Human Blood Exosomes: Isolation and Characterization Methods, Variability, and the Need for Standardized Protocols—A Review
Abstract
1. Introduction
1.1. Exosome Clinical Applications
1.2. Exosome Isolation Methods
1.3. Exosome Quantification Methods
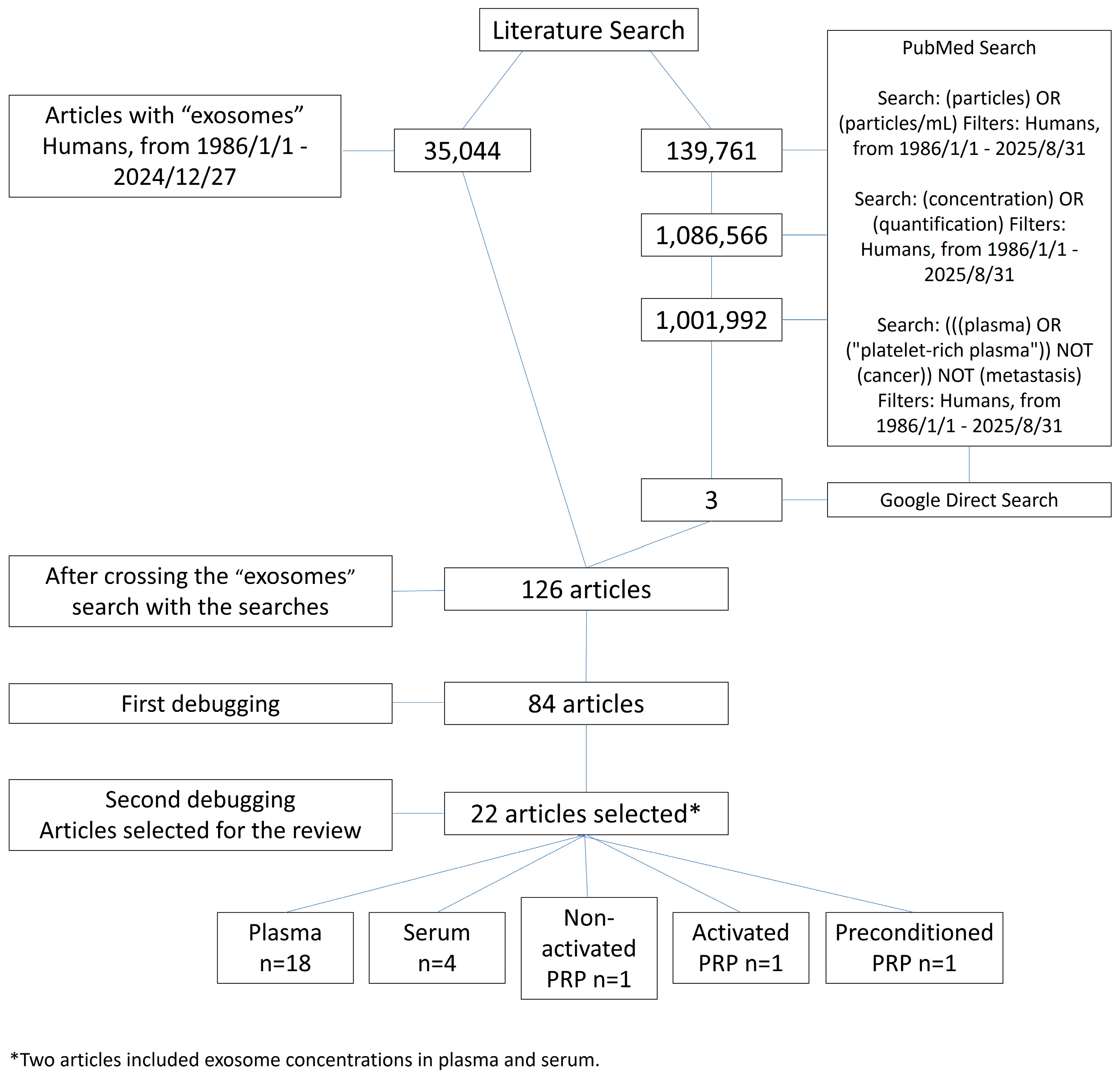
2. Materials and Methods
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| DLS | Dynamic light scattering |
| EM | Electron Microscopy |
| FCM | Flow cytometry |
| IAC | Immunoaffinity-based capture |
| miRNA | MicroRNA |
| MSC-Exo | Mesenchymal stem cell-derived exosome |
| MV | Microvesicle |
| NTA | Nanoparticle tracking analysis |
| PBM | Photobiomodulation |
| PRP | Platelet-rich plasma |
| PTBM | Photothermal biomodulation |
| SD | Standard deviation |
| SEC | Size-exclusion chromatography |
| SOPs | Standardized operating procedures |
| SP-IRIS | Single-particle interferometric reflectance imaging sensor |
| SPR | Surface plasmon resonance |
| TRPS | Tunable resistive pulse sensing |
| UC | Ultracentrifugation |
| UF | Ultrafiltration |
References
- Momen-Heravi, F.; Getting, S.J.; Moschos, S.A. Extracellular Vesicles and Their Nucleic Acids for Biomarker Discovery. Pharmacol. Ther. 2018, 192, 170–187. [Google Scholar] [CrossRef] [PubMed]
- Witwer, K.W.; Buzás, E.I.; Bemis, L.T.; Bora, A.; Lässer, C.; Lötvall, J.; Nolte-‘t Hoen, E.N.; Piper, M.G.; Sivaraman, S.; Skog, J.; et al. Standardization of Sample Collection, Isolation and Analysis Methods in Extracellular Vesicle Research. J. Extracell. Vesicles 2013, 2, 20360. [Google Scholar] [CrossRef]
- Doyle, L.; Wang, M. Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells 2019, 8, 727. [Google Scholar] [CrossRef]
- Corrado, C.; Raimondo, S.; Chiesi, A.; Ciccia, F.; De Leo, G.; Alessandro, R. Exosomes as Intercellular Signaling Organelles Involved in Health and Disease: Basic Science and Clinical Applications. Int. J. Mol. Sci. 2013, 14, 5338–5366. [Google Scholar] [CrossRef]
- Bobrie, A.; Colombo, M.; Raposo, G.; Théry, C. Exosome Secretion: Molecular Mechanisms and Roles in Immune Responses. Traffic 2011, 12, 1659–1668. [Google Scholar] [CrossRef]
- Palomar-Alonso, N.; Lee, M.; Kim, M. Exosomes: Membrane-Associated Proteins, Challenges and Perspectives. Biochem. Biophys. Rep. 2024, 37, 101599. [Google Scholar] [CrossRef]
- Johnstone, R.M.; Adam, M.; Hammond, J.R.; Orr, L.; Turbide, C. Vesicle Formation during Reticulocyte Maturation. Association of Plasma Membrane Activities with Released Vesicles (Exosomes). J. Biol. Chem. 1987, 262, 9412–9420. [Google Scholar] [CrossRef] [PubMed]
- van Niel, G.; Porto-Carreiro, I.; Simoes, S.; Raposo, G. Exosomes: A Common Pathway for a Specialized Function. J. Biochem. 2006, 140, 13–21. [Google Scholar] [CrossRef]
- Yuan, Y.-G.; Wang, J.-L.; Zhang, Y.-X.; Li, L.; Reza, A.M.M.T.; Gurunathan, S. Biogenesis, Composition and Potential Therapeutic Applications of Mesenchymal Stem Cells Derived Exosomes in Various Diseases. Int. J. Nanomed. 2023, 18, 3177–3210. [Google Scholar] [CrossRef] [PubMed]
- EL Andaloussi, S.; Lakhal, S.; Mäger, I.; Wood, M.J.A. Exosomes for Targeted SiRNA Delivery across Biological Barriers. Adv. Drug Deliv. Rev. 2013, 65, 391–397. [Google Scholar] [CrossRef]
