Role of Compensatory miRNA Networks in Cognitive Recovery from Heart Failure
Abstract
1. Introduction
2. Results
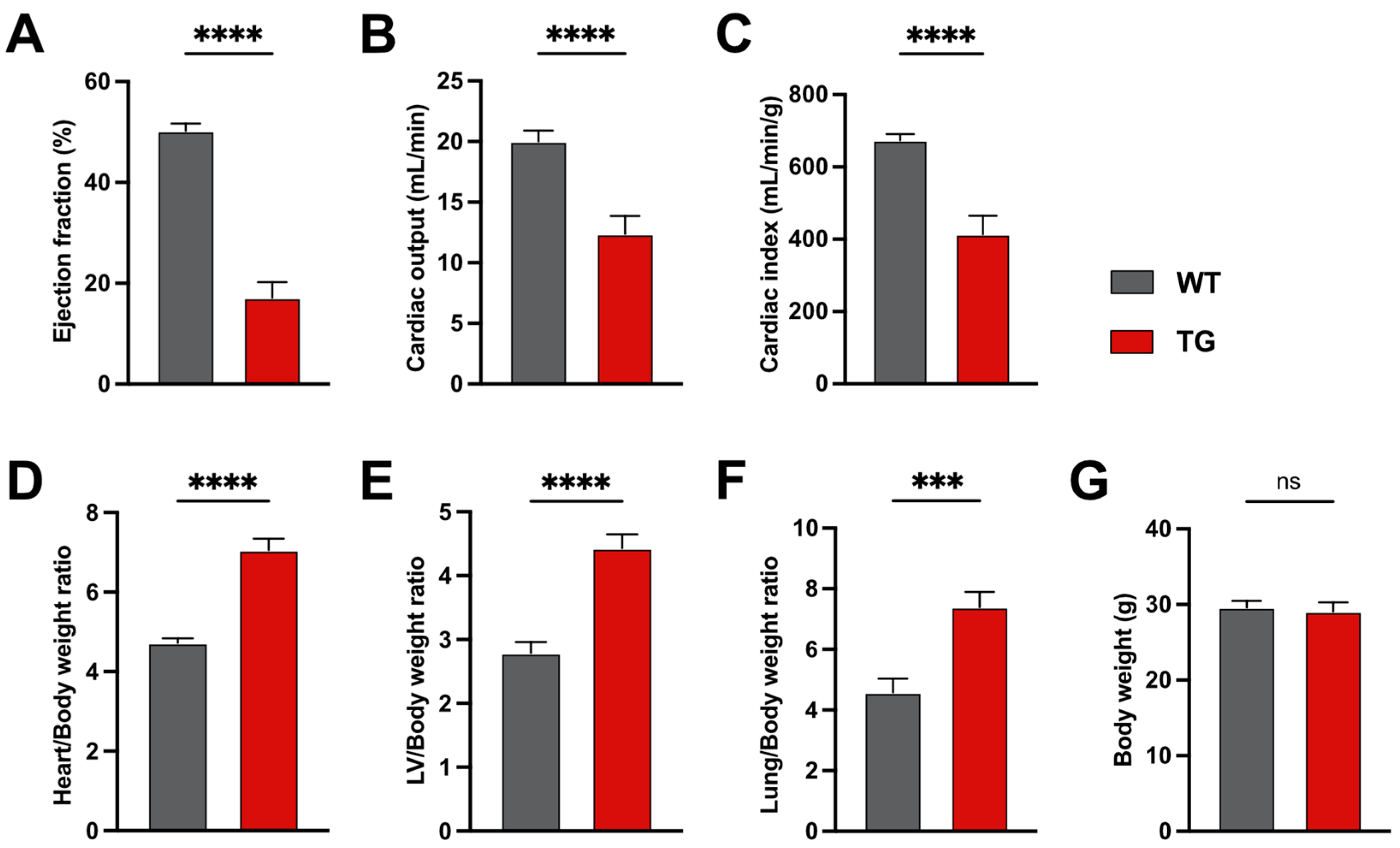
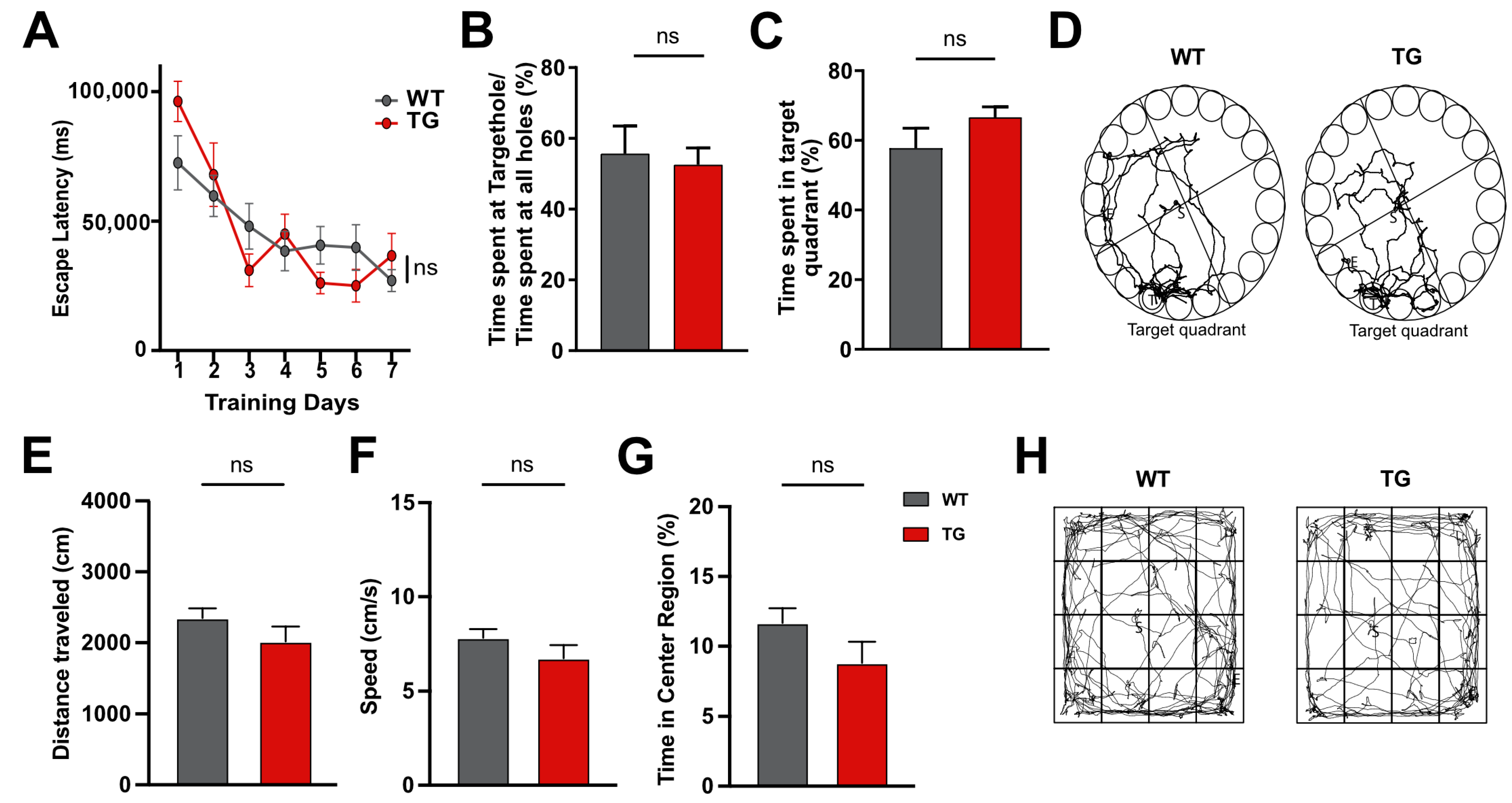
2.1. Six-Month-Old CamKIIδC Mice Exhibit No Memory Impairments Despite Heart Failure
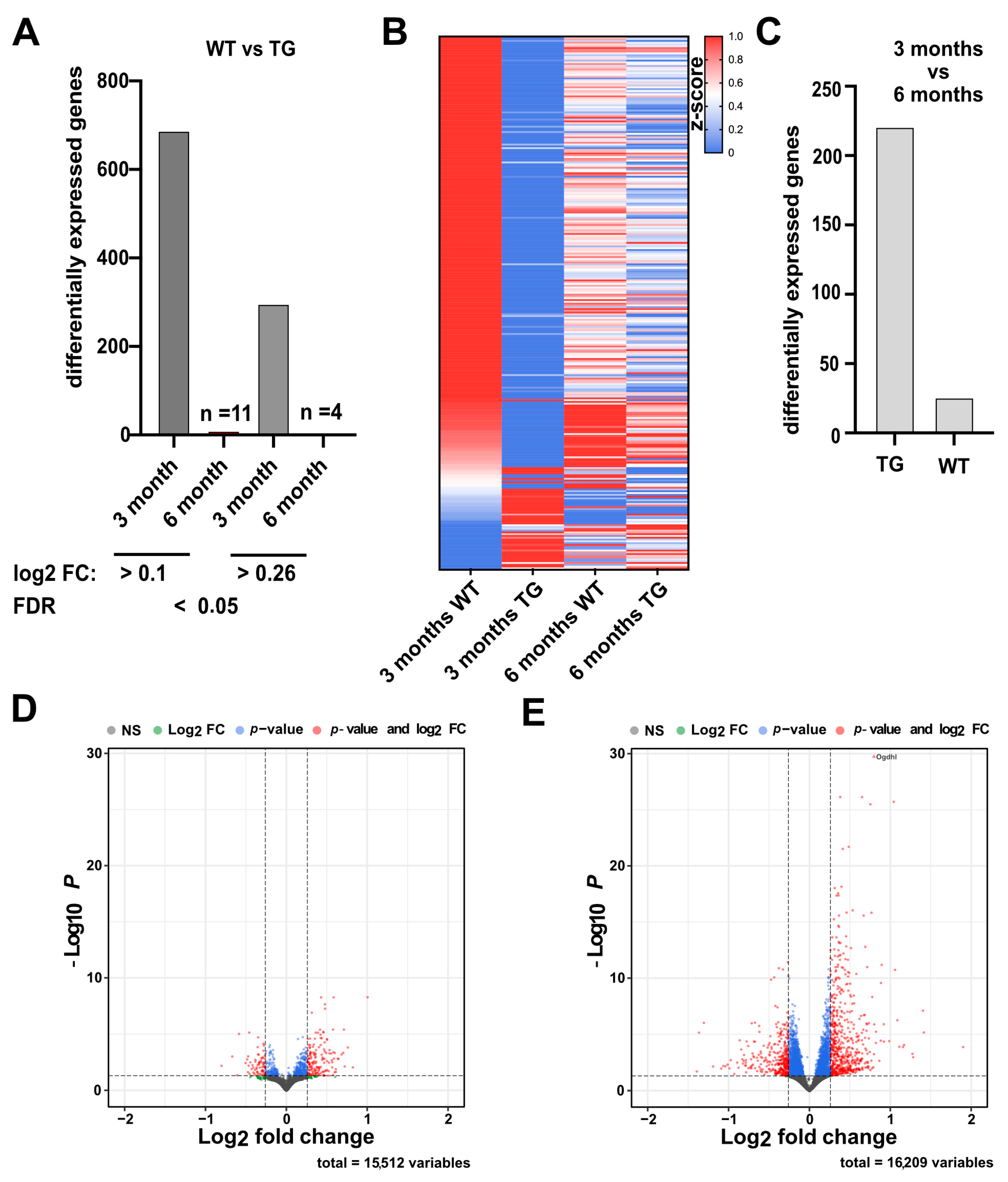
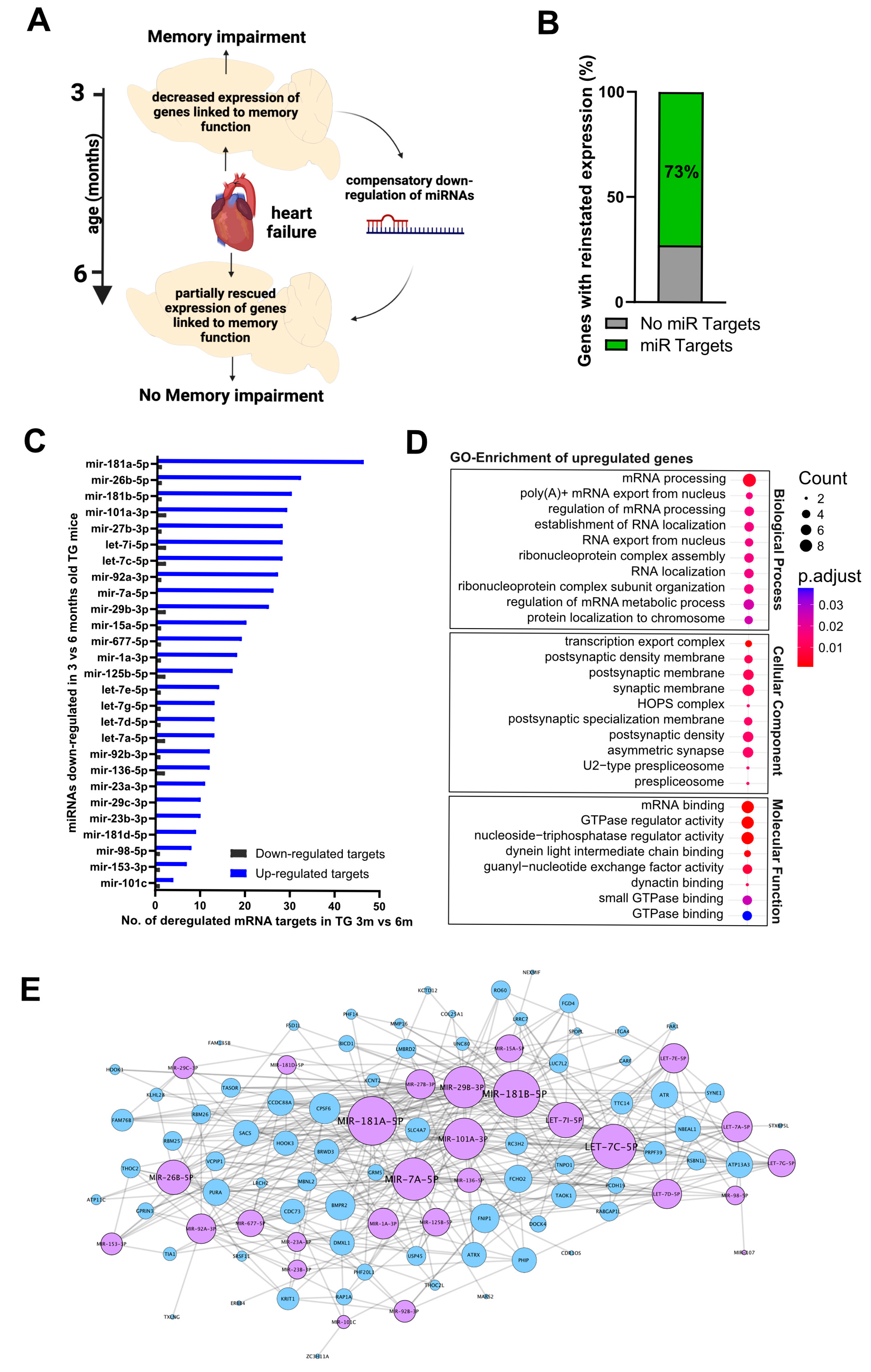
2.2. Hippocampal Gene Expression Reveals a Potential Compensatory Mechanism
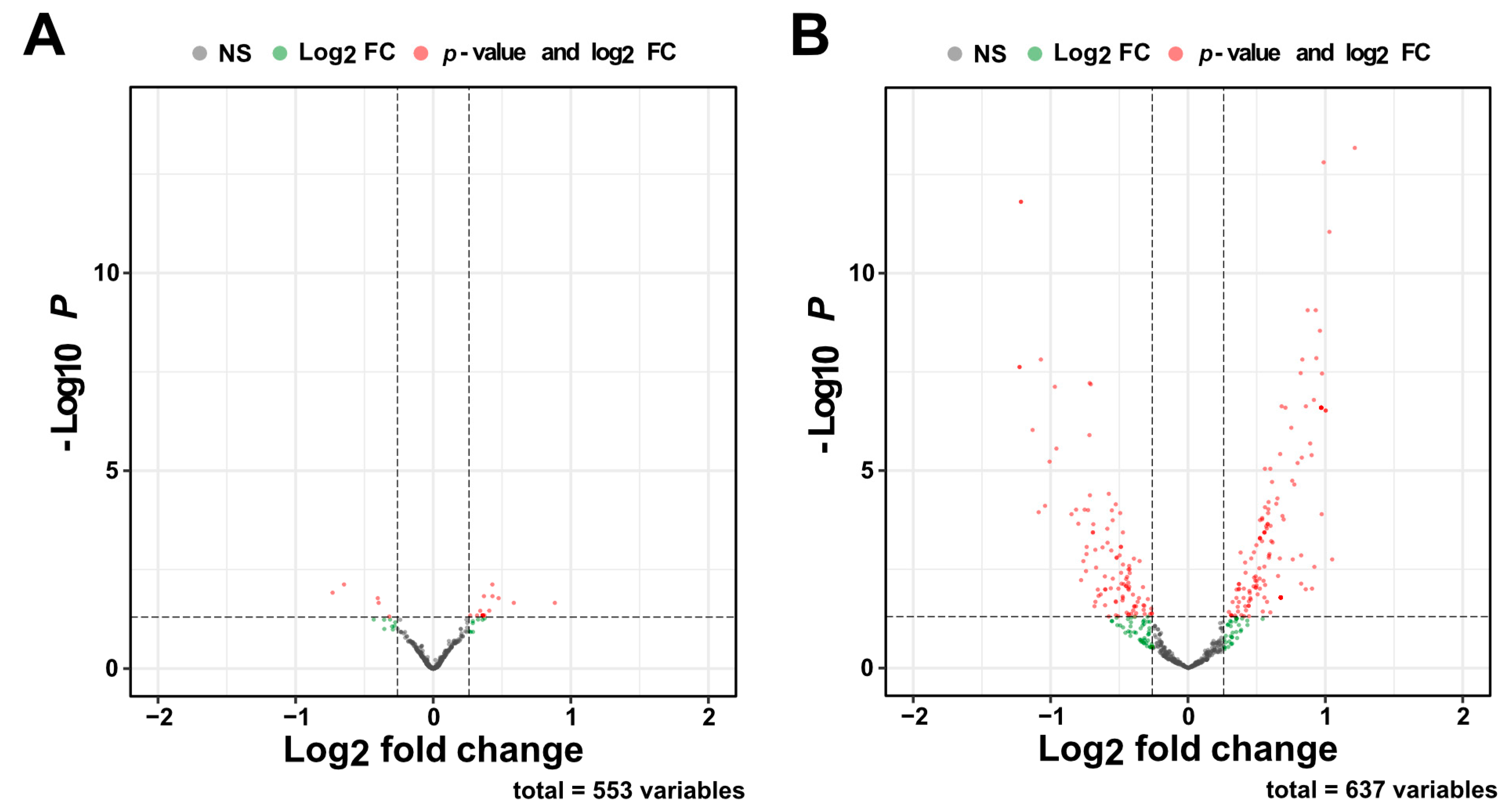
2.3. A Compensatory miRNA Network May Regulate Gene Expression Recovery
3. Discussion
4. Materials and Methods
4.1. Animals and Tissue Preparation
4.2. Echocardiography
4.3. Open Field Test and Barnes Maze Experiment
4.4. RNA Isolation and Sequencing
4.5. Bioinformatics Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Shahim, B.; Kapelios, C.J.; Savarese, G.; Lund, L.H. Global Public Health Burden of Heart Failure: An Updated Review. Card. Fail. Rev. 2023, 9, e11. [Google Scholar] [CrossRef] [PubMed]
- Carpenter, A.E.; Jones, T.R.; Lamprecht, M.R.; Clarke, C.; Kang, I.H.; Friman, O.; Guertin, D.A.; Chang, J.H.; Lindquist, R.A.; Moffat, J.; et al. CellProfiler: Image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006, 7, R100. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Yadav, S.K.; Palomares, J.A.; Park, B.; Joshi, S.H.; Ogren, J.A.; Macey, P.M.; Fonarow, G.C.; Harper, R.M.; Woo, M.A. Reduced regional brain cortical thickness in patients with heart failure. PLoS ONE 2015, 10, e0126595. [Google Scholar] [CrossRef] [PubMed]
- Forman, D.E.; Daniels, K.M.; Cahalin, L.P.; Zavin, A.; Allsup, K.; Cao, P.; Santhanam, M.; Joseph, J.; Arena, R.; Lazzari, A.; et al. Analysis of skeletal muscle gene expression patterns and the impact of functional capacity in patients with systolic heart failure. J. Card. Fail. 2014, 20, 422–430. [Google Scholar] [CrossRef]
