Re-Identification of Dollar Spot Pathogen on Creeping Bentgrass and Kentucky Bluegrass in South Korea
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation of the Pathogen from Hosts
2.2. Pathogenicity Tests in Pots
2.3. Pathogenicity Tests in Field
2.4. Koch’s Postulates Test
2.5. Apothecia Production and Morphological Examinations
2.6. DNA Extractions, PCR Amplification, and Sequencing
2.7. Alignment and Phylogenetic Analysis
2.8. Statistical Analyses
3. Results
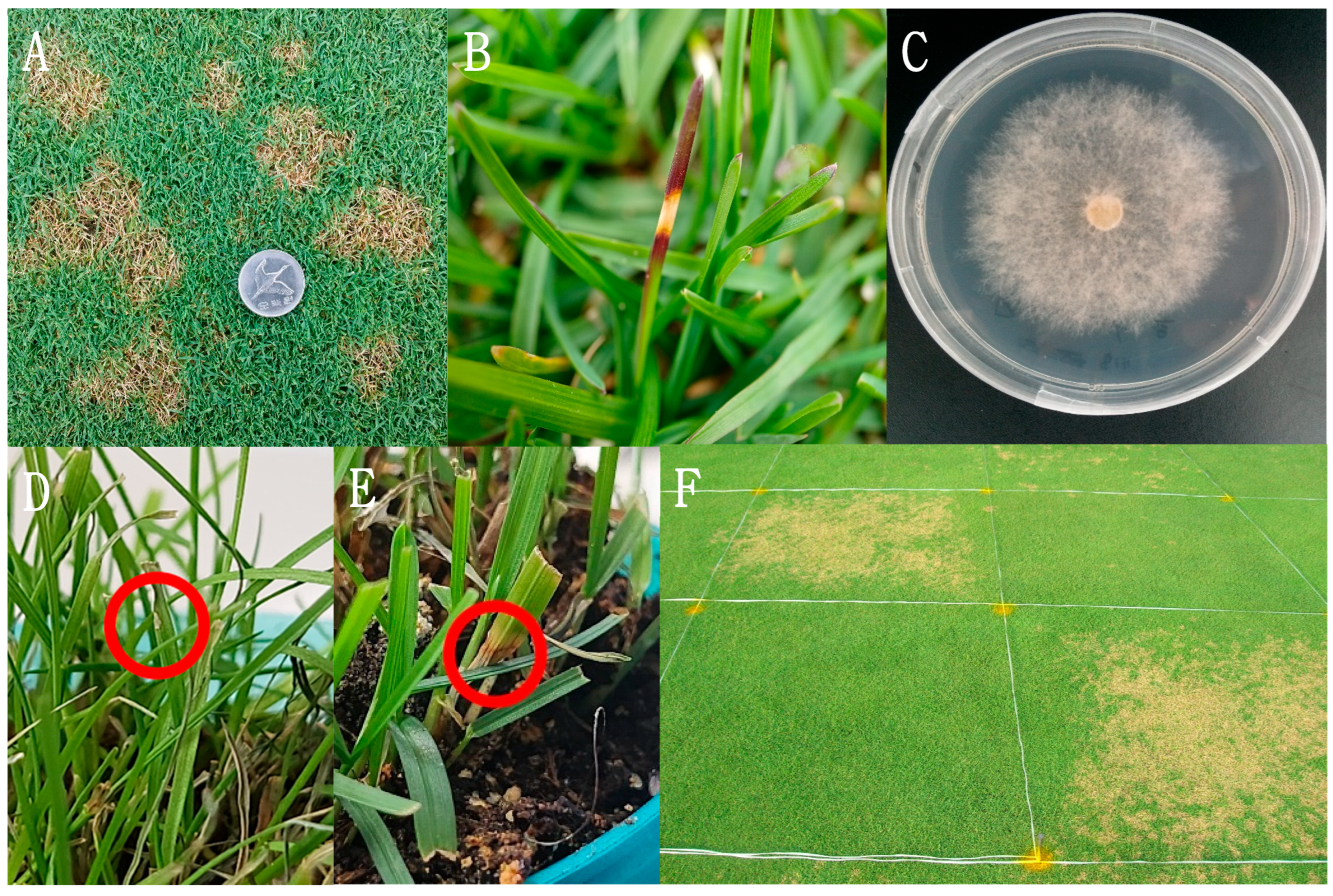
3.1. Isolation of the Pathogen and Pathogenicity Tests and Koch’s Postulates Test
3.2. Morphological Examinations and Apothecia Production
3.3. Molecular Re-Identification of Dollar Spot Pathogen and Phylogenetic Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Smiley, R.W.; Dernoden, P.H.; Clarke, B.B. Compendium of Turfgrass Diseases, 2nd ed.; American Phytopathological Society: St. Paul, MN, USA, 1992. [Google Scholar]
- Lee, J.H.; Min, G.Y.; Shim, G.Y.; Kim, D.S.; Sang, H.; Jung, G.; Kwak, Y.S. Toothpick-aided detection of Sclerotinia homoeocarpa in the turfgrass leaf canopy, thatch, and soil in relation to dollar spot infection centers. Weed Turfgrass Sci. 2015, 4, 376–382. [Google Scholar] [CrossRef]
- Smiley, R.W.; Dernoeden, P.P.; Clarke, B.B. Compendium of Turfgrass Diseases, 3rd ed.; American Phytopathological Society: St. Paul, MN, USA, 2005. [Google Scholar]
- Goodman, D.M.; Burpee, L.L. Biological control of dollar spot disease of creeping bentgrass. Phytopathology 1991, 81, 1438–1446. [Google Scholar] [CrossRef]
- Tredway, L.P.; Tomaso-Peterson, M.; Kerns, J.P.; Clarke, B.B. Compendium of Turfgrass Diseases, 4th ed.; American Phytopathological Society: St. Paul, MN, USA, 2023. [Google Scholar]
- Held, D.W.; Potter, D.A. Prospects for managing turfgrass pests with reduced chemical inputs. Annu. Rev. Entomol. 2012, 57, 329–354. [Google Scholar] [CrossRef]
- Bennett, F.T. Dollar spot disease of turf and its causal organism, Sclerotinia homoeocarpa n. sp. Ann. Appl. Biol. 1937, 24, 236–257. [Google Scholar] [CrossRef]
- Whetzel, H.H. The cypericolous and juncicolous species of Sclerotinia. Farlowia 1946, 2, 385–437. [Google Scholar] [CrossRef]
- Salgado-Salazar, C.; Beirn, L.A.; Ismaiel, A.; Boehm, M.J.; Carbone, I.; Putman, A.I.; Tredway, L.P.; Clarke, B.B.; Crouch, J.A. Clarireedia: A new fungal genus comprising four pathogenic species responsible for dollar spot disease of turfgrass. Fungal Biol. 2018, 122, 761–773. [Google Scholar] [CrossRef]
