Genome-Wide Association Study of Gluteus Medius Muscle Size in a Crossbred Pig Population
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Phenotypic Data
2.2. Genotypic Data
2.3. Estimation of Genetic Parameters
2.4. Genome-Wide Association Study
2.5. Functional Annotation of Significant Loci
3. Results
3.1. Phenotypic Variation and Heritability Estimates
3.2. Genome-Wide Association Study
3.3. Candidate Genes Search and Functional Annotation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lebret, B.; Candek-Potokar, M. Review: Pork quality attributes from farm to fork. Part II. Processed pork products. Animal 2022, 16, 100383. [Google Scholar] [CrossRef]
- van Milgen, J.; Noblet, J.; Dourmad, J.Y.; Labussière, E.; Garcia-Launay, F.; Brossard, L. Precision pork production: Predicting the impact of nutritional strategies on carcass quality. Meat Sci. 2012, 92, 182–187. [Google Scholar] [CrossRef]
- Candek-Potokar, M.; Lebret, B.; Gispert, M.; Font-i-Furnols, M. Challenges and future perspectives for the European grading of pig carcasses—A quality view. Meat Sci. 2024, 208, 109390. [Google Scholar] [CrossRef]
- Hwang, Y.H.; Bakhsh, A.; Lee, J.G.; Joo, S.T. Differences in Muscle Fiber Characteristics and Meat Quality by Muscle Type and Age of Korean Native Black Goat. Food Sci. Anim. Resour. 2019, 39, 988–999. [Google Scholar] [CrossRef] [PubMed]
- Gou, P.; Comaposada, J.; Arnau, J. NaCl content and temperature effects on moisture diffusivity in the muscle of pork ham. Meat Sci. 2003, 63, 29–34. [Google Scholar] [CrossRef]
- Arkfeld, E.K.; Wilson, K.B.; Overholt, M.F.; Harsh, B.N.; Lowell, J.E.; Hogan, E.K.; Klehm, B.J.; Bohrer, B.M.; Mohrhauser, D.A.; King, D.A.; et al. Pork loin quality is not indicative of fresh belly or fresh and cured ham quality. J. Anim. Sci. 2016, 94, 5155–5167. [Google Scholar] [CrossRef] [PubMed]
- He, Y.N.; Ma, J.W.; Zhang, F.; Hou, L.J.; Chen, H.; Guo, Y.M.; Zhang, Z.Y. Multi-breed genome-wide association study reveals heterogeneous loci associated with loin eye area in pigs. J. Appl. Genet. 2016, 57, 511–518. [Google Scholar] [CrossRef]
- Wang, H.Y.; Wang, X.Y.; Yang, Y.L.; Zhu, Y.X.; Wang, S.Y.; Chen, Q.; Yan, D.W.; Dong, X.X.; Li, M.L.; Lu, S.X. Genome-wide identification of quantitative trait loci and candidate genes for seven carcass traits in a four-way intercross porcine population. BMC Genom. 2024, 25, 582. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Wu, P.X.; Wang, K.; Chen, D.J.; Zhou, J.; Ma, J.D.; Li, M.Z.; Xiao, W.H.; Jiang, A.A.; Jiang, Y.Z.; et al. SNPs associated with body weight and backfat thickness in two pig breeds identified by a genome-wide association study. Genomics 2019, 111, 1583–1589. [Google Scholar] [CrossRef]