- Rademaker, M.T.; Pilbrow, A.P.; Ellmers, L.J.; Palmer, S.C.; Davidson, T.; Mbikou, P.; Scott, N.J.A.; Permina, E.; Charles, C.J.; Endre, Z.H.; et al. Acute Decompensated Heart Failure and the Kidney: Physiological, Histological and Transcriptomic Responses to Development and Recovery. J. Am. Heart Assoc. 2021, 10, e021312. [Google Scholar] [CrossRef]
- Islam, M.R.; Lbik, D.; Sakib, M.S.; Hofmann, M.; Berulava, T.; Jiménez Mausbach, M.; Cha, J.; Goldberg, M.; Vakhtang, E.; Schiffmann, C.; et al. Epigenetic gene expression links heart failure to memory impairment. EMBO Mol. Med. 2021, 13. Epub ahed of print. [Google Scholar] [CrossRef]
- Valinezhad Orang, A.; Safaralizadeh, R.; Kazemzadeh-Bavili, M. Mechanisms of miRNA-Mediated Gene Regulation from Common Downregulation to mRNA-Specific Upregulation. Int. J. Genom. 2014, 2014, 970607. [Google Scholar] [CrossRef]
- Guo, H.; Ingolia, N.T.; Weissman, J.S.; Bartel, D.P. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 2010, 466, 835–840. [Google Scholar] [CrossRef]
- Manakov, S.A.; Morton, A.; Enright, A.J.; Grant, S.G. A Neuronal Transcriptome Response Involving Stress Pathways is Buffered by Neuronal microRNAs. Front. Neurosci. 2012, 6, 156. [Google Scholar] [CrossRef]
- Daws, S.E.; Jamieson, S.; de Nijs, L.; Jones, M.; Snijders, C.; Klengel, T.; Joseph, N.F.; Krauskopf, J.; Kleinjans, J.; Vinkers, C.H.; et al. MicroRNA regulation of persistent stress-enhanced memory. Mol. Psychiatry 2020, 25, 965–976. [Google Scholar] [CrossRef]
- Maier, L.S.; Zhang, T.; Chen, L.; DeSantiago, J.; Brown, J.H.; Bers, D.M. Transgenic CaMKIIdeltaC overexpression uniquely alters cardiac myocyte Ca2+ handling: Reduced SR Ca2+ load and activated SR Ca2+ release. Circ. Res. 2003, 92, 904–911. [Google Scholar] [CrossRef] [PubMed]
