The Multifaceted Roles of CHROMR in Innate Immunity, Cancer, and Cholesterol Homeostasis
Abstract
1. Introduction
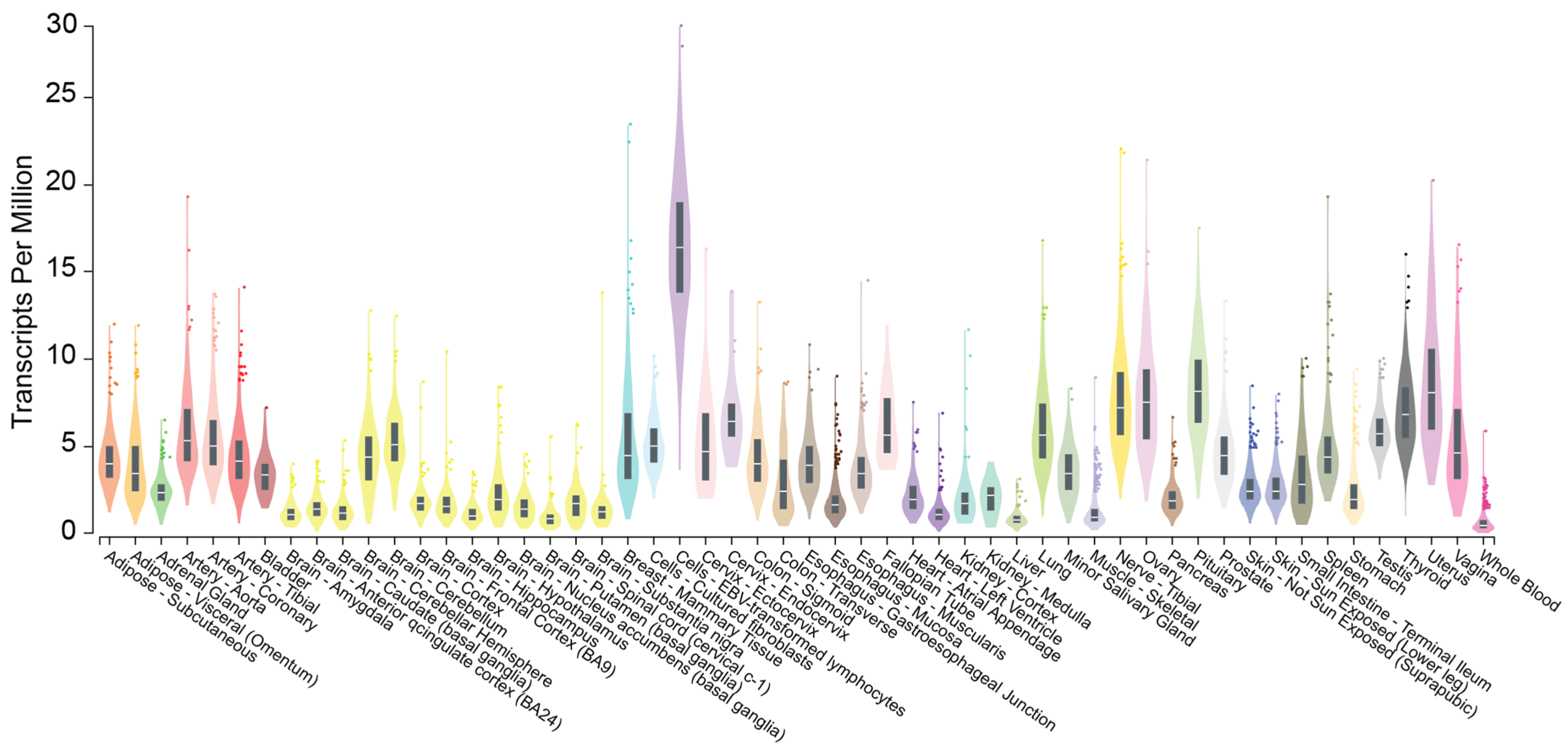
2. CHROMR: Nomenclature, Genomic Location, and Subcellular Localization
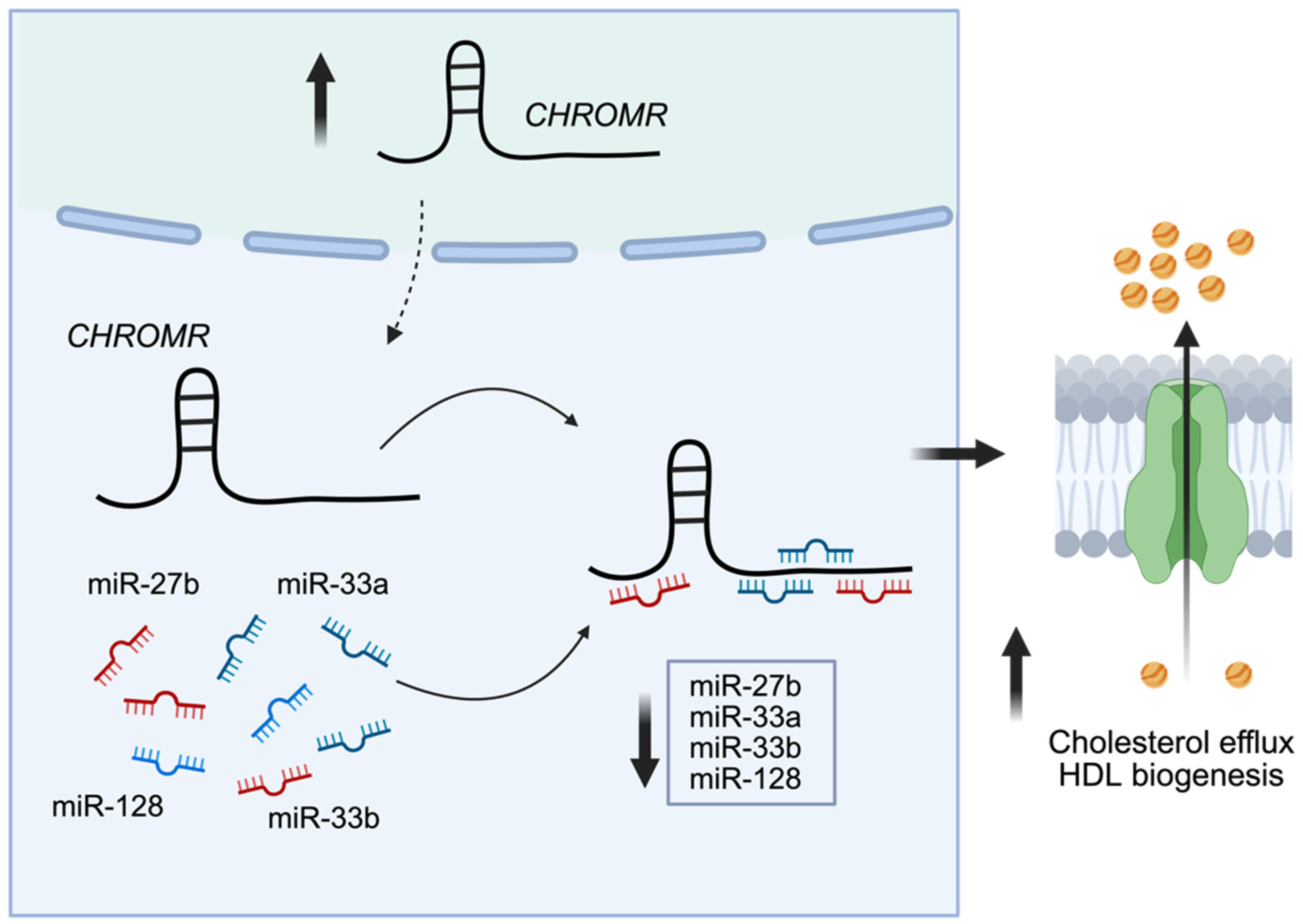
3. CHROMR in Cholesterol Homeostasis
4. CHROMR in Cancer
4.1. Diffuse Large B-Cell Lymphoma
4.2. Lung Adenocarcinoma
4.3. Stomach Adenocarcinoma and Glioma
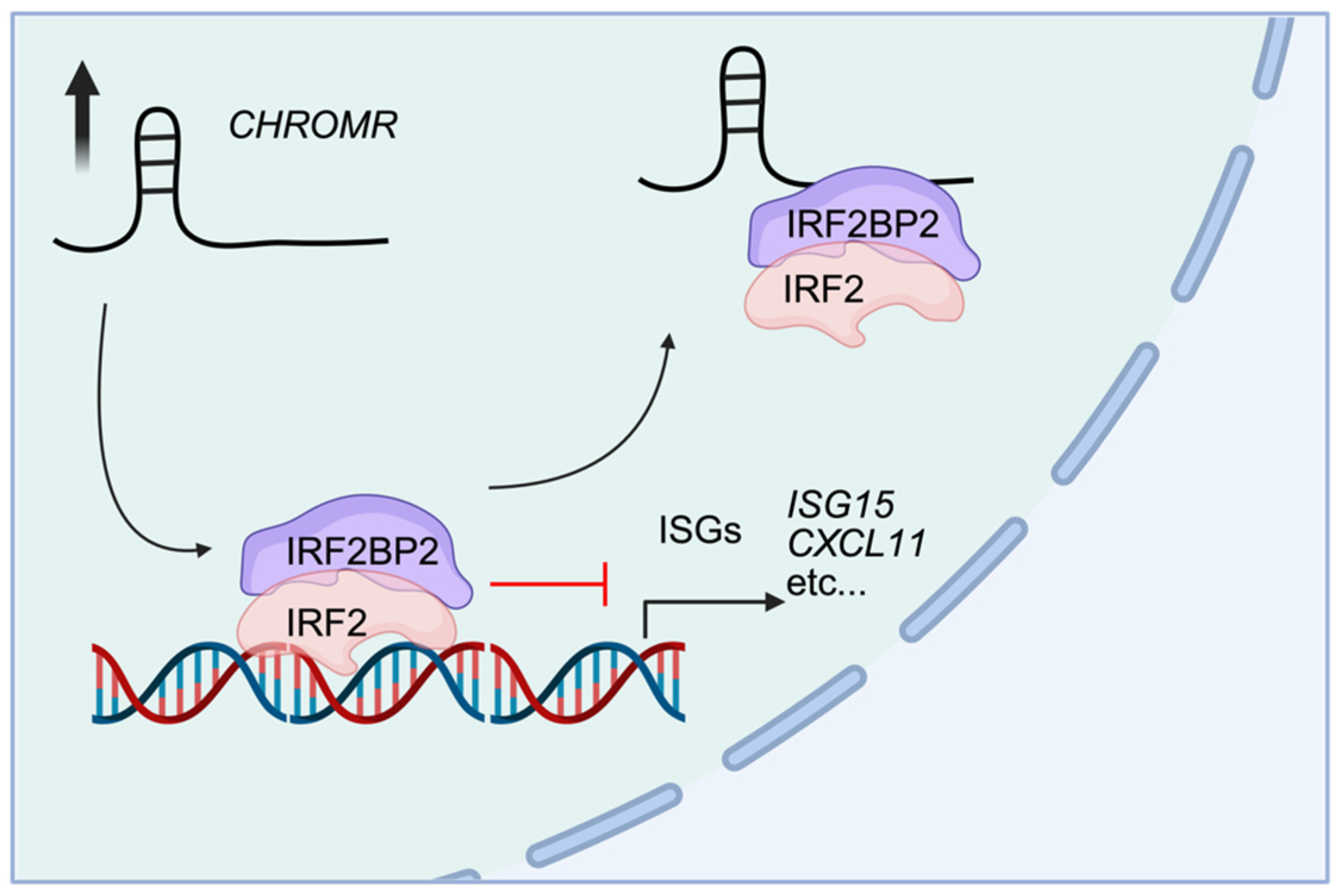
5. CHROMR in Innate Immunity
6. Perspective
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Rinn, J.L.; Chang, H.Y. Genome Regulation by Long Noncoding Rnas. Annu. Rev. Biochem. 2012, 81, 145–166. [Google Scholar] [CrossRef] [PubMed]
- Palazzo, A.F.; Lee, E.S. Non-Coding Rna: What Is Functional and What Is Junk? Front. Genet. 2015, 6, 2. [Google Scholar] [CrossRef]
- Schmitz, S.U.; Grote, P.; Herrmann, B.G. Mechanisms of Long Noncoding Rna Function in Development and Disease. Cell. Mol. Life Sci. 2016, 73, 2491–2509. [Google Scholar] [CrossRef]
- Bridges, M.C.; Daulagala, A.C.; Kourtidis, A. Lnccation: Lncrna Localization and Function. J. Cell Biol. 2021, 220, e202009045. [Google Scholar] [CrossRef] [PubMed]
- Krause, H.M. New and Prospective Roles for Lncrnas in Organelle Formation and Function. Trends Genet. 2018, 34, 736–745. [Google Scholar] [CrossRef]
- Goff, L.A.; Rinn, J.L. Linking Rna Biology to Lncrnas. Genome Res. 2015, 25, 1456–1465. [Google Scholar] [CrossRef]
- Satpathy, A.T.; Chang, H.Y. Long Noncoding Rna in Hematopoiesis and Immunity. Immunity 2015, 42, 792–804. [Google Scholar] [CrossRef]
- Wang, K.C.; Chang, H.Y. Molecular Mechanisms of Long Noncoding Rnas. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef] [PubMed]
- Guil, S.; Esteller, M. Cis-Acting Noncoding Rnas: Friends and Foes. Nat. Struct. Mol. Biol. 2012, 19, 1068–1075. [Google Scholar] [CrossRef]
- Joung, J.; Engreitz, J.M.; Konermann, S.; Abudayyeh, O.O.; Verdine, V.K.; Aguet, F.; Gootenberg, J.S.; Sanjana, N.E.; Wright, J.B.; Fulco, C.P.; et al. Genome-Scale Activation Screen Identifies a Lncrna Locus Regulating a Gene Neighbourhood. Nature 2017, 548, 343–346. [Google Scholar] [CrossRef]
- Kumar, V.; Westra, H.J.; Karjalainen, J.; Zhernakova, D.V.; Esko, T.; Hrdlickova, B.; Almeida, R.; Zhernakova, A.; Reinmaa, E.; Vosa, U.; et al. Human Disease-Associated Genetic Variation Impacts Large Intergenic Non-Coding Rna Expression. PLoS Genet. 2013, 9, e1003201. [Google Scholar] [CrossRef] [PubMed]
- Kopp, F.; Mendell, J.T. Functional Classification and Experimental Dissection of Long Noncoding Rnas. Cell 2018, 172, 393–407. [Google Scholar] [CrossRef]
- Congrains, A.; Kamide, K.; Oguro, R.; Yasuda, O.; Miyata, K.; Yamamoto, E.; Kawai, T.; Kusunoki, H.; Yamamoto, H.; Takeya, Y.; et al. Genetic Variants at the 9p21 Locus Contribute to Atherosclerosis through Modulation of Anril and Cdkn2a/B. Atherosclerosis 2012, 220, 449–455. [Google Scholar] [CrossRef] [PubMed]
- Holdt, L.M.; Beutner, F.; Scholz, M.; Gielen, S.; Gabel, G.; Bergert, H.; Schuler, G.; Thiery, J.; Teupser, D. Anril Expression Is Associated with Atherosclerosis Risk at Chromosome 9p21. Arter. Thromb. Vasc. Biol. 2010, 30, 620–627. [Google Scholar] [CrossRef]