- Li, X.; Corbett, A.L.; Taatizadeh, E.; Tasnim, N.; Little, J.P.; Garnis, C.; Daugaard, M.; Guns, E.; Hoorfar, M.; Li, I.T.S. Challenges and Opportunities in Exosome Research—Perspectives from Biology, Engineering, and Cancer Therapy. APL Bioeng. 2019, 3, 011503. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, N.; Whiteside, T.L.; Reichert, T.E. Challenges in Exosome Isolation and Analysis in Health and Disease. Int. J. Mol. Sci. 2019, 20, 4684. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, T.; Takahashi, Y.; Takakura, Y. Possibility of Exosome-Based Therapeutics and Challenges in Production of Exosomes Eligible for Therapeutic Application. Biol. Pharm. Bull. 2018, 41, 835–842. [Google Scholar] [CrossRef]
- Chung, I.-M.; Rajakumar, G.; Venkidasamy, B.; Subramanian, U.; Thiruvengadam, M. Exosomes: Current Use and Future Applications. Clin. Chim. Acta 2020, 500, 226–232. [Google Scholar] [CrossRef]
- Vitha, A.E.; Kollefrath, A.W.; Huang, C.-Y.C.; Garcia-Godoy, F. Characterization and Therapeutic Uses of Exosomes: A New Potential Tool in Orthopedics. Stem Cells Dev. 2019, 28, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Popowski, K.; Lutz, H.; Hu, S.; George, A.; Dinh, P.; Cheng, K. Exosome Therapeutics for Lung Regenerative Medicine. J. Extracell. Vesicles 2020, 9, 1785161. [Google Scholar] [CrossRef]
- Sanghani, A.; Andriesei, P.; Kafetzis, K.N.; Tagalakis, A.D.; Yu-Wai-Man, C. Advances in Exosome Therapies in Ophthalmology–From Bench to Clinical Trial. Acta Ophthalmol. 2022, 100, 243–252. [Google Scholar] [CrossRef]
- Kost, Y.; Muskat, A.; Mhaimeed, N.; Nazarian, R.S.; Kobets, K. Exosome Therapy in Hair Regeneration: A Literature Review of the Evidence, Challenges, and Future Opportunities. J. Cosmet. Dermatol. 2022, 21, 3226–3231. [Google Scholar] [CrossRef]
- Salarpour, S.; Barani, M.; Pardakhty, A.; Khatami, M.; Pal Singh Chauhan, N. The Application of Exosomes and Exosome-Nanoparticle in Treating Brain Disorders. J. Mol. Liq. 2022, 350, 118549. [Google Scholar] [CrossRef]
- Sheridan, C. Exosome Cancer Diagnostic Reaches Market. Nat. Biotechnol. 2016, 34, 359–360. [Google Scholar] [CrossRef]
- Reiss, A.B.; Ahmed, S.; Johnson, M.; Saeedullah, U.; De Leon, J. Exosomes in Cardiovascular Disease: From Mechanism to Therapeutic Target. Metabolites 2023, 13, 479. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Shang, X.; Guo, M.; Su, L.; Wang, J. Exosomes in the Diagnosis of Neuropsychiatric Diseases: A Review. Biology 2024, 13, 387. [Google Scholar] [CrossRef] [PubMed]
- Duan, L.; Lin, W.; Zhang, Y.; Jin, L.; Xiao, J.; Wang, H.; Pang, S.; Wang, H.; Sun, D.; Gong, Y.; et al. Exosomes in Autoimmune Diseases: A Review of Mechanisms and Diagnostic Applications. Clin. Rev. Allergy Immunol. 2025, 68, 5. [Google Scholar] [CrossRef]
- Crenshaw, B.J.; Sims, B.; Matthews, Q.L. Biological Function of Exosomes as Diagnostic Markers and Therapeutic Delivery Vehicles in Carcinogenesis and Infectious Diseases. In Nanomedicines; IntechOpen: London, UK, 2019. [Google Scholar]
- Xu, L.; Wu, L.-F.; Deng, F.-Y. Exosome: An Emerging Source of Biomarkers for Human Diseases. Curr. Mol. Med. 2019, 19, 387–394. [Google Scholar] [CrossRef]
- Cully, M. Exosome-Based Candidates Move into the Clinic. Nat. Rev. Drug Discov. 2021, 20, 6–7. [Google Scholar] [CrossRef]
- Jiang, L.; Gu, Y.; Du, Y.; Liu, J. Exosomes: Diagnostic Biomarkers and Therapeutic Delivery Vehicles for Cancer. Mol. Pharm. 2019, 16, 3333–3349. [Google Scholar] [CrossRef]
- Santos, P.; Almeida, F. Exosome-Based Vaccines: History, Current State, and Clinical Trials. Front. Immunol. 2021, 12, 711565. [Google Scholar] [CrossRef]
