungi and actinomycetes, and belongs to a crucial class of hydrolases with a global market valuation of approximately 500 million USD [10].
Numerous factors influence xylanase synthesis in all fermentation methods. The initial aspect refers to the origin of the microorganism. Efforts are being made to improve the production yields of bacteria, fungi, protozoa, algae, etc., which are frequently utilized in xylanase synthesis, by recombinant methodologies. Recombinant production of xylanase can improve the yields of enzyme activity [11,12]. Enhancing the capabilities of species through recombinant technologies is an efficient method; yet, it is labor-intensive and expensive.
The selection of the appropriate bioreactor configuration is another aspect influencing production efficiency in xylanase synthesis. The decision of the process type depends on the microorganism’s origin [13]. Numerous studies in the literature address xylanase production using batch or continuous reactors [14]; however, the predominant methods in recent years are deep culture and solid state fermentations [15,16]. The growing interest in solid-state fermentations is driven by the need to minimize process costs. Agricultural waste materials serve as a substrate source in solid-state fermentations. Utilizing agricultural waste for xylanase production with the proper microorganism is a highly effective method for cost reduction [17]. Nonetheless, similar to other bioprocesses, scaling up solid-state fermentation suffers significant costs, and reductions in yields obtained at the laboratory size may occur during the scale-up stages. It is widely recognized that 30–40% of the production expenses for commercial enzymes are attributed to the cost of the nutritional medium [18]. The fermentation profile of an organism is affected by nutritional and physiological factors, notably carbon and nitrogen sources, along with pH, temperature, agitation, dissolved oxygen, and inoculum density. Consequently, in xylanase production, it is imperative to establish appropriate media and culture conditions to attain optimal enzyme yield. Optimizing these growth parameters is crucial for maximizing industrial enzyme production, as improper optimization results in lower enzyme yields [19,20]. Therefore, the optimization of xylanase production is a multifaceted endeavor that encompasses the selection of microbial strains, fermentation methods, nutrient media composition, and the application of statistical optimization techniques. The integration of these elements, coupled with a focus on sustainability and commercial viability, positions xylanase production as a promising area of research with significant industrial implications.
Statistical approaches, particularly design of experiments (DoE), have been widely used to optimize enzyme production by systematically analyzing the effects of multiple variables. DoE helps reduce the number of experimental trials while improving efficiency, making it a powerful tool in biotechnological optimization [21,22]. However, many DoE techniques, including advanced experimental design methodologies, often require specialized software or tools that come with significant costs, which can be a disadvantage for researchers with limited budgets. Among various experimental design methodologies, factorial design and response surface methodology (RSM) have been extensively applied for optimizing medium composition in xylanase production, Table 1.

Table 1.
Statistical Optimization Methods for Different Microorganisms.
While DoE provides valuable insights, machine-learning-based regression models offer a more flexible and cost-effective alternative. Unlike DoE, which relies on controlled experiments, regression models utilize existing data to predict xylanase production under various conditions, minimizing the need for additional costly and time-consuming trials. Moreover, ML-based approaches, such as boosting techniques, can capture complex interactions between parameters more effectively than traditional statistical methods, ultimately enhancing accuracy and reducing prediction errors. Using machine learning, enzyme production processes can be optimized more efficiently, making these techniques a promising alternative to traditional statistical methods [21].
A significant study by Pensupa et al. [24] shows the effectiveness of machine learning models in optimizing biomass production through fermentation processes, revealing that the Matern 5/2 Gaussian process regression model achieved the lowest root mean squared error of 0.75 g/L and an R-squared value of 0.90. This highlights the power of complex statistical frameworks to examine complex datasets and augment predictive precision in fermentation processes, therefore enhancing the comprehension of ideal growing circumstances for microbial cultures, including xylanase producers.
Moreover, Wu et al. [25] highlighted the significance of machine learning in monitoring yeast fermentation by Raman spectroscopy, delivering real-time data that improve monitoring precision throughout fermentation activities. These strategies could similarly improve the monitoring of essential parameters in xylanase production, allowing dynamic modifications to optimize enzyme yield.
Jeong and Kim [26] integrated image processing into machine learning methodologies, concentrating on quantitative evaluations in fermentation processes. Their use of convolutional neural networks (CNNs) creates new opportunities for visually studying fermentation dynamics, which can be crucial in enhancing both the visual and operational sides of xylanase production by enabling real-time modifications based on measurable fermentation indicators.
Additionally, Bowler et al. [27] introduce a novel use of ultrasonic measurements integrated with machine learning to forecast alcohol content during beer fermentation. Their findings demonstrate that simple monitoring techniques can significantly improve fermentation control. The implementation of comparable simple measurement techniques in xylanase production may facilitate accurate adjustment of fermentation parameters, hence ensuring optimal conditions during the process.
A comparison between DoE and ML-based approaches highlights their respective advantages. In DoE methodologies, data collection typically occurs through controlled experiments, whereas ML approaches, particularly boosting techniques, can process datasets to improve accuracy and minimize prediction errors. While DoE relies on hypothesis-driven mathematical models, ML methods utilize iterative algorithms that continuously refine predictions. Beyond serving as a complement to DoE techniques, machine learning methods provide a significant advantage by optimizing processes without dependence on proprietary software, employing predictive capabilities directly from the acquired data.
This work evaluates the use of machine-learning-based predictive modeling on publicly available experimental datasets extracted from the literature. No new laboratory experiments were conducted. The aim is to explore the capability of ML models, particularly XGBoost, to replicate or improve upon the predictive patterns captured by DoE approaches.
This study focuses on predicting xylanase production using the XGBoost regressor. The model was trained on a dataset comprising various experimental conditions, including glucose, (NH4)HPO4, K2HPO4, KH2PO4, MgSO4, (NH4)2HPO4, urea, malt sprout, corn cobs, and wheat bran concentrations. The objective was to accurately predict xylanase activity and identify key factors influencing enzyme production. The results demonstrate that machine learning methods can effectively optimize protein production processes, providing a robust and scalable alternative to traditional statistical approaches. This study not only highlights the potential of machine learning in biotechnological optimization but also provides a foundation for future research in this rapidly developing field.
2. Materials and Methods
2.1. Dataset
The data from studies that optimized xylanase production through experimental design were analyzed in this study. Various factorial design levels and response surface methodology approaches (central composite design, Box–Behnken design, etc.) were used for media optimization in the assessed research. The independent variables and the levels that were utilized in these studies can be seen in Table 1.
All datasets used in this study were compiled from previously published experimental studies focused on xylanase production using various microorganisms and fermentation designs. These datasets represent secondary sources obtained through manual data extraction from the literature, and no original experiments or synthetic (simulated) data were produced.
Farliahati et al. [3] conducted a two-stage study to optimize the medium composition in xylanase production using Escherichia coli DH5α. In the first phase of this study, the most effective factors were determined using a factorial design with five factors. Within the scope of the study, the factors considered in 18 different experimental setups were glucose (10–20 g/L), (NH4)2HPO4 (2–10 g/L), K2HPO4 (5–18 g/L), KH2PO4 (1–6 g/L), and MgSO4 (0.5–3 g/L) (see Table A1). In the second part of the study, the concentration ranges of three effective factors, (NH4)2HPO4 (1–13 g/L), K2HPO4 (1.5–23.5 g/L), and MgSO4 (0.75–3.75 g/L), were altered, and optimization results were improved in 15 experimental sets using the response surface methodology. All the experiments were conducted in 250 mL baffled flasks with 50 mL working volume, and the pH was kept at 7.4. The incubation temperature was 37 °C, and all the flasks were agitated at 200 rpm for 18 h in an orbital shaker. The dataset is available in tabular form in Table A5.