- Holdt, L.M.; Stahringer, A.; Sass, K.; Pichler, G.; Kulak, N.A.; Wilfert, W.; Kohlmaier, A.; Herbst, A.; Northoff, B.H.; Nicolaou, A.; et al. Circular Non-Coding Rna Anril Modulates Ribosomal Rna Maturation and Atherosclerosis in Humans. Nat. Commun. 2016, 7, 12429. [Google Scholar] [CrossRef] [PubMed]
- Ishii, N.; Ozaki, K.; Sato, H.; Mizuno, H.; Saito, S.; Takahashi, A.; Miyamoto, Y.; Ikegawa, S.; Kamatani, N.; Hori, M.; et al. Identification of a Novel Non-Coding Rna, Miat, That Confers Risk of Myocardial Infarction. J. Human Genet. 2006, 51, 1087–1099. [Google Scholar] [CrossRef]
- Gao, W.; Zhu, M.; Wang, H.; Zhao, S.; Zhao, D.; Yang, Y.; Wang, Z.M.; Wang, F.; Yang, Z.J.; Lu, X.; et al. Association of Polymorphisms in Long Non-Coding Rna H19 with Coronary Artery Disease Risk in a Chinese Population. Mutat. Res. 2015, 772, 15–22. [Google Scholar] [CrossRef]
- Sallam, T.; Jones, M.; Thomas, B.J.; Wu, X.; Gilliland, T.; Qian, K.; Eskin, A.; Casero, D.; Zhang, Z.; Sandhu, J.; et al. Transcriptional Regulation of Macrophage Cholesterol Efflux and Atherogenesis by a Long Noncoding Rna. Nat. Med. 2018, 24, 304–312. [Google Scholar] [CrossRef]
- Sallam, T.; Jones, M.C.; Gilliland, T.; Zhang, L.; Wu, X.; Eskin, A.; Sandhu, J.; Casero, D.; Vallim, T.Q.; Hong, C.; et al. Feedback Modulation of Cholesterol Metabolism by the Lipid-Responsive Non-Coding Rna Lexis. Nature 2016, 534, 124–128. [Google Scholar] [CrossRef]
- Hennessy, E.J.; van Solingen, C.; Scacalossi, K.R.; Ouimet, M.; Afonso, M.S.; Prins, J.; Koelwyn, G.J.; Sharma, M.; Ramkhelawon, B.; Carpenter, S.; et al. The Long Noncoding Rna Chrome Regulates Cholesterol Homeostasis in Primate. Nat. Metab. 2019, 1, 98–110. [Google Scholar] [CrossRef]
- Paez, I.; Prado, Y.; Ubilla, C.G.; Zambrano, T.; Salazar, L.A. Atorvastatin Increases the Expression of Long Non-Coding Rnas Arsr and Chrome in Hypercholesterolemic Patients: A Pilot Study. Pharmaceuticals 2020, 13, 382. [Google Scholar] [CrossRef] [PubMed]
- van Solingen, C.; Cyr, Y.; Scacalossi, K.R.; de Vries, M.; Barrett, T.J.; de Jong, A.; Gourvest, M.; Zhang, T.; Peled, D.; Kher, R.; et al. Long Noncoding Rna Chromr Regulates Antiviral Immunity in Humans. Proc. Natl. Acad. Sci. USA 2022, 119, e2210321119. [Google Scholar] [CrossRef]
- Teimuri, S.; Hosseini, A.; Rezaenasab, A.; Ghaedi, K.; Ghoveud, E.; Etemadifar, M.; Nasr-Esfahani, M.H.; Megraw, T.L. Integrative Analysis of Lncrnas in Th17 Cell Lineage to Discover New Potential Biomarkers and Therapeutic Targets in Autoimmune Diseases. Mol. Ther. Nucleic Acids 2018, 12, 393–404. [Google Scholar] [CrossRef] [PubMed]
- Fawcett, K.A.; Demidov, G.; Shrine, N.; Paynton, M.L.; Ossowski, S.; Sayers, I.; Wain, L.V.; Hollox, E.J. Exome-Wide Analysis of Copy Number Variation Shows Association of the Human Leukocyte Antigen Region with Asthma in Uk Biobank. BMC Med. Genom. 2022, 15, 119. [Google Scholar] [CrossRef]
- Kok, K.H.; Lui, P.Y.; Ng, M.H.; Siu, K.L.; Au, S.W.; Jin, D.Y. The Double-Stranded Rna-Binding Protein Pact Functions as a Cellular Activator of Rig-I to Facilitate Innate Antiviral Response. Cell Host Microbe 2011, 9, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Baurecht, H.; Hotze, M.; Brand, S.; Buning, C.; Cormican, P.; Corvin, A.; Ellinghaus, D.; Ellinghaus, E.; Esparza-Gordillo, J.; Folster-Holst, R.; et al. Genome-Wide Comparative Analysis of Atopic Dermatitis and Psoriasis Gives Insight into Opposing Genetic Mechanisms. Am. J. Human Genet. 2015, 96, 104–120. [Google Scholar] [CrossRef]