- Li, Q.; Li, Y.; Shao, J.; Sun, J.; Hu, L.; Yun, X.; Liuqing, C.; Gong, L.; Wu, S. Exploring Regulatory Frameworks for Exosome Therapy: Insights and Perspectives. Health Care Sci. 2025, 4, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Dilsiz, N. A Comprehensive Review on Recent Advances in Exosome Isolation and Characterization: Toward Clinical Applications. Transl. Oncol. 2024, 50, 102121. [Google Scholar] [CrossRef]
- Li, P.; Kaslan, M.; Lee, S.H.; Yao, J.; Gao, Z. Progress in Exosome Isolation Techniques. Theranostics 2017, 7, 789–804. [Google Scholar] [CrossRef]
- Zeringer, E.; Barta, T.; Li, M.; Vlassov, A.V. Strategies for Isolation of Exosomes. Cold Spring Harb. Protoc. 2015, 2015, 319–323. [Google Scholar] [CrossRef] [PubMed]
- Hassanzadeh-Barforoushi, A.; Sango, X.; Johnston, E.L.; Haylock, D.; Wang, Y. Microfluidic Devices for Manufacture of Therapeutic Extracellular Vesicles: Advances and Opportunities. J. Extracell. Vesicles 2025, 14, e70132. [Google Scholar] [CrossRef]
- Comfort, N.; Cai, K.; Bloomquist, T.R.; Strait, M.D.; Ferrante, A.W., Jr.; Baccarelli, A.A. Nanoparticle Tracking Analysis for the Quantification and Size Determination of Extracellular Vesicles. J. Vis. Exp. 2021, e62447. [Google Scholar] [CrossRef]
- Cook, S.; Tang, V.A.; Lannigan, J.; Jones, J.C.; Welsh, J.A. Quantitative Flow Cytometry Enables End-to-End Optimization of Cross-Platform Extracellular Vesicle Studies. Cell Rep. Methods 2023, 3, 100664. [Google Scholar] [CrossRef]
- Koritzinsky, E.H.; Street, J.M.; Star, R.A.; Yuen, P.S.T. Quantification of Exosomes. J. Cell. Physiol. 2017, 232, 1587–1590. [Google Scholar] [CrossRef]
- Khan, M.A.; Anand, S.; Deshmukh, S.K.; Singh, S.; Singh, A.P. Determining the Size Distribution and Integrity of Extracellular Vesicles by Dynamic Light Scattering. In Cancer Biomarkers: Methods and Protocols; Springer: New York, NY, USA, 2022; pp. 165–175. [Google Scholar]
- Wu, Y.; Wang, Y.; Lu, Y.; Luo, X.; Huang, Y.; Xie, T.; Pilarsky, C.; Dang, Y.; Zhang, J. Microfluidic Technology for the Isolation and Analysis of Exosomes. Micromachines 2022, 13, 1571. [Google Scholar] [CrossRef]
- Vafadar, A.; Nayerain Jazi, N.; Eghtesadi, M.; Ehtiati, S.; Movahedpour, A.; Savardashtaki, A. Exosome Biosensors for Detection of Ovarian Cancer. Mol. Cell. Probes 2025, 84, 102051. [Google Scholar] [CrossRef]
- Deng, F.; Ratri, A.; Deighan, C.; Daaboul, G.; Geiger, P.C.; Christenson, L.K. Single-Particle Interferometric Reflectance Imaging Characterization of Individual Extracellular Vesicles and Population Dynamics. J. Vis. Exp. 2022, 179, e62988. [Google Scholar] [CrossRef] [PubMed]
- Kurian, T.K.; Banik, S.; Gopal, D.; Chakrabarti, S.; Mazumder, N. Elucidating Methods for Isolation and Quantification of Exosomes: A Review. Mol. Biotechnol. 2021, 63, 249–266. [Google Scholar] [CrossRef] [PubMed]
- Jeppesen, D.K.; Hvam, M.L.; Primdahl-Bengtson, B.; Boysen, A.T.; Whitehead, B.; Dyrskjøt, L.; Ørntoft, T.F.; Howard, K.A.; Ostenfeld, M.S. Comparative Analysis of Discrete Exosome Fractions Obtained by Differential Centrifugation. J. Extracell. Vesicles 2014, 3, 25011. [Google Scholar] [CrossRef]
- Liu, W.; Ma, Z.; Kang, X. Current Status and Outlook of Advances in Exosome Isolation. Anal. Bioanal. Chem. 2022, 414, 7123–7141. [Google Scholar] [CrossRef] [PubMed]
- Zarovni, N.; Corrado, A.; Guazzi, P.; Zocco, D.; Lari, E.; Radano, G.; Muhhina, J.; Fondelli, C.; Gavrilova, J.; Chiesi, A. Integrated Isolation and Quantitative Analysis of Exosome Shuttled Proteins and Nucleic Acids Using Immunocapture Approaches. Methods 2015, 87, 46–58. [Google Scholar] [CrossRef] [PubMed]