- Takao, T.; Toshihiro, H.; Koya, S. Re-identification of Clarireedia spp. causing dollar spot disease of turfgrasses in Japan. J. Jpn. Soc. Turfgrass Sci. 2022, 49, 71–77. [Google Scholar]
- Hu, J.; Zhou, Y.; Geng, J.; Dai, Y.; Ren, H.; Lamour, K. A new dollar spot disease of turfgrass caused by Clarireedia paspali. Mycol. Prog. 2019, 18, 1423–1435. [Google Scholar] [CrossRef]
- Bahri, B.A.; Parvathaneni, R.K.; Spratling, W.T.; Saxena, H.; Sapkota, S.; Raymer, P.L.; Martinez-Espinoza, A.D. Whole genome sequencing of Clarireedia aff. paspali reveals potential pathogenesis factors in Clarireedia species, causal agents of dollar spot in turfgrass. Front. Genet. 2023, 13, 1033437. [Google Scholar] [CrossRef]
- Stackhouse, T.; Bass, A.; Waliullah, S.; Ali, E.; Bahri, B.A.; Martinez-Espinoza, A. Probe-based loop-mediated isothermal amplification assay for rapid detection of two Clarireedia spp., the causal agent of dollar spot of turfgrass. Phytopathology 2024, 108, 3115–3122. [Google Scholar] [CrossRef]
- Huangwei, Z.; Yinglu, D.; Yuxin, Z.; Jian, H.; Kurt, L.; Zhimin, Y. Clarireedia hainanense: A new species is associated with dollar spot of turfgrass in Hainan, China. Plant Dis. 2022, 106, 996–1002. [Google Scholar]
- Hu, J.; Zhang, H.; Dong, Y.; Jiang, S.; Lamour, K.; Liu, J.; Chen, Y.; Yang, Z. Global distributions of Clarireedia species and their in vitro sensitivity profiles to fungicides. Agronomy 2021, 11, 2036. [Google Scholar] [CrossRef]
- Sang, H.; Hulvey, J.; Popko, J.T., Jr.; Lopes, J.; Swaminathan, A.; Chang, T.; Jung, G. A pleiotropic drug resistance transporter is involved in reduced sensitivity to multiple fungicide classes in Sclerotinia homoeocarpa (F.T. Bennett). Mol. Plant Pathol. 2015, 16, 251–261. [Google Scholar] [CrossRef]
- Sapkota, S.; Catching, K.E.; Raymer, P.L.; Martinez-Espinoza, A.D.; Bahri, B.A. New approaches to an old problem: Dollar spot of turfgrass. Phytopathology 2022, 112, 469–480. [Google Scholar] [CrossRef]
- Ok, C.H.; Popko, J.T., Jr.; Campbell-Nelson, K.; Jung, G. In vitro assessment of Sclerotinia homoeocarpa resistance to fungicides and plant growth regulators. Plant Dis. 2011, 95, 51–56. [Google Scholar] [CrossRef]
- Popko, J.T., Jr.; Ok, C.H.; Campbell-Nelson, K.; Jung, G. The association between in vitro propiconazole sensitivity and field efficacy of five New England Sclerotinia homoeocarpa populations. Plant Dis. 2012, 96, 552–561. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Jiang, S.; Zhao, Z.; Guan, J.; Dong, Y.; Hu, J.; Lamour, K.; Yin, S.; Yang, Z. Fungicide sensitivity of Clarireedia spp. isolates from golf courses in China. Crop Prot. 2021, 149, 105785. [Google Scholar] [CrossRef]