- Zhao, M.; Di, X.; Jin, X.; Tian, C.; Cong, S.; Liu, J.; Wang, K. Identification of Biomarkers for Sarcoidosis and Tuberculosis of the Lung Using Systematic and Integrated Analysis. Med. Sci. Monit. 2020, 26, e925438. [Google Scholar] [CrossRef]
- Bai, J.; Li, H.; Chen, X.; Chen, L.; Hu, Y.; Liu, L.; Zhao, Y.; Zuo, W.; Zhang, B.; Yin, C. Lncrna-Ac009948.5 Promotes Invasion and Metastasis of Lung Adenocarcinoma by Binding to Mir-186-5p. Front. Oncol. 2022, 12, 949951. [Google Scholar] [CrossRef]
- Liu, C.; Zhao, X.; Wang, Z.; Zhang, C.; Zheng, W.; Zhu, X.; Zhang, D.; Gong, T.; Zhao, H.; Li, F.; et al. Lncrna Chromr/Mir-27b-3p/Met Axis Promotes the Proliferation, Invasion, and Contributes to Rituximab Resistance in Diffuse Large B-Cell Lymphoma. J. Biol. Chem. 2024, 300, 105762. [Google Scholar] [CrossRef]
- Luo, L.; Li, L.; Liu, L.; Feng, Z.; Zeng, Q.; Shu, X.; Cao, Y.; Li, Z. A Necroptosis-Related Lncrna-Based Signature to Predict Prognosis and Probe Molecular Characteristics of Stomach Adenocarcinoma. Front. Genet. 2022, 13, 833928. [Google Scholar] [CrossRef]
- Sirvinskas, D.; Steponaitis, G.; Stakaitis, R.; Tamasauskas, A.; Vaitkiene, P.; Skiriute, D. Antisense Lncrna Chromr Is Linked to Glioma Patient Survival. Front. Mol. Biosci. 2023, 10, 1101953. [Google Scholar] [CrossRef]
- Wang, M.; Miao, Z.; Cen, H.; He, J.; Wei, C. Long Non-Coding Rna (Lncrna) Chromr Promotes the Expression of the Cnnm1 Gene by Adsorbing Hsa-Mir-1299 to Obtain Drug Resistance in Diffuse Large B Lymphoma Cells. Transl. Cancer Res. 2022, 11, 1362–1371. [Google Scholar] [CrossRef] [PubMed]
- Seal, R.L.; Tweedie, S.; Bruford, E.A. A Standardised Nomenclature for Long Non-Coding Rnas. IUBMB Life 2023, 75, 380–389. [Google Scholar] [CrossRef]
- Wright, M.W. A Short Guide to Long Non-Coding Rna Gene Nomenclature. Human Genom. 2014, 8, 7. [Google Scholar] [CrossRef]
- Consortium, G.T. The Genotype-Tissue Expression (Gtex) Project. Nat. Genet. 2013, 45, 580–585. [Google Scholar]
- Cabili, M.N.; Dunagin, M.C.; McClanahan, P.D.; Biaesch, A.; Padovan-Merhar, O.; Regev, A.; Rinn, J.L.; Raj, A. Localization and Abundance Analysis of Human Lncrnas at Single-Cell and Single-Molecule Resolution. Genome Biol. 2015, 16, 20. [Google Scholar] [CrossRef]
- Hubstenberger, A.; Courel, M.; Benard, M.; Souquere, S.; Ernoult-Lange, M.; Chouaib, R.; Yi, Z.; Morlot, J.B.; Munier, A.; Fradet, M.; et al. P-Body Purification Reveals the Condensation of Repressed Mrna Regulons. Mol. Cell 2017, 68, 144–157.e5. [Google Scholar] [CrossRef]
- Yang, Y.; Hsu, P.J.; Chen, Y.-S.; Yang, Y.-G. Dynamic Transcriptomic m6A Decoration: Writers, Erasers, Readers and Functions in Rna Metabolism. Cell Res. 2018, 28, 616–624. [Google Scholar] [CrossRef]
- Yang, L.-R.; Lin, Z.-Y.; Hao, Q.-G.; Li, T.-T.; Zhu, Y.; Teng, Z.-W.; Zhang, J. The Prognosis Biomarkers Based on M6a-Related Lncrnas for Myeloid Leukemia Patients. Cancer Cell Int. 2022, 22, 10. [Google Scholar] [CrossRef]
- Loganathan, T.; Doss, C.G. Non-Coding Rnas in Human Health and Disease: Potential Function as Biomarkers and Therapeutic Targets. Funct. Integr. Genom. 2023, 23, 33. [Google Scholar] [CrossRef]
- Goedeke, L.; Fernandez-Hernando, C. Regulation of Cholesterol Homeostasis. Cell. Mol. Life Sci. 2012, 69, 915–930. [Google Scholar] [CrossRef]