- Shivdasani, R.A. MicroRNAs: Regulators of gene expression and cell differentiation. Blood 2006, 108, 3646–3653. [Google Scholar] [CrossRef]
- van Wijk, N.; Zohar, K.; Linial, M. Challenging Cellular Homeostasis: Spatial and Temporal Regulation of miRNAs. Int. J. Mol. Sci. 2022, 23, 16152. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.R.; Kaurani, L.; Berulava, T.; Heilbronner, U.; Budde, M.; Centeno, T.P.; Elerdashvili, V.; Zafieriou, M.P.; Benito, E.; Sertel, S.M.; et al. A microRNA-signature that correlates with cognition and is a target against cognitive decline. EMBO Mol. Med. 2021; Online ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Farnsworth von Cederwald, B.; Josefsson, M.; Wåhlin, A.; Nyberg, L.; Karalija, N. Association of Cardiovascular Risk Trajectory With Cognitive Decline and Incident Dementia. Neurology 2022, 98, e2013–e2022. [Google Scholar] [CrossRef]
- van Gennip, A.C.E.; van Sloten, T.T.; Fayosse, A.; Sabia, S.; Singh-Manoux, A. Age at cardiovascular disease onset, dementia risk, and the role of lifestyle factors. Alzheimer’s Dement. 2024, 20, 1693–1702. [Google Scholar] [CrossRef]
- Liang, X.; Huang, Y.; Han, X. Associations between coronary heart disease and risk of cognitive impairment: A meta-analysis. Brain Behav. 2021, 11, e02108. [Google Scholar] [CrossRef]
- Blount, G.S.; Coursey, L.; Kocerha, J. MicroRNA Networks in Cognition and Dementia. Cells 2022, 11, 1882. [Google Scholar] [CrossRef]
- Mendell, J.T.; Olson, E.N. MicroRNAs in stress signaling and human disease. Cell 2012, 148, 1172–1187. [Google Scholar] [CrossRef]
- Ripa, R.; Dolfi, L.; Terrigno, M.; Pandolfini, L.; Savino, A.; Arcucci, V.; Groth, M.; Terzibasi Tozzini, E.; Baumgart, M.; Cellerino, A. MicroRNA miR-29 controls a compensatory response to limit neuronal iron accumulation during adult life and aging. BMC Biol. 2017, 15, 9. [Google Scholar] [CrossRef]