- Ghimire, B.; Aktaruzzaman, M.; Chowdhury, S.R.; Spratling, W.T.; Vermeer, C.B.; Buck, J.W.; Martinez-Espinoza, A.D.; Bahri, B.A. Sensitivity of Clarireedia spp. to benzimidazoles and dimethyl inhibitors fungicides and efficacy of biofungicides on dollar spot of warm season turfgrass. Front. Plant Sci. 2023, 14, 1155670. [Google Scholar] [CrossRef]
- Hu, J.; Yang, J.; Li, J.; Ma, Z.; Yao, W.; Ren, H.; Zhang, F.; Yang, G.; Sun, X.; Xiao, Y. Sensitivity of Sclerotinia homoeocarpa from turfgrass to thiophanate-methyl, iprodione and propiconazole. Chin. J. Pestic. Sci. 2017, 19, 694–700. [Google Scholar]
- Chang, S.W.; Chang, T.H.; Tredway, L.; Jung, G. Aggressiveness of Typhula ishikariensis isolates to cultivars of bentgrass species (Agrostis spp.) under controlled environment conditions. Plant Dis. 2006, 90, 951–956. [Google Scholar] [CrossRef]
- Rengwalska, M.; Simon, P.W. Laboratory evaluation of pink root and Fusarium basal rot resistance in garlic. Plant Dis. 1986, 70, 670–672. [Google Scholar] [CrossRef]
- Chang, S.W.; Chang, T.H.; Hong, J.K.; Park, J.H.; Jung, S.W. Vegetative compatibility grouping of Sclerotinia homoeocarpa isolates infecting turfgrass in South Korea. Asian J. Turfgrass Sci. 2012, 25, 171–176. [Google Scholar]
- Schmitt, I.; Crespo, A.; Divakar, P.K.; Fankhauser, J.D.; Herman-Sackett, E.; Kalb, K.; Nelsen, M.P.; Nelson, N.A.; Rivas Plata, E.; Shimp, A.D.; et al. New primers for promising single-copy genes in fungal phylogenetics and systematics. Persoonia 2009, 23, 35–40. [Google Scholar] [CrossRef] [PubMed]
- Carbone, I.; Kohn, L.M. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 1999, 91, 553–556. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Aynardi, B.A.; Jiménez-Gasco, M.M.; Uddin, W. Effects of isolates of Clarireedia jacksonii and Clarireedia monteithiana on severity of dollar spot in turfgrasses by host type. Eur. J. Plant Pathology. 2019, 155, 817–829. [Google Scholar] [CrossRef]
- Du, Y.; Li, J.; Chen, S.; Xia, Y.; Jin, K. Pathogenicity analysis and comparative genomics reveal the different infection strategies between the generalist Metarhizium anisopliae and the specialist Metarhizium acridum. Pest Manag. Sci. 2024, 80, 820–836. [Google Scholar] [CrossRef]
- Rep, M.; Kistler, H.C. The genomic organization of plant pathogenicity in Fusarium species. Curr. Opin. Plant Biol. 2010, 13, 420–426. [Google Scholar] [CrossRef]
- Weir, B.S.; Johnston, P.R.; Damm, U. The Colletotrichum gloeosporioides species complex. Stud. Mycol. 2012, 73, 115–180. [Google Scholar] [CrossRef]
