Abstract
Mass spectrometry (MS) remains under-utilized for the analysis of expressed proteins because it is inaccessible to the non-specialist, and sample-turnaround from service labs is slow. Here, we describe 3.5 min Liquid-Chromatography (LC)-MS and 16 min LC-MSMS methods which are tailored to validation and characterization of recombinant proteins in a high throughput structural biology pipeline. We illustrate the type and scope of MS data typically obtained from a 96-well expression and purification test for both soluble and integral membrane proteins (IMPs), and describe their utility in the selection of constructs for scale-up structural work, leading to cost and efficiency savings. We propose that value of MS data lies in how quickly it becomes available and that this can fundamentally change the way in which it is used.
1. Introduction
Worldwide efforts in structural genomics have led to the development of highly parallel methods of cloning, protein production and crystallization [1,2,3,4]. These technologies are also useful outside the context of structural genomics, in more focused structural projects and in research groups and facilities specializing in protein production. A common feature is the handling of diverse entities (clones, proteins) in parallel, and there is an important need for robust and informative measures to assess the quality and properties of the final purified protein. No less important is the ability to achieve a comparative assessment of different clones or methodologies, in order to select the best route to obtain the target protein.
In laboratories where it is available, mass spectrometry (MS) is used to confirm the identity, purity and structural homogeneity of purified proteins or to characterize post-translational modifications (PTMs) prior to entering crystallization trials [5]. Whilst the use of MS purely as a quality control tool is prudent, an important benefit of applying MS to the structural genomics pipeline lies in influencing the experimental process in real time. However, typical turnaround times of days to weeks usually mean that the analysis is after the fact, with little chance of influencing the flow of an experiment in real time. Figure 1 describes schematically our protein production and analysis pipeline and indicates the points of use and the impact of MS in the process.
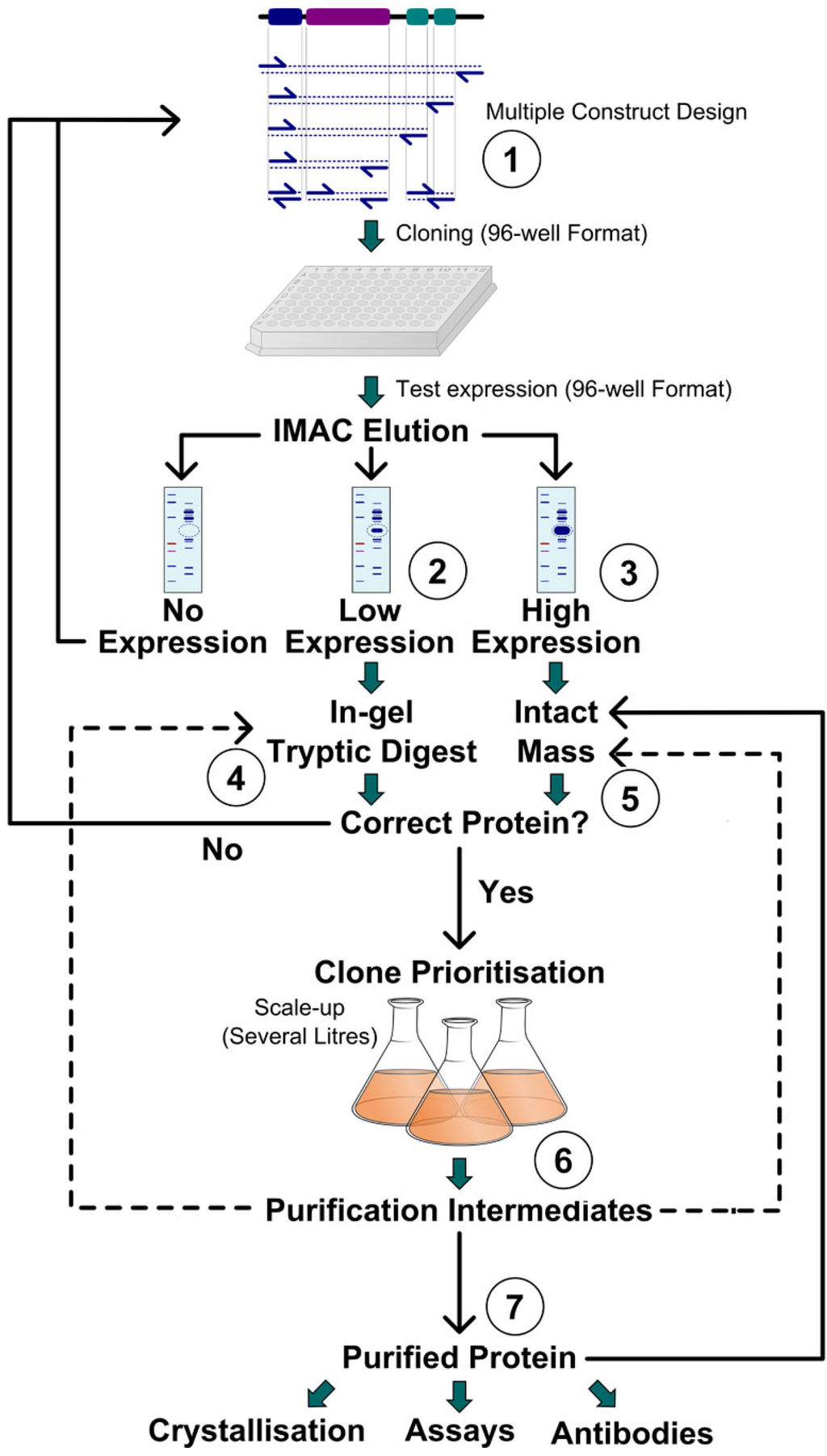
Figure 1.
Integration of mass spectrometry (MS) in the protein production pipeline.
We have applied high-throughput methods for intact mass analysis and in-gel tryptic digest MSMS analysis to the early, small scale (1–3 mL) test expression phase of our pipeline. Intact mass analysis can confirm expression of the target protein with the expected sequence, identify PTMs based on assignment of mass differences and quantify expression yield. This data is additional and complementary to SDS-PAGE analysis, allowing informed decision making, prior to scale-up. It is also complementary to DNA sequence data, in that it identifies features of the protein product, as well as detecting post-cloning effects including sequence changes and clone cross-contamination. In fact, in a high-volume operation where most of the constructs do not yield soluble protein, we have found that in-house MS is more practical than sequencing of hundreds of DNA constructs.
In intact mass analysis where the discrepancy between observed and expected mass is too large to be accounted for by modification, or where the target protein is not readily amenable to intact mass measurement (if, for example the target is larger than 80 kDa or is an IMP), tryptic digest MSMS analysis will confirm the identity of the target protein [6]. Although less informative than intact mass, most proteins are amenable to this analysis. IMPs express at lower abundance, have anomalous SDS-PAGE mobility and poor gel resolution, compounded by glycosylation when this is present [7]. Together, these factors render SDS-PAGE alone a particularly unreliable method for IMP identification following test expression. Western blots may be used when suitable antibodies are available; otherwise MSMS analysis is our method of choice. Rather than focusing on in-depth protein analysis, which is incompatible with high throughput and fast sample turnaround, these improvements allow high quality MS data to be generated quickly. Taken together, these methods allow low cost, high-throughput protein characterization of 96-well test expressions with rapid turnaround. We show typical data generated with these methods and we argue that the timely availability of MS protein characterization data at the earliest test expression stage of a structural genomics pipeline improves the efficiency and cost effectiveness of the entire pipeline.