- Imanbekova, M.; Suarasan, S.; Lu, Y.; Jurchuk, S.; Wachsmann-Hogiu, S. Recent Advances in Optical Label-Free Characterization of Extracellular Vesicles. Nanophotonics 2022, 11, 2827–2863. [Google Scholar] [CrossRef]
- Libregts, S.F.W.M.; Arkesteijn, G.J.A.; Németh, A.; Nolte-’t Hoen, E.N.M.; Wauben, M.H.M. Flow Cytometric Analysis of Extracellular Vesicle Subsets in Plasma: Impact of Swarm by Particles of Non-interest. J. Thromb. Haemost. 2018, 16, 1423–1436. [Google Scholar] [CrossRef]
- Anderson, W.; Lane, R.; Korbie, D.; Trau, M. Observations of Tunable Resistive Pulse Sensing for Exosome Analysis: Improving System Sensitivity and Stability. Langmuir 2015, 31, 6577–6587. [Google Scholar] [CrossRef]
- Daaboul, G.G.; Gagni, P.; Benussi, L.; Bettotti, P.; Ciani, M.; Cretich, M.; Freedman, D.S.; Ghidoni, R.; Ozkumur, A.Y.; Piotto, C.; et al. Digital Detection of Exosomes by Interferometric Imaging. Sci. Rep. 2016, 6, 37246. [Google Scholar] [CrossRef]
- Chen, W.; Li, Z.; Cheng, W.; Wu, T.; Li, J.; Li, X.; Liu, L.; Bai, H.; Ding, S.; Li, X.; et al. Surface Plasmon Resonance Biosensor for Exosome Detection Based on Reformative Tyramine Signal Amplification Activated by Molecular Aptamer Beacon. J. Nanobiotechnol. 2021, 19, 450. [Google Scholar] [CrossRef]
- Gao, J.; Li, A.; Hu, J.; Feng, L.; Liu, L.; Shen, Z. Recent Developments in Isolating Methods for Exosomes. Front. Bioeng. Biotechnol. 2023, 10, 1100892. [Google Scholar] [CrossRef]
- Yadav, A.; Sharma, A.; Moulick, M.; Gavande, P.V.; Nandy, A.; Xuan, Y.; Sen, C.K.; Ghatak, S. Labeling, Isolation and Characterization of Cell-Type-Specific Exosomes Derived from Mouse Skin Tissue. Nat. Protoc. 2025; online ahead of print. [Google Scholar] [CrossRef]
- Whittaker, T.E.; Nagelkerke, A.; Nele, V.; Kauscher, U.; Stevens, M.M. Experimental Artefacts Can Lead to Misattribution of Bioactivity from Soluble Mesenchymal Stem Cell Paracrine Factors to Extracellular Vesicles. J. Extracell. Vesicles 2020, 9, 1807674. [Google Scholar] [CrossRef]
- Raju, D.; Bathini, S.; Badilescu, S.; Ghosh, A.; Packirisamy, M. Microfluidic Platforms for the Isolation and Detection of Exosomes: A Brief Review. Micromachines 2022, 13, 730. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; Wu, Z.; Wang, Y.; Li, H. Regulating the Production and Biological Function of Small Extracellular Vesicles: Current Strategies, Applications and Prospects. J. Nanobiotechnol. 2021, 19, 422. [Google Scholar] [CrossRef] [PubMed]
- Erwin, N.; Serafim, M.F.; He, M. Enhancing the Cellular Production of Extracellular Vesicles for Developing Therapeutic Applications. Pharm. Res. 2023, 40, 833–853. [Google Scholar] [CrossRef]
- Kong, F.; Upadya, M.; Wong, A.S.W.; Dalan, R.; Dao, M. Isolating Small Extracellular Vesicles from Small Volumes of Blood Plasma Using Size Exclusion Chromatography and Density Gradient Ultracentrifugation: A Comparative Study. bioRxiv 2023. [Google Scholar] [CrossRef]
- Johnsen, K.B.; Gudbergsson, J.M.; Andresen, T.L.; Simonsen, J.B. What Is the Blood Concentration of Extracellular Vesicles? Implications for the Use of Extracellular Vesicles as Blood-Borne Biomarkers of Cancer. Biochim. Biophys. Acta (BBA)—Rev. Cancer 2019, 1871, 109–116. [Google Scholar] [CrossRef]
- Dragovic, R.A.; Gardiner, C.; Brooks, A.S.; Tannetta, D.S.; Ferguson, D.J.P.; Hole, P.; Carr, B.; Redman, C.W.G.; Harris, A.L.; Dobson, P.J.; et al. Sizing and Phenotyping of Cellular Vesicles Using Nanoparticle Tracking Analysis. Nanomedicine 2011, 7, 780–788. [Google Scholar] [CrossRef]