- Siddiqi, H.K.; Kiss, D.; Rader, D. Hdl-Cholesterol and Cardiovascular Disease: Rethinking Our Approach. Curr. Opin. Cardiol. 2015, 30, 536–542. [Google Scholar] [CrossRef]
- van Solingen, C.; Scacalossi, K.R.; Moore, K.J. Long Noncoding Rnas in Lipid Metabolism. Curr. Opin. Lipidol. 2018, 29, 224–232. [Google Scholar] [CrossRef] [PubMed]
- Gluba-Sagr, A.; Franczyk, B.; Rysz-Gorzynska, A.; Olszewski, R.; Rysz, J. The Role of Selected Lncrnas in Lipid Metabolism and Cardiovascular Disease Risk. Int. J. Mol. Sci. 2024, 25, 9244. [Google Scholar] [CrossRef]
- Yan, C.; Chen, J.; Chen, N. Long Noncoding Rna Malat1 Promotes Hepatic Steatosis and Insulin Resistance by Increasing Nuclear Srebp-1c Protein Stability. Sci. Rep. 2016, 6, 22640. [Google Scholar] [CrossRef]
- Liu, C.; Yang, Z.; Wu, J.; Zhang, L.; Lee, S.; Shin, D.J.; Tran, M.; Wang, L. Long Noncoding Rna H19 Interacts with Polypyrimidine Tract-Binding Protein 1 to Reprogram Hepatic Lipid Homeostasis. Hepatology 2018, 67, 1768–1783. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Cheng, M.; Niu, Y.; Chi, X.; Liu, X.; Fan, J.; Fan, H.; Chang, Y.; Yang, W. Identification of a Novel Human Long Non-Coding Rna That Regulates Hepatic Lipid Metabolism by Inhibiting Srebp-1c. Int. J. Biol. Sci. 2017, 13, 349–357. [Google Scholar] [CrossRef]
- Li, P.; Ruan, X.; Yang, L.; Kiesewetter, K.; Zhao, Y.; Luo, H.; Chen, Y.; Gucek, M.; Zhu, J.; Cao, H. A Liver-Enriched Long Non-Coding Rna, Lnclstr, Regulates Systemic Lipid Metabolism in Mice. Cell Metab. 2015, 21, 455–467. [Google Scholar] [CrossRef]
- Halley, P.; Kadakkuzha, B.M.; Faghihi, M.A.; Magistri, M.; Zeier, Z.; Khorkova, O.; Coito, C.; Hsiao, J.; Lawrence, M.; Wahlestedt, C. Regulation of the Apolipoprotein Gene Cluster by a Long Noncoding Rna. Cell Rep. 2014, 6, 222–230. [Google Scholar] [CrossRef]
- Qin, W.; Li, X.; Xie, L.; Li, S.; Liu, J.; Jia, L.; Dong, X.; Ren, X.; Xiao, J.; Yang, C.; et al. A Long Non-Coding Rna, Apoa4-as, Regulates Apoa4 Expression Depending on Hur in Mice. Nucleic Acids Res. 2016, 44, 6423–6433. [Google Scholar] [CrossRef]
- Nsengimana, J.; Samani, N.J.; Hall, A.S.; Balmforth, A.J.; Mangino, M.; Yuldasheva, N.; Maqbool, A.; Braund, P.; Burton, P.; Bishop, D.T.; et al. Enhanced Linkage of a Locus on Chromosome 2 to Premature Coronary Artery Disease in the Absence of Hypercholesterolemia. Eur. J. Human Genet. 2007, 15, 313–319. [Google Scholar] [CrossRef]
- North, K.E.; Martin, L.J.; Dyer, T.; Comuzzie, A.G.; Williams, J.T.; Framingham Heart, S. Hdl Cholesterol in Females in the Framingham Heart Study Is Linked to a Region of Chromosome 2q. BMC Genet. 2003, 4 (Suppl. S1), S98. [Google Scholar] [CrossRef] [PubMed]
- Rayner, K.J.; Suarez, Y.; Davalos, A.; Parathath, S.; Fitzgerald, M.L.; Tamehiro, N.; Fisher, E.A.; Moore, K.J.; Fernandez-Hernando, C. Mir-33 Contributes to the Regulation of Cholesterol Homeostasis. Science 2010, 328, 1570–1573. [Google Scholar] [CrossRef]
- Vickers, K.C.; Shoucri, B.M.; Levin, M.G.; Wu, H.; Pearson, D.S.; Osei-Hwedieh, D.; Collins, F.S.; Remaley, A.T.; Sethupathy, P. Microrna-27b Is a Regulatory Hub in Lipid Metabolism and Is Altered in Dyslipidemia. Hepatology 2013, 57, 533–542. [Google Scholar] [CrossRef]
- Wagschal, A.; Najafi-Shoushtari, S.H.; Wang, L.; Goedeke, L.; Sinha, S.; deLemos, A.S.; Black, J.C.; Ramirez, C.M.; Li, Y.; Tewhey, R.; et al. Genome-Wide Identification of Micrornas Regulating Cholesterol and Triglyceride Homeostasis. Nat. Med. 2015, 21, 1290–1297. [Google Scholar] [CrossRef] [PubMed]
