Transcriptomic and Lipidomic Analysis Reveals the Regulatory Network of Lipid Metabolism in Cannabis sativa
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. RNA Sequencing and Quantitative Real-Time PCR (qRT-PCR)
2.3. Lipid Extraction and Identification
2.4. Statistical Analysis and Data Visualization
3. Results
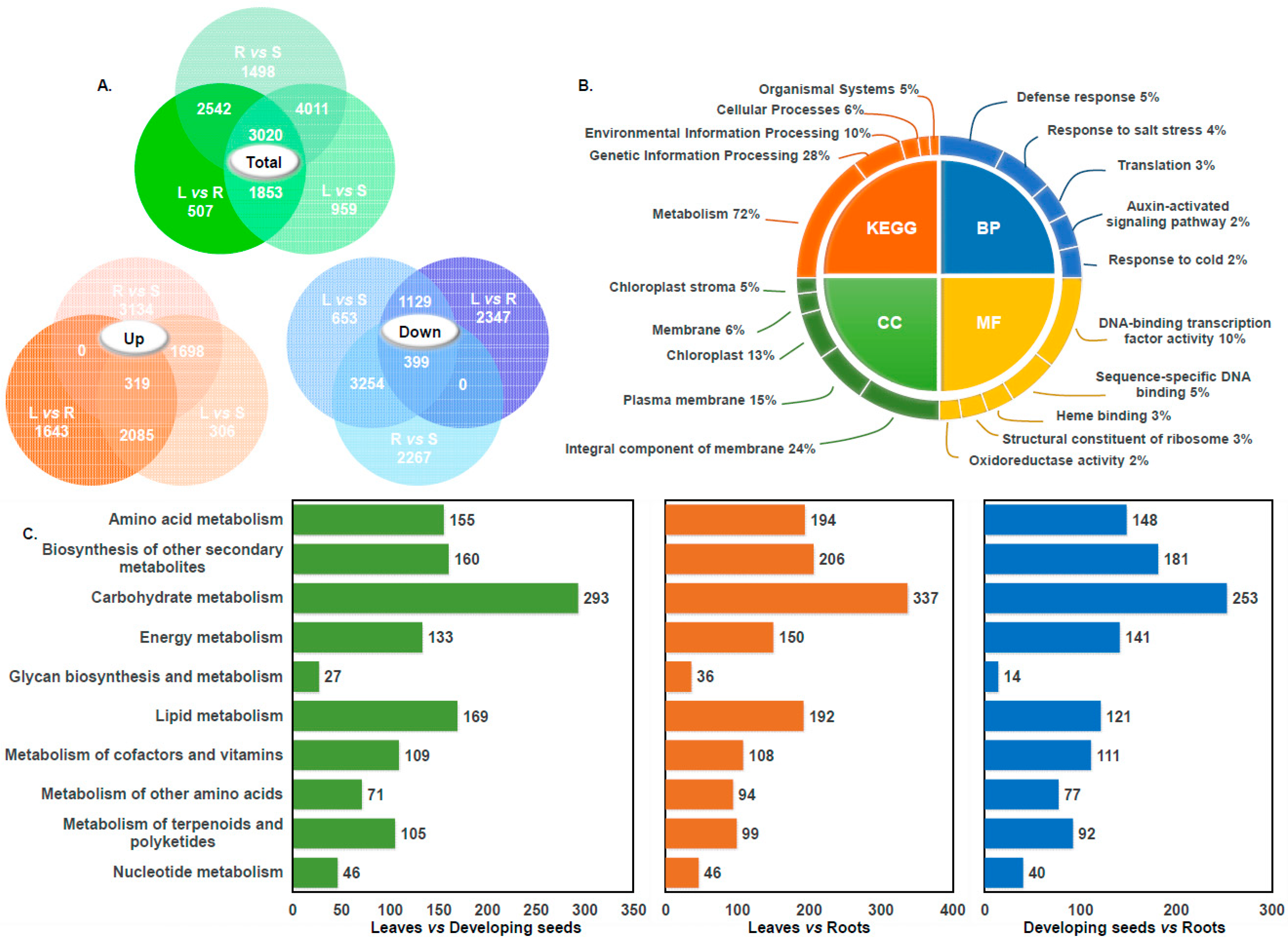
3.1. Transcriptomic Analysis of Hemp
3.2. The GO and KEGG Enrichment Analysis
3.3. Lipid Metabolism-Related Genes
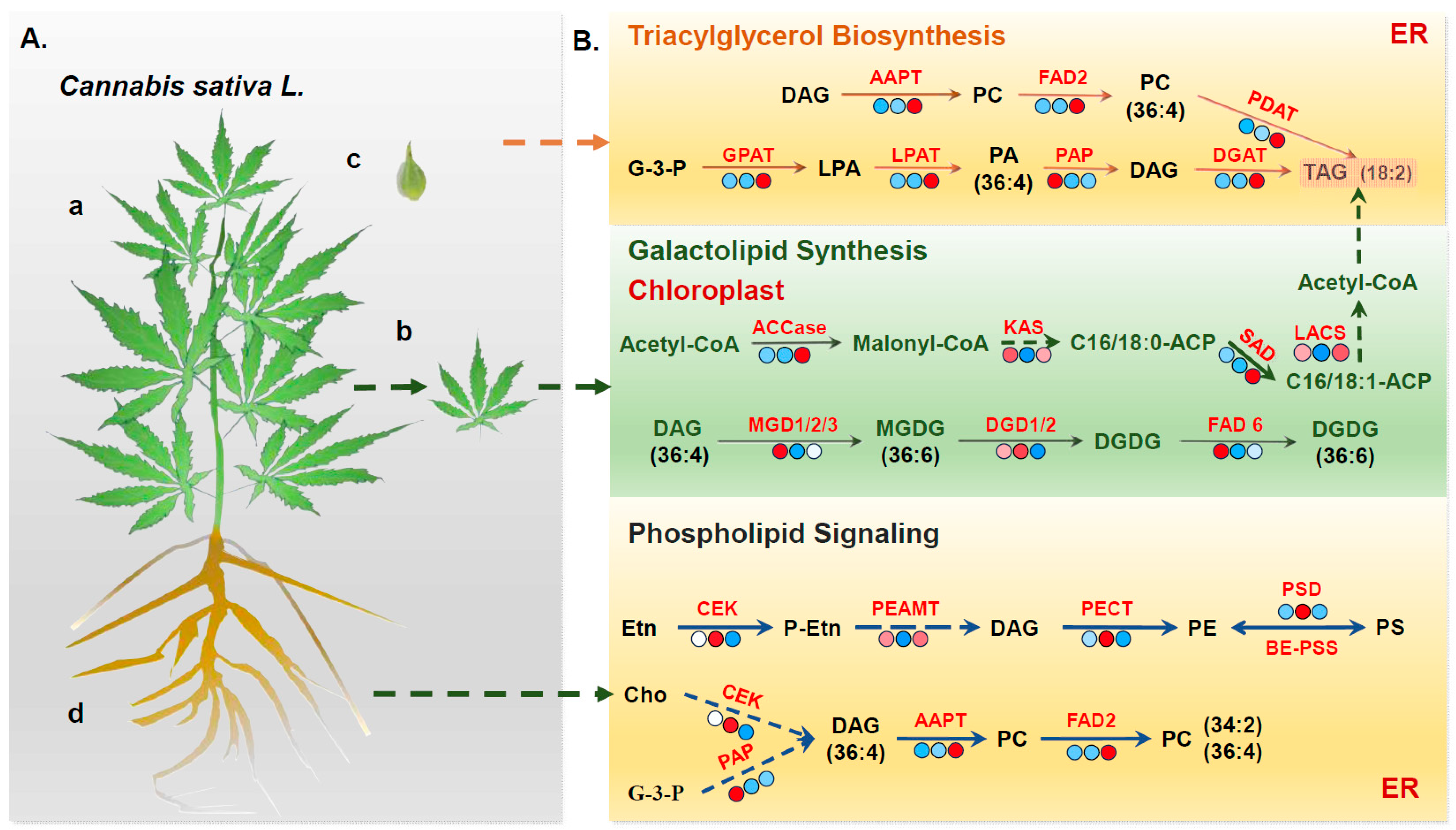
3.3.1. Genes Associated with Triglyceride Biosynthesis
3.3.2. Analysis of Genes Involved in Photosynthetic-Membrane Lipid Synthesis
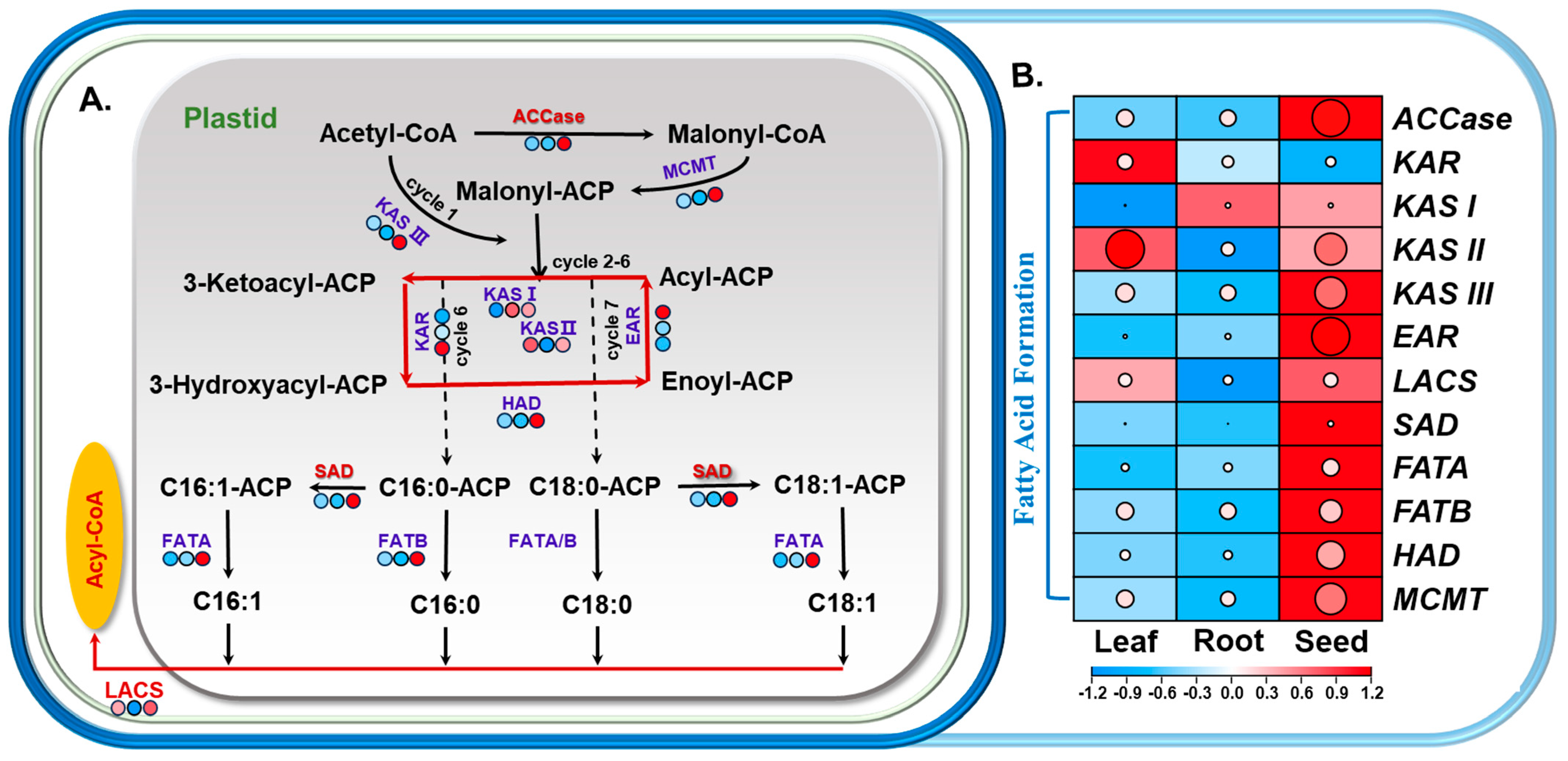
3.3.3. Analysis of DEGs in Fatty-Acid Metabolic Pathways
3.4. Lipidome of Hemp
3.5. Analysis of Molecular Species Types of Lipids in Different Tissue Sites
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Cai, S.; Zhang, Z.; Huang, S.; Bai, X.; Huang, Z.; Zhang, Y.; Huang, L.; Tang, W.; Haughn, G.; You, S.; et al. CannabisGDB: A comprehensive genomic database for Cannabis sativa L. Plant Biotechnol. J. 2021, 19, 857–859. [Google Scholar] [CrossRef]
- Farinon, B.; Molinari, R.; Costantini, L.; Merendino, N. The seed of industrial hemp Cannabis sativa: Nutritional quality and potential functionality for human health and nutrition. Nutrients 2020, 12, 1935. [Google Scholar] [CrossRef]
- Gonçalves, E.C.D.; Baldasso, G.M.; Bicca, M.A.; Paes, R.S.; Capasso, R.; Dutra, R.C. Terpenoids, cannabimimetic ligands, beyond the cannabis plant. Molecules 2020, 25, 1567. [Google Scholar] [CrossRef]
- Lynch, R.C.; Padgitt-Cobb, L.K.; Garfinkel, A.R.; Knaus, B.J.; Hartwick, N.T.; Allsing, N.; Aylward, A.; Bentz, P.C.; Carey, S.B.; Mamerto, A.; et al. Domesticated cannabinoid synthases amid a wild mosaic cannabis pangenome. Nature 2025, 643, 1001–1010. [Google Scholar] [CrossRef]