In another study conducted by Dobrev et al. [2], an optimization study was carried out considering the cost advantage provided by using cheaper and more accessible types of waste instead of xylan. In the deep culture experiments conducted with Aspergillus niger B03, 26 different setups were used with (NH4)2HPO4 (2.6–5.4 g/L), urea (0.9–2.1 g/L), malt sprout (6–18 g/L), corn cobs (12–24 g/L), and wheat bran (6–16 g/L) (Table A2). In the second part of the study, the concentrations required for the maximum xylanase amount were optimized in 14 different experimental setups using the important factors identified as (NH4)2HPO4 (2.6–20.4 g/L), urea (0.3–0.9 g/L), and malt sprout (0.4–10 g/L). The biosynthesis reactors were 500 mL flasks with 50 mL working volume. All flasks were kept at 28 °C in an orbital shaker, agitated at 200 rpm for 18 h (Table A6).
A solid culture fermentation study by Bacillus circulans was conducted by Bocchini et al. [4] using a 3 × 3 factorial design to optimize xylan concentrations (5–10 g/L), pH levels (8–9), and incubation times (24–72 h) for enhanced xylanase production. The concentrations of these three significant components were optimized at 27 different levels in the conducted studies. Xylanase production was performed in 125 mL Erlenmeyer flasks with a working volume of 20 mL. All the flasks were incubated for 12 h at 45 °C and agitated at 150 rpm. The detailed dataset of the experiments and the xylanase production values are listed in Table A3.
Pham et al. [23] modified the amounts of xylan (2.5–7.5 g/L), casein (1–2 g/L), and NH4Cl (0.3–1.3 g/L) in the culture medium for xylanase production from Bacillus sp. L-1018 using response surface techniques and used a two-stage optimization process. Initially, a full factorial design was applied to navigate toward the ideal region. The full factorial design technique simultaneously assesses the primary impacts of variables and their interactions (Table A4). Moreover, full factorial design effectively identifies the path of steepest ascent to reach within range of the optimal answer. It is thus especially suited for the preliminary phases. A 2 × 3 factorial design with three components at two levels necessitates eight experimental runs. In the second phase, 13 tests were conducted adopting response surface methodology to optimize concentrations of xylan (2.5–3.5 g/L) and casein (1.8–2.0 g/L) for maximum xylanase production. Xylanase production experiments were conducted in 250 mL Erlenmeyer flasks with 50 mL working volume. All the flasks were kept in a water bath, and agitation was supplied by a magnetic agitator at 250 rpm (Table A7).
For each dataset, the “DoE” values refer to reported predictions or interpolated outputs from factorial or response surface methods in the original publications, while the “XGBoost” values are generated by training a machine learning model on the same experimental input–output pairs. The XGBoost model was trained and tested independently from any statistical modeling carried out in the original study.
2.2. Regression Analysis for Xylanase Production Prediction
In this study, we used supervised machine learning regression techniques to predict xylanase production based on experimental input parameters such as nutrient concentrations, pH, and temperature. Among various regression algorithms, we selected extreme gradient boosting (XGBoost) due to its high accuracy, scalability, and robustness in modeling nonlinear interactions in structured datasets [28].
Figure 1 illustrates the workflow of building and applying a machine learning model for predicting xylanase activity based on experimental input variables. Initially, data obtained from laboratory experiments—including features such as xylan, casein, and glucose concentrations—are used to train a predictive model. During the training phase (top row), these features are paired with experimentally determined xylanase activity values to construct a supervised learning model.Once the model is trained, it can then be applied to new data points (bottom row), where the same set of input features is fed into the model to generate predicted xylanase activity values. This approach allows for the estimation of enzymatic performance without conducting additional experiments, thereby facilitating process optimization and decision making in a data-driven manner.
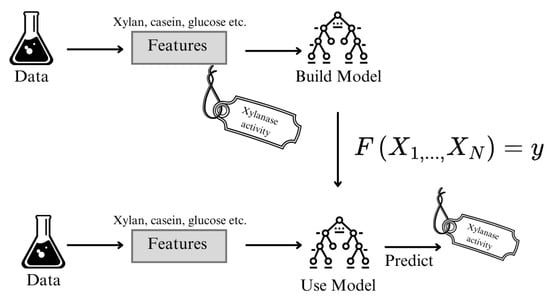
Figure 1.
Blok Diagram of the Xylanase Production Prediction using XGBoost.
Regression analysis aims to model the relationship between a set of input features and a continuous output variable y (xylanase activity in this case). Formally, it estimates a function such that
where represents random error or noise.
XGBoost is an open-source implementation of gradient boosted decision trees. It constructs an ensemble of regression trees where each new tree is trained to minimize the residual errors of the previous model [29]. The training process is guided by a regularized objective function:
Here, ℓ is typically the mean squared error (MSE):
and is a regularization term that penalizes overly complex models, thereby improving generalizability. At each iteration t, the model updates its prediction as
Figure 2 shows the block diagram of the XGBoost algorithm. In each booster iteration k, k = 1, 2, …, T and T represent the total number of trees, the function represents a regression tree trained to minimize the objective function , which consists of the loss of prediction and a regularization term. Here, denotes the set of parameters that define the structure of the regression tree . These parameters include split decisions, feature thresholds, leaf weights, and tree depth. They are used both for fitting the model and for computing the regularization term , which penalizes overly complex trees to improve generalization. This objective ensures that each tree improves the model by fitting the residuals while controlling model complexity.
XGBoost has demonstrated superior performance in various domains, including bioinformatics [30,31], chemical engineering [32,33], and biotechnology [34], particularly when data are tabular and heterogeneous [35]. Furthermore, its built-in feature importance mechanism helps identify the most predictive variables in a given dataset, although this does not imply causality or biological significance.
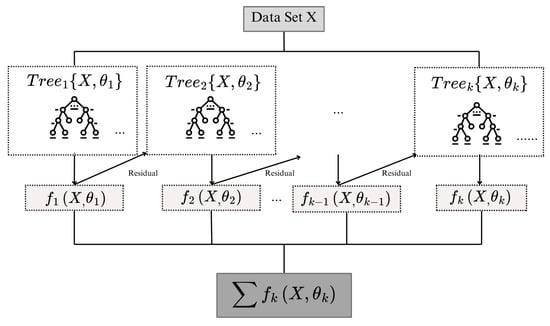
Figure 2.
Block diagram of the XGBoost learning process. Adapted from [36]. In each boosting iteration (k), the function ((X, )) represents a regression tree trained to minimize the objective function ().
In this study, XGBoost was trained using a diverse set of input variables collected from prior factorial and response surface experiments, allowing the model to capture complex interactions and generalize across different fermentation conditions.