- Najafi-Shoushtari, S.H.; Kristo, F.; Li, Y.; Shioda, T.; Cohen, D.E.; Gerszten, R.E.; Naar, A.M. Microrna-33 and the Srebp Host Genes Cooperate to Control Cholesterol Homeostasis. Science 2010, 328, 1566–1569. [Google Scholar] [CrossRef]
- Goedeke, L.; Rotllan, N.; Ramirez, C.M.; Aranda, J.F.; Canfran-Duque, A.; Araldi, E.; Fernandez-Hernando, A.; Langhi, C.; de Cabo, R.; Baldan, A.; et al. Mir-27b Inhibits Ldlr and Abca1 Expression but Does Not Influence Plasma and Hepatic Lipid Levels in Mice. Atherosclerosis 2015, 243, 499–509. [Google Scholar] [CrossRef]
- Xie, Y.; Wang, Y.; Gong, R.; Lin, J.; Li, X.; Ma, J.; Huo, L. Snhg7 Facilitates Hepatocellular Carcinoma Occurrence by Sequestering Mir-9-5p to Upregulate Cnnm1 Expression. Cancer Biother. Radiopharm. 2020, 35, 731–740. [Google Scholar] [CrossRef]
- Li, H.; Yin, C.; Zhang, B.; Sun, Y.; Shi, L.; Liu, N.; Liang, S.; Lu, S.; Liu, Y.; Zhang, J.; et al. Pttg1 Promotes Migration and Invasion of Human Non-Small Cell Lung Cancer Cells and Is Modulated by Mir-186. Carcinogenesis 2013, 34, 2145–2155. [Google Scholar] [CrossRef]
- Zhan, P.; Xi, G.M.; Zhang, B.; Wu, Y.; Liu, H.B.; Liu, Y.F.; Xu, W.J.; Zhu, Q.; Cai, F.; Zhou, Z.J.; et al. Ncapg2 Promotes Tumour Proliferation by Regulating G2/M Phase and Associates with Poor Prognosis in Lung Adenocarcinoma. J. Cell. Mol. Med. 2017, 21, 665–676. [Google Scholar] [CrossRef]
- Wang, F.; Liu, Y.; Chen, Y. Pituitary Tumor Transforming Gene-1 in Non-Small Cell Lung Cancer: Clinicopathological and Immunohistochemical Analysis. Biomed. Pharmacother. 2016, 84, 1595–1600. [Google Scholar] [CrossRef] [PubMed]
- Samani, N.J.; Burton, P.; Mangino, M.; Ball, S.G.; Balmforth, A.J.; Barrett, J.; Bishop, T.; Hall, A.; Group, B.H.F.F.H.S.R. A Genomewide Linkage Study of 1,933 Families Affected by Premature Coronary Artery Disease: The British Heart Foundation (Bhf) Family Heart Study. Am. J. Human Genet. 2005, 77, 1011–1020. [Google Scholar]
- Wang, J.; Ma, R.; Ma, W.; Chen, J.; Yang, J.; Xi, Y.; Cui, Q. Lncdisease: A Sequence Based Bioinformatics Tool for Predicting Lncrna-Disease Associations. Nucleic Acids Res. 2016, 44, e90. [Google Scholar] [CrossRef] [PubMed]
- Mero, I.L.; Gustavsen, M.W.; Saether, H.S.; Flam, S.T.; Berg-Hansen, P.; Sondergaard, H.B.; Jensen, P.E.; Berge, T.; Bjolgerud, A.; Muggerud, A.; et al. Oligoclonal Band Status in Scandinavian Multiple Sclerosis Patients Is Associated with Specific Genetic Risk Alleles. PLoS ONE 2013, 8, e58352. [Google Scholar] [CrossRef]
- Cyr, Y.; Gourvest, M.; Ciabattoni, G.O.; Zhang, T.; Newman, A.A.; Zahr, T.; Delbare, S.; Schlamp, F.; Dittmann, M.; Moore, K.J.; et al. Lncrna Carinh Regulates Expression and Function of Innate Immune Transcription Factor Irf1 in Macrophages. Life Sci. Alliance 2025, 8, e202403021. [Google Scholar] [CrossRef]
- Barriocanal, M.; Carnero, E.; Segura, V.; Fortes, P. Long Non-Coding Rna Bst2/Bispr Is Induced by Ifn and Regulates the Expression of the Antiviral Factor Tetherin. Front. Immunol. 2014, 5, 655. [Google Scholar] [CrossRef]
- Mariotti, B.; Servaas, N.H.; Rossato, M.; Tamassia, N.; Cassatella, M.A.; Cossu, M.; Beretta, L.; van der Kroef, M.; Radstake, T.; Bazzoni, F. The Long Non-Coding Rna Nrir Drives Ifn-Response in Monocytes: Implication for Systemic Sclerosis. Front. Immunol. 2019, 10, 100. [Google Scholar] [CrossRef]
