Abstract
Proteins are the largest defined molecular component of marine organic nitrogen, and hydrolysable amino acids, the building blocks of proteins, are important components of particulate nitrogen in marine sediments. In oceanic systems, the largest contributors are phytoplankton proteins, which have been tracked from newly produced bloom material through the water column to surface sediments in the Bering Sea, but it is not known if proteins buried deeper in sediment systems can be identified with confidence. Electrophoretic gel protein extraction methods followed by proteomic mass spectrometry and database searching were used as the methodology to identify buried phytoplankton proteins in sediments from the 8–10 cm section of a Bering Sea sediment core. More peptides and proteins were identified using an SDS-PAGE tube gel than a standard 1D flat gel or digesting the sediment directly with trypsin. The majority of proteins identified correlated to the marine diatom, Thalassiosira pseudonana, rather than bacterial protein sequences, indicating an algal source not only dominates the input, but also the preserved protein fraction. Abundant RuBisCO and fucoxanthin chlorophyll a/c binding proteins were identified, supporting algal sources of these proteins and reinforcing the proposed mechanisms that might protect proteins for long time periods. Some preserved peptides were identified in unexpected gel molecular weight ranges, indicating that some structural changes or charge alteration influenced the mobility of these products during electrophoresis isolation. Identifying buried photosystem proteins suggests that algal particulate matter is a significant fraction of the preserved organic carbon and nitrogen pools in marine sediments.
1. Introduction
Much of the ocean is influenced by nitrogen limitation [], and thus, understanding marine protein cycling is important for tracking the global organic nitrogen cycle. Solid state NMR has provided evidence that the majority of organic nitrogen in dissolved and particulate marine organic matter contains amide bonds as found in proteins [,,]. Protein building blocks, such as total hydrolysable amino acids (THAAs, total amino acids that can be extracted using 6 N HCl), are found to account for up to 30%–40% of particulate nitrogen in marine sediments [,,,]. In addition to proteins representing the largest fraction of organic nitrogen, within the unique amino acid sequence, proteins also can provide functional and phylogenetic information on the organisms from which they were produced, potentially making peptides unique to taxonomic groups useful biomarkers. Identifying intact proteins and peptides buried in marine sediments would give valuable information towards understanding marine biogeochemical cycles and reconstructing algal/microbial populations [,]. The complex matrix effects of sediments [,,], however, have made extracting buried proteins for further analysis a challenge [,,]. New bioseparation methods are needed to successfully extract proteins from deep sediments for subsequent analyses.
Gel electrophoresis is the classic method for protein separation and visualization, including sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and related approaches that separate proteins based primarily on molecular weight []. The SDS-PAGE method has been a standard technique for protein analysis in the fields of biochemistry, cell biology and medical sciences for decades [,,,,,,]. Modified SDS-PAGE methods have been used to purify proteins from cell cultures before mass spectrometry analysis [,] and to separate proteins from soil trichloroacetic acid (TCA) extracts [,]. Recent studies by Moore et al. [,] found that intact algal proteins and peptides could be tracked and identified from the marine water column and extracted from surface sediments in the Bering Sea using modified SDS-PAGE tube gels where sediment-buffer mixtures were loaded directly to the gels for separation. In-gel protein extractions were then performed, and peptides were identified using proteomic mass spectrometry and database searching. It is unknown if these methods could be applied to identify algal proteins that have potentially been buried in marine sediments for extensive periods of time.
The goal of this study was to test sediment gel-based extraction methods with tube and 1D flat gels on field-collected deep marine sediments of the Bering Sea to determine if algal proteins identified previously in surface sediments could be identified after long-term burial. These methods were compared with direct sediment trypsin digest as a control.
2. Experimental Section
2.1. Sediment Collection
The Bering Sea is known to be one of the world’s most productive ecosystems [,,], resulting, in part, in the high export of primary production material to sediments after bloom termination [,,]. These factors, along with the identification of algal proteins in Bering Sea surface sediments, make down core sediments from this region ideal for identifying buried proteins. Bering Sea sediment was collected from the outer shelf during the Bering Sea Ecosystem Study (BEST) spring 2009 cruise on April 9, 2009, using a multi-corer (see []). Sediment was collected before the spring phytoplankton bloom to ensure that primary production exported to the sediment was from the previous year. The location of the sediment core was Latitude 59.9004 N, Longitude 171.5952 W, depth 101 m, temperature −0.15 °C. The sediment core was sliced into multiple sections, and the 8 to 10 cm section was used for the analysis of buried proteins. Sediments were frozen and stored at −70 °C until analysis.
2.2. Gel Electrophoresis Protein Extraction
To extract buried proteins from sediment, 1.5 g wet weight of 8 to 10 cm sediment was treated with 500 μL of extraction buffer in a 2 mL Eppendorf tube and pulse sonicated for 1 min on ice (Bronson sonicating microprobe, 20 kHz). The extraction buffer consisted of 7 M urea, 2 M thiourea, 0.01 M Tris-HCl, 1 mM EDTA, 10% v/v glycerol, 2% CHAPS, 0.2% w/v ampholytes (Fluka BioChemika, high resolution pH 3–10, 40% in water) and 2 mM Tributyl-phosphine []. The entire sediment + extraction buffer mixture was then loaded onto gel-prep cell tubes for isolation and molecular weight separation. The gel composition was 17% acrylamide/Bis, 0.125 Tris-HCl. The amount of protein material loaded onto the gel was determined by measuring the concentration of THAAs in the sediment buffer mixture as a proxy for total protein (buffer extraction efficiency was 12%). Gels were covered with running buffer (0.25 M Tris, 0.192 M glycine, 0.1% SDS, pH 8.3) and run at 180 V until the ion front traveled 7 cm or 2 cm from the top in separate gels. After electrophoresis, the 7 cm gel section was cut into five molecular weight ranges for digestion and analysis (<10, 10–25, 25–50, 50–100 and >100 kDa) based on external MW standards (Bio-Rad Kalaidoscope) in identical gels. In addition, 1-dimensional pre-cast 12% Bis-Tris flat gels (Invitrogen NuPAGE Novex) were loaded with 45 µL of sediment + buffer (the same ratio of sediment and extraction buffer as the above tube gels). Gel lanes were covered with agarose overlay and running buffer and run at 180 V until the ion front traveled 2 cm down the 1D gel. The top 2 cm of the ion front gels were excised for digestion analysis. All gels were washed three times with nanopure water to remove excess SDS prior to digestion.
A control gel was run with multiple lanes loaded with sonicated diatom cells. Approximately 0.75 g (wet weight) of cultured Thalassiosira weissflogii cells were sonicated in 1 mL of 44 mM ammonium bicarbonate for 1 min on ice (Bronson sonicating microprobe, 20 kHz). Total hydrolysable amino acid (THAA) analysis was performed on the sonicated T. weissflogii/ammonium bicarbonate mixture as a proxy for protein material for gel loading. Six lanes of a 1-dimensional pre-cast 12% Bis-Tris flat gel (Invitrogen NuPAGE Novex) were loaded with sonicated cellular material containing 40 µg of THAAs per lane. Gel lanes were covered with agarose overlay and running buffer and run at 180 V until the ion front traveled 10 cm down the gel. The gel was washed three times with DI water, and each gel lane was cut separately into molecular weight sections (<10, 10–25, 25–50, 50–100 and >100 kDa) based on internal molecular weight standards run in a separate gel lane. Three gel lanes were taken for trypsin digestions and proteomic analysis, and three gel lanes were taken for electroelution and THAA analysis. The electroelution molecular weight sections were separately placed in the electroelution cell (Bio-Rad, Hercules, CA, USA), covered with running buffer and run at 100 V for 1 h to elute protein material out of the gel section. The eluted protein was collected in an elution chamber with a 1 kDa molecular weight membrane, so that all material above 1 kDa was collected.