2.3. Performance Evaluation
To assess the performance of the regression model, we used three common evaluation metrics: root mean squared error (RMSE), mean absolute percentage error (MAPE), and coefficient of determination (R2). These metrics were selected to provide a comprehensive assessment of predictive accuracy from multiple perspectives. RMSE penalizes larger errors more heavily and is sensitive to outliers, which is important when high deviation points can influence process reliability. MAPE expresses the average prediction error as a percentage, allowing for easier interpretation across different output scales. Finally, measures how well the variation in the dependent variable is explained by the model, offering an overall goodness-of-fit evaluation.
By combining these three metrics, we aim to ensure a balanced and interpretable performance comparison between the XGBoost and DoE approaches.
RMSE measures the average error between the actual and predicted values. A lower RMSE indicates better model performance.
MAPE calculates the average percentage error between actual and predicted values, making it useful for understanding relative error.
explains how well the model fits the data. A value closer to 1 indicates a better fit.
In this study, we analyzed these three metrics in detail to evaluate the model’s performance comprehensively. We calculated RMSE, MAPE, and R² for both training and test datasets to compare how well the model generalizes. Additionally, we performed an evaluation using the predictions obtained from the design of experiments (DoE) approach, allowing us to assess the prediction accuracy of DoE-based models.
3. Results
In all regression experiments conducted within this study, the dataset was divided into training and testing sets using an 80–20% split. Performance metrics were calculated accordingly for both sets. It is important to note that while the XGBoost model was trained and evaluated on both training and test data, the design of experiments (DoE) approach does not involve a learning-based prediction process. Therefore, for DoE-based results, predictions on the training data were not derived from a model but rather interpolated or estimated through statistical fitting within the experimental design boundaries.
As a result, relatively higher error metrics for the training set in the DoE results—compared to XGBoost—are expected and do not indicate model underperformance. In contrast, performance on the test set provides a more objective and fair comparison between the two approaches, as it reflects the ability to generalize to unseen data. Accordingly, emphasis in the comparison of model effectiveness is placed primarily on the test set results throughout the following sections.
3.1. Regression for the Experiments Performed by Full Factorial Experimental Design
The figures below present the results of regression analyses performed using the XGBoost model on datasets generated through full factorial experimental designs. In each plot, the X-axis represents the experimentally observed xylanase activity, while the Y-axis shows the corresponding predicted values. Dots are the symbols for train values and stars are the symbols of test values. Blue dots indicate the predictions made by the XGBoost regression model, whereas red dots represent the estimations obtained via the design of experiments (DoE) approach. The black dashed diagonal line serves as a reference, illustrating the ideal case where predicted values perfectly match the experimental results.
Farliahati et al. [3] conducted a two-stage study using recombinant Escherichia coli DH5 to optimize xylanase production. In the first stage, five variables—glucose (10–20 g/L), (NH4)HPO4 (2–10 g/L), K2HPO4 (5–18 g/L), KH2PO4 (1–6 g/L), and MgSO4 (0.5–3 g/L)—were investigated under 18 experimental conditions across 115 levels to maximize xylanase yield. As shown in Figure 3a, most data points lie close to the diagonal reference line, indicating strong agreement between the XGBoost model predictions and the experimental outcomes. However, a few discrepancies are observed, notably around the value of 2.2, which corresponds to a prediction from the DoE method. These deviations are likely due to experimental uncertainties rather than model inaccuracy. Such uncertainties may stem from factors common in bioprocess experiments, including variability in complex medium components (e.g., yeast extract), measurement errors in enzymatic activity assays (often due to sensitivity to time, temperature, or reagent stability), fluctuations in environmental conditions during fermentation (such as pH or aeration), and biological variation among cultures (e.g., inoculum density or age). The variation observed along the primary line results from the wide range of data intervals in the experimental design. The chosen range of 5–18 g/L for K2HPO4 indicates that the experimental values deviate from the expected values. The detailed experimental data used in this analysis are provided in Table A5. Such significant variations in the quantities utilized in fermentation media can only be noticed for complex substrates. The research of Farliahati et al. [3] indicates that glucose’s purity as a substrate permits a more limited operational range for K2HPO4. Within the parameters of the study, this phenomenon was also noted, resulting in a refinement of the outcome ranges and initiating the secondary optimization phase utilizing the RSM methodology (Table A5).
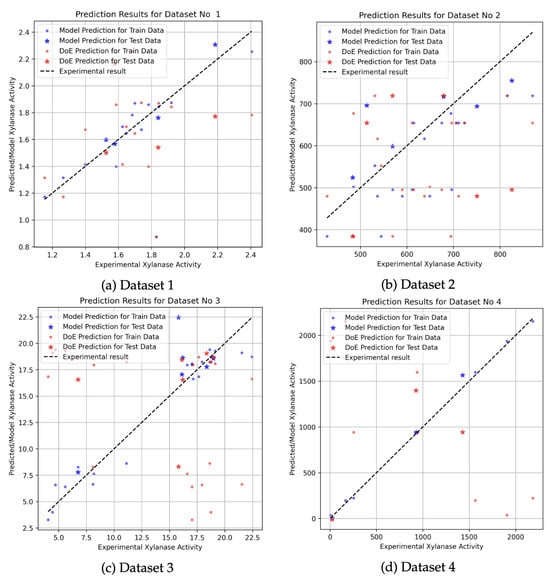
Figure 3.
Prediction results for full-factorial datasets using XGBoost and DoE approaches. Blue dots indicate predictions made by the XGBoost model, and red dots represent estimations from the DoE approach. Dot symbols correspond to predictions on the training data, while star symbols indicate predictions on the test data. The black dashed line shows the ideal prediction line ().
Dobrev et al. [2] conducted a study to optimize the nutrient medium for xylanase production using Aspergillus niger B03 cultivated on agricultural wastes. The experimental design included a range of components such as (NH4)2HPO4, urea, malt sprout, corn cobs, and wheat bran. Although the XGBoost regression model was trained on the same dataset (detailed in Table 2), the prediction performance was comparatively lower than that of other datasets. As shown in Figure 3b, the model predictions exhibit noticeable deviations from the reference line at several points, particularly at higher activity values. The factorial design employed in this study provides a broad range of variable combinations, which is beneficial for model training, but the observed deviations suggest that refined modeling with lower boundaries may be required to improve accuracy in such complex media formulations.

Table 2.
XGBoost Results.
In another study, Bocchini et al. [4] optimized xylanase production by Bacillus circulans D1 using a combination of full factorial design and Box–Behnken design (BBD). The optimization focused on three key variables: xylan concentration, pH, and cultivation time, across 27 experimental conditions (see Table A3). As shown in Figure 3c, the predictions obtained from the DoE and XGBoost models demonstrate a more pronounced divergence compared to other datasets. In particular, the DoE predictions show significant deviations from the experimental values, while the XGBoost regression model offers relatively closer estimates to the actual measurements. However, despite being more consistent than the DoE approach, the XGBoost model also exhibits variability and does not fully capture the experimental outcomes with high precision. These findings suggest that while the regression model performs better overall, the complexity of the underlying biological interactions in this dataset may require more advanced modeling strategies or a larger sample size to improve prediction accuracy.
The fermentation study conducted by Pham et al. [23] investigated xylanase production by Bacillus sp. I-1018 by optimizing three critical parameters: xylan, casein, and ammonium chloride concentrations. In Figure 3d, detailed in Table A7, the model prediction data are situated near the black dashed line, whereas the DOE predictions exhibit a wider dispersion. The increased frequency of the red points signifies a bigger variance in the predictions; however, a lower variance in the model’s prediction data implies that the model produces successful outcomes.