- Kolodziej, F.; McDonagh, B.; Burns, N.; Goljanek-Whysall, K. MicroRNAs as the Sentinels of Redox and Hypertrophic Signalling. Int. J. Mol. Sci. 2022, 23, 14716. [Google Scholar] [CrossRef]
- Rusu-Nastase, E.G.; Lupan, A.M.; Marinescu, C.I.; Neculachi, C.A.; Preda, M.B.; Burlacu, A. MiR-29a Increase in Aging May Function as a Compensatory Mechanism Against Cardiac Fibrosis Through SERPINH1 Downregulation. Front. Cardiovasc. Med. 2022, 8, 810241. [Google Scholar] [CrossRef]
- Rodriguez-Ortiz, C.J.; Baglietto-Vargas, D.; Martinez-Coria, H.; LaFerla, F.M.; Kitazawa, M. Upregulation of miR-181 decreases c-Fos and SIRT-1 in the hippocampus of 3xTg-AD mice. J. Alzheimers Dis. 2014, 42, 1229–1238. [Google Scholar] [CrossRef] [PubMed]
- Ansari, A.; Maffioletti, E.; Milanesi, E.; Marizzoni, M.; Frisoni, G.B.; Blin, O.; Richardson, J.C.; Bordet, R.; Forloni, G.; Gennarelli, M.; et al. miR-146a and miR-181a are involved in the progression of mild cognitive impairment to Alzheimer’s disease. Neurobiol. Aging 2019, 82, 102–109. [Google Scholar] [CrossRef]
- Khanna, A.; Muthusamy, S.; Liang, R.; Sarojini, H.; Wang, E. Gain of survival signaling by down-regulation of three key miRNAs in brain of calorie-restricted mice. Aging 2011, 3, 223–236. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, Y.B.; Lu, Y.; Yue, S.; Xu, L.J.; Xiong, X.X.; White, R.E.; Sun, X.; Giffard, R.G. miR-181 regulates GRP78 and influences outcome from cerebral ischemia in vitro and in vivo. Neurobiol. Dis. 2012, 45, 555–563. [Google Scholar] [CrossRef]
- Moon, J.M.; Xu, L.; Giffard, R.G. Inhibition of microRNA-181 reduces forebrain ischemia-induced neuronal loss. J. Cereb. Blood Flow. Metab. 2013, 33, 1976–1982. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.J.; Ouyang, Y.B.; Xiong, X.; Stary, C.M.; Giffard, R.G. Post-stroke treatment with miR-181 antagomir reduces injury and improves long-term behavioral recovery in mice after focal cerebral ischemia. Exp. Neurol. 2015, 26, 1–7. [Google Scholar] [CrossRef]