- Hurgobin, B.; Tamiru-Oli, M.; Welling, M.T.; Doblin, M.S.; Bacic, A.; Whelan, J.; Lewsey, M.G. Recent advances in Cannabis sativa genomics research. New Phytol. 2021, 230, 73–89. [Google Scholar] [CrossRef]
- Karus, M.; Vogt, D. European hemp industry: Cultivation, processing and product lines. Euphytica 2004, 140, 7–12. [Google Scholar] [CrossRef]
- Liu, Y.; Li, S.; Shen, M.; Guo, F.; Li, M.; Cai, S.; Huang, J.; Wu, J.; Li, X.; Peng, L.; et al. Integration of transcriptome and metabolome provides insights into metabolites and pathways associated with antiproliferative activity of cannabis flower extracts. Ind. Crops Prod. 2025, 223, 120239. [Google Scholar] [CrossRef]
- Nie, J.; Ma, W.; Ma, X.; Zhu, D.; Li, X.; Wang, C.; Xu, G.; Chen, C.; Luo, D.; Xie, S.; et al. Integrated Transcriptomic and Metabolomic Analysis Reveal the Dynamic Process of Bama Hemp Seed Development and the Accumulation Mechanism of α-Linolenic Acid and Linoleic Acid. J. Agric. Food Chem. 2024, 72, 10862–10878. [Google Scholar] [CrossRef] [PubMed]
- Kim, O.Y.; Song, J. Important roles of linoleic acid and α-linolenic acid in regulating cognitive impairment and neuropsychiatric issues in metabolic-related dementia. Life Sci. 2024, 337, 122356. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Shanklin, J. Triacylglycerol metabolism, function, and accumulation in plant vegetative tissues. Annu. Rev. Plant Biol. 2016, 67, 179–206. [Google Scholar] [CrossRef]
- Yang, Y.; Benning, C. Functions of triacylglycerols during plant development and stress. Curr. Opin. Biotechnol. 2018, 49, 191–198. [Google Scholar] [CrossRef]
- Shomo, Z.D.; Mahboub, S.; Vanviratikul, H.; McCormick, M.; Tulyananda, T.; Roston, R.L.; Warakanont, J. All members of the arabidopsis DGAT and PDAT acyltransferase families operate during high and low temperatures. Plant Physiol. 2024, 195, 685–697. [Google Scholar] [CrossRef]
- Melero, A.; Jiménez-Rojo, N. Cracking the membrane lipid code. Curr. Opin. Cell Biol. 2023, 83, 102203. [Google Scholar] [CrossRef]