3.2. Regression for the Experiments Performed by RSM
Figure 4a illustrates the results obtained from an optimization study conducted by Farliahati et al. [3] using response surface methodology (RSM) with three independent variables (dataset is available in Table A5). In this case, the predictions of the XGBoost model (represented by blue dots) exhibit greater stability from the reference line when compared to the predictions made by the DoE approach (red dots). The clustering of DoE predictions around the reference line indicates a high degree of accuracy and alignment with the experimental data, particularly in the mid-range xylanase activity levels. At higher activity values, both methods begin to diverge slightly, which may be attributed to experimental uncertainties or limitations in model generalizability. These findings suggest that although both models can track the general trend of the data, the XGBoost model provides more precise and stable predictions across the experimental range.
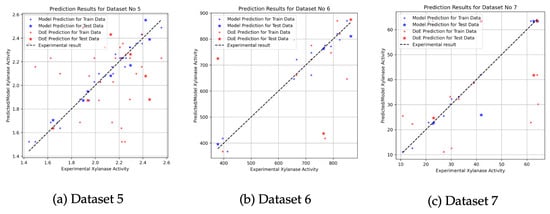
Figure 4.
Prediction results for RSM-based datasets using XGBoost and DoE approaches. Blue dots represent predictions from the XGBoost model, while red dots correspond to the DoE estimations. Dot symbols indicate training data predictions; star symbols indicate test data predictions. The black dashed line represents the ideal prediction line ().
A similar evaluation was conducted on the dataset reported by Dobrev et al. [2] (see Table A6), who also utilized RSM for optimization purposes. As shown in the Figure 4b, both the XGBoost and DoE models show substantial correlation with the experimental values, especially at elevated xylanase activity levels. However, the XGBoost model exhibits noticeable variability in the mid-range activity region, where deviations from the reference line become more prominent. While predictions at lower activity values are largely consistent across both models, minor overestimations and underestimations occur in the higher range. The XGBoosting model displays superior consistency, as evidenced by the denser clustering of blue points near the reference line. Overall, both methods successfully capture the general behavior of the process, but the XGBoosting model outperforms in terms of accuracy and robustness across all activity intervals.
Further evaluation was performed using the dataset derived from the work of Pham et al. [23] (see Table A7), which also employed an RSM-based experimental design. The scatter plot (Figure 4c) shows that the predicted values from both models closely align with the experimental xylanase activity measurements, indicating strong agreement with the reference line. Despite this overall alignment, more noticeable deviations are observed at the upper end of the activity spectrum, particularly for the DOE model. In contrast, the XGBoosting predictions maintain a more stable correspondence with the experimental data across the full range of activity levels. These results can be improved by further refinement of the ML-based model to enhance accuracy, particularly in boundary conditions or under extreme parameter settings.
The comparative analysis of experimental and predicted xylanase activity values across these three datasets offers valuable insights into the relative performance of machine learning and classical statistical approaches. While both methods capture the main trends in the data, the DoE approach demonstrates more stable performance, especially in the test datasets and at activity extremes. The results indicated variability in the performance of the XGBoost model across various datasets. Datasets with narrow experimental ranges, such as Dataset 4, demonstrated higher predictive accuracy, indicating that machine learning models are more effective within well-defined input spaces. In contrast, datasets characterized by broader or more heterogeneous parameter ranges (e.g., Dataset 2 and Dataset 3) demonstrated elevated prediction errors, likely related to increased complexity and noise that constrained the model’s generalizability. The observations emphasize the significance of dataset characteristics, such as size, homogeneity, and noise levels, in influencing the performance of machine learning models.Therefore The XGBoost model, though promising, shows increased variability and may require further tuning, extended training datasets, or hybrid modeling strategies to improve prediction reliability.
Taken together, the results suggest that integrating the strengths of both methodologies—leveraging the predictive power of machine learning and the structural rigor of statistical design—may offer a more comprehensive and accurate framework for modeling and optimizing xylanase production processes (Table 3).

Table 3.
DoE Results.
The feature importance values obtained from XGBoost for each dataset indicate the relative contribution of each input variable to the model’s predictive performance. It is important to clarify that these importance scores do not imply any underlying biological or chemical significance. Instead, they reflect how frequently and effectively each feature is used by the XGBoost algorithm to reduce prediction error during training.
For instance, in Dataset 1, the feature NH4HPO4 dominated the model with an importance score of 0.775, suggesting it played a major role in the decision paths constructed by the algorithm. Similarly, in Dataset 3, cultivation time (h) had a remarkably high importance of 0.964, indicating that XGBoost found this feature highly predictive within the context of the dataset.
Conversely, some features with known biochemical relevance may appear with low importance scores simply because they did not provide significant predictive value in the context of the model structure and available data. For example, in Dataset 4 features like X2 (casein) g/L and X3 (NH4Cl) g/L had minimal contribution to the model outcome (0.008 and 0.001, respectively), not due to their irrelevance in a biological sense but rather due to their limited utility in reducing model error (Table 4).

Table 4.
Feature contribution scores generated by XGBoost across six experimental datasets. The values reflect model-based importance, not causal or biological relationships.
Overall, these results provide a data-driven insight into how XGBoost utilizes features within each specific dataset. They are valuable for model interpretation and optimization but should not be over-interpreted in terms of causal or mechanistic implications without further experimental validation.
4. Discussion
Recent research underscores the essential impact of machine learning techniques in optimizing xylanase production within the field of biotechnology. Xylanase is a multifunctional enzyme used in several industries including pulp and paper, biofuels, food processing, textiles, and animal feed. Due to the enzyme’s extensive applications, effective production methods are essential, especially for industrial scale, where variables such as temperature, pH, and working volumes critically affect yield.
This work illustrates the potential benefits of machine learning (ML), specifically the XGBoost algorithm, in forecasting xylanase production utilizing datasets initially created through conventional design of experiments (DoE). In this study, XGBoost was selected as the machine learning model due to the characteristics of the available datasets. Specifically, the datasets contained a limited number of observations and were structured in a tabular format, making them less suitable for artificial neural networks (ANNs) or other conventional classifiers that typically require large volumes of data to perform effectively.
Prior to finalizing the modeling approach, we conducted preliminary experiments using artificial neural networks (ANN) and other conventional machine learning algorithms. However, these models showed suboptimal predictive performance, primarily attributed to overfitting on the relatively small and heterogeneous datasets [37,38]. While overfitting was a major concern, the decision to exclude these models was also informed by their limited generalization ability, sensitivity to small sample sizes, and inadequate capacity to capture complex nonlinear interactions effectively. In contrast, XGBoost was chosen for its well-documented robustness on small-to-medium-sized tabular datasets, its built-in regularization mechanisms, and its strong predictive performance observed in our initial evaluations.
Therefore, ANN and other conventional classifiers were not pursued further in the main analysis of this study. Our findings indicate that machine learning may match or even exceed traditional statistical methods in specific cases, particularly when the relationship between inputs and outputs is complex or nonlinear. Xylanase production processes often exhibit complex and nonlinear interactions among operational parameters such as pH, temperature, and substrate concentration. While DoE captures these interactions up to second-order polynomial levels, tree-based methods like XGBoost can flexibly partition the input space and approximate more intricate nonlinear patterns without predefining a functional form.