2.3. In Gel Protein Digestion
Gel molecular weight sections and 2 cm gels were cut into 2 × 2 mm slices to increase the surface area for enzyme and chemical access. Pieces were covered together with 100 mM ammonium bicarbonate and rinsed for 15 min to hydrate the gel sections and then rinsed for 15 min in acetonitrile to dehydrate the gel sections and remove detergents and other chemical interferences. The rinse cycle was repeated five times, and the gel sections were then dried by speed-vac for 45 min. Reduction, alkylation and digestion followed the standard procedure by Schevchenko et al. []. Digests were dried, and volumes were adjusted to give a final protein concentration of 1 µg protein/10 µL based on THAA concentration recoveries.
2.4. Direct Digestion of Sediment
As a control, 8–10 cm sediment was digested directly with trypsin by combining 100 mg of sediment with 300 µL of 6 M urea and 50 mM ammonium bicarbonate. The sediment was then sonicated (Bronson sonicating probe, 20 kHz for 60 s on ice). The pH was raised by adding 18 µL 1.5 M Tris-HCl (pH 8.8). To reduce sulfide bonds in proteins, 7.5 µL TCEP (2,2',2'(-phosphanetriyltripropanoic acid) was added to the sediment, vortexed and incubated for 1 h (37 °C). Proteins were then alkylated by adding 60 µL of 200 mM iodoacetic acid (IAM) and incubated in the dark for 1 h. After the addition of and incubation of 60 µL of dithiothreitol (1 h room temperature), the urea was diluted with the addition of 2.4 mL 25 mM ammonium bicarbonate, 600 µL HPLC-grade methanol and 1 µg of sequencing grade trypsin. The trypsin incubation was completed overnight at room temperature. Samples were centrifuged (14,000× g, 20 min) and the digest with buffer removed. The sediments were then washed 3 times with 1 mL 25 mM ammonium bicarbonate, centrifuged and the extracts combined. The volume was reduced to ~10 µL, and 200 µL of 5% acetonitrile and 0.1% trifluoroacetic acid were added before desalting the peptides using C18 micro prep desalting centrifuge columns (NEST Group, Southboro, MA, USA). Sample pH was adjusted to <2 using small additions of 10% TFA and was desalted using the protocol provided by manufacturer.
2.5. Mass Spectrometry and Database Searching
Protein identification was accomplished via shotgun proteomics with samples introduced into the ion trap (LTQ Velos) mass spectrometer (Thermo Fisher, Waltham, MA, USA) by NanoAcquity high performance liquid chromatography (HPLC, Waters, Milford, MA, USA). New analytical and trapping columns were packed in-house prior to batch analyses of Bering Sea samples to prevent sample carry over. Analytical columns were made using 11 cm 75 µm i.d. fused silica capillaries packed with C18 particles (Magic C18AQ, 100 A, 5 µm; Michrom, Bioresources, Billerica, MA, USA) preceded by a 2 cm, 100 µm i.d. trapping column (Magic C18AQ, 200 A, 5 µm; Michrom). Samples were loaded onto the trapping column with a flow rate of 4 µL min−1 for 7 min and then entered the analytical column at a flow rate of 250 nL min−1 (total run time: 100 min). Peptides were eluted using an acidified (formic acid, 0.1% v/v) water-acetonitrile linear gradient (5%–35% acetonitrile in 60 min) and ionized in atmospheric pressure before entering the mass spectrometer. Ions that entered the ion trap were surveyed (MS1), and the fourteen most intense ions from scans having either +2, +3, +4 or +5 charge states were selected for collision-induced dissociation (CID) and tandem mass spectral (MS2) detection. Sample digests were analyzed using full scan (m/z 350–2,000), followed by gas phase fractionation with repeat analyses over multiple narrow, but overlapping, mass to charge ranges (e.g., m/z 350–444, 444–583, 583–825, 825–1600; []).
Mass spectra were interpreted and searched using an in-house copy of SEQUEST on a Beowolf-style computer cluster with 800 dedicated processing cores with 22 terabytes of storage [,]. All data searches were performed with no assumption of proteolytic enzyme (i.e., unconstrained search), specifically to allow for the identification of the maximal number of protein degradation products. Fixed modifications were set for 57 Da on cysteine, resulting from the IAM alkylation step, and 16 Da on methionine via oxidation. Each tandem mass spectrum was then searched against a protein sequence database to correlate predicted peptide fragmentation patterns with observed sample ions. To objectively validate peptide and protein identifications, two statistical evaluations using PeptideProphet and ProteinProphet were used to provide probability-based scores [,]. Probability thresholds for positive identifications of proteins and peptides were strictly set at 90% confidence on ProteinProphet and PeptideProphet for SEQUEST search results. Based on the databases used, the false discovery rate was calculated to be 0.5% for database searches. Mass spectra from all samples were searched against a database containing the proteomes of Thalassiosira pseudonana (marine diatom), Prochlorococcus marinus (marine cyanobacterium) and Pelagibacter ubique (marine bacterium belonging to the SAR11 clade). Identified sedimentary peptides possessed amino acid sequences that matched peptide sequences within the proteomes of the above mentioned species. This database was chosen after extensive comparison revealed that larger databases, including the NCBI non-redundant database containing over 11.9 million protein sequences and the Global Ocean Survey Combined Assembly Protein database [,], did not enhance the number of proteins identified, added limited species diversity to identified proteins and had 95% functional agreement with proteins identified from Bering Sea sediment with the smaller database [].
2.6. Total Hydrolysable Amino Acid Analysis
Individual amino acids were identified and quantified by gas chromatography (GC) and GC mass spectrometry (GC/MS) using the EZFaast method (Phenomenex, Torrance, CA, USA), which uses derivatization of AAs with propyl chloroformate and propanol for sensitive detection (see [] for a comparison of the methods). Briefly, sediment samples, sediment buffer mixtures, T. weissflogii buffer mixtures and gel section electroelutions were hydrolyzed for 4 h at 110 °C [,] with 6 M analytical-grade HCl and L-methylleucine as the recovery standard. Following hydrolysis and derivatization, amino acids were quantified using an Agilent 6890 capillary GC with samples injected at 250 °C and separated via a DB-5MS (0.25 mm ID, 30 m) column with H2 as the carrier gas. The oven was ramped from an initial temperature of 110–280 °C at 10 °C per minute followed by a 5 min hold. For amino acid identification, the GC was coupled to an Agilent 5973N mass spectrometer run under the same conditions with helium as the carrier gas and acquisition of spectra over the 50–600 Da range. Bovine serum albumin (BSA) was analyzed in parallel to correct for responses among individual amino acids and calculation of molar ratios.