- Zhuang, Z.W.; Li, S.Y.; Ding, R.R.; Yang, M.; Zheng, E.Q.; Yang, H.Q.; Gu, T.; Xu, Z.; Cai, G.Y.; Wu, Z.F.; et al. Meta-analysis of genome-wide association studies for loin muscle area and loin muscle depth in two Duroc pig populations. PLoS ONE 2019, 14, e0218263. [Google Scholar] [CrossRef] [PubMed]
- Browning, B.L.; Zhou, Y.; Browning, S.R. A One-Penny Imputed Genome from Next-Generation Reference Panels. Am. J. Hum. Genet. 2018, 103, 338–348. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.L.; Zhang, H.H.; Tang, Z.S.; Yin, D.; Fu, Y.H.; Yuan, X.H.; Li, X.Y.; Liu, X.L.; Zhao, S.H. HIBLUP: An integration of statistical models on the BLUP framework for efficient genetic evaluation using big genomic data. Nucleic Acids Res. 2023, 51, 3501–3512. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.B.; Zhang, Z.W. GAPIT Version 3: Boosting Power and Accuracy for Genomic Association and Prediction. Genom. Proteom. Bioinf. 2021, 19, 629–640. [Google Scholar] [CrossRef]
- Huang, M.; Liu, X.L.; Zhou, Y.; Summers, R.M.; Zhang, Z.W. BLINK: A package for the next level of genome-wide association studies with both individuals and markers in the millions. Gigascience 2019, 8, giy154. [Google Scholar] [CrossRef]
- Bland, J.M.; Altman, D.G. Multiple significance tests: The Bonferroni method. BMJ 1995, 310, 170. [Google Scholar] [CrossRef]
- Zeng, H.N.; Zhang, W.J.; Lin, Q.; Gao, Y.H.; Teng, J.Y.; Xu, Z.T.; Cai, X.D.; Zhong, Z.M.; Wu, J.; Liu, Y.Q.; et al. PigBiobank: A valuable resource for understanding genetic and biological mechanisms of diverse complex traits in pigs. Nucleic Acids Res. 2024, 52, D980–D989. [Google Scholar] [CrossRef]
- Matskova, L.; Zheng, S.X.; Kashuba, E.; Ernberg, I.; Aspenström, P. MTSS1: Beyond the integration of actin and membrane dynamics. Cell Mol. Life Sci. 2024, 81, 472. [Google Scholar] [CrossRef]
- Poch, C.M.; Foo, K.S.; De Angelis, M.T.; Jennbacken, K.; Santamaria, G.; Bähr, A.; Wang, Q.D.; Reiter, F.; Hornaschewitz, N.; Zawada, D.; et al. Migratory and anti-fibrotic programmes define the regenerative potential of human cardiac progenitors. Nat. Cell Biol. 2022, 24, 659–671. [Google Scholar] [CrossRef]
- Zhou, F.C.; Quan, J.P.; Ruan, D.L.; Qiu, Y.B.; Ding, R.R.; Xu, C.E.; Ye, Y.; Cai, G.Y.; Liu, L.Q.; Zhang, Z.B.; et al. Identification of Candidate Genes for Economically Important Carcass Cutting in Commercial Pigs through GWAS. Animal 2023, 13, 3243. [Google Scholar] [CrossRef]