- Lehmann, S.K.C.; Park, B.; Derkow, K.; Rosenberger, K.; Baumgart, J.; Trimbuch, T.; Eom, G.; Hinz, M.; Kaul, D.; Habbel, P.; et al. An unconventional role for miRNA: Let-7 activates Toll-like receptor 7 and causes neurodegeneration. Nat. Neurosci. 2012, 15, 827–835. [Google Scholar] [CrossRef]
- Roshan, R.; Shridhar, S.; Sarangdhar, M.A.; Banik, A.; Chawla, M.; Garg, M.; Singh, V.P.; Pillai, B. Brain-specific knockdown of miR-29 results in neuronal cell death and ataxia in mice. RNA 2014, 20, 1287–1297. [Google Scholar] [CrossRef]
- Vetere, G.; Barbato, C.; Pezzola, S.; Frisone, P.; Aceti, M.; Ciotti, M.; Cogoni, C.; Ammassari-Teule, M.; Ruberti, F. Selective inhibition of miR-92 in hippocampal neurons alters contextual fear memory. Hippocampus 2014, 24, 1458–1465. [Google Scholar] [CrossRef]
- Ma, R.; Wang, M.; Gao, S.; Zhu, L.; Yu, L.; Hu, D.; Zhu, L.; Huang, W.; Zhang, W.; Deng, J.; et al. miR-29a Promotes the Neurite Outgrowth of Rat Neural Stem Cells by Targeting Extracellular Matrix to Repair Brain Injury. Stem Cells Dev. 2020, 29, 599–614. [Google Scholar] [CrossRef]
- Swahari, V.; Nakamura, A.; Hollville, E.; Stroud, H.; Simon, J.M.; Ptacek, T.S.; Beck, M.V.; Flowers, C.; Guo, J.; Plestant, C.; et al. MicroRNA-29 is an essential regulator of brain maturation through regulation of CH methylation. Cell Rep. 2021, 35, 108946. [Google Scholar] [CrossRef]
- Roshan-Milani, S.; Sattari, P.; Ghaderi-Pakdel, F.; Naderi, R. miR-23b/TAB3/NF-κB/p53 axis is involved in hippocampus injury induced by cerebral ischemia-reperfusion in rats: The protective effect of chlorogenic acid. Biofactors 2022, 48, 908–917. [Google Scholar] [CrossRef]