3. Results and Discussion
Using the tube gel method run for 7 cm, 21 peptides were identified combined from all of the gel molecular weight range sections, correlating to 11 proteins (Table 1). From the 2 cm tube gel, 2 cm flat gel and direct digest methods, 3, 1 and 1 peptides were identified correlating to 3, 1 and 1 proteins, respectively. The majority of identified peptides and proteins correlated to T. pseudonana chloroplast proteins, including the RuBisCO large subunit and three fucoxanthin chlorophyll a/c binding proteins (FCPs). The proteins with the most identified peptides were the RuBisCO large subunit and the histone H4 protein.

Table 1.
Proteins (in bold) and associated peptides identified using each extraction method (DD = direct digest). Correlated species, molecular weight (MW), cellular location (CL) and percent sequence coverage (%) are given for each protein. For cellular location: C = chloroplast; M = mitochondria; N = nucleus; S = secretory.
| Method | Protein (Annotation)/Peptides | Species | MW | Location | % |
|---|---|---|---|---|---|
| 7 cm Tube Gel | RuBisCO large subunit | T. pseudonana | 54,324.7 | C | 13.3 |
| (gi|118411104|ref|YP_874498.1|) | |||||
| DTDVLALFR | |||||
| FLNCLEGINR | |||||
| MGYWDAAYTVK | |||||
| MSGVDHIHAGTVVGK | |||||
| VALEAMVLAR | |||||
| YESGVIPYAK | |||||
| Histone H4 | T. pseudonana | 11,383.6 | N | 43.7 | |
| (jgi|Thaps3|3184|fgenesh1_pg.C_ chr_3000118) | |||||
| DNIQGITKPAIR | |||||
| ISGLIYEETR | |||||
| VFLENVIR | |||||
| VLRDNIQGITKPAIR | |||||
| ATP-CF1-Beta | T. pseudonana | 51,143.1 | C | 5.3 | |
| (gi|118411134|ref|YP_874528.1|) | |||||
| FTQAGSEVSALLGR | |||||
| IGLFGGAGVGK | |||||
| P2A | T. pseudonana | 56,408.1 | C | 4.7 | |
| (gi|118411113|ref|YP_874507.1|) | |||||
| YQWDSGYFQQEIER | |||||
| TSLESDGVFR | |||||
| FCP-1 | T. pseudonana | 22,628.0 | C | 6.2 | |
| (jgi|Thaps3|268127|estExt_thaps1_ ua_kg.C_chr_10394) | |||||
| IAQLAFLGQVVTR | |||||
| FCP-2 | T. pseudonana | 21,807.2 | C | 6.8 | |
| (jgi|Thaps3|38667|e_gw1.22.164.1) | |||||
| IAQLAFLGNIITR | |||||
| FCP-3 | T. pseudonana | 20,354.4 | C | 6.8 | |
| (jgi|Thaps3|38494|e_gw1.20.149.1) | |||||
| ISQLAFLGQIVTR | |||||
| P2-10P | T. pseudonana | 7,388.0 | C | 21.2 | |
| (gi|118411116|ref|YP_874510.1|) | |||||
| LGEILRPLNAEYGK | |||||
| Putative cysteine synthase | T. pseudonana | 37,867.3 | M | 4.6 | |
| (jgi|Thaps3|38294|e_gw1.19a.45.1) | |||||
| MENLNPGGTGKDRAAR | |||||
| DNA-RNA-P-Gamma | P. marinus | 72,277.0 | S | 1.7 | |
| (gi|33862040|ref|NP_893601.1|) | |||||
| FATSDLNDLYR | |||||
| ATP-PBS | P. marinus | 49,496.0 | S | 3.1 | |
| (gi|33862213|ref|NP_893774.1|) | |||||
| DVSGEGVQQALLK | |||||
| 2 cm Tube Gel | FCP-2 | T. pseudonana | 21,807.2 | C | 7.3 |
| (jgi|Thaps3|38667|e_gw1.22.164.1) | |||||
| DIEGTGNEFVGDFR | |||||
| P2RC-D2 | T. pseudonana | 39,064.0 | C | 3.7 | |
| (gi|118411148|ref|YP_874542.1|) | |||||
| AAEDPEFETFYTK | |||||
| H-DNA-BP | P. marinus | 9,768.0 | S | 24.2 | |
| (gi|33861877|ref|NP_893438.1|) | |||||
| TDVSLVVDAAIETIVDSVVEGK | |||||
| 1D Gel | H-DNA-BP | P. marinus | 9,768.0 | S | 24.2 |
| (gi|33861877|ref|NP_893438.1|) | |||||
| TDVSLVVDAAIETIVDSVVEGK | |||||
| DD | H-DNA-BP | P. marinus | 9,768.0 | S | 24.2 |
| (gi|33861877|ref|NP_893438.1|) | |||||
| TDVSLVVDAAIETIVDSVVEGK |
* ATP-CF1-Beta = ATP synthase CF1 beta chain; P2A = Photosystem II chlorophyll A core antenna apoprotein; FCP = fucoxanthin chlorophyll a/c binding protein; P2-10P = Photosystem II 10-kDa phosphoprotein; DNA-RNA-P-Gamma = DNA-directed RNA polymerase subunit gamma; ATP-PBS = ATP-dependent protease ATP-binding subunit; P2RC-D2 = Photosystem II reaction center protein D2; H-DNA-BP = histone-like DNA-binding protein.
The 7 cm tube gel molecular weight section with the largest number of peptides and proteins identified was the >100 kDa section (Table 2). The 7 cm tube gel >100 kDa section was also the only section to contain peptides and proteins from four cellular locations (chloroplast, nucleus, cytoplasm and mitochondria). All other 7-cm tube gel sections contained only chloroplast and nucleus peptides and proteins or no identifications in the case of the <10 kDa section. The majority of identified peptides in the 7 cm tube gel correlated to proteins that had a molecular weight that was outside the molecular weight range of the gel section in which the peptide was identified. Of these peptides identified in unexpected gel molecular weight sections, five correlated to proteins that were less than 6.5 kDa different from their identified gel molecular weight section. Five proteins were identified in multiple 7 cm tube gel molecular weight sections, and from these proteins, the same peptides were often identified in multiple gel sections.

Table 2.
Proteins (in bold) and associated peptides identified from each 7 cm tube gel molecular weight section method. Correlated species, molecular weight (MW), cellular location (CL) and percent sequence coverage (%) are given for each protein. Molecular weights in bold represent proteins that are outside the expected gel molecular weight section; molecular weights in bold and italic represent proteins that are less than 6.5 kDa outside the expected gel molecular weight section. For cellular location: C = chloroplast; M = mitochondria; N = nucleus; S = secretory.