- Wang, B.B.; Li, P.H.; Hou, L.M.; Zhou, W.D.; Tao, W.; Liu, C.X.; Liu, K.Y.; Niu, P.P.; Zhang, Z.P.; Li, Q.; et al. Genome-wide association study and genomic prediction for intramuscular fat content in Suhuai pigs using imputed whole-genome sequencing data. Evol. Appl. 2022, 15, 2054–2066. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Xu, Q.; Tang, C. RGS proteins and their roles in cancer: Friend or foe? Cancer Cell Int. 2023, 23, 81. [Google Scholar] [CrossRef]
- Yowe, D.; Weich, N.; Prabhudas, M.; Poisson, L.; Errada, P.; Kapeller, R.; Yu, K.; Faron, L.; Shen, M.H.; Cleary, J.; et al. RGS18 is a myeloerythroid lineage-specific regulator of G-protein-signalling molecule highly expressed in megakaryocytes. Biochem. J. 2001, 359, 109–118. [Google Scholar] [CrossRef]
- Kimple, A.J.; Garland, A.L.; Cohen, S.P.; Setola, V.; Willard, F.S.; Zielinski, T.; Lowery, R.G.; Tarran, R.; Siderovski, D.P. RGS21, A Regulator of Taste and Mucociliary Clearance? Laryngoscope 2014, 124, E56–E63. [Google Scholar] [CrossRef]
- Schroer, A.B.; Branyan, K.W.; Gross, J.D.; Chantler, P.D.; Kimple, A.J.; Vandenbeuch, A.; Siderovski, D.P. The stability of tastant detection by mouse lingual chemosensory tissue requires Regulator of G protein Signaling-21 (RGS21). Chem. Senses 2021, 46, bjab048. [Google Scholar] [CrossRef]
- von Buchholtz, L.; Elischer, A.; Tareilus, E.; Gouka, R.; Kaiser, C.; Breer, H.; Conzelmann, S. RGS21 is a novel regulator of G protein signalling selectively expressed in subpopulations of taste bud cells. Eur. J. Neurosci. 2004, 19, 1535–1544. [Google Scholar] [CrossRef]
- Óvilo, C.; Benítez, R.; Núñez, Y.; Peiró-Pastor, R.; García, F.; De Mercado, E.; Gómez-Izquierdo, E.; García-Casco, J.; López-Bote, C.; Muñoz, M. Expression and structural analysis of taste receptor genes in Iberian and Duroc pigs. Genet. Sel. Evol. 2025, 57, 22. [Google Scholar] [CrossRef]
- Zhang, Z.; Xiao, Q.; Zhang, Q.Q.; Sun, H.; Chen, J.C.; Li, Z.C.; Xue, M.; Ma, P.P.; Yang, H.J.; Xu, N.Y.; et al. Genomic analysis reveals genes affecting distinct phenotypes among different Chinese and western pig breeds. Sci. Rep. 2018, 8, 13352. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.H.; Xu, J.Y.; Tang, Z.S.; Wang, L.; Yin, D.; Fan, Y.; Zhang, D.D.; Deng, F.; Zhang, Y.P.; Zhang, H.H.; et al. A gene prioritization method based on a swine multi-omics knowledgebase and a deep learning model. Commun. Biol. 2020, 3, 502. [Google Scholar] [CrossRef] [PubMed]