- Mørk, M.; Pedersen, S.; Botha, J.; Lund, S.M.; Kristensen, S.R. Preanalytical, Analytical, and Biological Variation of Blood Plasma Submicron Particle Levels Measured with Nanoparticle Tracking Analysis and Tunable Resistive Pulse Sensing. Scand. J. Clin. Lab. Invest. 2016, 76, 349–360. [Google Scholar] [CrossRef]
- Soares Martins, T.; Catita, J.; Martins Rosa, I.; da Cruz e Silva, O.A.B.; Henriques, A.G. Exosome Isolation from Distinct Biofluids Using Precipitation and Column-Based Approaches. PLoS ONE 2018, 13, e0198820. [Google Scholar] [CrossRef]
- Connolly, K.D.; Wadey, R.M.; Mathew, D.; Johnson, E.; Rees, D.A.; James, P.E. Evidence for Adipocyte-Derived Extracellular Vesicles in the Human Circulation. Endocrinology 2018, 159, 3259–3267. [Google Scholar] [CrossRef]
- Zhang, X.; Takeuchi, T.; Takeda, A.; Mochizuki, H.; Nagai, Y. Comparison of Serum and Plasma as a Source of Blood Extracellular Vesicles: Increased Levels of Platelet-Derived Particles in Serum Extracellular Vesicle Fractions Alter Content Profiles from Plasma Extracellular Vesicle Fractions. PLoS ONE 2022, 17, e0270634. [Google Scholar] [CrossRef] [PubMed]
- Jamaly, S.; Ramberg, C.; Olsen, R.; Latysheva, N.; Webster, P.; Sovershaev, T.; Brækkan, S.K.; Hansen, J.-B. Impact of Preanalytical Conditions on Plasma Concentration and Size Distribution of Extracellular Vesicles Using Nanoparticle Tracking Analysis. Sci. Rep. 2018, 8, 17216. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Bruce, T.F.; Huang, S.; Marcus, R.K. Isolation and Quantitation of Exosomes Isolated from Human Plasma via Hydrophobic Interaction Chromatography Using a Polyester, Capillary-Channeled Polymer Fiber Phase. Anal. Chim. Acta 2019, 1082, 186–193. [Google Scholar] [CrossRef]
- Božič, D.; Sitar, S.; Junkar, I.; Štukelj, R.; Pajnič, M.; Žagar, E.; Kralj-Iglič, V.; Kogej, K. Viscosity of Plasma as a Key Factor in Assessment of Extracellular Vesicles by Light Scattering. Cells 2019, 8, 1046. [Google Scholar] [CrossRef]
- Mohammad, S.; Hutchinson, K.A.; da Silva, D.F.; Bhattacharjee, J.; McInnis, K.; Burger, D.; Adamo, K.B. Circulating Small Extracellular Vesicles Increase after an Acute Bout of Moderate-Intensity Exercise in Pregnant Compared to Non-Pregnant Women. Sci. Rep. 2021, 11, 12615. [Google Scholar] [CrossRef]
- Gelibter, S.; Marostica, G.; Mandelli, A.; Siciliani, S.; Podini, P.; Finardi, A.; Furlan, R. The Impact of Storage on Extracellular Vesicles: A Systematic Study. J. Extracell. Vesicles 2022, 11, e12162. [Google Scholar] [CrossRef]
- Woud, W.W.; Hesselink, D.A.; Hoogduijn, M.J.; Baan, C.C.; Boer, K. Direct Detection of Circulating Donor-Derived Extracellular Vesicles in Kidney Transplant Recipients. Sci. Rep. 2022, 12, 21973. [Google Scholar] [CrossRef] [PubMed]
- Weber, B.; Henrich, D.; Schindler, C.R.; Marzi, I.; Leppik, L. Release of Exosomes in Polytraumatized Patients: The Injury Pattern Is Reflected by the Surface Epitopes. Front. Immunol. 2023, 14, 1107150. [Google Scholar] [CrossRef] [PubMed]
- Lichá, K.; Pastorek, M.; Repiská, G.; Celec, P.; Konečná, B. Investigation of the Presence of DNA in Human Blood Plasma Small Extracellular Vesicles. Int. J. Mol. Sci. 2023, 24, 5915. [Google Scholar] [CrossRef]
- Yim, K.H.W.; Krzyzaniak, O.; Al Hrout, A.; Peacock, B.; Chahwan, R. Assessing Extracellular Vesicles in Human Biofluids Using Flow-Based Analyzers. Adv. Healthc. Mater. 2023, 12, 2301706. [Google Scholar] [CrossRef]