- Kulkarni, S.; Lied, A.; Kulkarni, V.; Rucevic, M.; Martin, M.P.; Walker-Sperling, V.; Anderson, S.K.; Ewy, R.; Singh, S.; Nguyen, H.; et al. Ccr5as Lncrna Variation Differentially Regulates Ccr5, Influencing Hiv Disease Outcome. Nat. Immunol. 2019, 20, 824–834. [Google Scholar] [CrossRef]
- Agarwal, S.; Vierbuchen, T.; Ghosh, S.; Chan, J.; Jiang, Z.; Kandasamy, R.K.; Ricci, E.; Fitzgerald, K.A. The Long Non-Coding Rna Lucat1 Is a Negative Feedback Regulator of Interferon Responses in Humans. Nat. Commun. 2020, 11, 6348. [Google Scholar] [CrossRef]
- Childs, K.S.; Goodbourn, S. Identification of Novel Co-Repressor Molecules for Interferon Regulatory Factor-2. Nucleic Acids Res. 2003, 31, 3016–3026. [Google Scholar] [CrossRef]
- Keller, M.D.; Pandey, R.; Li, D.; Glessner, J.; Tian, L.; Henrickson, S.E.; Chinn, I.K.; Monaco-Shawver, L.; Heimall, J.; Hou, C.; et al. Mutation in Irf2bp2 Is Responsible for a Familial Form of Common Variable Immunodeficiency Disorder. J. Allergy Clin. Immunol. 2016, 138, 544–550.e4. [Google Scholar] [CrossRef] [PubMed]
- Udemgba, C.; Pillay, B.; Shafer, S.; Alberstadt, A.; Abers, M.; Gilliaux, O.; Chen, K.; Rae, W.; Hanitsch, L.; Von Bernuth, H.; et al. Irf2bp2 Deficiency: An Important Form of Common Variable Immunodeficiency with Inflammation. J. Allergy Clin. Immunol. 2025, in press. [Google Scholar] [CrossRef]
- Anim, M.; Sogkas, G.; Camacho-Ordonez, N.; Schmidt, G.; Elsayed, A.; Proietti, M.; Witte, T.; Grimbacher, B.; Atschekzei, F. Novel Hypermorphic Variants in Irf2bp2 Identified in Patients with Common Variable Immunodeficiency and Autoimmunity. Clin. Immunol. 2024, 266, 110326. [Google Scholar] [CrossRef]
- Kumar, A.; Das, S.; Agrawal, A.; Mukhopadhyay, I.; Ghosh, B. Genetic Association of Key Th1/Th2 Pathway Candidate Genes, Irf2, Il6, Ifngr2, Stat4 and Il4ra, with Atopic Asthma in the Indian Population. J. Human Genet. 2015, 60, 443–448. [Google Scholar] [CrossRef]
- Arruda, L.C.; Lorenzi, J.C.; Sousa, A.P.; Zanette, D.L.; Palma, P.V.; Panepucci, R.A.; Brum, D.S.; Barreira, A.A.; Covas, D.T.; Simoes, B.P.; et al. Autologous Hematopoietic Sct Normalizes Mir-16, -155 and -142-3p Expression in Multiple Sclerosis Patients. Bone Marrow Transplant. 2015, 50, 380–389. [Google Scholar] [CrossRef]
- Gao, P.S.; Leung, D.Y.; Rafaels, N.M.; Boguniewicz, M.; Hand, T.; Gao, L.; Hata, T.R.; Schneider, L.C.; Hanifin, J.M.; Beaty, T.H.; et al. Genetic Variants in Interferon Regulatory Factor 2 (Irf2) Are Associated with Atopic Dermatitis and Eczema Herpeticum. J. Investig. Dermatol. 2012, 132, 650–657. [Google Scholar] [CrossRef]
- Chen, L.; Liu, C.; Xiang, X.; Qiu, W.; Guo, K. Mir-155 Promotes an Inflammatory Response in Hacat Cells Via the Irf2bp2/Klf2/Nf-Kappab Pathway in Psoriasis. Int. J. Mol. Med. 2024, 54, 91. [Google Scholar] [CrossRef] [PubMed]
- Mattick, J.S.; Amaral, P.P.; Carninci, P.; Carpenter, S.; Chang, H.Y.; Chen, L.L.; Chen, R.; Dean, C.; Dinger, M.E.; Fitzgerald, K.A.; et al. Long Non-Coding Rnas: Definitions, Functions, Challenges and Recommendations. Nat. Rev. Mol. Cell Biol. 2023, 24, 430–447. [Google Scholar] [CrossRef]
- Bajimaya, S.; Frankl, T.; Hayashi, T.; Takimoto, T. Cholesterol Is Required for Stability and Infectivity of Influenza a and Respiratory Syncytial Viruses. Virology 2017, 510, 234–241. [Google Scholar] [CrossRef]