- Nakamura, Y. Plant phospholipid diversity: Emerging functions in metabolism and protein–lipid interactions. Trends Plant Sci. 2017, 22, 1027–1040. [Google Scholar] [CrossRef]
- Kobayashi, K. Role of membrane glycerolipids in photosynthesis, thylakoid biogenesis and chloroplast development. J. Plant Res. 2016, 129, 565–580. [Google Scholar] [CrossRef]
- Yu, L.; Zhou, C.; Fan, J.; Shanklin, J.; Xu, C. Mechanisms and functions of membrane lipid remodeling in plants. Plant J. 2021, 107, 37–53. [Google Scholar] [CrossRef]
- Cook, R.; Lupette, J.; Benning, C. The role of chloroplast membrane lipid metabolism in plant environmental responses. Cells 2021, 10, 706. [Google Scholar] [CrossRef] [PubMed]
- LaBrant, E.; Barnes, A.C.; Roston, R.L. Lipid transport required to make lipids of photosynthetic membranes. Photosynth. Res. 2018, 138, 345–360. [Google Scholar] [CrossRef] [PubMed]
- Zafar, S.; Li, Y.-L.; Li, N.-N.; Zhu, K.-M.; Tan, X.-L. Recent advances in enhancement of oil content in oilseed crops. J. Biotechnol. 2019, 301, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Kaushal, C.; Sachdev, M.; Parekh, M.; Gowrishankar, H.; Jain, M.; Sankaranarayanan, S.; Pathak, B. Transcriptional engineering for value enhancement of oilseed crops: A forward perspective. Front. Genome Ed. 2025, 6, 1488024. [Google Scholar] [CrossRef]
- Khan, A.; Awan, A.A.; Yasin, M.; Ramzan, A.; Cheema, M.W.A.; Jan, A. Edible Oilseeds: Historical Perspectives, Recent Advances, and Future Directions. Food Science and Nutrition; IntechOpen: London, UK, 2025; p. 156. [Google Scholar] [CrossRef]
- Spano, M.; Di Matteo, G.; Rapa, M.; Ciano, S.; Ingallina, C.; Cesa, S.; Menghini, L.; Carradori, S.; Giusti, A.M.; Di Sotto, A.; et al. Commercial hemp seed oils: A multimethodological characterization. Appl. Sci. 2020, 10, 6933. [Google Scholar] [CrossRef]