- Choi, Y.H.; Mageswari, A.; Choi, H.R.; Lee, J.S.; Lee, D.S.; Hong, S.B. Re-identification of Fusarium sambucinum species complex strains in Korea and their literature review. Res. Plant Dis. 2023, 29, 118–129. [Google Scholar] [CrossRef]
- Dučkena, L.; Bimšteine, G.; Bankina, B.; Skinderskis, E.; Roga, A.; Frîdmanis, D. Morphological diversity and mycelial compatibility of Botrytis pseudocinerea and Botrytis cinerea isolated in Latvia. Proc. Latv. Acad. Sci. Sect. B 2024, 78, 197–205. [Google Scholar] [CrossRef]
- Thao, L.D.; Choi, H.; Choi, Y.; Mageswari, A.; Lee, D.; Kim, D.H.; Shin, H.D.; Choi, H.; Ju, H.J.; Hong, S.B. Re-identification of Colletotrichum gloeosporioides species complex isolates in Korea and their host plants. Plant Pathol. J. 2024, 40, 16–29. [Google Scholar] [CrossRef] [PubMed]
- Manawasinghe, I.S.; Phillips, A.J.L.; Xu, J.; Balasuriya, A.; Hyde, K.D.; Stępień, Ł.; Harischandra, D.L.; Karunarathna, A.; Yan, J.; Weerasinghe, J.; et al. Defining a species in fungal plant pathology: Beyond the species level. Fungal Divers. 2021, 109, 267–282. [Google Scholar] [CrossRef]
- Dallacorte, L.V.; Bosse, M.A.; Capelin, D.; Paladini, M.V.; Cattani, F.; Remor, M.B.; Lima, J.D.; Perboni, A.T.; Marchese, J.A. Economic versus technical efficiency in using ASM combined with fungicides to elicit wheat varieties with different disease susceptibilities. Heliyon 2023, 9, e17012. [Google Scholar] [CrossRef] [PubMed]
- Devyatkina, T.F.; Chigorin, S.S.; Silaev, A.I.; Bochkarev, D.V. Effectiveness of fungicides in reducing harmfulness of biotrophic pathogens on spring rape. Agrar. Sci. J. 2024, 4–10. [Google Scholar] [CrossRef]
- Mikaberidze, A.; Gokhale, C.S.; Bargués-Ribera, M.; Verma, P. Economic analysis of disease and control of multi-field epidemics in agriculture. bioRxiv 2025. [CrossRef]


| Isolate Location | Host | Isolate | Isolated Year |
|---|---|---|---|
| Icheon, Gyeonggi | Kentucky bluegrass | KKBD-02, KKBD-36 | 2022 |
| Incheon | creeping bentgrass | KCBD-01 | 2020 |
| Hoengseong, Gangwon | creeping bentgrass | KCBD-02, KCBD-03, KCBD-04 | 2022 |
| Kentucky bluegrass | KKBD-01 | 2022 |
| Gene | Primer | Sequence (5′-3′) | Annealing Temperature (°C) | References |
|---|---|---|---|---|
| Mcm7 | Mcm7-709for | ACIMGIGTITCVGAYGTHAARCC | 56 | [26] |
| Mcm7-1348rev | GAYTTDGCIACICCIGGRTCWCCCAT | |||
| Cam | CAL-228F | GAGTTCAAGGAGGCCTTCTCCC | 55 | [27] |
| CAL-737R | CATCTTTCTGGCCATCATGG | |||
| ITS | ITS4 | TCCTCCGCTTATTGATATGC | 55 | [28] |
| ITS5 | GGAAGTAAAAGTCGTAACAAGG |
| Fungal Species | Isolate ID | Locale | Year | GenBank Number | |
|---|---|---|---|---|---|
| CaM | ITS | ||||