| Gel Section (kDa) | Protein/Peptides | Species | MW | CL | % |
|---|---|---|---|---|---|
| >100 | RuBisCO large subunit | T. pseudonana | 54,324.7 | C | 7.1 |
| MSGVDHIHAGTVVGK | |||||
| VALEAMVLAR | |||||
| FLNCLEGINR | |||||
| Histone H4 | T. pseudonana | 11,383.6 | N | 22.3 | |
| DNIQGITKPAIR | |||||
| VFLENVIR | |||||
| VLRDNIQGITKPAIR | |||||
| FCP-2 | T. pseudonana | 21,807.2 | C | 6.8 | |
| IAQLAFLGNIITR | |||||
| ATP-CF1-Beta | T. pseudonana | 51,143.1 | C | 3.0 | |
| FTQAGSEVSALLGR | |||||
| Putative cysteine synthase | T. pseudonana | 37,867.3 | M | 4.6 | |
| MENLNPGGTGKDRAAR | |||||
| DNA-RNA-P-Gamma | P. marinus | 72,277.0 | S | 1.7 | |
| FATSDLNDLYR | |||||
| ATP-PBS | P. marinus | 49,496.0 | S | 3.1 | |
| DVSGEGVQQALLK | |||||
| 50 to 100 | RuBisCO large subunit | T. pseudonana | 54,324.7 | C | 9.2 |
| DTDVLALFR | |||||
| MSGVDHIHAGTVVGK | |||||
| YESGVIPYAK | |||||
| MGYWDAAYTVK | |||||
| Histone H4 | T. pseudonana | 11,383.6 | N | 21.4 | |
| ISGLIYEETR | |||||
| DNIQGITKPAIR | |||||
| ATP-CF1-Beta | T. pseudonana | 51,143.1 | C | 3.0 | |
| FTQAGSEVSALLGR | |||||
| FCP-2 | T. pseudonana | 21,807.2 | C | 6.8 | |
| IAQLAFLGNIITR | |||||
| 25 to 50 | Histone H4 | T. pseudonana | 11,383.6 | N | 17.5 |
| ISGLIYEETR | |||||
| VFLENVIR | |||||
| P2A | T. pseudonana | 56,408.1 | C | 4.7 | |
| YQWDSGYFQQEIER | |||||
| TSLESDGVFR | |||||
| ATP-CF1-Beta | T. pseudonana | 51,143.1 | C | 3.0 | |
| FTQAGSEVSALLGR | |||||
| FCP-2 | T. pseudonana | 21,807.2 | C | 6.8 | |
| IAQLAFLGNIITR | |||||
| P2-10P | T. pseudonana | 7,388.0 | C | 21.2 | |
| LGEILRPLNAEYGK | |||||
| 10 to 25 | P2-10P | T. pseudonana | 7,388.0 | C | 21.2 |
| LGEILRPLNAEYGK | |||||
| Histone H4 | T. pseudonana | 11,383.6 | N | 29.1 | |
| ISGLIYEETR | |||||
| VFLENVIR | |||||
| DNIQGITKPAIR | |||||
| ATP-CF1-Beta | T. pseudonana | 51,143.1 | C | 5.3 | |
| IGLFGGAGVGK | |||||
| FTQAGSEVSALLGR | |||||
| FCP-1 | T. pseudonana | 22,628.0 | C | 6.2 | |
| IAQLAFLGQVVTR | |||||
| FCP-2 | T. pseudonana | 21,807.2 | C | 6.8 | |
| IAQLAFLGNIITR | |||||
| FCP-3 | T. pseudonana | 20,354.4 | C | 6.8 | |
| ISQLAFLGQIVTR | |||||
| <10 | no IDs |
* ATP-CF1-Beta = ATP synthase CF1 beta chain; P2A = Photosystem II chlorophyll A core antenna apoprotein; FCP = fucoxanthin chlorophyll a/c binding protein; P2-10P = Photosystem II 10 kDa phosphoprotein; DNA-RNA-P-Gamma = DNA-directed RNA polymerase subunit gamma; ATP-PBS = ATP-dependent protease ATP-binding subunit; P2RC-D2 = Photosystem II reaction center protein D2; H-DNA-BP = histone-like DNA-binding protein.
The concentration of THAAs in the 8 to 10 cm sediments, 0.74 mg/g sediment (dry weight), was approximately 23% less than surface sediments of the same sediment core collected before the spring phytoplankton bloom and 36% less than surface sediments collected two months later after the spring phytoplankton bloom (both surface sediment samples analyzed by Moore et al. []). The number of proteins identified in 8 to 10 cm sediments was 56% less than surface sediments from the same core and 79% less than surface sediment collected two months later after the spring phytoplankton bloom. The THAA distribution was very similar between the three sediment samples (Figure 1). The amount of THAAs varied between the molecular weight sections of the T. weissflogii gel, with relatively large amounts in the >100 kDa section and small amounts in the 50–100 kDa section (Figure 2).
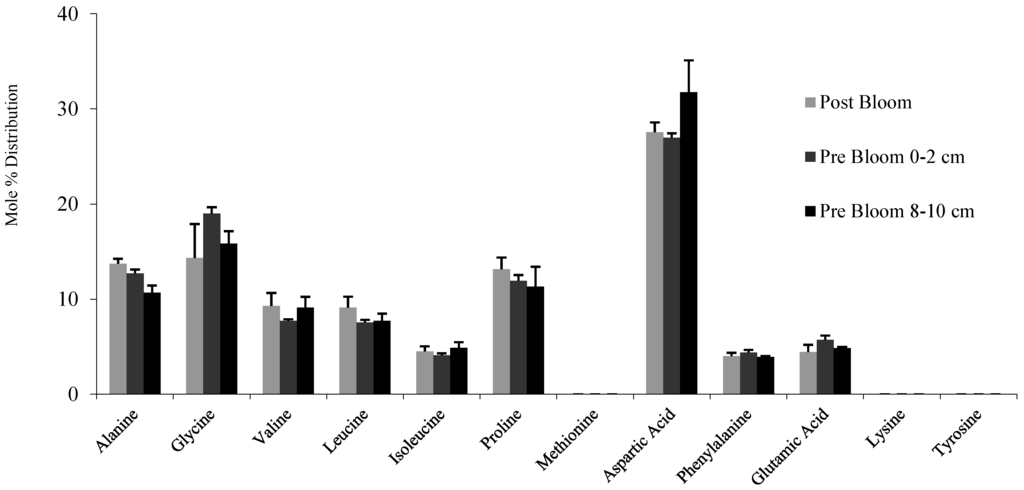
Figure 1.
Amino acid distribution and total hydrolysable amino acid (THAA) concentration (µg THAA/g sediment DW) of post spring phytoplankton bloom Bering Sea surface sediment, pre-bloom surface sediment (0–2 cm) and deeper pre-bloom sediment (8–10 cm).
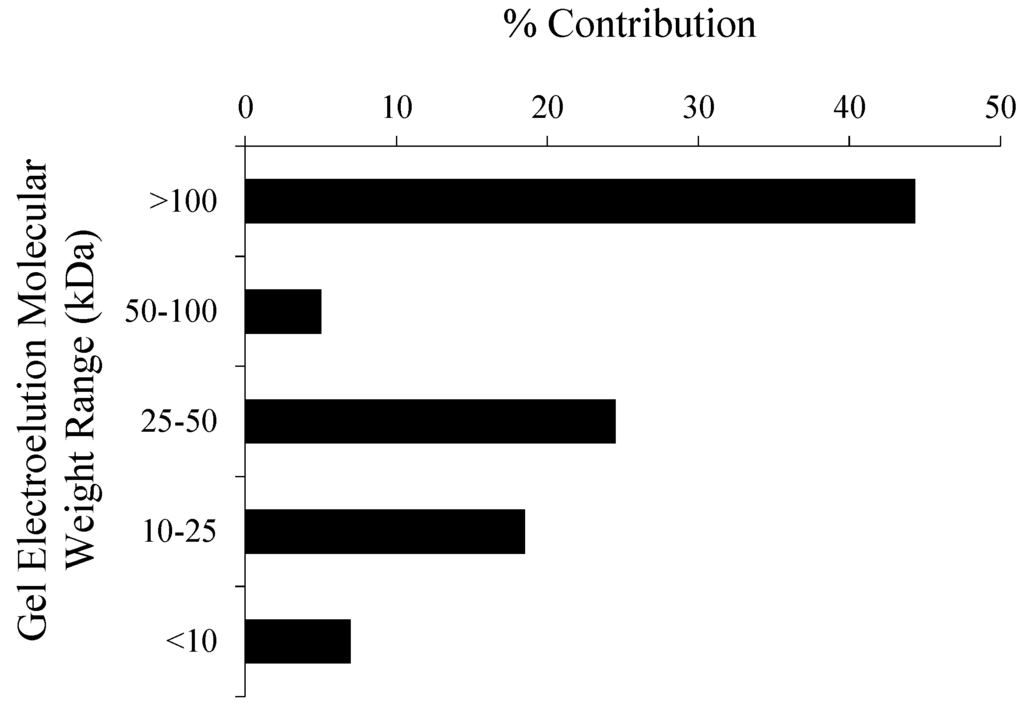
Figure 2.