- Yan, B.; Chang, C.; Gu, Y.; Sui, Y.; Zheng, N.; Fang, Y.; Zhang, Y.; Zhang, M.; Xu, J.; Zhang, L. Lipidomic remodeling in Cannabis sativa L. under cold tolerance. Ind. Crops Prod. 2025, 224, 120346. [Google Scholar] [CrossRef]
- Holčapek, M.; Liebisch, G.; Ekroos, K. Lipidomic analysis. Anal. Chem. 2018, 90, 4249–4257. [Google Scholar] [CrossRef]
- Beisson, F.; Koo, A.J.K.; Ruuska, S.; Schwender, J.; Pollard, M.; Thelen, J.J.; Paddock, T.; Salas, J.J.; Savage, L.; Milcamps, A.; et al. Arabidopsis genes involved in acyl lipid metabolism. A 2003 census of the candidates, a study of the distribution of expressed sequence tags in organs, and a web-based database. Plant Physiol. 2003, 132, 681–697. [Google Scholar] [CrossRef]
- Ge, S.; Liu, D.; Chu, M.; Liu, X.; Wei, Y.; Che, X.; Zhu, L.; He, L.; Xu, J. Dynamic and adaptive membrane lipid remodeling in leaves of sorghum under salt stress. Crop J. 2022, 10, 1557–1569. [Google Scholar] [CrossRef]
- Liu, H.; Xin, W.; Wang, Y.; Zhang, D.; Wang, J.; Zheng, H.; Yang, L.; Nie, S.; Zou, D. An integrated analysis of the rice transcriptome and lipidome reveals lipid metabolism plays a central role in rice cold tolerance. BMC Plant Biol. 2022, 22, 91. [Google Scholar] [CrossRef]
- Pfaff, J.; Denton, A.K.; Usadel, B.; Pfaff, C. Phosphate starvation causes different stress responses in the lipid metabolism of tomato leaves and roots. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2020, 1865, 158763. [Google Scholar] [CrossRef]
- Zhao, X.; Wei, Y.; Zhang, J.; Yang, L.; Liu, X.; Zhang, H.; Shao, W.; He, L.; Li, Z.; Zhang, Y.; et al. Membrane Lipids’ Metabolism and Transcriptional Regulation in Maize Roots Under Cold Stress. Front. Plant Sci. 2021, 12, 639132. [Google Scholar] [CrossRef] [PubMed]