The high-throughput protein production pipeline (Figure 1; see references [1,8] for more detail) starts with parallel cloning (1 in Figure 1) of multiple truncated constructs of the target gene (in one or more expression vectors and hosts) and testing the yield of the recombinant proteins from small-scale (1–3 mL) cultures; After IMAC purification, SDS-PAGE and coomassie staining, clones are classified as no expression, low expression (2 in Figure 1), where band of correct size is a minor component among contaminating proteins; and high expression (3 in Figure 1), where a band of correct size is the major species. Intact mass analysis of eluates from low-expression clones rarely identifies the target protein, but eluates from highly expressed clones allow quick identification of the target protein, as well as information on its integrity, possible PTM, and homogeneity (5 in Figure 1). Bands of low-expressing clones can be identified by MS/MS of tryptic digests of the gel bands (4 in Figure 1). All this information is integrated into decisions on prioritization of correct clones for large-scale expression, for elimination or re-cloning of incorrect clones, and (when partial proteolysis is evident), the design of new constructs. Large scale (1–24 L) expression followed by multi-step purification allows to recover proteins from low and high-expression clones. In a structural genomics pipeline, there are often no specific assays for the activity of the target protein, so the purification intermediates are monitored through purification by presence of gel bands of the correct size. When purifying low-expressing or membrane proteins there is often uncertainty about the identity of gel bands; tryptic digest and MS/MS (6 in Figure 1) are used to provide definitive identification. Intact MS analysis is also used during purification to monitor enzymatic treatments (tag removal, dephosphorylation) and to differentiate fractions with minute differences (e.g., phosphorylation states) in high-resolution chromatography. Finally, intact MS is used for quality control and characterization of all purified protein batches (7 in Figure 1), as well as MS/MS to map PTMs, when necessary.
2. Experimental Section
2.1. Protein Samples
Proteins were purified from recombinant E. coli or Baculovirus-infected insect cells using a variety of procedures, often starting with immobilized metal affinity chromatography (IMAC) [1,4,8,9,10,11]. Protein samples should be free of detergents and polymers such as PEG or DNA. Otherwise, when the protein is of sufficient purity (>70%) and concentration (>0.2 mg/mL), a range of salt and pH values are tolerated. Small-scale test expression in E. coli (1-mL cultures) or insect cells (3-mL cultures) was performed as described [1,9,10]. The proteins were eluted from 50 µL of Ni-IDA for soluble proteins or TALON beads for IMPs using 50–100 µL of elution buffer (50 mM HEPES, pH 7.5, 10% glycerol, 500 mM NaCl, 1 mM TCEP and 300–500 mM imidazole).
2.2. Electrospray Mass Spectrometry-Time of Flight (ESI-TOF) Intact Mass Analysis
Purified proteins (0.5–1 mg/mL) were diluted 1:50 in 0.1% (v/v) formic acid (50 µL final volume), in round bottomed 96-well microtiter plates (Agilent part number 5042–1385). It is important not to exceed this amount of protein, as column overloading reduces performance and leads to sample cross-contamination. When analyzing nickel eluates from small-scale test expression, 5 µL of each sample was transferred using a hand held 8-channel 10 µL automatic pipette to wells containing 45 µL 0.1% (v/v) formic acid. Reversed-phase chromatography was performed in-line prior to mass spectrometry using an Agilent 1100 HPLC system (Agilent Technologies Inc., Palo Alto, CA, USA). 50 µL was injected from each well onto a 2.1 mm × 12.5 mm Zorbax 5µm 300SB-C3 guard column housed in a column oven set at 40 °C. The solvent system used consisted of 0.1% (v/v) formic acid in LC-MS grade water (Millipore, solvent A) and 0.1% (v/v) formic acid in methanol (LC-MS grade, Chromasolv, solvent B). Chromatography was performed as follows: Initial conditions were 90% A and 10% B and a flow rate of 1.0 mL/min. After 15 s at 10% B, a linear gradient from 10% B to 80% B was applied over 45 s, followed by 80% B to 95% B over 3 s. Elution continued isocratically at 95% B for 1 min 12 s followed by equilibration at initial conditions for a further 45 s. Protein intact mass was determined using an MSD-TOF electrospray ionization orthogonal time-of-flight mass spectrometer (Agilent Technologies Inc., Palo Alto, CA, USA). The instrument was configured with the standard ESI source and operated in positive ion mode. The ion source was operated with the capillary voltage at 4000 V, nebulizer pressure at 60 psig, drying gas at 350 °C and drying gas flow rate at 12 L/min. The instrument ion optic voltages were as follows: Fragmentor 250 V, skimmer 60 V and octopole RF 250 V.
2.3. Intact Mass Data Analysis
LC-MS data files were imported into the data analysis program Masshunter Qualitative Analysis v 6.0 (Agilent, Santa Clara, CA, USA). Total ion chromatograms were overlaid and spectra summed over the elution time of interest. All m/z spectra were simultaneously deconvoluted between minimum and maximum expected masses (±1 kDa) using the maximum entropy algorithm. Peak masses were compared to the expected average mass generated from a local database (Beehive from Molsoft). Mass shifts corresponding to known PTMs were tentatively identified using the Unimod database [12].
2.4. In-Gel Tryptic Digestion
SDS-PAGE gel bands selected for analysis were cut using 1 mm × 4 mm gel cutting tips (Genecatcher, Webscientific, Crewe, UK) attached to a 1-mL pipettor, and transferred to a 96-well PCR plate. Gel plugs were covered with 200 µL 10% (v/v) methanol, the plate sealed and stored at 4 °C until ready for analysis. Using a hand held 12-channel 200 µL automatic pipette, the storage solution was removed and the gel plugs were dehydrated by immersion in 200 µL acetonitrile and allowed to stand for 30 s. The acetronitrile was then removed (taking care that the gel plugs remain in situ) and replaced with 200 µL of 0.1 mM DTT, 100 mM (NH4) HCO3 pH 8.0. The plate was then sealed, transferred to a PCR thermal cycler and incubated at 56 °C for 40 min. The DTT solution was removed, the gel plugs dehydrated as previously described, replaced with 200 µL of 0.1 mM iodoacetamide, 100 mM (NH4) HCO3 pH 8.0, the plate sealed and incubated in the dark at room temperature for 20 min. The iodoacetamide solution was removed, the gel plugs were dehydrated once more with acetonitrile and then 50 µL of trypsin solution was added (2.5 µg/mL sequencing grade trypsin (Sigma) in 25 mM (NH4) HCO3 pH 8.0). The plate was resealed and incubated 37 °C overnight. About 45 µL of digest supernatant was transferred to a 0.5 mL round bottomed polypropylene 96-well microtiter plate (Agilent part No. 5042–1385). Digest supernatants which appeared mid to dark blue (i.e., still heavily Coomassie blue stained) were diluted by addition of 200 µL 2% (v/v) acetonitrile, 0.1% (v/v) formic acid. The plate was sealed using autosampler compatible PP/PE sealing film (Kinesis, UK) with an adhesive free zone corresponding to each well, and loaded on to the LC-MSMS system. When necessary, digests were concentrated by drying using a rotary evaporator at 60 °C for 2 h and re-suspended in 5 µL 2% (v/v) acetonitrile, 0.1% (v/v) formic acid.