In the analyzed datasets, XGBoost demonstrated comparable or superior test set performance to DoE in several cases, particularly in datasets with complex or nonlinear relationships. While design of experiments (DoE) methodologies remain effective for structured experimental planning and hypothesis testing, their limited capacity to generalize beyond the training data can be restrictive in certain contexts. Conversely, machine learning models such as XGBoost offer increased flexibility by identifying hidden patterns in the data, which proved beneficial in our study. These findings are in line with the work of Zhai et al. [39], who applied ML models to predict volatile fatty acid concentrations in anaerobic sludge fermentation, achieving an of up to 0.949.
Our research aligns with the findings of Pensupa et al. [24], who employed Gaussian process regression to forecast biomass output from Yarrowia lipolytica fermentation and identified 14 critical predictors. Our study demonstrates that the integration of data mining and machine learning can provide significant insights into fermentation performance, especially when utilizing secondary or heterogeneous datasets.
While XGBoost did not consistently outperform the DoE approach across all datasets, it demonstrated comparable or better performance in several cases, highlighting its potential as a robust, data-driven alternative for modeling bioprocess outcomes.
In summary, ML models like XGBoost provide a data-driven enhancement to traditional design of experiments, enabling improved generalization and predictive accuracy in complex fermentation systems. Subsequent efforts should incorporate feature interpretation methodologies such as SHAP, examine ensemble and hybrid models, and analyze time-series dynamics to enhance prediction and process optimization in biotechnological applications. Moreover, the incorporation of machine learning—especially via regression models and advanced data analysis methodologies—offers considerable potential for enhancing xylanase production. Assessing critical variables influencing enzyme activity might improve efficiency and cost-effectiveness, thereby matching with overreaching objectives in biotechnology including sustainability and food security.
5. Conclusions
This study highlights the effectiveness of XGBoost in predicting experimental outcomes compared to traditional DoE methods. The results show that machine learning models can significantly improve prediction accuracy and reduce error metrics, making them suitable for complex, data-driven experimental processes. However, the limitations of purely data-driven methods should not be overlooked, as they require extensive and high-quality datasets for optimal performance.
Although XGBoost demonstrated strong predictive capacity in several datasets, it did not consistently outperform DoE in all cases. The comparative advantage of each method appeared to depend on the dataset size, experimental design type, and variability in input parameters.
While DoE methods remain valuable for structured experimental design, incorporating machine learning techniques such as XGBoost can enhance predictive power and efficiency. Future research could explore hybrid approaches that leverage the strengths of both methods, ensuring a balance between statistical rigor and predictive accuracy. Additionally, expanding the dataset and implementing feature selection techniques could further improve model generalization and reliability in real-world applications.
By integrating advanced machine learning techniques with established statistical methods, researchers can achieve more precise and reliable experimental predictions, ultimately enhancing decision-making processes in various scientific and industrial applications.
Author Contributions
Conceptualization, T.K.-G.; methodology, B.E.K.-G.; software, B.E.K.-G.; validation, B.E.K.-G. and T.K.-G.; formal analysis, B.E.K.-G.; investigation, B.E.K.-G.; resources, M.A.E.; data curation, M.A.E.; writing—original draft preparation, M.A.E., B.E.K.-G. and T.K.-G.; writing—review and editing, T.K.-G.; visualization, M.A.E.; supervision, T.K.-G.; project administration, T.K.-G. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the Scientific Research Projects Coordination Unit of İzmir Demokrasi University. Project No: TEZ-MHF/2501.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The Python code for the XGBoost-based regression analysis, along with sample data and result generation tools, is available at: https://github.com/basakesin/xylanase-model (accessed on 28 March 2025).
Acknowledgments
During the preparation of this manuscript, the authors used GPT-4o and Quillbot Premium for the purposes of language editing and text fluency. The authors have reviewed and edited the output and take full responsibility for the content of this publication.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| AI | Artificial intelligence |
| ANN | Artificial neural network |
| BBD | Box–Behnken design |
| CCD | Central composite design |
| CNN | Convolutional neural network |
| DoE | Design of experiments |
| GPR | Gaussian process regression |
| IU/mL | International units per milliliter |
| MAPE | Mean absolute percentage error |
| ML | Machine learning |
| MLR | Multiple linear regression |
| R2 | Coefficient of determination |
| RMSE | Root mean squared error |
| RSM | Response surface methodology |
| SHAP | Shapley additive explanations |
| SVR | Support vector regression |
| VFAs | Volatile fatty acids |
| XGBoost | Extreme gradient boosting |
Appendix A

Table A1.
Dataset No: 1.
Table A1.
Dataset No: 1.
| Glucose (g/L) | (NH4)HPO4 (g/L) | K2HPO4 (g/L) | KH2PO4 (g/L) | MgSO4 (g/L) | Experimental Xylanase Activity | Predicted Xylanase Activity |
|---|---|---|---|---|---|---|
| 20 | 10 | 18 | 6 | 3 | 1.699 | 1.870 |
| 10 | 2 | 18 | 6 | 3 | 1.586 | 1.398 |
| 10 | 10 | 18 | 1 | 3 | 1.782 | 1.859 |
| 10 | 10 | 18 | 6 | 0.5 | 1.843 | 1.843 |
| 10 | 10 | 5 | 1 | 0.5 | 1.738 | 1.673 |
| 20 | 10 | 5 | 1 | 3 | 1.647 | 1.645 |
| 20 | 2 | 5 | 1 | 0.5 | 1.266 | 1.314 |
| 15 | 6 | 11.5 | 3.5 | 1.75 | 1.829 | 0.874 |
| 20 | 2 | 18 | 6 | 0.5 | 1.578 | 1.540 |
| 10 | 2 | 18 | 1 | 0.5 | 1.683 | 1.782 |
| 15 | 6 | 11.5 | 3.5 | 1.75 | 1.920 | 1.874 |
| 10 | 2 | 5 | 1 | 3 | 1.153 | 1.172 |
| 20 | 2 | 18 | 1 | 3 | 1.525 | 1.501 |
| 20 | 10 | 18 | 1 | 0.5 | 2.406 | 2.254 |
| 20 | 10 | 5 | 6 | 0.5 | 2.185 | 2.167 |
| 20 | 2 | 5 | 6 | 3 | 1.399 | 1.414 |
| 10 | 10 | 5 | 6 | 3 | 1.840 | 1.772 |
| 10 | 2 | 5 | 6 | 0.5 | 1.625 | 1.695 |

Table A2.
Dataset No: 2.
Table A2.