- Bekele, B.; Andargie, M.; Gallach, M.; Beyene, D.; Tesfaye, K. Decoding gene expression dynamics during seed development in sesame (Sesamum indicum L.) through RNA-Seq analysis. Genomics 2025, 117, 110997. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xiao, L.; Cao, H.; Cao, Y.; Zhang, L. Phylogenomics and functional analysis of glycerol-3-phosphate acyltransferase (GPAT) genes: A critical role in lipid biosynthesis. Physiol. Plant. 2024, 176, e14509. [Google Scholar] [CrossRef] [PubMed]
- Mi, C.; Zhang, Y.; Zhao, Y.; Lin, L. Mechanisms of low nighttime temperature promote oil accumulation in brassica napus L. based on in-depth transcriptome analysis. Physiol. Plant. 2024, 176, e14372. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, C.F.; Weusthuis, R.A.; D’Adamo, S.; Wijffels, R.H. Effect of single and combined expression of lysophosphatidic acid acyltransferase, glycerol-3-phosphate acyltransferase, and diacylglycerol acyltransferase on lipid accumulation and composition in neochloris oleoabundans. Front. Plant Sci. 2019, 10, 1573. [Google Scholar] [CrossRef] [PubMed]
- Bhatt-Wessel, B.; Jordan, T.W.; Miller, J.H.; Peng, L. Role of DGAT enzymes in triacylglycerol metabolism. Arch. Biochem. Biophys. 2018, 655, 1–11. [Google Scholar] [CrossRef]
- Wang, L.; Xu, S.; Zhou, M.; Hu, H.; Li, J. The role of DGAT1 and DGAT2 in tumor progression via fatty acid metabolism: A comprehensive review. Int. J. Biol. Macromol. 2024, 278, 134835. [Google Scholar] [CrossRef]
- Xu, J.; Francis, T.; Mietkiewska, E.; Giblin, E.M.; Barton, D.L.; Zhang, Y.; Zhang, M.; Taylor, D.C. Cloning and characterization of an acyl-CoA-dependent diacylglycerol acyltransferase 1 (DGAT1) gene from tropaeolum majus, and a study of the functional motifs of the DGAT protein using site-directed mutagenesis to modify enzyme activity and oil content. Plant Biotechnol. J. 2008, 6, 799–818. [Google Scholar] [CrossRef]
- Nakamura, Y. Headgroup biosynthesis of phosphatidylcholine and phosphatidylethanolamine in seed plants. Prog. Lipid Res. 2021, 82, 101091. [Google Scholar] [CrossRef]