- Marić, I.; Žiberna, K.; Kolenc, A.; Maličev, E. Platelet Activation and Blood Extracellular Vesicles: The Influence of Venepuncture and Short Blood Storage. Blood Cells Mol. Dis. 2024, 106, 102842. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Cheng, L.M. Application of EXODUS System Combined with Allosteric DNA Nanoswitches in the Detection of MiR-107 among Plasma Exosomes of Parkinson’s Disease Patients. Zhonghua Yu Fang Yi Xue Za Zhi 2024, 58, 1191–1196. [Google Scholar] [CrossRef]
- Małys, M.S.; Aigner, C.; Schulz, S.M.; Schachner, H.; Rees, A.J.; Kain, R. Isolation of Small Extracellular Vesicles from Human Sera. Int. J. Mol. Sci. 2021, 22, 4653. [Google Scholar] [CrossRef]
- Helwa, I.; Cai, J.; Drewry, M.D.; Zimmerman, A.; Dinkins, M.B.; Khaled, M.L.; Seremwe, M.; Dismuke, W.M.; Bieberich, E.; Stamer, W.D.; et al. A Comparative Study of Serum Exosome Isolation Using Differential Ultracentrifugation and Three Commercial Reagents. PLoS ONE 2017, 12, e0170628. [Google Scholar] [CrossRef] [PubMed]
- Rui, S.; Yuan, Y.; Du, C.; Song, P.; Chen, Y.; Wang, H.; Fan, Y.; Armstrong, D.G.; Deng, W.; Li, L. Comparison and Investigation of Exosomes Derived from Platelet-Rich Plasma Activated by Different Agonists. Cell Transplant. 2021, 30, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Cordero, L.; Domingo, J.C.; Sánchez-Vizcaíno Mengual, E.; Pinto, H. Autologous Platelet-Rich Plasma Exosome Quantification after Two Thermo-Photobiomodulation Protocols with Different Fluences. J. Photochem. Photobiol. 2025, 29, 100267. [Google Scholar] [CrossRef]
- Lee, K.W.A.; Chan, L.K.W.; Hung, L.C.; Phoebe, L.K.W.; Park, Y.; Yi, K.-H. Clinical Applications of Exosomes: A Critical Review. Int. J. Mol. Sci. 2024, 25, 7794. [Google Scholar] [CrossRef]
- Nguyen, V.V.T.; Witwer, K.W.; Verhaar, M.C.; Strunk, D.; van Balkom, B.W.M. Functional Assays to Assess the Therapeutic Potential of Extracellular Vesicles. J. Extracell. Vesicles 2020, 10, e12033. [Google Scholar] [CrossRef]
- Li, B.; Wu, W.; Xu, W.; Qian, H.; Ji, C. Advances of Extracellular Vesicles Isolation and Detection Frontier Technology: From Heterogeneity Analysis to Clinical Application. J. Nanobiotechnol. 2025, 23, 678. [Google Scholar] [CrossRef]
- Willis, G.R.; Kourembanas, S.; Mitsialis, S.A. Toward Exosome-Based Therapeutics: Isolation, Heterogeneity, and Fit-for-Purpose Potency. Front. Cardiovasc. Med. 2017, 4, 63. [Google Scholar] [CrossRef] [PubMed]
- Vergauwen, G.; Tulkens, J.; Pinheiro, C.; Avila Cobos, F.; Dedeyne, S.; De Scheerder, M.; Vandekerckhove, L.; Impens, F.; Miinalainen, I.; Braems, G.; et al. Robust Sequential Biophysical Fractionation of Blood Plasma to Study Variations in the Biomolecular Landscape of Systemically Circulating Extracellular Vesicles across Clinical Conditions. J. Extracell. Vesicles 2021, 10, e12122. [Google Scholar] [CrossRef]
- Shu, S.L.; Allen, C.L.; Benjamin-Davalos, S.; Koroleva, M.; MacFarland, D.; Minderman, H.; Ernstoff, M.S. A Rapid Exosome Isolation Using Ultrafiltration and Size Exclusion Chromatography (REIUS) Method for Exosome Isolation from Melanoma Cell Lines. In Melanoma: Methods and Protocols; Springer: New York, NY, USA, 2021; pp. 289–304. [Google Scholar]
- Guo, J.; Wu, C.; Lin, X.; Zhou, J.; Zhang, J.; Zheng, W.; Wang, T.; Cui, Y. Establishment of a Simplified Dichotomic Size-exclusion Chromatography for Isolating Extracellular Vesicles toward Clinical Applications. J. Extracell. Vesicles 2021, 10, e12145. [Google Scholar] [CrossRef]
- Zhu, J.; Zhang, J.; Ji, X.; Tan, Z.; Lubman, D.M. Column-Based Technology for CD9-HPLC Immunoaffinity Isolation of Serum Extracellular Vesicles. J. Proteome Res. 2021, 20, 4901–4911. [Google Scholar] [CrossRef]