| Ciboria aestivalis | CBS 119.47 | Australia | 1947 | KF545281 | KF545326 |
| Clarireedia bennettii | CBS 309.37 | United Kingdom | 1937 | MF964321 | MF964270 |
| Clarireedia bennettii | CBS 311.37 | United Kingdom | 1937 | MF964272 | MF964323 |
| Clarireedia homoeocarpa | HP-50 | NJ, USA | N/A | KF545247 | KF545291 |
| Clarireedia homoeocarpa | MB-01 | OH, USA | 2001 | KF545244 | KF545290 |
| Clarireedia homoeocarpa | D19 | OH, USA | 2002 | KF545252 | KF545298 |
| Clarireedia homoeocarpa | SH44 | Canada | 2000 | KF545251 | KF545299 |
| Clarireedia homoeocarpa | RCCPG-1 | NC, USA | 2003 | KF545253 | KF545297 |
| Clarireedia homoeocarpa | 236-941 | NJ, USA | 2013 | KF545248 | KF545296 |
| Clarireedia homoeocarpa | A4 | OH, USA | 2001 | KF545243 | KF545295 |
| Clarireedia homoeocarpa | SH80 | Canada | 2000 | KF545245 | KF545294 |
| Clarireedia jacksonii | CBS 510.89 | Netherlands | 1989 | KF545261 | KF545289 |
| Clarireedia jacksonii | LEF17T-21 | Italy | 2008 | KF545250 | KF545293 |
| Clarireedia jacksonii | LWC-10 | NC, USA | 2003 | MF964269 | MF964320 |
| Clarireedia jacksonii | MAFF 235854 | Japan | 1987 | KF545242 | KF545301 |
| Clarireedia jacksonii | MAFF 235856 | Japan | 1987 | KF545246 | KF545302 |
| Clarireedia jacksonii | MAFF 235858 | Japan | 1988 | MF964273 | MF964324 |
| Clarireedia monteithiana | TEKP-2 | HI, USA | 2008 | KF545259 | KF545304 |
| Clarireedia monteithiana | BC-14 | NC, USA | 2008 | KF545255 | KF545307 |
| Clarireedia monteithiana | MAFF 236938 | Japan | 1991 | KF545258 | KF545305 |
| Clarireedia paspali | Hawaii2 | GA, USA | 2021 | MZ620646 | MZ578438 |
| Clarireedia paspali | Hawaii3 | GA, USA | 2021 | MZ620644 | MZ578439 |
| Clarireedia paspali | KCBD-01 | Incheon, Republic of Korea | 2020 | LC867002 | LC867009 |
| Clarireedia paspali | KCBD-02 | Icheon, Republic of Korea | 2022 | LC867003 | LC867010 |
| Clarireedia paspali | KCBD-03 | Hoengseong, Republic of Korea | 2022 | LC867004 | LC867011 |
| Clarireedia paspali | KCBD-04 | Hoengseong, Republic of Korea | 2022 | LC867005 | LC867012 |
| Clarireedia paspali | KKBD-01 | Hoengseong, Republic of Korea | 2022 | LC867006 | LC867013 |
| Clarireedia paspali | KKBD-02 | Hoengseong, Republic of Korea | 2022 | LC867007 | LC867014 |
| Clarireedia sp. | KKBD-36 | Icheon, Republic of Korea | 2022 | LC867008 | LC867015 |
| Monilinia vaccinii-corymbosi | SSI-1 | NJ, USA | 2009 | MF964274 | MF964325 |
| Sclerotinia matthiolae | CBS 111.17 | Switzerland | N/A | MF964263 | MF964314 |
| Sclerotinia minor | CBS 112.17 | Netherlands | N/A | MF964264 | MF964315 |
| Sclerotinia sclerotiorum | SS1 | NJ, USA | 2009 | KF545279 | KF545320 |