- Xiong, H.L.; Zhang, V.; Zhao, Z.Y. Investigation of single nucleotide polymorphisms in differentially expressed genes and proteins reveals the genetic basis of skeletal muscle growth differences between Tibetan and Large White pigs. Anim. Biosci. 2024, 37, 2021–2032. [Google Scholar] [CrossRef]
- Chen, Z.Y.; Wu, X.Q.; Zheng, D.B.; Wang, Y.L.; Chai, J.; Zhang, T.H.; Wu, P.X.; Wei, M.H.; Zhou, T.; Long, K.R.; et al. Single-Nucleus RNA Sequencing Reveals Cellular Transcriptome Features at Different Growth Stages in Porcine Skeletal Muscle. Cells 2025, 14, 37. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.Y.; Liu, S.Q.; Zhang, S.; Wang, Y.Z.; Zhou, Y.B.; Shan, T.Z. Single-nucleus transcriptomics reveal the cytological mechanism of conjugated linoleic acids in regulating intramuscular fat deposition. Elife 2025, 13, RP99790. [Google Scholar] [CrossRef]
- Wang, L.Y.; Zhou, Y.B.; Wang, Y.Z.; Shan, T.Z. Integrative cross-species analysis reveals conserved and unique signatures in fatty skeletal muscles. Sci. Data 2024, 11, 290. [Google Scholar] [CrossRef]
- Lindstrand, A.; Grigelioniene, G.; Nilsson, D.; Pettersson, M.; Hofmeister, W.; Anderlid, B.M.; Kant, S.G.; Ruivenkamp, C.A.; Gustavsson, P.; Valta, H.; et al. Different mutations in PDE4D associated with developmental disorders with mirror phenotypes. J. Med. Genet. 2014, 51, 45–54. [Google Scholar] [CrossRef]
- Himes, B.E.; Hunninghake, G.M.; Baurley, J.W.; Rafaels, N.M.; Sleiman, P.; Strachan, D.P.; Wilk, J.B.; Willis-Owen, S.A.; Klanderman, B.; Lasky-Su, J.; et al. Genome-wide association analysis identifies PDE4D as an asthma-susceptibility gene. Am. J. Hum. Genet. 2009, 84, 581–593. [Google Scholar] [CrossRef]
- Xu, R.Y.; Song, J.L.; Ruze, R.; Chen, Y.; Yin, X.P.; Wang, C.C.; Zhao, Y.P. SQLE promotes pancreatic cancer growth by attenuating ER stress and activating lipid rafts-regulated Src/PI3K/Akt signaling pathway. Cell Death Dis. 2023, 14, 497. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.Y.; Zhang, Y.W.; Zhang, B.; Zhong, H.A.; Lu, Y.F.; Zhang, H. Candidate gene screening for lipid deposition using combined transcriptomic and proteomic data from Nanyang black pigs. BMC Genom. 2021, 22, 441. [Google Scholar] [CrossRef] [PubMed]
- Ha, J.; Kwon, S.; Hwang, J.H.; Park, D.H.; Kim, T.W.; Kang, D.G.; Yu, G.E.; Park, H.C.; An, S.M.; Kim, C.W. Squalene epoxidase plays a critical role in determining pig meat quality by regulating adipogenesis, myogenesis, and ROS scavengers. Sci. Rep. 2017, 7, 16740. [Google Scholar] [CrossRef]