- Yu, Z.; Liu, X.; Wu, M.; Shi, S.; Fu, Q.; Jia, J.; Chen, G. Untouched Isolation Enables Targeted Functional Analysis of Tumour-cell-derived Extracellular Vesicles from Tumour Tissues. J. Extracell. Vesicles 2022, 11, e12214. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Li, D.; Wang, M.; Xu, Z.; Chen, X.; Liu, Q.; Sun, W.; Li, J.; Gong, Y.; Liu, D.; et al. Exposure to Blue Light Stimulates the Proangiogenic Capability of Exosomes Derived from Human Umbilical Cord Mesenchymal Stem Cells. Stem Cell Res. Ther. 2019, 10, 358. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Su, J.; Ma, K.; Li, H.; Fu, X.; Zhang, C. Photobiomodulation Promotes Hair Regeneration in Injured Skin by Enhancing Migration and Exosome Secretion of Dermal Papilla Cells. Wound Repair Regen. 2022, 30, 245–257. [Google Scholar] [CrossRef] [PubMed]
- Bagheri, H.S.; Mousavi, M.; Rezabakhsh, A.; Rezaie, J.; Rasta, S.H.; Nourazarian, A.; Avci, Ç.B.; Tajalli, H.; Talebi, M.; Oryan, A.; et al. Low-Level Laser Irradiation at a High Power Intensity Increased Human Endothelial Cell Exosome Secretion via Wnt Signaling. Lasers Med. Sci. 2018, 33, 1131–1145. [Google Scholar] [CrossRef]
- Chang, C.-Y.; Aviña, A.E.; Chang, C.-J.; Lu, L.-S.; Chong, Y.-Y.; Ho, T.Y.; Yang, T.-S. Exploring the Biphasic Dose-Response Effects of Photobiomodulation on the Viability, Migration, and Extracellular Vesicle Secretion of Human Adipose Mesenchymal Stem Cells. J. Photochem. Photobiol. B 2024, 256, 112940. [Google Scholar] [CrossRef]
| Isolation Method | Disadvantages |
|---|---|
| High speed can damage exosomes [42]. UC can cause mechanical damage, making it difficult to maintain the bioactivity and morphological integrity of exosomes [50]. |
| Exosomes with moderate purity. Exosomes can be lost due to membrane damage [32], which can impair their ability to bind to and communicate with target cells [51]. |
| Co-precipitates with non-exosomal contaminants, including proteins and polymeric materials [43]. Common contaminants include residual proteins, lipids, and polymers from isolation methods, which can retain biological activity, leading to inaccurate conclusions about exosome-mediated processes [52]. |
| IAC can impair exosomes’ functional capacity [44]. |
| A less sensitive method with less pure isolated exosomes due to the complexity of biological samples, the size overlap between exosomes and other EVs, and the heterogeneity of exosomes [11,53]. |
| Quantification Method | Disadvantages |
| The method also measures non-exosomal contaminants. NTA cannot distinguish EVs from other particles, such as lipoproteins [45]. |
| Smaller vesicles are counted as single particles when the concentration of smaller vesicles is high in the sample and the scattering or fluorescence signal exceeds the detection limit [46]. |
| Insensitivity to smaller exosomes and smaller vesicles, which are counted as single particles [47]. |
| This technique cannot analyze heterogeneous exosome populations [37]. |
| It has difficulty discriminating between specific and non-specific interactions and mass-sensitive and sensor area limitations [49]. |
| It has a detection limit of 3.94 × 109 for CD81 and 5.07 × 109 particles/mL for CD63 [48]. |
| Concept | Search Terms Used |
|---|---|
| #1 Exosomes/EVs | “extracellular vesicles” OR “exosomes” OR related terms |
| #2 Sample type | “plasma” OR “serum” OR “platelet-rich plasma” |
| #3 Quantification | “concentration” OR “quantification” |
| #4 Particles | “particles” OR “particles/mL” |