- Lu, J.; Zhou, N.; Yang, P.; Deng, L.; Liu, G. MicroRNA-27a-3p Downregulation Inhibits Inflammatory Response and Hippocampal Neuronal Cell Apoptosis by Upregulating Mitogen-Activated Protein Kinase 4 (MAP2K4) Expression in Epilepsy: In Vivo and In Vitro Studies. Med. Sci. Monit. 2019, 25, 8499–8508. [Google Scholar] [CrossRef]
- Wang, Z.; Yuan, Y.; Zhang, Z.; Ding, K. Inhibition of miRNA-27b enhances neurogenesis via AMPK activation in a mouse ischemic stroke model. FEBS Open Bio 2019, 9, 859–869. [Google Scholar] [CrossRef]
- Neumann, E.; Brandenburger, T.; Santana-Varela, S.; Deenen, R.; Köhrer, K.; Bauer, I.; Hermanns, H.; Wood, J.N.; Zhao, J.; Werdehausen, R. MicroRNA-1-associated effects of neuron-specific brain-derived neurotrophic factor gene deletion in dorsal root ganglia. Mol. Cell Neurosci. 2016, 75, 36–43. [Google Scholar] [CrossRef]
- He, J.; Zhao, J.; Peng, X.; Shi, X.; Zong, S.; Zeng, G. Molecular Mechanism of MiR-136-5p Targeting NF-κB/A20 in the IL-17-Mediated Inflammatory Response after Spinal Cord Injury. Cell. Physiol. Biochem. 2017, 44, 1224–1241. [Google Scholar] [CrossRef]
- Cui, Y.; Xiao, Z.; Han, J.; Sun, J.; Ding, W.; Zhao, Y.; Chen, B.; Li, X.; Dai, J. MiR-125b orchestrates cell proliferation, differentiation and migration in neural stem/progenitor cells by targeting Nestin. BMC Neurosci. 2012, 13, 116. [Google Scholar] [CrossRef] [PubMed]
- Marcuzzo, S.; Bonanno, S.; Kapetis, D.; Barzago, C.; Cavalcante, P.; D’Alessandro, S.; Mantegazza, R.; Bernasconi, P. Up-regulation of neural and cell cycle-related microRNAs in brain of amyotrophic lateral sclerosis mice at late disease stage. Mol. Brain 2015, 8, 5. [Google Scholar] [CrossRef] [PubMed]
- Qiao, J.; Zhao, J.; Chang, S.; Sun, Q.; Liu, N.; Dong, J.; Chen, Y.; Yang, D.; Ye, D.; Liu, X.; et al. MicroRNA-153 improves the neurogenesis of neural stem cells and enhances the cognitive ability of aged mice through the notch signaling pathway. Cell Death Differ. 2020, 27, 808–825. [Google Scholar] [CrossRef] [PubMed]