2.5. LC-MSMS Analysis of Digests
Reversed-phase chromatography was performed in-line prior to MSMS using a Dionex U3000 nano HPLC system (Thermo, Waltham, MA, USA). For soluble proteins, 1 µL was injected from each well on to a 200 µm × 5 cm Pepswift PS-DVB monolithic column (Thermo, Waltham, MA, USA) housed in a column oven set at 60 °C, and equipped with a 1:97 flow splitter. For IMPs, 5 µL was injected. The solvent system used consisted of 2% (v/v) acetonitrile, 0.1% (v/v) formic acid in LC-MS grade water (solvent A) and 80% (v/v) acetonitrile, 0.1% (v/v) formic acid in LC-MS grade water (solvent B). Chromatography was performed as follows: Initial conditions were 0% B and a post-splitter flow rate of 2.5 µL/min. A linear gradient was developed over 5 min to 15% B and a further 2 min to 40% B. Isocratic elution at 90% B proceeded for 1 min, followed by isocratic elution at 0% B for 6 min to equilibrate the column at the initial conditions. MSMS was performed using a Bruker Esquire HTC ESI-ion trap mass spectrometer fitted with a standard ion source in positive ion mode. Nebulizer pressure was 16 psi, drying gas flow was 5 L/min and drying gas temperature was 300 °C. Voltages were capillary −4 k V, skimmer 40 V, capillary exit 145 V. Scan range was 200–2000 m/z. Data-dependent peptide fragmentation was achieved in auto MSn mode. Three precursor ions were fragmented per MS1 scan. Precursors were actively excluded after 2 spectra, and released after 20 s. Each LC-MS sample run was preceded by a blank run to identify whether significant sample carryover had occurred. The blank was spiked with 1 fmol BSA tryptic digest. This was sufficient to obtain a Mascot hit for BSA reliably without interfering with the blank’s original purpose. This standard/blank injection enabled the LC-MSMS performance to be monitored continuously throughout the batch. If no signal was observed because of low amounts of peptides, the digests were concentrated and re-analyzed.
2.6. MSMS Data Analysis
Automated compound selection, peptide deconvolution and .mgf file generation was performed using DA software (Bruker). Fragment ion files were exported from DA to Biotools batch processing software (Bruker) using an automation script. Fragment ion searches were done in-house on a 1 CPU Mascot server (Matrix Science, London, UK) with the following search parameters: Global modifications carbamidomethyl (C), variable modifications oxidation (M), peptide mass tolerance 1.5 Da (C13 = 1), fragment mass tolerance 1.3 Da, missed cleavages 4. Three databases were searched sequentially without taxonomic restrictions: (i) in-house construct database, 2.5 × 107 residues; (ii) Uniprot, 1.9 × 108 residues; (iii) Mascot contaminants, 1.3 × 105 residues. Total search time was less than 5 min and always less than the LC-MSMS cycle time of 16 min. Mascot search results including server hyperlinks were exported in Biotools Batch Result (.btr) format and imported into Excel where they were manually annotated and assigned one of the following categories: (i) target protein only; (ii) target protein with contaminants; (iii) contaminants predominate over target; (iv) contaminants only; (v) incorrect target; (vi) no protein detected. Data was submitted to Scarab, a laboratory information management system (LIMS by MolSoft LLC) and users notified by email.
3. Results and Discussion
3.1. MS Can be Integrated in a High-Throughput Structural Biology Pipeline
Our pipeline of protein production for crystallization typically ends, after 1–4 chromatographic steps, in purified proteins with yields in the milligram range. Each sample, of which only a small amount (e.g., 0.5–1 µg) is needed, is analyzed by ESI-TOF mass spectrometry to check for protein identity, homogeneity and PTMs. Rapid access to mass spectroscopy allows scientists to retrieve information that supplements that from SDS-PAGE on a similar time scale and can immediately inform the next experimental steps. Implementing multi-user access to the MS facility potentially risks instrumentation misuse and data misinterpretation. We have eliminated most of these problems using some simple measures: (1) providing strict guidelines for sample preparation; in particular, the avoidance of detergents and polymers (PEG, DNA) and of sample overloading; (2) performing data storage and data analysis on computers which are not driving the LC-MS instrument, so users have unobstructed access; (3) integration with an in-house relational database via the LIMS (Scarab) so each data point can be reliably evaluated with regard to the expected mass, gel images and the particulars of each experiment and; (4) having in place a focal person for more sophisticated analysis, troubleshooting, training and maintenance. With these measures in place, we have been able to sustain universal access to >50 users to intact mass analysis. MSMS analysis of tryptic peptides is still performed by expert users. To achieve high levels of access and rapid turnaround, we have streamlined the entire process, routinely achieving readout within 3–4 days for up to 2 × 96 samples per week.
3.2. Intact Mass Analysis Can Guide Small-Scale Test Expression of Multiple Clones
The above setup and throughput enable us to extend MS analysis to an earlier stage in the protein production process, namely the parallel small-scale expression testing of multiple expression clones. This is routinely performed in a 96-well format, which will cover ~5–10 target proteins each sampling 8–12 different N- and C-terminal truncated variants, as part of our multi-construct approach to optimize recombinant expression and solubility [1,4]. This analysis poses additional challenges as the proteins are only partially purified (after one step of affinity purification), the yields are variable and may be limiting, and the proteins are eluted in a complex solution with a high concentration of imidazole. However, where possible, such early-stage analysis can provide valuable information on the selection of constructs before embarking on large-scale purifications.
An example of summary intact mass analysis for a subset of constructs from a 96-well test expression is shown in Table 1 (the full dataset can be found in Supplementary Material Section 5, along with the complete SDS-PAGE analysis in Section 6). LC-MS acquisition time was 5.6 h (overnight) and data analysis time was approximately 3 h. Spectra were obtained for constructs which were scored as high or medium expressers by Coomassie Blue staining following SDS-PAGE analysis (Figure 2 elutions). Low expressers or non-homogeneous samples did not produce protein spectra therefore bands from these elutions were submitted for MSMS analysis (see Section 3.4). Deviation from the expected theoretical mass was less than 1 Da and as small as 0.02 Da. This degree of mass accuracy was sufficient to confirm correct target protein expression, correct construct expression and that no non-isobaric mutations were present. Subsidiary peaks were always observed which corresponded to multiple sodium adducts (+22 Da). Subsidiary peaks which did not conform to this mass deviation were accounted for as PTMs, such as +178 Da (gluconylation [13]) which are known to occur in the expression system used. In some instances, no peak corresponded to the expected unmodified mass, but could be accounted for by single or multiple PTMs where 100% modification had occurred.