- Sanders, D.W.; Jumper, C.C.; Ackerman, P.J.; Bracha, D.; Donlic, A.; Kim, H.; Kenney, D.; Castello-Serrano, I.; Suzuki, S.; Tamura, T.; et al. SARS-Cov-2 Requires Cholesterol for Viral Entry and Pathological Syncytia Formation. eLife 2021, 10, e65962. [Google Scholar] [CrossRef]
- Winkler, R.; Gillis, E.; Lasman, L.; Safra, M.; Geula, S.; Soyris, C.; Nachshon, A.; Tai-Schmiedel, J.; Friedman, N.; Le-Trilling, V.T.K.; et al. M6a Modification Controls the Innate Immune Response to Infection by Targeting Type I Interferons. Nat. Immunol. 2019, 20, 173–182. [Google Scholar] [CrossRef] [PubMed]
- McFadden, M.J.; Horner, S.M. N6-Methyladenosine Regulates Host Responses to Viral Infection. Trends Biochem. Sci. 2021, 46, 366–377. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Y.; Gu, J.; Su, T.; Gu, X.; Feng, Y. The Role of Rna M6a Methylation in Lipid Metabolism. Front. Endocrinol. 2022, 13, 866116. [Google Scholar] [CrossRef]
- He, R.-Z.; Jiang, J.; Luo, D.-X. The Functions of N6-Methyladenosine Modification in Lncrnas. Genes Dis. 2020, 7, 598–605. [Google Scholar] [CrossRef]
- Liu, J.; Yue, Y.; Han, D.; Wang, X.; Fu, Y.; Zhang, L.; Jia, G.; Yu, M.; Lu, Z.; Deng, X.; et al. A Mettl3-Mettl14 Complex Mediates Mammalian Nuclear Rna N6-Adenosine Methylation. Nat. Chem. Biol. 2014, 10, 93–95. [Google Scholar] [CrossRef]
- Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Pan, T.; Yang, Y.G.; et al. N6-Methyladenosine in Nuclear Rna Is a Major Substrate of the Obesity-Associated Fto. Nat. Chem. Biol. 2011, 7, 885–887. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.; Dahl, J.A.; Niu, Y.; Fedorcsak, P.; Huang, C.-M.; Li, C.J.; Vagbo, C.B.; Shi, Y.; Wang, W.-L.; Song, S.-H.; et al. Alkbh5 Is a Mammalian Rna Demethylase That Impacts Rna Metabolism and Mouse Fertility. Mol. Cell 2013, 49, 18–29. [Google Scholar] [CrossRef]
- Kumari, R.; Ranjan, P.; Suleiman, Z.G.; Goswami, S.K.; Li, J.; Prasad, R.; Verma, S.K. Mrna Modifications in Cardiovascular Biology and Disease: With a Focus on M6a Modification. Cardiovasc. Res. 2022, 118, 1680–1692. [Google Scholar] [CrossRef]
- Zaccara, S.; Ries, R.J.; Jaffrey, S.R. Reading, Writing and Erasing Mrna Methylation. Nat. Rev. Mol. Cell Biol. 2019, 20, 608–624. [Google Scholar] [CrossRef]
- Yang, D.; Qiao, J.; Wang, G.; Lan, Y.; Li, G.; Guo, X.; Xi, J.; Ye, D.; Zhu, S.; Chen, W.; et al. N6-Methyladenosine Modification of Lincrna 1281 Is Critically Required for Mesc Differentiation Potential. Nucleic Acids Res. 2018, 46, 3906–3920. [Google Scholar] [CrossRef]
- Fu, Y.; Zhuang, X. m6A-Binding Ythdf Proteins Promote Stress Granule Formation. Nat. Chem. Biol. 2020, 16, 955–963. [Google Scholar] [CrossRef] [PubMed]
- Petri, B.J.; Klinge, C.M. M6a Readers, Writers, Erasers, and the M6a Epitranscriptome in Breast Cancer. J. Mol. Endocrinol. 2023, 70, e220110. [Google Scholar] [CrossRef]
- Patil, D.P.; Chen, C.K.; Pickering, B.F.; Chow, A.; Jackson, C.; Guttman, M.; Jaffrey, S.R. m6A Rna Methylation Promotes Xist-Mediated Transcriptional Repression. Nature 2016, 537, 369–373. [Google Scholar] [CrossRef]
- Porman, A.M.; Roberts, J.T.; Duncan, E.D.; Chrupcala, M.L.; Levine, A.A.; Kennedy, M.A.; Williams, M.M.; Richer, J.K.; Johnson, A.M. A Single N6-Methyladenosine Site Regulates Lncrna Hotair Function in Breast Cancer Cells. PLoS Biol. 2022, 20, e3001885. [Google Scholar] [CrossRef] [PubMed]