Dataset No: 2.
| (NH4)2HPO4 (g/L) | Urea (g/L) | Malt Sprout (g/L) | Corn Cobs (g/L) | Wheat Bran (g/L) | Experimental Xylanase Activity | Predicted Xylanase Activity |
|---|---|---|---|---|---|---|
| 2.6 | 0.9 | 6 | 12 | 16 | 428.12 | 384.32 |
| 5.4 | 0.9 | 6 | 12 | 6 | 568.79 | 479.86 |
| 2.6 | 2.1 | 6 | 12 | 6 | 649.09 | 479.86 |
| 5.4 | 2.1 | 6 | 12 | 16 | 544.56 | 384.32 |
| 2.6 | 0.9 | 18 | 12 | 6 | 589.81 | 479.86 |
| 5.4 | 0.9 | 18 | 12 | 16 | 483.24 | 384.32 |
| 2.6 | 2.1 | 18 | 12 | 16 | 484.91 | 384.32 |
| 5.4 | 2.1 | 18 | 12 | 6 | 536.66 | 479.86 |
| 2.6 | 0.9 | 6 | 24 | 6 | 569.31 | 495.54 |
| 5.4 | 0.9 | 6 | 24 | 16 | 750.47 | 718.72 |
| 2.6 | 2.1 | 6 | 24 | 16 | 869.88 | 718.72 |
| 5.4 | 2.1 | 6 | 24 | 6 | 513.49 | 495.54 |
| 2.6 | 0.9 | 18 | 24 | 16 | 825.29 | 718.72 |
| 5.4 | 0.9 | 18 | 24 | 6 | 695.87 | 495.54 |
| 2.6 | 2.1 | 18 | 24 | 6 | 611.15 | 495.54 |
| 5.4 | 2.1 | 18 | 24 | 16 | 815.58 | 718.72 |
| 5.4 | 1.5 | 12 | 18 | 11 | 723.41 | 654.25 |
| 2.6 | 1.5 | 12 | 18 | 11 | 678.61 | 654.25 |
| 4 | 2.1 | 12 | 18 | 11 | 674.75 | 654.25 |
| 4 | 0.9 | 12 | 18 | 11 | 614.37 | 654.25 |
| 4 | 1.5 | 18 | 18 | 11 | 710.87 | 654.25 |
| 4 | 1.5 | 6 | 18 | 11 | 705.62 | 654.25 |
| 4 | 1.5 | 12 | 24 | 11 | 694.44 | 677.02 |
| 4 | 1.5 | 12 | 12 | 11 | 484.97 | 501.98 |
| 4 | 1.5 | 12 | 18 | 16 | 637.56 | 616.27 |
| 4 | 1.5 | 12 | 18 | 6 | 531.08 | 552.45 |

Table A3.
Dataset No: 3.
Table A3.
Dataset No: 3.
| Xylan (g/L) | pH | Cultivation Time (h) | Experimental Xylanase Activity | Predicted Xylanase Activity |
|---|---|---|---|---|
| 5 | 8 | 24 | 11.11 | 8.62 |
| 5 | 8 | 48 | 16.20 | 18.45 |
| 5 | 8 | 72 | 15.81 | 16.54 |
| 5 | 8.5 | 24 | 8.17 | 7.62 |
| 5 | 8.5 | 48 | 17.04 | 17.96 |
| 5 | 8.5 | 72 | 16.12 | 16.56 |
| 5 | 9 | 24 | 6.75 | 8.26 |
| 5 | 9 | 48 | 21.54 | 19.11 |
| 5 | 9 | 72 | 18.65 | 18.22 |
| 7.5 | 8 | 24 | 6.75 | 8.31 |
| 7.5 | 8 | 48 | 18.63 | 19.39 |
| 7.5 | 8 | 72 | 22.45 | 18.73 |
| 7.5 | 8.5 | 24 | 8.10 | 6.63 |
| 7.5 | 8.5 | 48 | 18.73 | 18.22 |
| 7.5 | 8.5 | 72 | 17.04 | 18.07 |
| 7.5 | 9 | 24 | 4.70 | 6.58 |
| 7.5 | 9 | 48 | 18.36 | 18.69 |
| 7.5 | 9 | 72 | 18.97 | 19.05 |
| 10 | 8 | 24 | 5.59 | 6.39 |
| 10 | 8 | 48 | 18.75 | 18.69 |
| 10 | 8 | 72 | 19.12 | 19.28 |
| 10 | 8.5 | 24 | 4.44 | 3.99 |
| 10 | 8.5 | 48 | 17.64 | 16.84 |
| 10 | 8.5 | 72 | 16.60 | 17.94 |
| 10 | 9 | 24 | 4.05 | 3.27 |
| 10 | 9 | 48 | 17.14 | 16.63 |
| 10 | 9 | 72 | 17.93 | 18.24 |

Table A4.
Dataset No: 4.
Table A4.
Dataset No: 4.
| X1 (Xylan) (g/L) | X2 (Casein) (g/L) | X3 (NH4Cl) (g/L) | Observed XA (nkat/mL) | Predicted XA (nkat/mL) |
|---|---|---|---|---|
| 2.5 | 1 | 0.3 | 1428.00 | 1397.80 |
| 7.5 | 1 | 0.3 | 5.30 | 36.10 |
| 2.5 | 2 | 0.3 | 1905.50 | 1936.34 |
| 7.5 | 2 | 0.3 | 253.70 | 222.86 |
| 2.5 | 1 | 1.3 | 1565.10 | 1595.94 |
| 7.5 | 1 | 1.3 | 22.10 | −8.73 |
| 2.5 | 2 | 1.3 | 2184.90 | 2154.10 |
| 7.5 | 2 | 1.3 | 166.10 | 196.60 |
| 5.0 | 1.5 | 0.8 | 925.40 | 941.30 |
| 5.0 | 1.5 | 0.8 | 942.60 | 941.30 |
| 5.0 | 1.5 | 0.8 | 938.40 | 941.30 |

Table A5.
Dataset No: 5.
Table A5.
Dataset No: 5.
| (NH4)2HPO4 (g/L) | K2HPO4 (g/L) | MgSO4 (g/L) | Experimental Xylanase Activity | Predicted Xylanase Activity |
|---|---|---|---|---|
| 10 | 7 | 3 | 1.871 | 1.880 |
| 10 | 7 | 3 | 1.898 | 1.880 |
| 10 | 18 | 1.5 | 2.423 | 2.489 |
| 10 | 18 | 3 | 2.292 | 2.322 |
| 1 | 12.5 | 2.25 | 1.443 | 1.523 |
| 7 | 12.5 | 0.75 | 2.456 | 2.431 |
| 7 | 12.5 | 2.25 | 2.219 | 2.230 |
| 7 | 12.5 | 2.25 | 2.219 | 2.230 |
| 10 | 18 | 3 | 2.290 | 2.322 |
| 10 | 7 | 1.5 | 2.296 | 2.263 |
| 7 | 12.5 | 3.75 | 1.992 | 2.029 |
| 4 | 7 | 3 | 1.705 | 1.636 |
| 7 | 12.5 | 2.25 | 2.257 | 2.230 |
| 4 | 7 | 3 | 1.645 | 1.636 |
| 13 | 12.5 | 2.25 | 2.141 | 2.157 |
| 10 | 7 | 1.5 | 2.169 | 2.263 |
| 7 | 1.5 | 2.25 | 1.625 | 1.686 |
| 7 | 1.5 | 2.25 | 1.683 | 1.686 |
| 7 | 23.5 | 2.25 | 2.345 | 2.354 |
| 7 | 12.5 | 3.75 | 2.042 | 2.029 |
| 4 | 18 | 3 | 2.130 | 2.079 |
| 4 | 7 | 1.5 | 1.937 | 1.873 |
| 7 | 23.5 | 2.25 | 2.395 | 2.354 |
| 4 | 18 | 3 | 2.081 | 2.079 |
| 4 | 18 | 1.5 | 2.031 | 2.099 |
| 1 | 12.5 | 2.25 | 1.496 | 1.523 |
| 7 | 12.5 | 0.75 | 2.390 | 2.431 |
| 10 | 18 | 1.5 | 2.555 | 2.489 |
| 4 | 7 | 1.5 | 1.948 | 1.873 |
| 4 | 18 | 1.5 | 2.150 | 2.099 |
| 7 | 12.5 | 2.25 | 2.207 | 2.230 |
| 7 | 12.5 | 2.25 | 2.232 | 2.230 |
| 13 | 12.5 | 2.25 | 2.249 | 2.157 |
| 7 | 12.5 | 2.25 | 2.224 | 2.230 |

Table A6.