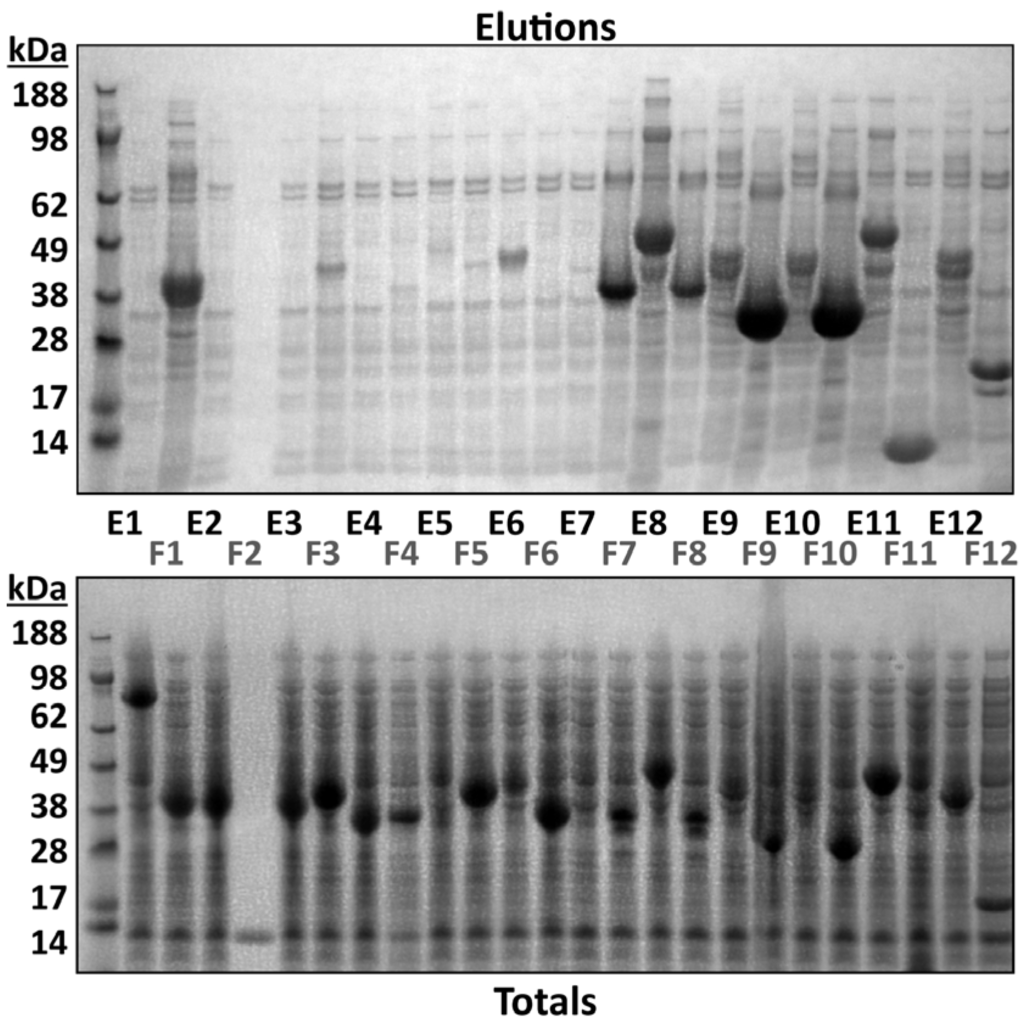
Figure 2.
SDS-PAGE analysis of both the IMAC elutions (15 μL) and total cell lysates (3 μL) for a subset of the 96-well test expression. These samples include examples of high (F10), medium (E6) and low (F3) expressers.

Table 1.
Combined SDS-PAGE, intact mass and tryptic digest MSMS data for 24 representative 1 mL test expressions. Corresponding gel images for IMAC elutions and total protein are shown in Figure 2.
| Sample Description | Intact Mass Analysis | MSMS Analysis Ni Elution | MSMS Analysis Total Protein | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Construct | N or C Terminal Tag | Elution Yield (SDS PAGE) | Plate Well | Expected Mass (Da) | Observed Mass (Da) | Delta Mass | Relative Intensity × 103 | Comments | MOWSE Score | Identity | MOWSE Score | Identity |
| HDAC6 (478–1215) | N | 0: None | E01 | 81,248.4 | 519 | HDAC6 | ||||||
| HDAC6 (478–844) | N | 0: None | E02 | 42,797.7 | 543 | HDAC6 | ||||||
| HDAC6 (478–835) | N | 0: None | E03 | 41,753.5 | 571 | HDAC6 | ||||||
| HDAC6 (478–801) | N | 0: None | E04 | 37,878 | 220 | HDAC6 | ||||||
| HDAC7 (483–903) | N | 0: None | E05 | 48,005.4 | 314 | HDAC7 | ||||||
| HDAC8 (1–286) | N | 3: Medium | E06 | 44,294.4 | not found | 122 | HDAC8 | 564 | HDAC8 | |||
| No construct | ||||||||||||
| NDEL1 (1–345) | N | 5: High | E08 | 41,900.7 | 44,011.52 | 2110.82 | 310 | Unidentified | 1341 | NDEL1 | 1466 | NDEL1 |
| NDEL1 (1–321) | N | 3: Medium | E09 | 38,777.1 | 697 | NDEL1 | 391 | NDEL1 | ||||
| No construct | ||||||||||||
| NDEL1 (1–310) | N | 5: High | E10 | 37,477.7 | not found | 1071 | NDEL1 | 1241 | NDEL1 | |||
| NDEL1 (13–345) | N | 5: High | E11 | 40,724.5 | 42,836 | 2111.5 | 7 | Unidentified | 1487 | NDEL1 | 1306 | NDEL1 |
| NDEL1 (13–321) | N | 5: High | E12 | 37,600.9 | not found | 1089 | NDEL1 | 61 | NDEL1 | |||
| NDEL1 (13–310) | N | 5: High | F01 | 36,301.5 | not found | 1272 | NDEL1 | |||||
| No construct | ||||||||||||
| SIRT2 (34–389) | N | 1: Low | F03 | 42,571.9 | not found | 299 | SIRT2 | 1011 | SIRT2 | |||
| SIRT2 (34–356) | N | 1: Low | F04 | 39,173.1 | not found | 144 | SIRT2 | 951 | SIRT2 | |||
| SIRT2 (38–389) | N | 1: Low | F05 | 42,068.3 | not found | 191 | SIRT2 | 1158 | SIRT2 | |||
| SIRT2 (38–356) | N | 0: None | F06 | 38,669.6 | 841 | SIRT2 | ||||||
| cobB (1–279) | N | 5: High | F07 | 34,016.9 | not found | 802 | COB1 | 718 | COB1 | |||
| cobB (1–274) | N | 5: High | F08 | 33,617.4 | 33,618.47 | 1.07 | 260 | Intact mass | 837 | COB1 | 785 | COB1 |
| cobB (40–279) | N | 5: High | F09 | 29,140.1 | 29,140.27 | 0.17 | 4100 | Intact mass | 756 | COB1 | 609 | COB1 |
| cobB (40–274) | N | 5: High | F10 | 28,740.6 | 28,741.33 | 0.73 | 2200 | Intact mass | 662 | COB1 | 655 | COB1 |
| RPS27A (1–76) | N | 5: High | F11 | 11,117.6 | 11,118.01 | 0.41 | 26,000 | Intact mass | 206 | RPS27A | no hit | |
| SSBP1 (17–148) | N | 5: High | F12 | 17,879.1 | 17,879.64 | 0.54 | 3600 | Intact mass | 834 | SSBP1 | 726 | SSBP1 |

Table 2.