- Khyzha, N.; Khor, M.; DiStefano, P.V.; Wang, L.; Matic, L.; Hedin, U.; Wilson, M.D.; Maegdefessel, L.; Fish, J.E. Regulation of Ccl2 Expression in Human Vascular Endothelial Cells by a Neighboring Divergently Transcribed Long Noncoding RNA. Proc. Natl. Acad. Sci. USA 2019, 116, 16410–16419. [Google Scholar] [CrossRef]
- Wang, Y.; Song, F.; Zhang, B.; Zhang, L.; Xu, J.; Kuang, D.; Li, D.; Choudhary, M.N.K.; Li, Y.; Hu, M.; et al. The 3d Genome Browser: A Web-Based Browser for Visualizing 3d Genome Organization and Long-Range Chromatin Interactions. Genome Biol. 2018, 19, 151. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Wang, X. Prediction of Functional Microrna Targets by Integrative Modeling of Microrna Binding and Target Expression Data. Genome Biol. 2019, 20, 18. [Google Scholar] [CrossRef]
- Lorenz, R.; Bernhart, S.H.; Honer Zu Siederdissen, C.; Tafer, H.; Flamm, C.; Stadler, P.F.; Hofacker, I.L. Viennarna Package 2.0. Algorithms Mol. Biol. 2011, 6, 26. [Google Scholar] [CrossRef]
- van Heesch, S.; Witte, F.; Schneider-Lunitz, V.; Schulz, J.F.; Adami, E.; Faber, A.B.; Kirchner, M.; Maatz, H.; Blachut, S.; Sandmann, C.L.; et al. The Translational Landscape of the Human Heart. Cell 2019, 178, 242–260.e29. [Google Scholar] [CrossRef]
- Prensner, J.R.; Abelin, J.G.; Kok, L.W.; Clauser, K.R.; Mudge, J.M.; Ruiz-Orera, J.; Bassani-Sternberg, M.; Moritz, R.L.; Deutsch, E.W.; van Heesch, S. What Can Ribo-Seq, Immunopeptidomics, and Proteomics Tell Us About the Noncanonical Proteome? Mol. Cell. Proteom. 2023, 22, 100631. [Google Scholar] [CrossRef]
- Prensner, J.R.; Enache, O.M.; Luria, V.; Krug, K.; Clauser, K.R.; Dempster, J.M.; Karger, A.; Wang, L.; Stumbraite, K.; Wang, V.M.; et al. Noncanonical Open Reading Frames Encode Functional Proteins Essential for Cancer Cell Survival. Nat. Biotechnol. 2021, 39, 697–704. [Google Scholar] [CrossRef] [PubMed]


| Cardiovascular disease | |||
| Pathway | Direct target(s) | Effects on | Ref. |
| Cholesterol homeostasis | miR-27b, miR-33a, miR-33b, miR-128 | ABCA1, ABCB11, ANGPTL3, ATP8B1, CPT1A, CROT, GPAM, HNF4A, OSBPL6 | [20] |
| Cancer | |||
| Type | Direct target(s) | Effects on | Ref. |
| B-cell lymphoma | miR-27b-3p, miR-1299 | CNNM1, MET | [29,32] |
| HDAC3 | CD20 | [29] | |
| Lung adenocarcinoma | miR-186-5p | NCAPG2 | [28] |
| Innate immunity | |||
| Pathway | Direct target(s) | Effects on | Ref. |
| Viral response | IRF2BP2 | Interferon-stimulated genes | [22] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Blaustein, E.R.; van Solingen, C. The Multifaceted Roles of CHROMR in Innate Immunity, Cancer, and Cholesterol Homeostasis. Non-Coding RNA 2025, 11, 44. https://doi.org/10.3390/ncrna11030044
Blaustein ER, van Solingen C. The Multifaceted Roles of CHROMR in Innate Immunity, Cancer, and Cholesterol Homeostasis. Non-Coding RNA. 2025; 11(3):44. https://doi.org/10.3390/ncrna11030044
Chicago/Turabian StyleBlaustein, Emma R., and Coen van Solingen. 2025. "The Multifaceted Roles of CHROMR in Innate Immunity, Cancer, and Cholesterol Homeostasis" Non-Coding RNA 11, no. 3: 44. https://doi.org/10.3390/ncrna11030044
APA StyleBlaustein, E. R., & van Solingen, C. (2025). The Multifaceted Roles of CHROMR in Innate Immunity, Cancer, and Cholesterol Homeostasis. Non-Coding RNA, 11(3), 44. https://doi.org/10.3390/ncrna11030044