Dataset No: 6.
Table A6.
Dataset No: 6.
| (NH4)2HPO4 (g/L) | Urea (g/L) | Malt Sprout (g/L) | Experimental Xylanase Activity | Predicted Xylanase Activity |
|---|---|---|---|---|
| 0.4 | 0.3 | 0.4 | 413.45 | 367.28 |
| 2.6 | 0.3 | 0.4 | 395.83 | 417.70 |
| 0.4 | 0.9 | 0.4 | 764.69 | 724.98 |
| 2.6 | 0.9 | 0.4 | 770.09 | 775.39 |
| 0.4 | 0.3 | 10 | 666.32 | 721.08 |
| 2.6 | 0.3 | 10 | 791.83 | 771.49 |
| 0.4 | 0.9 | 10 | 815.09 | 819.69 |
| 2.6 | 0.9 | 10 | 850.41 | 870.11 |
| 0.4 | 0.6 | 5.2 | 720.35 | 746.87 |
| 2.6 | 0.6 | 5.2 | 823.81 | 797.29 |
| 1.5 | 0.3 | 5.2 | 656.61 | 646.49 |
| 1.5 | 0.9 | 5.2 | 864.53 | 874.65 |
| 1.5 | 0.3 | 0.4 | 378.05 | 436.73 |
| 1.5 | 0.3 | 10 | 719.73 | 661.02 |

Table A7.
Dataset No: 7.
Table A7.
Dataset No: 7.
| A: Wheat Bran (g/L) | B: Yeast Extract + Peptone (g/L) | C: Temperature (°C) | Observed Xylanase Activity (IU/mL) | Predicted Xylanase Activity (IU/mL) |
|---|---|---|---|---|
| 10 | 10 | 25 | 64.44 | 63.38 |
| 2 | 10 | 20 | 10.83 | 11.11 |
| 2 | 10 | 30 | 27.04 | 25.41 |
| 18 | 2 | 25 | 41.61 | 41.81 |
| 2 | 18 | 25 | 21.05 | 22.85 |
| 18 | 18 | 25 | 29.64 | 30.09 |
| 10 | 2 | 30 | 30.14 | 32.24 |
| 10 | 18 | 20 | 14.60 | 12.54 |
| 10 | 18 | 30 | 41.93 | 41.76 |
| 10 | 10 | 25 | 64.08 | 63.38 |
| 10 | 2 | 20 | 33.01 | 33.17 |
| 18 | 10 | 20 | 23.04 | 24.67 |
| 2 | 2 | 25 | 22.68 | 22.23 |
| 10 | 10 | 25 | 64.03 | 63.38 |
| 10 | 10 | 25 | 61.60 | 63.38 |
| 18 | 10 | 30 | 38.95 | 38.67 |
| 10 | 10 | 25 | 62.73 | 63.38 |
References
- Collins, T.; Gerday, C.; Feller, G. Xylanases, xylanase families and extremophilic xylanases. FEMS Microbiol. Rev. 2005, 29, 3–23. [Google Scholar] [CrossRef] [PubMed]
- Dobrev, G.T.; Pishtiyski, I.G.; Stanchev, V.S.; Mircheva, R. Optimization of nutrient medium containing agricultural wastes for xylanase production by Aspergillus niger B03 using optimal composite experimental design. Bioresour. Technol. 2007, 98, 2671–2678. [Google Scholar] [CrossRef] [PubMed]
- Farliahati, M.R.; Ramanan, R.N.; Mohamad, R.; Puspaningsih, N.N.T.; Ariff, A.B. Enhanced production of xylanase by recombinant Escherichia coli DH5α through optimization of medium composition using response surface methodology. Ann. Microbiol. 2010, 60, 279–285. [Google Scholar] [CrossRef]
- Bocchini, D.A.; Alves-Prado, H.F.; Baida, L.C.; Roberto, I.C.; Gomes, E.; Da Silva, R. Optimization of xylanase production by Bacillus circulans D1 in submerged fermentation using response surface methodology. Process. Biochem. 2002, 38, 727–731. [Google Scholar] [CrossRef]
- Limkar, M.B.; Pawar, S.V.; Rathod, V.K. Statistical optimization of xylanase and alkaline protease co-production by Bacillus spp using Box-Behnken Design under submerged fermentation using wheat bran as a substrate. Biocatal. Agric. Biotechnol. 2019, 17, 455–464. [Google Scholar] [CrossRef]
- Patel, K.; Dudhagara, P. Optimization of xylanase production by Bacillus tequilensis strain UD-3 using economical agricultural substrate and its application in rice straw pulp bleaching. Biocatal. Agric. Biotechnol. 2020, 30, 101846. [Google Scholar] [CrossRef]
- Prade, R.A. Xylanases: From biology to biotechnology. Biotechnol. Genet. Eng. Rev. 1996, 13, 101–132. [Google Scholar] [CrossRef]
- Sharma, D.; Sahu, S.; Singh, G.; Arya, S.K. An eco-friendly process for xylose production from waste of pulp and paper industry with xylanase catalyst. Sustain. Chem. Environ. 2023, 3, 100024. [Google Scholar] [CrossRef]
- Garai, D.; Kumar, V. Response surface optimization for xylanase with high volumetric productivity by indigenous alkali tolerant Aspergillus candidus under submerged cultivation. 3 Biotech 2013, 3, 127–136. [Google Scholar] [CrossRef]
- Yıldırım, A.; İlhan Ayışığı, E.; Düzel, A.; Mayfield, S.P.; Sargın, S. Optimization of culture conditions for the production and activity of recombinant xylanase from microalgal platform. Biochem. Eng. J. 2023, 197, 108967. [Google Scholar] [CrossRef]
- Bhardwaj, N.; Kumar, B.; Verma, P. A detailed overview of xylanases: An emerging biomolecule for current and future prospective. Bioresour. Bioprocess. 2019, 6, 40. [Google Scholar] [CrossRef]
- Huang, K.; Chu, Y.; Qin, X.; Zhang, J.; Bai, Y.; Wang, Y.; Luo, H.; Huang, H.; Su, X. Recombinant production of two xylanase-somatostatin fusion proteins retaining somatostatin immunogenicity and xylanase activity in Pichia pastoris. Appl. Microbiol. Biotechnol. 2021, 105, 4167–4175. [Google Scholar] [CrossRef] [PubMed]
- Kallel, F.; Driss, D.; Chaari, F.; Zouari-Ellouzi, S.; Chaabouni, M.; Ghorbel, R.; Chaabouni, S.E. Statistical optimization of low-cost production of an acidic xylanase by Bacillus mojavensis UEB-FK: Its potential applications. Biocatal. Agric. Biotechnol. 2016, 5, 1–10. [Google Scholar] [CrossRef]
- Abdella, A.; Segato, F.; Wilkins, M.R. Optimization of process parameters and fermentation strategy for xylanase production in a stirred tank reactor using a mutant Aspergillus nidulans strain. Biotechnol. Rep. 2020, 26, e00457. [Google Scholar] [CrossRef]
- Ajijolakewu, A.K.; Leh, C.P.; Abdullah, W.N.W.; Lee, C.K. Optimization of production conditions for xylanase production by newly isolated strain Aspergillus niger through solid state fermentation of oil palm empty fruit bunches. Biocatal. Agric. Biotechnol. 2017, 11, 239–247. [Google Scholar] [CrossRef]