Commonly observed deviations from theoretical protein mass in test expression and scale-up. (Deconvolution artefacts are identified by presence of the wrong (+11 or +44) sodium mass interval).
| Delta Mass (Da) | Tentative Interpretation |
|---|---|
| +(22) n | Sodium adduct and proton loss |
| −89 | N-terminal methionine loss and acetylation |
| −131 | N-terminal methionine loss |
| +178 | Gluconoylation [13] |
| +256 | Phosphogluconylation |
| +(80) n | Phosphorylation |
| −18 | Pyroglutamic acid from N-terminal glutamine |
| +42 | Acetylation |
| +(16) n | Oxidation |
| +14 | Methylation |
| <−131 | (1) N-terminal or C-terminal truncation |
| <−131 | (2) N-terminal and C-terminal truncation |
| <−131 | (3) Different construct |
| <−131 | (4) Different protein |
| +305 | Glutathione |
| +56 | Nickel adduct and 2 proton loss |
| +227 | Biotinylation |
| +1216 | Glycan core |
| +1216 + (161) n | Glycosylation envelope |
| various | Point mutation |
| + theoretical mass | (1) Deconvolution artefact |
| + theoretical mass | (2) Dimer |
| − theoretical mass/2 | Deconvolution artefact |
| +29 | S-nitroyslation |
| +70 | N-pyruvic acid 2-iminyl |
| +119 | Cysteinylation |
| +454 | FMN |
| +48 (n) | Selenomethionine |
| −14 | Lysine demethylation |
| −28 | Arginine-Lysine substitution |
Intact mass analysis of integral membrane proteins would be highly desirable, especially since their size cannot be reliably estimated by SDS-PAGE. The literature on MS characterization of IMPs is substantial (reviewed by Whitelegge [14]), yet intact mass analysis of these proteins is not routine. Sharma [15] describes intact mass measurement of small photosystem II components using LC-MS with acetonitrile as the organic solvent. In our laboratory, we have been unable to generate membrane protein spectra using this solvent. Other authors [16,17,18,19,20] have used acetone precipitation prior to removing detergent prior to LC-MS. This procedure requires relatively a large amount of protein, which is seldom available in an IMP structural genomics context and never available from test expressions. Moreover, this method is not easily adapted for high throughput. We have previously described methods in which detergents are separated from IMPs via in-line LC-MS [21]. However, these methods require more prolonged elution protocols which need to be varied based on the specifics of the protein and the detergents used. Furthermore, they are not easy to integrate on an instrument devoted to high-throughput standardized analysis. Hence, we have found it more useful to use MS/MS as a means of identifying membrane proteins. This is of crucial importance as the expression levels of membrane proteins are 1–2 orders of magnitude lower than those of soluble proteins, and there is an operational need to identify protein bands in the complex mixes of contaminating proteins during the first steps of purification.
3.3. Intact Mass Analysis Is Used to Reveal Expected/Unexpected Post-Translational Modifications
PTM characterization by pin-pointing the location of these modifications within a construct sequence is not a high throughput technique. Nor is it trivial with the limited material available from small scale test expression. Since speed is critical to high throughput sample preparation, acquisition and data analysis, no attempt is made to do this. Relying instead on intact mass measurement alone, the presence or absence of all commonly observed PTMs may be reliably and routinely detected. An intact mass spectrum from a test expression showing typical PTMs is shown in Figure 3. Protein intact mass spectra from high expressers typically show protein peaks with signal to noise of greater than 200:1 and mass accuracy better than ±1 Da. This degree of mass accuracy allows subsidiary peaks such as sodium adducts and PTMs to be readily distinguished. Although Unimod [12] and DeltaMass [22] websites provide extensive lists of known protein mass variants, caution must be used in assigning these to the target protein because nearly all these mass variants are extremely rare, and many are chemical or isotopic derivatives, not possible outside a specific experimental context. More useful is a knowledge of which PTMs are known to occur in the expression system being used and with what frequency. Table 2 lists 19 PTMs observed by us, with a further eight mass differences with their likely interpretation. In some instances, verification of these mass differences is possible. Phosphorylation, for example, can be verified by an additional intact mass measurement following enzymatic removal of phosphate groups (e.g., lambda phosphatase) [23,24,25]. N-terminal modifications will be lost following TEV cleavage of an N-terminal tag. Histidine tag-associated modifications such as gluconoylation will also be lost following TEV cleavage. Intact mass measurements can be used to monitor deliberate modifications of the protein, such as biotinylation [26], reductive methylation [27], glycosylation [28], incorporation of SeMet or isotopically-labelled amino acids, or the removal of the tag by TEV cleavage. In all cases where a mass discrepancy cannot be unequivocally accounted for, the expression clone is (re)-sequenced. Peaks with double the calculated mass, require careful interpretation. The denaturing LC-MS conditions described do not always result in total denaturation. Some structure can still be retained and consequently, though rarely, dimers and other structures may be observed. More commonly, maximum entropy deconvolution may generate artifactual peaks of double or half the protein mass. In this case, the sodium adducts normally associated with these peaks will appear to have double or half the proper sodium mass shift (+44 Da or +11 Da). So called top-down MSMS techniques [29] are now available for protein identification and mapping of post translational modifications using the latest generation of high end mass spectrometers. However, it should be borne in mind that no MSMS technique generates full sequence coverage; hence, the possibility that PTMs will be missed. In contrast, intact mass measurement allows detects the presence of all PTMs wherever their location in the sequence, allowing confirmation of the entire covalent structure.
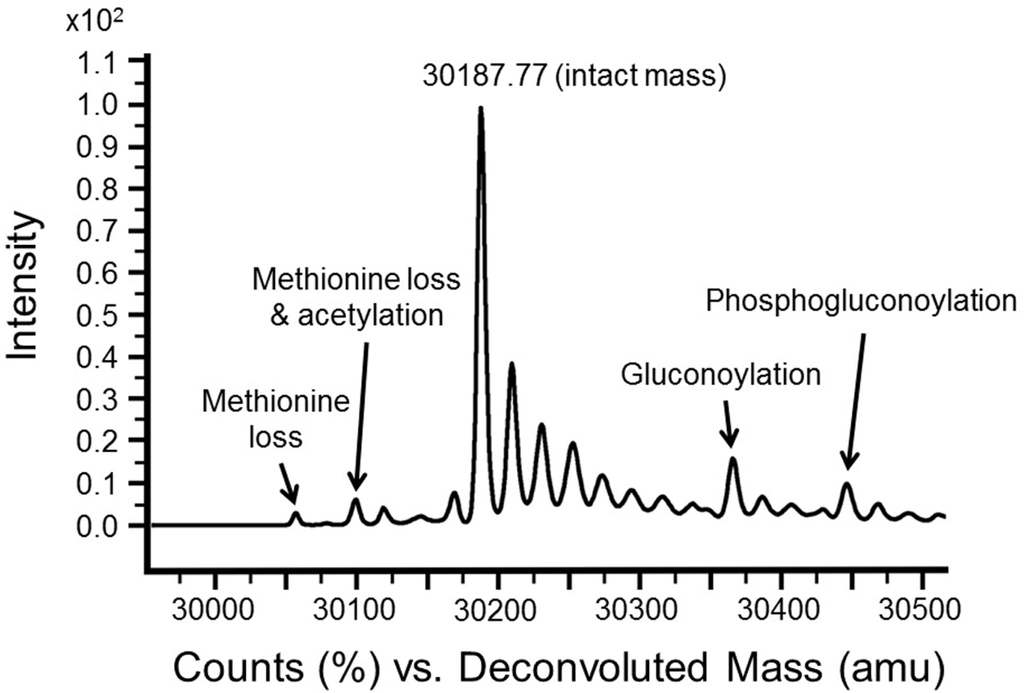
Figure 3.