The percent contribution of total hydrolysable amino acids (THAAs) electroeluted from each gel molecular weight section (>100, 50–100, 25–50, 10–25, <10 kDa) of 1D SDS-PAGE lanes loaded with sonicated diatom (Thalassiosira weissflogii) cell material.
Trypsin digestions were found to be more effective in all of the gel methods than the direct digest method. In each of the gel methods, trypsin peptides were identified as the most abundant HPLC/MS peaks in the sample, particularly in the 7 cm gel >100 kDa section (Figure 3A). The most abundant HPLC/MS peaks in the direct digest were m/z 519.27 at multiple retention times, possibly representing sample matrix material (Figure 3B). The trypsin peaks were much lower in abundance in the direct digest, with many fewer peptides identified than the gel methods, as well.
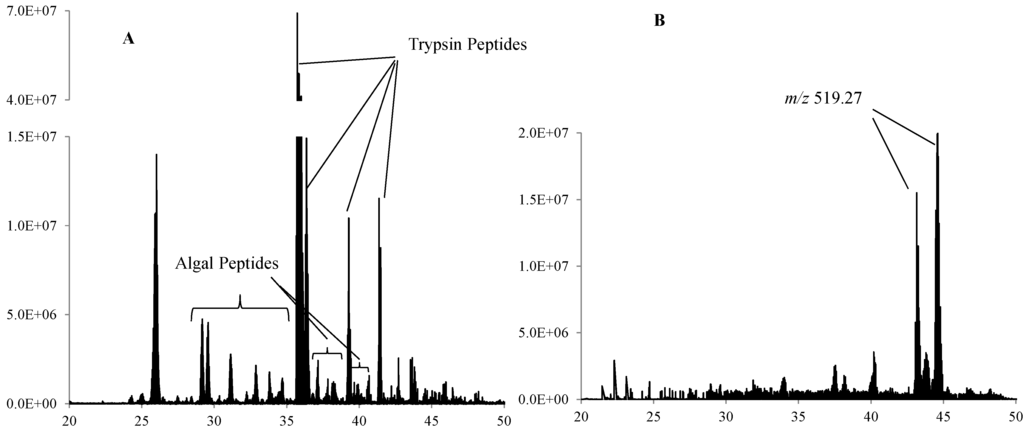
Figure 3.
LC/MS chromatograms of (A) Bering Sea 8–10 cm sediment 7 cm tube gel >100 kDa section trypsin digest and (B) Bering Sea 8–10 cm sediment direct trypsin digest.
The identification of algal peptides and proteins in the deep marine sediment core section demonstrates that gel electrophoresis can be an effective method for isolating proteins present in complex matrices, but further improvements can be made in the total recovery of material. The results obtained by loading sediment buffer mixture directly to the gel interface supports previous findings by Moore et al. [] that an electric field applied directly to sediment particles enhances protein extraction. The number of proteins identified was increased by running the gel for a longer period of time, allowing the ion front to move further down the gel (7 cm vs. 2 cm) and the electric field to mobilize proteins out of the sediment. Some brown colored sedimentary material traveled into the gel, as well, but the separation of protein from matrix was sufficient for peptides to be digested out of the gel sections with trypsin and identified via highly sensitive proteomic mass spectrometry and database searching (Figure 4, []). Protein function and amino acid sequence can be conserved across many species and taxonomic groups. Identified peptide sequences that correlated with T. pseudonana indicate that the peptide was likely conserved among species and not directly produced by T. pseudonana, but by some other algal source.
The difficult identification of peptides and proteins using the direct digest method indicates that the complex sedimentary matrix includes a suite of interferences, including mineral surfaces and organic materials, which impact protein digestion, extraction and identification. The flat gels loaded with sediment buffer mixture could not be reproducibly run over time frames for the ion front to travel greater than 2 cm down the gel. The is likely due to the fact that the electrophoresis current mobilized charged matrix particles, clays and other minerals, which were too large to move into the gel itself. The mobilized matrix particles were then either pushed down the edge of the gel or the gel was torn by the material pushing against it. The sediment material pushed against the tube gel, as well, but the thicker tube gel was sturdy enough to consistently withstand the pressure of the charged matrix material and allow proteins to be mobilized into the gel and matrix particles to be held at the top.
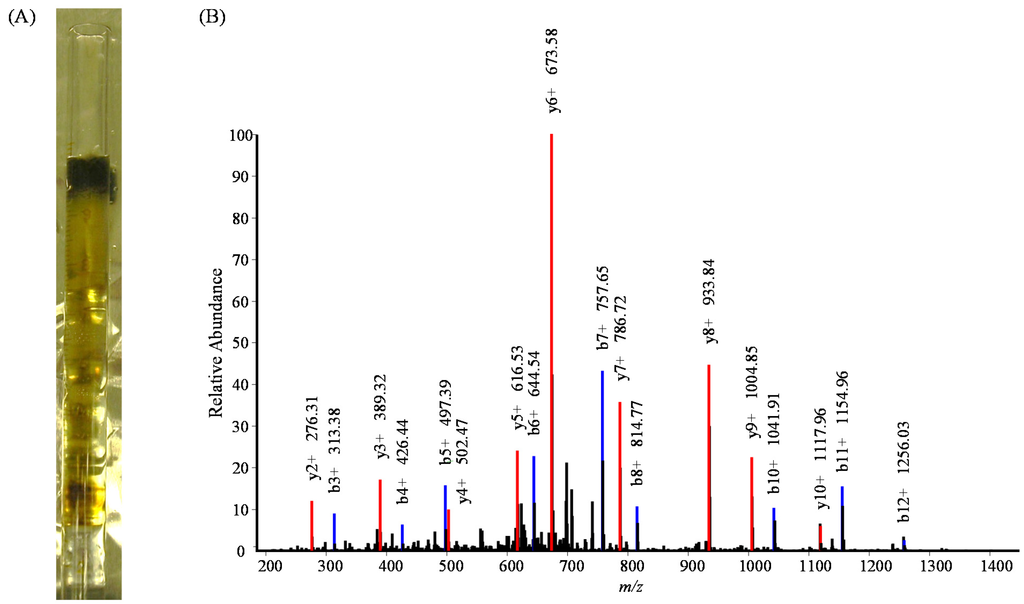
Figure 4.
(A) Tube gel with sediment slurry loaded directly to the top of the gel and run until the ion front traveled 7 cm down the gel; (B) tandem mass spectrum of peptide IAQLAFLGNIITR, MH+ 1,429.8526, m/z 715.4299, a tryptic peptide from fucoxanthin chlorophyll a/c binding protein 2 identified in molecular weight section 10–25 kDa.