- Krüger, D.M.; Pena, T.; Liu, S.; Park, T.; Kaurani, L.; Pradhan, R.; Huang, Y.; Rischer, S.L.; Burkhardt, S.; Schütz, A.; et al. The plasma miRNAome in ADNI: Signatures to aid the detection of at-risk individuals. Alzheimer’s Dement. 2024; online ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Wang, A.; Xia, C.; Lin, Q.; Chen, C. A single nucleotide polymorphism in primary-microRNA-146a reduces the expression of mature microRNA-146a in patients with Alzheimer’s disease and is associated with the pathogenesis of Alzheimer’s disease. Mol. Med. Rep. 2015, 12, 4037–4042. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef]
- Peng, J.Y.; Chen, Y.H.; Yen, J.H.; Huang, W.M.; Chen, C.N. Effects of Exercise Training on Cognitive Function in Individuals With Heart Failure: A Meta-Analysis. Phys. Ther. 2023, 103, pzad027. [Google Scholar] [CrossRef]
- Goldberg, M.; Islam, R.M.; Kerimolgu, C.; Lacelin, C.; Burkhart, S.; Krüger, D.M.; Marquardt, T.; Malchow, B.; Schmitt, A.; Falkai, P.; et al. Exercise as a model to identify microRNAs linked to human cognition: A role for microRNA-409 and microRNA-501. Transl. Psychiatry 2021, 11, 514. [Google Scholar] [CrossRef]
- Han, Z.; Zhang, L.; Ma, M.; Keshavarzi, M. Effects of MicroRNAs and Long Non-coding RNAs on Beneficial Action of Exercise on Cognition in Degenerative Diseases: A Review. Mol. Neurobiol. 2025, 62, 485–500. [Google Scholar] [CrossRef]
- Bahari-Javan, S.; Maddalena, A.; Kerimoglu, C.; Wittnam, J.; Held, T.; Bähr, M.; Burkhardt, S.; Delalle, I.; Kügler, S.; Fischer, A.; et al. HDAC1 Regulates Fear Extinction in Mice. J. Neurosci. 2012, 32, 5062–5073. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 1 June 2025).