- Wang, B.; Tontonoz, P. Phospholipid remodeling in physiology and disease. Annu. Rev. Physiol. 2019, 81, 165–188. [Google Scholar] [CrossRef]
- Chen, D.; Yuan, X.; Liang, L.; Liu, K.; Ye, H.; Liu, Z.; Liu, Y.; Huang, L.; He, W.; Chen, Y.; et al. Overexpression of acetyl-CoA carboxylase increases fatty acid production in the green alga chlamydomonas reinhardtii. Biotechnol. Lett. 2019, 41, 1133–1145. [Google Scholar] [CrossRef]
- Meng, Q.; Liang, H.; Gao, H. Roles of multiple KASIII homologues of Shewanella oneidensis in initiation of fatty acid synthesis and in cerulenin resistance. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2018, 1863, 1153–1163. [Google Scholar] [CrossRef]
- Xie, Z.; Mi, Y.; Kong, L.; Gao, M.; Chen, S.; Chen, W.; Meng, X.; Sun, W.; Chen, S.; Xu, Z. Cannabis sativa: Origin and history, glandular trichome development, and cannabinoid biosynthesis. Hortic. Res. 2023, 10, uhad150. [Google Scholar] [CrossRef] [PubMed]
- Liang, M.; Zhang, X.; Dong, Q.; Li, H.; Guo, S.; Luan, H.; Jia, P.; Yang, M.; Qi, G. Metabolomics and transcriptomics provide insights into lipid biosynthesis in the embryos of walnut (Juglans regia L.). Plants 2023, 12, 538. [Google Scholar] [CrossRef]
- Shang, X.; Cheng, C.; Ding, J.; Guo, W. Identification of candidate genes from the SAD gene family in cotton for determination of cottonseed oil composition. Mol. Genet. Genom. MGG 2017, 292, 173–186. [Google Scholar] [CrossRef]
- Yang, J.; Chen, B.; Manan, S.; Li, P.; Liu, C.; She, G.; Zhao, S.; Zhao, J. Critical metabolic pathways and SAD/FADs, WRI1s, and DGATs cooperate for high-oleic acid oil production in developing oil tea (Camellia oleifera) seeds. Hortic. Res. 2022, 9, uhac087. [Google Scholar] [CrossRef]