Intact mass spectrum from nickel elution of test expression SPIN1 (21–262) (theoretical mass 30,189.2 Da) showing four most frequently observed PTMs. Sodium adducts are unlabeled.
A commonly-used alternative to ESI-TOF is Matrix-Assisted Laser Desorption/Ionization (MALDI)-MS. Accurate mass determination of short peptides by MALDI compares favorably with ESI. However, the sensitivity, resolution and mass accuracy of MALDI instruments decreases rapidly for larger analytes such as intact proteins, such that accurate mass measurement of proteins greater than 15 kDa is generally not possible. While ESI protein mass measurements have a typical accuracy of ±10 ppm, similar measurements by MADLI have an accuracy of ±1000 ppm; hence, the information gained is of limited value and unsuitable for construct and PTM identification. The poor high mass performance of MALDI arises from fundamental differences in ion formation in comparison to ESI and how this affects the efficiency of micro channel plate ion detectors common to both types. Large, singly charged MALDI ions acquire much lower velocity during TOF acceleration than the equivalent multiply charged ESI ion. The impact velocity as ions strike the detector determines the efficiency at which secondary electrons are generated and hence the amplitude of the detector current [30]. ESI instruments require calibration only up to m/z 2700 because all charge states for a denatured protein fall within this range. In contrast, MALDI instruments require calibration up to an m/z value beyond the mass of the protein being measured. Monoisotopic masses are unresolved and few, if any, good high mass calibrants exist. Typically, BSA or IgG are used, despite the fact that these calibrants are heterogenous and display broad peaks. Moreover, MALDI analysis of proteins requires a non-homogeneous matrix such as ferrulic acid, needing manual laser positioning and is thus not suitable for automation and high throughput.
3.4. MSMS Data Is Used to Confirm Identity of Target Proteins
A second level of investigation after intact mass and SDS-PAGE analyses is the LC-MSMS of excised gel bands, usually performed for a subset of the 96-well constructs where (i) intact mass of a low expresser is not conclusive or there are multiple bands of similar mobility to the target protein in SDS-PAGE. Analysis of a full 96-well plate would take 60 h, but in practice, only those bands that have not been confirmed by intact mass analysis need be submitted. Typically, the turnaround time is therefore less than 48 h. MSMS analysis of a complete 96-well plate excepting IMPs is rarely justified, since successful identification based on intact mass analysis obviates the need for MSMS. Results for a batch analysis of gel bands from the corresponding subset of constructs (see Section 3.2) are shown in Table 1 (the full dataset can be found in Supplementary Material Section 5). For example, the identities of samples E06–F07 were not confirmed by intact mass. Expression levels ranged from low to high, but in all cases, target identities were confirmed by MSMS. Additionally, samples E08 and E11 each have an intact mass discrepancy of +2111 Da, while MSMS analysis shows that this is the target protein. Re-sequencing of these clones revealed a frameshift near the C-terminus which was previously overlooked, leading to read-through beyond the intended stop codon. This was remedied by re-cloning. In addition, it can be seen from the gel (Figure 2) that the purified proteins in samples E09, E10, E12 and F01 appear as doublet bands. Samples F03, F04 and F05 show weak levels of expression, requiring further optimization before scale-up.
Typical “hits” for proteins comprised of MOWSE scores of several hundred, well above the significance threshold for databases of this size. For successful target protein identifications, hits occurred in both construct and Uniprot searches. Uniprot hits only occurred for host contaminant proteins. Table 3 lists commonly expressed proteins observed by MSMS from bacterial and insect expression systems. Peptide coverage for IMPs is lower than for soluble proteins because tryptic cleavage sites are rare in membrane spanning regions and these are less accessible to proteases in general [31]. For IMPs, we therefore compensate with five-fold greater volume digest for LC-MSMS, or if necessary 40-fold. Less stringent criteria for identification of known protein targets are needed than in a proteomics context; hence, a single high scoring peptide match is often sufficient for IMP identification. While both detergent (used for IMP extraction) and high imidazole have a negative impact on intact mass analyses, they do not interfere with either SDS-PAGE purification or subsequent tryptic digestion and LC-MSMS. Similarly, partially degraded proteins or those which have aggregated and are insoluble are readily amenable to in-gel digestion and LC-MSMS. Figure 2 also includes analyses of total protein from IMAC purification. We do not routinely analyze this material, but here it is instructive to see that very high levels of expression are observed for nearly all constructs. The barrier to successful purification appears to be protein solubility, something which must be borne in mind during construct design.

Table 3.
Highly expressed contaminating proteins commonly observed by tryptic digest MSMS analysis of gel bands from E. coli and insect cell test expression.
| Expression System | Contaminant Protein | |
|---|---|---|
| E.coli | 50S ribosomal protein L2 | |
| 60 kDa chaperonin | ||
| Bifunctional polymyxin resistance protein ArnA | ||
| Catabolite gene activator | ||
| Chaperone protein htpG | ||
| Chloramphenicol acetyltransferase | ||
| Triosephosphate isomerase | ||
| Insect cell | Host origin | 40S ribosomal protein S16 |
| 40S ribosomal protein S3 | ||
| 40S ribosomal protein S3a | ||
| Eukaryotic translation initiation factor 3 subunit A | ||
| Guanine nucleotide-binding protein subunit beta-like protein | ||
| Heat shock 70 kDa protein cognate 4 | ||
| Tubulin alpha-1 chain | ||
| Tubulin beta-1 chain | ||
| Histone H2A | ||
| Histone H2B | ||
| Histone H4 | ||
| Viral origin | Early 39 kDa protein | |
| Major capsid protein | ||
| Probable endochitinase | ||
3.5. MS Is Complementary to SDS-PAGE in a Structural Biologist’s Toolkit
The value of intact mass and MSMS analyses is that information is gained over and above that from SDS-PAGE at minimal cost in terms of time and money. Foremost, it can confirm that the over-expressed protein is indeed the target. For low to medium expressers as judged by SDS-PAGE, one might expect these constructs would not normally be pursued for scale up. However, MSMS confirmation of target protein, for even some of these constructs, would help guide the next round of construct boundary optimization. Additional information from high/medium expressers is confirmation that the observed mass matches the theoretical mass of the construct. We assume from this that the construct is correct, the protein sequence is correct and that at this stage construct DNA sequencing is not prioritized. If present, PTMs are evident in the intact mass spectrum. They fall into two categories: N-terminal processing, which may or may not involve the affinity tag, and modifications which may affect protein activity or the likelihood of crystallization such as phosphorylation or glycosylation. Intact mass spectrometry is uniquely capable of assessing sample heterogeneity that is not discernible by SDS-PAGE, such as a partial proteolysis of a few amino acids or multiple phosphorylation states.