- Naganuma, T.; Sato, Y.; Sassa, T.; Ohno, Y.; Kihara, A. Biochemical characterization of the very long-chain fatty acid elongase ELOVL7. FEBS Lett. 2011, 585, 3337–3341. [Google Scholar] [CrossRef]
- Crespo-Piazuelo, D.; Criado-Mesas, L.; Revilla, M.; Castelló, A.; Noguera, J.L.; Fernández, A.I.; Ballester, M.; Folch, J.M. Identification of strong candidate genes for backfat and intramuscular fatty acid composition in three crosses based on the Iberian pig. Sci Rep. 2020, 10, 13962. [Google Scholar] [CrossRef]
- Passols, M.; Llobet-Cabau, F.; Sebastià, C.; Castelló, A.; Valdés-Hernández, J.; Criado-Mesas, L.; Sánchez, A.; Folch, J.M. Identification of genomic regions, genetic variants and gene networks regulating candidate genes for lipid metabolism in pig muscle. Animal 2023, 17, 101033. [Google Scholar] [CrossRef]
- Zhang, J.J.; Zhang, Y.F.; Gong, H.F.; Cui, L.L.; Ma, J.W.; Chen, C.Y.; Ai, H.S.; Xiao, S.J.; Huang, L.S.; Yang, B. Landscape of Loci and Candidate Genes for Muscle Fatty Acid Composition in Pigs Revealed by Multiple Population Association Analysis. Front Genet. 2019, 10, 1067. [Google Scholar] [CrossRef]
- Zhang, W.C.; Zhang, J.J.; Cui, L.L.; Ma, J.W.; Chen, C.Y.; Ai, H.S.; Xie, X.H.; Li, L.; Xiao, S.J.; Huang, L.S.; et al. Genetic architecture of fatty acid composition in the muscle revealed by genome-wide association studies on diverse pig populations. Genet. Sel. Evol. 2016, 48, 5. [Google Scholar] [CrossRef]
- Yang, B.; Zhang, W.C.; Zhang, Z.Y.; Fan, Y.; Xie, X.H.; Ai, H.S.; Ma, J.W.; Xiao, S.J.; Huang, L.S.; Ren, J. Genome-Wide Association Analyses for Fatty Acid Composition in Porcine Muscle and Abdominal Fat Tissues. PLoS ONE 2013, 8, e65554. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.Y.; Qi, X.L.; Hu, M.Y.; Lin, R.Y.; Hou, Y.; Wang, Z.X.; Zhou, H.H.; Zhao, Y.X.; Luan, Y.; Zhao, S.H.; et al. Transcriptome Analysis of Adipose Tissue Indicates That the cAMP Signaling Pathway Affects the Feed Efficiency of Pigs. Genes 2018, 9, 336. [Google Scholar] [CrossRef]
- Cook, E.C.L.; Nelson, J.K.; Sorrentino, V.; Koenis, D.; Moeton, M.; Scheij, S.; Ottenhoff, R.; Bleijlevens, B.; Loregger, A.; Zelcer, N. Identification of the ER-resident E3 ubiquitin ligase RNF145 as a novel LXR-regulated gene. PLoS ONE 2017, 12, e0172721. [Google Scholar] [CrossRef]
- Jiang, Y.; You, Q.X.; Mu, F.X.; Xiang, S.Q.; Zhang, N. Endoplasmic reticulum stress and unfolded protein response play roles in recurrent pregnancy loss: A bioinformatics study. J. Reprod. Immunol. 2025, 168, 104446. [Google Scholar] [CrossRef]