The identification of peptides correlating to proteins that occurred in unexpected gel molecular weight sections (based on their known intact molecular weights) suggests that some change to protein structure or perhaps the charge has taken place. Peptides identified in smaller than expected molecular weight ranges could be due to partial hydrolysis []. Covalent modifications [,,], hydrophobic interactions [,] and sequestration in potential energy fields [] have been proposed as mechanisms for enhanced protein preservation, which could influence the gel migration of proteins to be in larger than expected molecular weight sections. The denaturing conditions of the SDS-PAGE gels used in this study indicate that charge alteration could also play a role in protein mobility. Given the physical effect of mobilized sample matrix on the gels themselves, it is certainly possible that the complex mixture of charged matrix material could also impact protein migration in the gels.
Proteins in lower core sediments have likely been exposed to more degradation processes than in surface sediments. These identified proteins represent material that may have been preserved after burial [,,,,] or mixed from the surface into deeper sediments by bioturbators [,,]. Thus, it is expected that fewer proteins, with potentially lower sequence coverage, would be identified in the Bering Sea 8- to 10-cm core section than in surface sediments observed by Moore et al. [,]. Age estimation of these sediments can be uncertain, because the surface sedimentary mixed layer can extend from 0 to 16.5 cm on the outer Bering Sea shelf []. However, given a sedimentation rate of 30 to 40 cm/1,000 y for the Bering Sea outer shelf and slope [], one could estimate that the 8–10-cm sediment is conservatively 200 to 330 years old.
The proteins that were identified in the deeper sediments were also found in the water column particulate phase and surface sediments before and after the spring bloom []. The RuBisCO large subunit, histone H4 and fucoxanthin chlorophyll a/c binding proteins were all identified after 53 days of shipboard degradation experiments, indicating that these proteins can indeed survive water column recycling long enough to be buried in sediments and are present in sufficient quantities to be detected using tandem mass spectrometry []. Furthermore, eight out of the twelve total proteins identified correlating to T. pseudonana were found to be high abundance proteins in this species based on proteomic analysis []. This indicates that the identified algal proteins in 8- to 10-cm sediments signify buried primary production and exported organic nitrogen and carbon from the water column.
It has been estimated that only approximately 1% of the originally produced organic matter from the marine upper water column is transferred to the deep biosphere [,,]. The identification of buried algal proteins (RuBisCO, FCPs, photosystem proteins, etc.) in marine sediment suggests that primary production proteins could be preserved over long periods of time. FCPs are important light harvesting complex proteins in diatoms and other marine algae [,,], which along with other chloroplast proteins, could not have been produced in the sediments by bacteria and, thus, were transported and preserved from the water column. This would be expected, since bacteria make up a small fraction of the sedimentary buried carbon pool []. The greater decrease in protein identifications than THAAs between 8 to 10 cm sediments and surface sediments [] suggests that intact proteins are present, but below detection limits. Since proteins are the functional biomacromolecules in all organisms, they could be useful biomarker molecules for reconstructing past phytoplankton communities and biogeochemistry if peptides unique to specific taxonomic groups are detected and tracked.
4. Conclusions
The sediment gel method successfully extracted buried intact proteins and/or peptides from 8–10 cm Bering Sea marine sediment for identification by proteomic mass spectrometry. More peptides were identified using the sediment tube gel method than the 1D flat gel or direct digest of the sediment. The majority of identified peptides and proteins correlated with diatom protein sequences, indicating that algal proteins survived water column recycling and burial in sediment. Identification of peptides in unexpected gel molecular weight sections suggests that modifications may have changed the protein’s structure or gel mobility. Identifying buried proteins in marine sediments suggests that these functional biomacromolecules could be preserved over long time periods and be useful biomarkers for general algal growth. This unique method allows extraction of proteins from complex environmental matrices using conventional electrophoresis in a modified approach and may also be applied to other complex environmental samples, such as terrestrial sediments and soils, giving greater understanding of protein biogeochemistry.
Supplemental Information
Supplementary materials can be accessed at: http://www.mdpi.com/2227-9075/1/4/176/s1.
Protein identification information for each 7 cm tube gel sections (>100 kDa, 50–100 kDa, 25–50 kDa, 10–25 kDa, <10 kDa), 2 cm tube gel and direct digest (DD). Information includes: Protein, annotation name, accession number, peptide sequence, species, database matching score, molecular weight (MW), cellular location and percent sequence coverage. For cellular location: C = chloroplast; M = mitochondria; N = nucleus; S = secretory.
Acknowledgments
This work was supported by the National Science Foundation grants through the Chemical Oceanography Program to H. Rodger Harvey (HRH) (OCE-1233589), David R. Goodlett (DRG) and Brook L. Nunn (OCE-0825790, OCE-1233014), with samples obtained as part of the NSF Bering Ecosystem Study Program (BEST) to HRH (ARC-0732667). Computational analysis was conducted on a National Institute of Health sponsored computer cluster award to DRG (1S10RR023044-01, 1U54AI57141-01). This is Marine Organic Geochemistry and Ecology Laboratory-Ocean Earth and Atmospheric Sciences (MOGEL-OEAS) contribution No. 2011-3 of Old Dominion University.