| Sclerotinia sclerotiorum | SS5 | NJ, USA | 2009 | KF545280 | KF545319 |
| Sclerotinia sulcata | CBS 303.31 | MD, USA | 2017 | MF964266 | MF964317 |
| Isolate | Disease Severity y | |||
|---|---|---|---|---|
| Creeping Bentgrass | Kentucky Bluegrass | |||
| Pot | Field | Pot | Field | |
| KKBD-01 | ++ | N/A z | + | N/A |
| KKBD-02 | ++ | N/A | + | N/A |
| KKBD-36 | ++ | ++++ | + | N/A |
| KCBD-01 | +++ | N/A | + | N/A |
| KCBD-02 | ++ | N/A | + | N/A |
| KCBD-03 | +++ | N/A | + | N/A |
| KCBD-04 | ++ | N/A | + | N/A |
| Salgado-Salazar (2018) [9] w | Present Isolate x | ||
|---|---|---|---|
| Species | Growth (mm) y | Isolate | Growth (mm) y |
| C. jacksonii | 80 a z | KKBD-01 | 77.9 a |
| C. homoeocarpa | 40 b | KKBD-02 | 41.7 b |
| C. monteithiana | 80 a | KKBD-36 | 79.8 a |
| C. bennettii | 80 a | KCBD-01 | 78.0 a |
| KCBD-02 | 74.2 a | ||
| KCBD-03 | 80.1 a | ||
| KCBD-04 | 76.9 a | ||
| Clarireedia jacksonii [9] | C. homoeocarpa [9] | KKBD-01, KKBD-36, KCBD-01, KCBD-02, and KCBD-04 | KKBD-02 | KCBD-03 | |
|---|---|---|---|---|---|
| Cultural characteristics | Colonies are fast growing, cottony, front white to off-white with light brown spots, back white to off-white, later collapsing and turning tan to brown. Colony reaches 8 cm radial growth after 6 days at 25 °C under continuous light on PDA + ascorbic acid. Colonies > 15 days old form thick, flat, black stroma on PDA + ascorbic acid. Hyphae septate, hyaline. | Thalli at first aerial, white to off-white, later collapsing and turning brown, tan, olive, or gray, sometimes slightly pink. Colonies on PDA raised, aerial mycelium white to offwhite, collapsing and turning brown, tan, olive, or gray, with undulate margins. The colony reaches 4 cm radial growth after 6 days at 25 °C under continuous light on PDA + ascorbic acid. Colonies > 15 days old do not form a dark stroma on PDA + ascorbic acid. Hyphae septate, hyaline. | Colonies fast growing, cottony, front white to off-white with light brown spots, back white to off-white, later collapsing and turning tan to brown. Colony reaches 8 cm radial growth after 3 days at 25 °C under continuous light on PDA + ascorbic acid. Colonies > 15 days old form thick, flat, black stroma on PDA + ascorbic acid. Hyphae septate, hyaline. | Thalli at first aerial, white to off-white, later collapsing and turning brown, tan, olive, or gray, sometimes slightly pink. Colonies on PDA raised, aerial mycelium white to off-white, collapsing and turning brown, tan, olive, or gray, with undulate margins. Colony reaches 4 cm radial growth after 3 days