- Zhu, S.L.; Zhang, J.W.; Wang, W.; Jiang, X.; Chen, Y.Q. Blockage of NDUFB9-SCD1 pathway inhibits adipogenesis Blockage of NDUFB9-SCD1 pathway inhibits adipogenesis. J. Physiol. Biochem. 2022, 78, 377–388. [Google Scholar] [CrossRef] [PubMed]
- Yoon, A.; Tammen, S.A.; Park, S.; Han, S.N.; Choi, S.W. Genome-wide hepatic DNA methylation changes in high-fat diet-induced obese mice. Nutr. Res. Pract. 2017, 11, 105–113. [Google Scholar] [CrossRef] [PubMed]
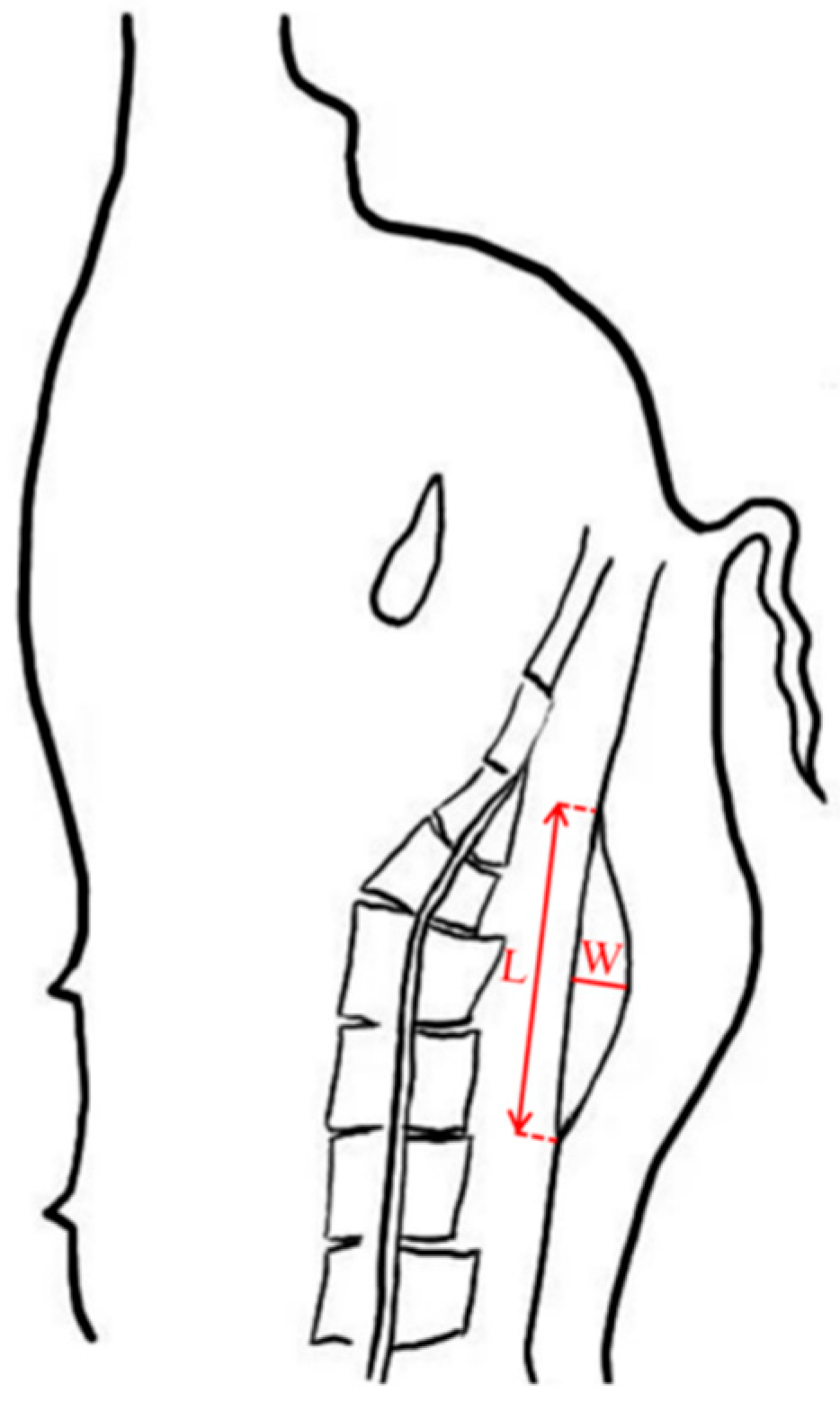
| Trait | Mean ± (SD) | Min | Max | CV (%) | h2 ± (SE) | p |
|---|---|---|---|---|---|---|
| GML (mm) | 103.99 ± 17.74 | 26.86 | 153.83 | 17.06 | 0.43 ± 0.10 | 1.22 × 10−5 |
| GMW (mm) | 20.45 ± 5.37 | 4.04 | 35.84 | 26.29 | 0.40 ± 0.10 | 1.13 × 10−4 |
| GMA (mm2) | 1086.60 ± 400.56 | 127.10 | 2364.20 | 36.86 | 0.46 ± 0.10 | 6.26× 10−6 |
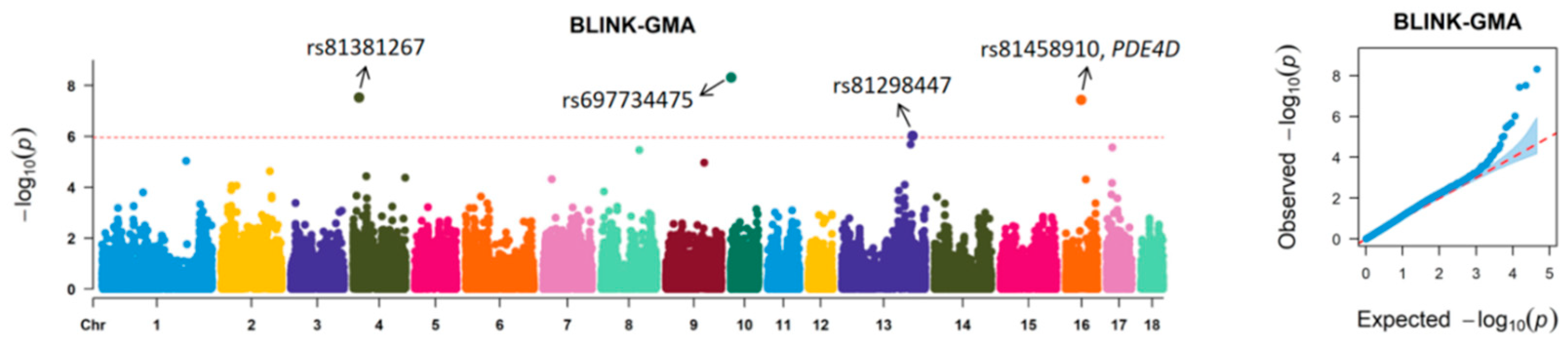
| SSC | Position | SNP ID | p | EPV (%) | Distance (kb) | Nearest Gene |
|---|---|---|---|---|---|---|
| 4 | 14,866,061 | rs81381267 | 2.98 × 10−8 | 11.30 | 67.27 | MTSS1 |
| 10 | 1,441,934 | rs697734475 | 4.84 × 10−9 | 6.21 | −72.29 | RGS21 |
| 13 | 176,619,148 | rs81298447 | 9.39 × 10−7 | 9.51 | −139.70 | ROBO1 |
| 16 | 39,179,599 | rs81458910 | 3.64 × 10−8 | 13.41 | within | PDE4D |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, Y.; Bai, C.; Fei, J.; Ke, J.; Chen, C.; Zhang, X.; Liu, W.; Li, J.; Liang, S.; Sun, B.; et al. Genome-Wide Association Study of Gluteus Medius Muscle Size in a Crossbred Pig Population. Vet. Sci. 2025, 12, 730. https://doi.org/10.3390/vetsci12080730
He Y, Bai C, Fei J, Ke J, Chen C, Zhang X, Liu W, Li J, Liang S, Sun B, et al. Genome-Wide Association Study of Gluteus Medius Muscle Size in a Crossbred Pig Population. Veterinary Sciences. 2025; 12(8):730. https://doi.org/10.3390/vetsci12080730
Chicago/Turabian StyleHe, Yu, Chunyan Bai, Junwen Fei, Juan Ke, Changyi Chen, Xiaoran Zhang, Wuyang Liu, Jing Li, Shuang Liang, Boxing Sun, and et al. 2025. "Genome-Wide Association Study of Gluteus Medius Muscle Size in a Crossbred Pig Population" Veterinary Sciences 12, no. 8: 730. https://doi.org/10.3390/vetsci12080730
APA StyleHe, Y., Bai, C., Fei, J., Ke, J., Chen, C., Zhang, X., Liu, W., Li, J., Liang, S., Sun, B., & Sun, H. (2025). Genome-Wide Association Study of Gluteus Medius Muscle Size in a Crossbred Pig Population. Veterinary Sciences, 12(8), 730. https://doi.org/10.3390/vetsci12080730