Author Contributions
The author contributions are as follows: EKM—sample analysis and data interpretations; HRH—sample collection and data interpretation; JFF—sample analysis; DRG—laboratory and instrument support; BLN—proteomic analysis and data interpretation. EKM, HRH, and BLN wrote the paper. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References and Notes
- Falkowski, P.G. Evolution of the nitrogen cycle and its influence of the biological sequestration of CO2 in the ocean. Nature 1997, 387, 272–275. [Google Scholar]
- McCarthy, M.; Pratum, T.; Hedges, J.; Benner, R. Chemical composition of dissolved organic nitrogen in the ocean. Nature 1997, 390, 150–154. [Google Scholar]
- Knicker, H. Solid-State 2-D double cross polarization magic angle spinning N-15 C-13 NMR spectroscopy on degraded algal residues. Org. Geochem. 2000, 31, 337–340. [Google Scholar]
- Zang, X.; Nguyen, R.T.; Harvey, H.R.; Knicker, H.; Hatcher, P.G. Preservation of proteinaceous material during the degradation of the green alga Botryococcus braunii: A solidstate 2D 15N 13C NMR Spectroscopy Study. Geochim. Cosmochim. Acta 2001, 65, 3299–3305. [Google Scholar]
- Cowie, G.L.; Hedges, J.I. Sources and reactivities of amino acids in a coastal marine environment. Limnol. Oceanogr. 1992, 37, 703–724. [Google Scholar]
- Grutters, M.; van Raaphorst, W.; Helder, W. Total hydrolysable amino acid mineralisation in sediments across the northeastern Atlantic continental slope (Goban Spur). Deep-Sea Res. I 2001, 48, 811–832. [Google Scholar]
- Vandewiele, S.; Cowie, G.; Soetaert, K.; Middelburg, J.J. Amino acid biogeochemistry and organic matter degradation state across the Pakistan margin oxygen minimum zone. Deep-Sea Res. II 2009, 56, 318–334. [Google Scholar]
- Fernandes, L.; Garg, A.; Borole, D.V. Amino acid biogeochemistry and bacterial contribution to sediment organic matter along the western margin of the Bay of Bengal. Deep-Sea Res. I 2014, 83, 81–92. [Google Scholar]
- Nunn, B.L.; Timperman, A.T. Marine proteomics. MEPS 2007, 332, 281–289. [Google Scholar]
- Morris, M.M.; Nunn, B.L.; Frazar, C.; Goodlett, D.R.; Ting, Y.S.; Rocap, G. Comparative metaproteomics reveals ocean-scale shifts in microbial nutrient utilization and energy transduction. ISME 2010, 4, 673–685. [Google Scholar]
- Zang, X.; van Heemst, J.D.H.; Dria, K.J.; Hatcher, P.G. Encapsulation of protein in humic acid from a histosol as an explanation for the occurrence of organic nitrogen in soil and sediment. Org. Geochem. 2000, 31, 679–695. [Google Scholar]
- Nguyen, R.T.; Harvey, H.R. Preservation of protein in marine systems: Hydrophobic and other noncovalent associations as major stabilizing forces. Geochim. Cosmochim. Acta 2001, 65, 1467–1480. [Google Scholar]
- Nguyen, R.T.; Harvey, H.R. Preservation via macromolecular associations during Botryococcus braunii decay: Proteins in the Pula Kerogen. Org. Geochem. 2003, 34, 1391–1403. [Google Scholar]
- Ogunseitan, O.A. Direct extraction of proteins from environmental samples. J. Microbiol. Meth. 1993, 17, 273–281. [Google Scholar]
- Nunn, B.L.; Keil, R.G. A comparison of non- hydrolytic methods for extracting amino acids and proteins from coastal marine sediments. Mar. Chem. 2006, 98, 31–42. [Google Scholar]
- Moore, E.K.; Nunn, B.L.; Faux, J.F.; Goodlett, D.R.; Harvey, H.R. Evaluation of electrophoretic protein extraction and database-driven protein identification from marine sediments. Limnol. Oceanogr.Meth. 2012, 10, 353–366. [Google Scholar]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar]
- Reisfeld, R.A.; Lewis, U.J.; Williams, D.E. Disc electrophoresis of basic proteins and peptides on polyacrylamide gels. Nature 1962, 195, 281–283. [Google Scholar]
- Laver, W.G. Structural studies on the protein subunits from three strains of influenza virus. J. Mol. Biol. 1964, 9, 109–124. [Google Scholar]
- Shapiro, A.L.; Vinuela, E.; Maizel, J.V., Jr. Molecular weight estimation of polypeptide chains by electrophoresis in SDS-polyacrylamide gels. Biochem. Biophys. Res. Commun. 1967, 28, 815–820. [Google Scholar]
- Fairbanks, G.; Steck, T.L.; Wallach, D.F.H. Electrophoretic analysis of the major proteins of the human erythrocyte membrane. Biochemistry 1971, 10, 2606–2617. [Google Scholar]
- Maizel, J.V. SDS polyacrylamide gel electrophoresis. Trends Biochem. Sci. 2000, 25, 590–592. [Google Scholar]
- Pederson, T. Turning a PAGE: The overnight sensation of SDS-polycrylamide gel electrophoresis. FASEB J. 2008, 22, 949–953. [Google Scholar]
- Wang, D.Z.; Xie, Z.X.; Zhang, S.F. Marine metaproteomics: Current status and future directions. J. Proteom. 2014, 97, 27–35. [Google Scholar]
- Tran, J.C.; Doucette, A.A. Multiplexed size separation of intact proteins in solution phase for mass spectrometry. Anal. Chem. 2009, 81, 6201–6209. [Google Scholar]
- Botelho, D.; Wall, M.J.; Vieira, D.B.; Fitzsimmons, S.; Liu, F.; Doucette, A. Top-Down and bottom-up proteomics of SDS-containing solutions following mass-based separation. J. Proteome Res. 2010, 9, 2863–2870. [Google Scholar]
- Chourey, K.; Jansson, J.; VerBerkmoes, N.; Shah, M.; Chavarria, K.L.; Tom, L.M.; Brodie, E.L.; Hettich, R.L. Direct cellular lysis/protein extraction protocol for soil metaproteomics. J. Proteome Res. 2010, 9, 6615–6622. [Google Scholar]
- Bastida, F.; Hernández, T.; Garcia, C. Metaproteomics of soils from semiarid environment: Functional and phylogenetic information obtained with different protein extraction methods. J. Proteomics 2014, 101, 31–42. [Google Scholar]
- Moore, E.K.; Brook, B.L.; Goodlett, D.R.; Harvey, H.R. Identifying and tracking proteins through the marine water column: Insights into the inputs and preservation mechanisms of protein in sediments. Geochim. Cosmochim. Acta 2012, 83, 324–359. [Google Scholar]
- Sambrotto, R.N.; Niebauer, H.J.; Goering, J.J.; Iverson, R.L. Relationships among vertical mixing, nitrate uptake, and phytoplankton growth during spring bloom in the S-E Bering Sea. Cont. Shelf Res. 1986, 5, 161–198. [Google Scholar]
- McRoy, C.P. Global maximum of primary production in the North Bering Sea. E.O.S. Comm. 1987, 68, 172. [Google Scholar]
- Walsh, J.J.; McRoy, C.P.; Coachman, L.K.; Goering, J.J.; Nihoul, J.J.; Whitledge, T.E.; Blackburn, T.H.; Parker, P.L.; Wirick, C.D.; Shuert, P.G.; et al. Carbon and nitrogen cycling within the Bering Chukchi Seas: Source regions for organic matter effecting AOU demands of the Arctic Ocean. Progr. Oceanogr. 1989, 22, 277–359. [Google Scholar]
- Highsmith, R.C.; Coyle, K.O. High productivity of northern Bering Sea benthic amphipods. Nature 1990, 862, 862–864. [Google Scholar]
- Chen, M.; Huang, Y.P.; Cai, P.G.; Guo, L.D. Particulate organic carbon export fluxes in the Canada Basin and Bering Sea as derived from 234Th/238U disequilibria. Arctic 2003, 56, 32–44. [Google Scholar]