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 3. [Google Scholar] [CrossRef]
- Griffiths-Jones, S. miRBase: The microRNA sequence database. Methods Mol. Biol. 2006, 342, 129–138. [Google Scholar]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 13, 673–691. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Friedländer, M.R.; Mackowiak, S.D.; Li, N.; Chen, W.; Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2011, 40, 37–52. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Risso, D.; Ngai, J.; Speed, T.P.; Dudoit, S. Normalization of RNA-seq data using factor analysis of control genes or samples. Nat. Biotechnol. 2014, 32, 896. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics J. Integr. Biol. 2012, 16, 284–286. [Google Scholar] [CrossRef]
- Blighe, K.; Rana, S.; Lewis, M. EnhancedVolcano: Publication-Ready Volcano Plots with Enhanced Colouring and Labeling. Available online: https://github.com/kevinblighe/EnhancedVolcano (accessed on 1 June 2025).
- Teng, X.; Chen, X.; Xue, H.; Tang, Y.; Zhang, P.; Kang, Q.; Hao, Y.; Chen, R.; Zhao, Y.; He, S. NPInter v4.0: An integrated database of ncRNA interactions. Nucleic Acids Res. 2020, 48, 160–165. [Google Scholar] [CrossRef]
- Liu, Z.P.; Wu, C.; Miao, H.; Wu, H. RegNetwork: An integrated database of transcriptional and post-transcriptional regulatory networks in human and mouse. Database J. Biol. Databases Curatio 2015, 2015, bav095. [Google Scholar] [CrossRef]
- Gong, J.; Shao, D.; Xu, K.; Lu, Z.; Lu, Z.; Yang, Y.T.; Zhang, Q.C. RISE: A database of RNA interactome from sequencing experiments. Nucleic Acids Res. 2018, 46, D194–D201. [Google Scholar] [CrossRef]
- Karagkouni, D.; Paraskevopoulou, M.D.; Chatzopoulos, S.; Vlachos, I.S.; Tastsoglou, S.; Kanellos, I.; Papadimitriou, D.; Kavakiotis, I.; Maniou, S.; Skoufos, G.; et al. DIANA-TarBase v8: A decade-long collection of experimentally supported miRNA-gene interactions. Nucleic Acids Res. 2018, 46, D239–D245. [Google Scholar] [CrossRef] [PubMed]
- Tong, Z.; Cui, Q.; Wang, J.; Zhou, Y. TransmiR v2.0: An updated transcription factor-microRNA regulation database. Nucleic Acids Res. 2019, 47, D253–D258. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Gil, D.P.; Law, J.N.; Murali, T.M. The PathLinker app: Connect the dots in protein interaction networks. F1000Research 2017, 6, 58. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gisa, V.; Islam, M.R.; Lbik, D.; Hofmann, R.M.; Pena, T.; Krüger, D.M.; Burkhardt, S.; Schütz, A.-L.; Sananbenesi, F.; Toischer, K.; et al. Role of Compensatory miRNA Networks in Cognitive Recovery from Heart Failure. Non-Coding RNA 2025, 11, 45. https://doi.org/10.3390/ncrna11030045
Gisa V, Islam MR, Lbik D, Hofmann RM, Pena T, Krüger DM, Burkhardt S, Schütz A-L, Sananbenesi F, Toischer K, et al. Role of Compensatory miRNA Networks in Cognitive Recovery from Heart Failure. Non-Coding RNA. 2025; 11(3):45. https://doi.org/10.3390/ncrna11030045
Chicago/Turabian StyleGisa, Verena, Md Rezaul Islam, Dawid Lbik, Raoul Maximilian Hofmann, Tonatiuh Pena, Dennis Manfred Krüger, Susanne Burkhardt, Anna-Lena Schütz, Farahnaz Sananbenesi, Karl Toischer, and et al. 2025. "Role of Compensatory miRNA Networks in Cognitive Recovery from Heart Failure" Non-Coding RNA 11, no. 3: 45. https://doi.org/10.3390/ncrna11030045
APA StyleGisa, V., Islam, M. R., Lbik, D., Hofmann, R. M., Pena, T., Krüger, D. M., Burkhardt, S., Schütz, A.-L., Sananbenesi, F., Toischer, K., & Fischer, A. (2025). Role of Compensatory miRNA Networks in Cognitive Recovery from Heart Failure. Non-Coding RNA, 11(3), 45. https://doi.org/10.3390/ncrna11030045