The two techniques should be considered as complementary within the structural biology pipeline. Gel analysis is superior to LC-MS for visualization of proteins in complex mixtures, large proteins, complexes and IMPs. In our laboratory, LC-MS analysis is performed on IMAC elutions following SDS-PAGE analysis, using surplus material which would otherwise be discarded. Where intact mass analysis has been unsuccessful and gel analysis indicates that the construct protein has been expressed, in-gel digestion and LC-MSMS analysis determines protein identification. Although less informative than intact mass analysis, LC-MSMS is far more sensitive and is applicable to heavily or unusually modified proteins, truncated proteins, very large proteins and IMPs [32]. Once again, it is done using surplus material which would otherwise be discarded. While this information is clearly useful, it will not actually be used in a structural genomics pipeline unless it is available quickly. Using the method described here, intact mass data for a 96-well test expression plate can be available the following day. A complete MSMS dataset can be available in less than 60 h. For soluble proteins, partial datasets available in around 30 h are sufficient when intact mass data constitutes the remainder. Technical discussion of the high-throughput in-gel digestion, LC-MSMS and data analysis methods is provided in the Supplementary Material.
3.6. MS Can Help Detect Mistakes and Mitigate Downstream Decision
Structural genomics is expensive, and costly mistakes (e.g., scaling up the “wrong” protein) and unexpected surprises (e.g., unplanned incorporation of chemical groups/amino acids) need to be avoided. The purpose of test expression is to identify from a series of constructs those which are actually expressed in a soluble form and those which express with highest yield. Over-expression coupled with apparent molecular weight calculated from SDS-PAGE relative mobility is often used to “confirm” the presence of the target protein. However, expression from a construct bearing vector is only one of many reasons why the host may over-express a protein. For example, antibiotic resistance proteins (see Table 3) are commonly over-expressed by bacteria grown in the presence of antibiotics. Moreover, the apparent molecular weight of these proteins is unpredictable due to different extents of degradation which is also common amongst highly expressed proteins. Our experience in a laboratory where data capture of construct design, cloning and expression are tightly coordinated through the use of the LIMS (Scarab) shows that mis-identification of over-expressed proteins from a gel is quite common, and the risk is much greater with IMPs. While the correct protein may be expressed, there is a risk that mutation has occurred during the cloning process (e.g., fidelity of polymerase, mismatch mistakes in primers) resulting in incorrect amino acid sequence of protein (substitution/insertion/deletion). Obtaining the correct accurate mass value for the protein obviates the need for DNA sequencing of the construct. It is known that proteins with significant heterogeneity, such as those carrying multiple phosphorylations or a diverse glycosylation pattern, are less likely to crystallize. This information may not be evident from gel analysis alone.
4. Conclusions
Although LC-MS is widely used for downstream characterisation of purified proteins, 1 mL test expression analysis involves low protein loading and high imidazole and represents a significant analytical challenge. Intact mass measurements are not always successful, but when they do succeed, the information gained at the earliest possible stage in our pipeline is significantly more informative than gel analysis alone. When they fail due to low expression, these constructs are intrinsically of less interest, although MSMS may still be deployed. Intact mass and tryptic digest MSMS analyses are powerful analytical tools generating unambiguous data. They can inform the structural biologist much about an expressed construct: Protein identification, confirmation of primary structure, heterogeneity, identity of any PTMs and numerical quantitation of expression yield. These tools can be seamlessly integrated into a protein production and crystallization pipeline to reduce costs and improve its overall efficiency and reliability.
Supplementary Materials
Supplementary materials can be accessed at: http://www.mdpi.com/2227-9075/1/4/159/s1.
Acknowledgments
The authors are grateful for support by the SGC, a registered charity (number 1097737) that receives funds from AbbVie, BAYER Pharma AG, Boehringer Ingelheim, the Canada Foundation for Innovation, the Canadian Institutes for Health Research, Genome Canada, GlaxoSmithKline, Janssen, Lilly Canada, the Novartis Research Foundation, the Ontario Ministry of Economic Development and Innovation, Pfizer, Takeda, and the Wellcome Trust (092809/Z/10/Z).
Author Contributions
Chalk wrote the manuscript and Chalk and Berridge developed methods and performed MS analyses. Gileadi, Burgess-Brown and Yue provided discussion and critical reading and writing of the manuscript. Shrestha and Mahajan performed test expressions and purifications. Strain-Damerell prepared figures and contributed to the formatting and editing of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Savitsky, P.; Bray, J.; Cooper, C.D.O.; Marsden, B.D.; Mahajan, P.; Burgess-Brown, N.A.; Gileadia, O. High-throughput production of human proteins for crystallization: The SGC experience. J. Struct. Biol. 2010, 172, 3–13. [Google Scholar] [CrossRef]
- Edwards, A. Large-Scale Structural Biology of the Human Proteome. Annu. Rev. Biochem. 2009, 78, 541–568. [Google Scholar] [CrossRef]
- Terwilliger, T.C.; Stuart, D.; Yokoyama, S. Lessons from Structural Genomics. Annu. Rev. Biophys. 2009, 38, 371–383. [Google Scholar] [CrossRef]
- Graslund, S.; Sagemark, J.; Berglund, H.; Dahlgren, L.G.; Flores, A.; Hammarström, M.; Johansson, I.; Kotenyova, T.; Nilsson, M.; Nordlund, P.; et al. The use of systematic N- and C-terminal deletions to promote production and structural studies of recombinant proteins. Protein Expr. Purif. 2008, 58, 210–221. [Google Scholar] [CrossRef]
- Cohen, S.L.; Chait, B.T. Mass spectrometry as a tool for protein crystallography. Annu. Rev. Biophys. Biomol. Struct. 2001, 30, 67–85. [Google Scholar] [CrossRef]
- Rosenfeld, J.; Capdevielle, J.; Guillemot, J.C.; Ferrara, P. In-gel digestion of proteins for internal sequence analysis after one-or two-dimensional gel electrophoresis. Anal. Biochem. 1992, 203, 173–179. [Google Scholar] [CrossRef]
- Rath, A.; Glibowicka, M.; Nadeau, V.G.; Chen, G.; Deber, C.M. Detergent binding explains anomalous SDS-PAGE migration of membrane proteins. Proc. Natl. Acad. Sci. USA 2009, 106, 1760–1765. [Google Scholar] [CrossRef]
- Graslund, S.; Nordlund, P.; Weigelt, J.; Hallberg, B.M.; Bray, J.; Gileadi, O.; Knapp, S.; Oppermann, U.; Arrowsmith, C.; Hui, R.; et al. Protein production and purification. Nat. Methods 2008, 5, 135–146. [Google Scholar] [CrossRef]
- Burgess-Brown, N.A.; Mahajan, P.; Strain-Damerell, C.; Gileadi, O.; Gräslund, S. Medium-throughput production of recombinant human proteins: Protein production in E. coli. Methods Mol. Biol. 2014, 1091, 73–94. [Google Scholar]
- Mahajan, P.; Strain-Damerell, C.; Gileadi, O.; Burgess-Brown, N.A. Medium-throughput production of recombinant human proteins: Protein production in insect cells. Methods Mol. Biol. 2014, 1091, 95–121. [Google Scholar]
- Strain-Damerell, C.; Mahajan, P.; Gileadi, O.; Burgess-Brown, N.A. Medium-throughput production of recombinant human proteins: Ligation-independent cloning. Methods Mol. Biol. 2014, 1091, 55–72. [Google Scholar]
- UNIMOD: Protein modifications for mass spectrometry. Available online: http://www.unimod.org (accessed on 16 September 2014).