at 25 °C under continuous light on PDA + ascorbic acid. Colonies > 15 days old do not form a dark stroma on PDA + ascorbic acid. | Colonies are fast growing, cottony, and front white to off-white with light brown spots and back white to off-white, later collapsing and turning brown. The colony reaches 8 cm radial growth after 3 days at 25 °C under continuous light on PDA + ascorbic acid. Colonies > 15 days old do not form a dark stroma on PDA + ascorbic acid. |
| Apothecia characteristics | Apothecia arising from a substratal stroma, cupulate to discoid, brown, cinnamon, or light orange, receptacle pubescent. Apothecia 2.73 × 1.91 mm arising from dark, substratal stroma. Ascospores and conidia have not been observed. | Hyphae septate, hyaline. Apothecia 0.5–1.5 mm in diameter, arising from a dark substratal stroma, cupulate to discoid, brown, cinnamon, or light orange, receptacle pubescent. Ascus 162.9 × 12.5 μm, on average. Ascospores hyaline, oblong to elliptical, mostly unicellular, occasionally with a medium septum, 20.7 × 8.3 μm. Conidia are not observed. Microconidia spherical, hyaline, 2.0 mm in diameter, formed in cream-colored pustules [7]. | Apothecia 0.5–2.0 mm in diameter, arising from a substrate stroma, cupulate to discoid, brown, cinnamon, or light orange, receptacle pubescent.Ascospores and conidia were not observed. | Apothecia, ascospores, and conidia were not observed. | Apothecia 0.5–2.0 mm in diameter, arising from a substrate stroma, cupulate to discoid, brown, cinnamon, or light orange, receptacle pubescent.Ascospores and conidia were not observed. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jee, J.U.; Ryu, J.H.; Lee, J.H.; Chang, S.W.; Chun, S.C. Re-Identification of Dollar Spot Pathogen on Creeping Bentgrass and Kentucky Bluegrass in South Korea. Processes 2025, 13, 1694. https://doi.org/10.3390/pr13061694
Jee JU, Ryu JH, Lee JH, Chang SW, Chun SC. Re-Identification of Dollar Spot Pathogen on Creeping Bentgrass and Kentucky Bluegrass in South Korea. Processes. 2025; 13(6):1694. https://doi.org/10.3390/pr13061694
Chicago/Turabian StyleJee, Jae Uk, Ju Hyun Ryu, Jeong Ho Lee, Seog Won Chang, and Se Chul Chun. 2025. "Re-Identification of Dollar Spot Pathogen on Creeping Bentgrass and Kentucky Bluegrass in South Korea" Processes 13, no. 6: 1694. https://doi.org/10.3390/pr13061694
APA StyleJee, J. U., Ryu, J. H., Lee, J. H., Chang, S. W., & Chun, S. C. (2025). Re-Identification of Dollar Spot Pathogen on Creeping Bentgrass and Kentucky Bluegrass in South Korea. Processes, 13(6), 1694. https://doi.org/10.3390/pr13061694