- Lovvorn, J.R.; Cooper, L.W.; Brooks, M.L.; de Ruyck, C.C.; Bump, J.K.; Grebmeier, J.M. Organic matter pathways to zooplankton and benthos under pack ice in late winter and open water in late summer in the north-central Bering Sea. MEPS 2005, 291, 135–150. [Google Scholar]
- Wiese, F.K.; Wiseman, W.J., Jr.; van Pelt, T.I. Bering sea linkages introduction. Deep-Sea Res. Pt. II 2012, 65–70, 2–5. [Google Scholar]
- Kan, J.; Hanson, T.E.; Ginter, J.M.; Wang, K.; Chen, F. Metaproteomic analysis of Chesapeake Bay bacterial communities. Saline Syst. 2005, 1, 1–13. [Google Scholar]
- Shevchenko, A.; Wilm, M.; Vorm, O.; Mann, M. Mass spectrometric sequencing of proteins from silver stained polyacrylamide gels. Anal. Chem. 1996, 68, 850–858. [Google Scholar]
- Nunn, B.L.; Aker, J.R.; Shaffer, S.A.; Tsai, Y.; Strzepek, R.F.; Boyd, P.W.; Freeman, T.L.; Brittnacher, M.; Malmström, L.; Goodlett, D.R. Deciphering diatom biochemical pathways via whole-cell proteomics. Aquat. Microb. Ecol. 2009, 55, 241–253. [Google Scholar]
- Eng, J.K.; McCormack, A.L.; Yates, J.R. An approach to correlate tandem mass spectral data of peptides with amino acid-sequences in a protein database. J. Am. Soc. Mass Spectrom. 1994, 5, 976–989. [Google Scholar]
- Eng, J.K.; Fischer, B.; Grossmann, J.; MacCoss, M.J. A fast SEQUEST cross correlation algorithm. J. Proteome. Res. 2008, 7, 4598–4602. [Google Scholar]
- Keller, A.; Nesvizhskii, A.I.; Kolker, E.; Aebersold, R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 2002, 74, 5383–5392. [Google Scholar]
- Nesvizhskii, A.I.; Keller, A.; Kolker, E.; Aebersold, R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 2003, 75, 4646–4658. [Google Scholar]
- Rusch, D.B.; Halpern, A.L.; Sutton, G.; Heidelberg, K.B.; Williamson, S.; Yooseph, S.; Wu, D.; Eisen, J.A.; Hoffman, J.M.; Remington, K.; et al. The Sorcerer II Global Ocean Sampling expedition: Northwest Atlantic through Eastern Tropical Pacific. PLoS Biol. 2007, 5, e77. [Google Scholar]
- Yooseph, S.; Sutton, G.; Rusch, D.B.; Halpern, A.L.; Williamson, S.J.; Remington, K.; Eisen, J.A.; Heidelberg, K.B.; Manning, G.; Li, W.; et al. The Sorcerer II Global Ocean Sampling expedition: Expanding the universe of protein families. PLoS Biol. 2007, 5, 432–466. [Google Scholar]
- Waldhier, M.C.; Dettmer, K.; Gruber, M.A.; Oefner, P.J. Comparison of derivatization and chromatographic methods for GC–MS analysis of amino acid enantiomers in physiological samples. J. Chromatogr. B 2010, 878, 1103–1112. [Google Scholar]
- Cheng, C.N.; Shufeldt, R.C.; Stevenson, F.J. Amino acid analysis of soils and sediments: Extraction and desalting. Soil Biol. Biochem. 1975, 7, 143–151. [Google Scholar]
- Cowie, G.L.; Hedges, J.I. Improved amino acid quantification in environmental-samples —charge-matched recovery standards and reduced analysis time. Mar. Chem. 1992b, 37, 223–238. [Google Scholar]
- Mass spectra data files can be downloaded online at: https://chorusproject.org/anonymous/download/experiment/52779a6b839e496e803de596edbff228
- Pantoja, S.; Lee, C. Molecular weight distribution of proteinaceous material in Long Island Sound sediments. Limnol. Oceanogr. 1999, 44, 1323–1330. [Google Scholar]
- Cronin, J.R.; Morris, R.J. Rapid formation of humic material from diatom debris. In Coastal Upwelling, Its Sediment Record; Suess, E.T., Ed.; Plenum: New York, NY, USA, 1981; pp. 485–496. [Google Scholar]
- Benner, R.; Pakulski, J.D.; McCarthy, M.; Hedges, J.I.; Hatcher, P.G. Bulk chemical characteristics of dissolved organic matter in the Ocean. Science 1992, 255, 1561–1564. [Google Scholar]
- Bennett, R.H.; Hulbert, M.H.; Curry, K.J.; Curry, A.; Douglas, J. Organic matter sequestered in potential energy fields predicted by 3-D clay microstructure model: Direct observations of organo-clay micro- and nanofabric. Marine Geol. 2012, 315, 108–114. [Google Scholar]
- Knicker, H.; Hatcher, P.G. Survival of protein in an organic-rich sediment; possible protection by encapsulation in organic matter. Naturwissenschaften 1997, 84, 231–234. [Google Scholar]
- Hedges, J.I.; Keil, R.G. Organic geochemical perspectives on estuarine processes: Sorption reactions and consequences. Mar. Chem. 1999, 65, 55–65. [Google Scholar]
- Arnarson, T.S.; Keil, R.G. Influence of organic-mineral aggregates on microbial degradation of the dinoflagellate Scrippsiella. trochoidea. Geochim. Cosmochim. Acta 2005, 69, 2111–2117. [Google Scholar]
- De Leeuw, J.W.; Versteegh, G.J.M.; van Bergen, P.F. Biomacromolecules of plants and algae and their fossil analogues. Plant Ecol. 2006, 189, 209–233. [Google Scholar]
- Meysman, F.J.R.; Middelburg, J.J.; Heip, C.H.R. Bioturbation: A fresh look at Darwin’s last idea. Trends Ecol. Evol. 2006, 21, 688–695. [Google Scholar]
- Waldbusser, G.G.; Marinelli, R.L. Evidence of infaunal effects on porewater advection and biogeochemistry in permeable sediments: A proposed infaunal functional group framework. J. Mar. Res. 2009, 67, 503–532. [Google Scholar]
- Zonneveld, K.A.F.; Versteegh, G.J.M.; Kasten, S.; Eglinton, T.I.; Emeis, K.C.; Huguet, C.; Koch, B.P.; de Lange, G.J.; de Leeuw, J.W.; Middelburg, J.J.; et al. Selective preservation of organic matter in marine environments; processes and impact on the sedimentary record. Biogeosciences 2010, 7, 483–511. [Google Scholar]
- Oguri, K.; Harada, N.; Tadai, O. Excess 21°Pb and 137Cs concentrations, mass accumulation rates, and sedimentary processes on the Bering Sea continental shelf. Deep-Sea Res. II 2012, 61–64, 193–204. [Google Scholar]
- Gershanovich, D.E. New data on geomorphology and recent sediments of the bering sea and gulf of Alaska. Marin. Geol. 1968, 6, 281–296. [Google Scholar]
- Moore, E.K.; Nunn, B.L.; Faux, J.F.; Goodlett, D.R.; Harvey, H.R. Protein recycling in the Bering Sea algal incubations. MEPS 2014. in review. [Google Scholar]
- Suess, E. Particulate organic carbon flux in the oceans–surface productivity and oxygen utilization. Nature 1980, 288, 260–263. [Google Scholar]
- Hedges, J.I.; Keil, R.G. Sedimentary organic matter preservation: An assessment and speculative synthesis. Mar. Chem. 1995, 49, 81–115. [Google Scholar]
- Middelburg, J.J.; Meysman, F.J.R. Burial at Sea. Science 2007, 316, 1294–1294. [Google Scholar]
- Grossman, A.R.; Bhaya, D.; Apt, K.E.; Kehoe, D.M. Light-Harvesting complexes in oxygenic photosynthesis: Diversity, control, and evolution. Annu. Rev. Genet. 1995, 29, 231–288. [Google Scholar]
- Lang, M.; Kroth, P.G. Diatom fucoxanthin chlorophyll a/c-binding protein (FCP) and land plant light-harvesting proteins use a similar pathway for thylakoid membrane insertion. J. Biol. Chem. 2001, 276, 7985–7991. [Google Scholar]
- Kallmeyer, J.; Pockalny, R.; Adhikari, R.R.; Smith, D.C.; D’Hondt, S. Global distribution of microbial abundance and biomass in subseafloor sediment. Proc. Natl. Acad. Sci. USA 2012, 109, 16213–16216. [Google Scholar]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).