- Geoghegan, K.F.; Dixon, H.B.; Rosner, P.J.; Hoth, L.R.; Lanzetti, A.J.; Borzilleri, K.A.; Marr, E.S.; Pezzullo, L.H.; Martin, L.B.; LeMotte, P.K.; et al. Spontaneous alpha-N-6-phosphogluconoylation of a “His tag” in Escherichia coli: The cause of extra mass of 258 or 178 Da in fusion proteins. Anal. Biochem. 1999, 267, 169–184. [Google Scholar] [CrossRef]
- Whitelegge, J.P. Integral membrane proteins and bilayer proteomics. Anal. Chem. 2013, 85, 2558–2568. [Google Scholar] [CrossRef]
- Sharma, J.; Panico, M.; Barber, J.; Morris, H.R. Purification and determination of intact molecular mass by electrospray ionization mass spectrometry of the photosystem II reaction center subunits. J. Biol. Chem. 1997, 272, 33153–33157. [Google Scholar] [CrossRef]
- Hufnagel, P.; Schweiger, U.; Eckerskorn, C.; Oesterhelt, D. Electrospray ionization mass spectrometry of genetically and chemically modified bacteriorhodopsins. Anal. Biochem. 1996, 243, 46–54. [Google Scholar] [CrossRef]
- Le Coutre, J.; Whitelegge, J.P.; Gross, A.; Turk, E.; Wright, E.M.; Kaback, H.R.; Faull, K.F. Proteomics on full-length membrane proteins using mass spectrometry. Biochemistry 2000, 39, 4237–4242. [Google Scholar] [CrossRef]
- Whitelegge, J.P.; Gundersen, C.B.; Faull, K.F. Electrospray-ionization mass spectrometry of intact intrinsic membrane proteins. Protein Sci. 1998, 7, 1423–1430. [Google Scholar] [CrossRef]
- Le Coutre, J.; Lee, J.C.; Engel, C.K.; Privé, G.G.; Faull, K.F.; Kaback, H.R. Toward the bilayer proteome, electrospray ionization-mass spectrometry of large, intact transmembrane proteins. Proc. Natl. Acad. Sci. USA 1999, 96, 10695–10698. [Google Scholar] [CrossRef]
- Whitelegge, J.P.; Zhang, H.; Aguilera, R.; Taylor, R.M.; Cramer, W.A. Full subunit coverage liquid chromatography electrospray ionization mass spectrometry (LCMS+) of an oligomeric membrane protein: Cytochrome b(6)f complex from spinach and the cyanobacterium Mastigocladus laminosus. Mol. Cell. Proteomics 2002, 1, 816–827. [Google Scholar] [CrossRef]
- Berridge, G.; Chalk, R.; D’Avanzo, N.; Dong, L.; Doyle, D.; Kim, J.I.; Xia, X.; Burgess-Brown, N.; Deriso, A.; Carpenter, E.P. High-performance liquid chromatography separation and intact mass analysis of detergent-solubilized integral membrane proteins. Anal. Biochem. 2011, 410, 272–280. [Google Scholar] [CrossRef]
- DeltaMass. A database of protein post-translational modifications. Available online: http://www.abrf.org/index.cfm/dm.home (accessed on 15 September 2014).
- Edmonds, C.G.; Smith, R.D. [22] Electrospray ionization mass spectrometry. In Methods in Enzymology; James, A.M., Ed.; Academic Press: Amsterdam, The Netherlands, 1990; pp. 412–431. [Google Scholar]
- Gibson, B.W.; Cohen, P. [26] Liquid Secondary Ion Mass Spectrometry of Phosphorylated and Sulfated Peptides and Proteins. In Methods in Enzymology; James, A.M., Ed.; Academic Press: Amsterdam, The Netherlands, 1990; pp. 480–501. [Google Scholar]
- Holmes, C.F.; Tonks, N.K.; Major, H.; Cohen, P. Analysis of the in vivo phosphorylation state of protein phosphatase inhibitor-2 from rabbit skeletal muscle by fast-atom bombardment mass spectrometry. BBA-Mol. Cell Res. 1987, 929, 208–219. [Google Scholar]
- Keates, T.; Cooper, C.D.; Savitsky, P.; Allerston, C.K.; Phillips, C.; Hammarström, M.; Daga, N.; Berridge, G.; Mahajan, P.; Burgess-Brown, N.A.; et al. Expressing the human proteome for affinity proteomics: Optimising expression of soluble protein domains and in vivo biotinylation. New Biotechnol. 2012, 29, 515–525. [Google Scholar] [CrossRef]
- Kim, Y.; Quartey, P.; Li, H.; Volkart, L.; Hatzos, C.; Chang, C.; Nocek, B.; Cuff, M.; Osipiuk, J.; Tan, K.; et al. Large-scale evaluation of protein reductive methylation for improving protein crystallization. Nat. Methods 2008, 5, 853–854. [Google Scholar] [CrossRef]
- Chaikuad, A.; Froese, D.S.; Berridge, G.; von Delft, F.; Oppermann, U.; Yue, W.W. Conformational plasticity of glycogenin and its maltosaccharide substrate during glycogen biogenesis. Proc. Natl. Acad. Sci. USA 2011, 108, 21028–21033. [Google Scholar] [CrossRef]
- Loo, J.A.; Edmonds, C.G.; Smith, R.D. Tandem mass spectrometry of very large molecules: Serum albumin sequence information from multiply charged ions formed by electrospray ionization. Anal. Chem. 1991, 63, 2488–2499. [Google Scholar] [CrossRef]
- Twerenbold, D.; Gerber, D.; Gritti, D.; Gonin, Y.; Netuschill, A.; Rossel, F.; Schenker, D.; Vuilleumier, J.L. Single molecule detector for mass spectrometry with mass independent detection efficiency. Proteomics 2001, 1, 66–69. [Google Scholar] [CrossRef]
- Wu, C.C.; Yates, J.R. The application of mass spectrometry to membrane proteomics. Nat. Biotechnol. 2003, 21, 262–267. [Google Scholar] [CrossRef]
- Wilm, M.; Shevchenko, A.; Houthaeve, T.; Breit, S.; Schweigerer, L.; Fotsis, T.; Mann, M. Femtomole sequencing of proteins from polyacrylamide gels by nano-electrospray mass spectrometry. Nature 1996, 379, 466–469. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).