- Siwach, R.; Sharma, S.; Khan, A.A.; Kumar, A.; Agrawal, S. Optimization of xylanase production by Bacillus sp. MCC2212 under solid-state fermentation using response surface methodology. Biocatal. Agric. Biotechnol. 2024, 57, 103085. [Google Scholar] [CrossRef]
- Liao, H.; Ying, W.; Li, X.; Zhu, J.; Xu, Y.; Zhang, J. Optimized production of xylooligosaccharides from poplar: A biorefinery strategy with sequential acetic acid/sodium acetate hydrolysis followed by xylanase hydrolysis. Bioresour. Technol. 2022, 347, 126683. [Google Scholar] [CrossRef]
- Liu, C.; Sun, Z.T.; Du, J.H.; Wang, J. Response surface optimization of fermentation conditions for producing xylanase by Aspergillus niger SL-05. J. Ind. Microbiol. Biotechnol. 2008, 35, 703–711. [Google Scholar] [CrossRef]
- Iram, A.; Cekmecelioglu, D.; Demirci, A. Optimization of the fermentation parameters to maximize the production of cellulases and xylanases using DDGS as the main feedstock in stirred tank bioreactors. Biocatal. Agric. Biotechnol. 2022, 45, 102514. [Google Scholar] [CrossRef]
- Ezeilo, U.R.; Wahab, R.A.; Mahat, N.A. Optimization studies on cellulase and xylanase production by Rhizopus oryzae UC2 using raw oil palm frond leaves as substrate under solid state fermentation. Renew. Energy 2020, 156, 1301–1312. [Google Scholar] [CrossRef]
- Uhoraningoga, A.; Kinsella, G.K.; Henehan, G.T.; Ryan, B.J. The goldilocks approach: A review of employing design of experiments in prokaryotic recombinant protein production. Bioengineering 2018, 5, 89. [Google Scholar] [CrossRef] [PubMed]
- Adhyaru, D.N.; Bhatt, N.S.; Modi, H.A.; Divecha, J. Insight on xylanase from Aspergillus tubingensis FDHN1: Production, high yielding recovery optimization through statistical approach and application. Biocatal. Agric. Biotechnol. 2016, 6, 51–57. [Google Scholar] [CrossRef]
- Pham, P.L.; Taillandier, P.; Delmas, M.; Strehaiano, P. Optimization of a culture medium for xylanase production by Bacillus sp. using statistical experimental designs. World J. Microbiol. Biotechnol. 1997, 14, 185–190. [Google Scholar] [CrossRef]
- Pensupa, N.; Treebuppachartsakul, T.; Pechprasarn, S. Machine learning models using data mining for biomass production from Yarrowia lipolytica fermentation. Fermentation 2023, 9, 239. [Google Scholar] [CrossRef]
- Wu, D.; Xu, Y.; Xu, F.; Shao, M.; Huang, M. Machine learning algorithms for in-line monitoring during yeast fermentations based on Raman spectroscopy. Vib. Spectrosc. 2024, 132, 103672. [Google Scholar] [CrossRef]
- Jeong, J.; Kim, S. Application of machine learning for quantitative analysis of industrial fermentation using image processing. Food Sci. Biotechnol. 2025, 34, 373–381. [Google Scholar] [CrossRef]
- Bowler, A.; Escrig, J.; Pound, M.; Watson, N. Predicting alcohol concentration during beer fermentation using ultrasonic measurements and machine learning. Fermentation 2021, 7, 34. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. Xgboost: A scalable tree boosting system. In Proceedings of the 22nd Acm Sigkdd International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar]
- Nielsen, D. Tree Boosting with Xgboost-Why Does Xgboost Win “Every” Machine Learning Competition? Master’s Thesis, NTNU, Taipei City, Taiwan, 2016. [Google Scholar]
- Zhang, D.; Chen, H.D.; Zulfiqar, H.; Yuan, S.S.; Huang, Q.L.; Zhang, Z.Y.; Deng, K.J. iBLP: An XGBoost-Based Predictor for Identifying Bioluminescent Proteins. Comput. Math. Methods Med. 2021, 2021, 6664362. [Google Scholar] [CrossRef]
- Dimitrakopoulos, G.N.; Vrahatis, A.G.; Plagianakos, V.; Sgarbas, K. Pathway analysis using XGBoost classification in Biomedical Data. In Proceedings of the 10th Hellenic Conference on Artificial Intelligence, Patras, Greece, 9–12 July 2018; pp. 1–6. [Google Scholar] [CrossRef]
- Yang, A.; Sun, S.; Mi, H.; Wang, W.; Liu, J.; Kong, Z.Y. Interpretable feedforward neural network and XGBoost-based algorithms to predict CO2 solubility in ionic liquids. Ind. Eng. Chem. Res. 2024, 63, 8293–8305. [Google Scholar] [CrossRef]
- Al-Jamimi, H.A.; BinMakhashen, G.M.; Saleh, T.A. From data to clean water: XGBoost and Bayesian optimization for advanced wastewater treatment with ultrafiltration. Neural Comput. Appl. 2024, 36, 18863–18877. [Google Scholar] [CrossRef]
- Xie, H.; Deng, Y.m.; Li, J.y.; Xie, K.h.; Tao, T.; Zhang, J.f. Predicting the risk of primary Sjögren’s syndrome with key N7-methylguanosine-related genes: A novel XGBoost model. Heliyon 2024, 10, e31307. [Google Scholar] [CrossRef] [PubMed]
- Shwartz-Ziv, R.; Armon, A. Tabular data: Deep learning is not all you need. Inf. Fusion 2022, 81, 84–90. [Google Scholar] [CrossRef]
- Guo, R.; Zhao, Z.; Wang, T.; Liu, G.; Zhao, J.; Gao, D. Degradation state recognition of piston pump based on ICEEMDAN and XGBoost. Appl. Sci. 2020, 10, 6593. [Google Scholar] [CrossRef]
- Grinsztajn, L.; Oyallon, E.; Varoquaux, G. Why do tree-based models still outperform deep learning on typical tabular data? Adv. Neural Inf. Process. Syst. 2022, 35, 507–520. [Google Scholar] [CrossRef]
- Stevanović, S.; Dashti, H.; Milošević, M.; Al-Yakoob, S.; Stevanović, D. Comparison of ANN and XGBoost surrogate models trained on small numbers of building energy simulations. PLoS ONE 2024, 19, e0312573. [Google Scholar] [CrossRef]
- Zhai, S.; Chen, K.; Yang, L.; Li, Z.; Yu, T.; Chen, L.; Zhu, H. Applying machine learning to anaerobic fermentation of waste sludge using two targeted modeling strategies. Sci. Total Environ. 2024, 916, 170232. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).