- Lyu, J.; Gao, R.; Guo, Z. Galactosyldiacylglycerols: From a photosynthesis-associated apparatus to structure-defined in vitro assembling. J. Agric. Food Chem. 2021, 69, 8910–8928. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; He, L.; Zhao, C.; Wang, F.; Yan, B.; Gao, Y.; Li, Z.; Yang, K.; Xu, J. Biochemical and Transcriptional Regulation of Membrane Lipid Metabolism in Maize Leaves under Low Temperature. Front. Plant Sci. 2017, 8, 2053. [Google Scholar] [CrossRef] [PubMed]
- Heemskerk, J.W.; Storz, T.; Schmidt, R.R.; Heinz, E. Biosynthesis of digalactosyldiacylglycerol in plastids from 16:3 and 18:3 plants. Plant Physiol. 1990, 93, 1286–1294. [Google Scholar] [CrossRef]
- Hölzl, G.; Dörmann, P. Chloroplast lipids and their biosynthesis. Annu. Rev. Plant Biol. 2019, 70, 51–81. [Google Scholar] [CrossRef] [PubMed]
- Hou, Q.; Ufer, G.; Bartels, D. Lipid signalling in plant responses to abiotic stress. Plant Cell Environ. 2016, 39, 1029–1048. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, B.; Chang, C.; Sui, Y.; Zheng, N.; Fang, Y.; Zhang, Y.; Zhang, M.; He, D.; Zhang, L. Transcriptomic and Lipidomic Analysis Reveals the Regulatory Network of Lipid Metabolism in Cannabis sativa. Foods 2025, 14, 2809. https://doi.org/10.3390/foods14162809
Yan B, Chang C, Sui Y, Zheng N, Fang Y, Zhang Y, Zhang M, He D, Zhang L. Transcriptomic and Lipidomic Analysis Reveals the Regulatory Network of Lipid Metabolism in Cannabis sativa. Foods. 2025; 14(16):2809. https://doi.org/10.3390/foods14162809
Chicago/Turabian StyleYan, Bowei, Chuanyi Chang, Yue Sui, Nan Zheng, Yuyan Fang, Yuanye Zhang, Ming Zhang, Dan He, and Liguo Zhang. 2025. "Transcriptomic and Lipidomic Analysis Reveals the Regulatory Network of Lipid Metabolism in Cannabis sativa" Foods 14, no. 16: 2809. https://doi.org/10.3390/foods14162809
APA StyleYan, B., Chang, C., Sui, Y., Zheng, N., Fang, Y., Zhang, Y., Zhang, M., He, D., & Zhang, L. (2025). Transcriptomic and Lipidomic Analysis Reveals the Regulatory Network of Lipid Metabolism in Cannabis sativa. Foods, 14(16), 2809. https://doi.org/10.3390/foods14162809