| Combined search | #1 AND #2 AND #3 AND #4 |
| Filters applied | “human”/excluded: “cancer” and “metastasis” |
| Author Year | Sample Type | N | Isolation Method | Quantification Method | Mean Concentration (Particles/mL) |
|---|---|---|---|---|---|
| Božič [65] 2019 | Plasma | 3 | – | FCM | 3.50 × 106 |
| Woud [68] 2022 | Plasma (donor group) | 36 | NA | IFCM | 1.26 × 108 |
| Yim [71] 2023 | Plasma | 6 | – | nFCM | 1.50 × 1010 |
| Marić [72] 2024 | Plasma | 20 | UC | NTA | 3.95 × 1010 |
| Wang [73] 2024 | Plasma (control group) | – | EXODUS | NTA | 4.82 × 1010 |
| Kong [56] 2023 | Plasma | 5 | No isolation | NTA | 1.78 × 1011 |
| 5 | SEC-PF | NTA | 9.02 × 1010 * | ||
| 5 | DGUC-SEC | NTA | 1.51 × 109 * | ||
| Weber [69] 2023 | Plasma (healthy donors) | 10 | SEC | NTA | 1.15 × 109 |
| Lichá [70] 2023 | Plasma without Dnase | 4 | IDGUC | NTA | 8.80 × 108 |
| Zhang [62] 2022 | Plasma | 5 | UC | NTA | 2.38 × 1010 |
| Mohammad [66] 2021 | Plasma (controls) | 9 | Differential DUC | NTA | 8.11 × 1010 |
| Bendix [57] 2019 | Plasma | – | All isolation methods combined | NTA | ~2.00 × 1010 |
| – | All UC techniques | NTA | ~5.00 × 1010 | ||
| Wang [64] 2019 | Plasma | – | HIC | NTA | 1.15 × 109 |
| Soares [60] 2018 | Plasma | – | TEI | NTA | 8.30 × 108 |
| ExoQ | NTA | 7.80 × 108 | |||
| ExoS | NTA | 9.90 × 108 | |||
| Connolly [61] 2018 | PFP | 7 | UC | NTA | 1.01 × 1011 |
| 7 | MB | NTA | 3.10 × 1011 | ||
| 7 | SPD | NTA | 2.50 × 1010 | ||
| Jamaly [63] 2018 | PPP | 10 | HSC | NTA | 1.80 × 1010 |
| Mørk [59] 2016 | PFP | 20 | SEC | NTA | 4.50 × 108 * |
| PFP (ERI) | 20 | SEC | NTA | 6.70 × 1011 | |
| Dragovic [58] 2011 | PPP | – | UC | NTA | 1.20 × 1010 |
| Gelibter [67] 2022 | Fresh plasma | 3 | SEC | TRPS | 4.36 × 1010 |
| Mørk [59] 2016 | PFP | 20 | SEC | TRPS | 9.05 × 108 |
| PFP (ERI) | 20 | SEC | TRPS | 1.70 × 109 |
| Author Year | Sample Type | N | Isolation Method | Quantification Method | Mean Concentration (Particles/mL) |
|---|---|---|---|---|---|
| Zhang [62] 2022 | Serum | 6 | UC | NTA | 4.23 × 1010 |
| Malys [74] 2021 | Serum | 3 | Exo-spinTM | NTA | 4.98 × 1010 |
| 3 | DUC | NTA | 9.90 × 1010 | ||
| Soares [60] 2018 | Serum | – | TEI | NTA | 5.30 × 108 |
| ExoQ | NTA | 5.40 × 108 | |||
| ExoS | NTA | 6.90 × 108 | |||
| Helwa [75] 2017 | Serum (ID) | 6 | miRCURY | NTA | 1.62 × 1011 * |
| 6 | TEIR | NTA | 2.13 × 1011 * | ||
| 6 | UC | NTA | 8.45 × 108 * | ||
| 6 | ExoQuick | NTA | 1.79 × 1011 * |
| Author Year | Sample Type | N | Isolation Method | Quantification Method | Mean Concentration (Particles/mL) |
|---|---|---|---|---|---|
| Cordero [77] 2025 | Preconditioned autologous PRP (blue light 467 nm, 1.0 J/cm2, 37 °C) | 3 | UC | NTA | 2.99 × 1011 * |
| Preconditioned autologous PRP (blue light 467 nm, 2.0 J/cm2, 37 °C) | 3 | UC | NTA | 2.53 × 1011 * | |
| Rui [76] 2021 | PRP with saline solution | 3 | UC | Nanoflow analysis | 7.52 × 109 * |
| PRP activated with calcium gluconate | 3 | UC | Nanoflow analysis | 5.85 × 1010 * | |
| PRP activated with thrombin | 3 | UC | Nanoflow analysis | 4.87 × 1010 * | |
| PRP activated with thrombin and calcium gluconate | 3 | UC | Nanoflow analysis | 7.16 × 1010 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sánchez-Vizcaíno Mengual, E.; Cordero, L.; Pinto, H. Human Blood Exosomes: Isolation and Characterization Methods, Variability, and the Need for Standardized Protocols—A Review. Biomedicines 2025, 13, 2970. https://doi.org/10.3390/biomedicines13122970
Sánchez-Vizcaíno Mengual E, Cordero L, Pinto H. Human Blood Exosomes: Isolation and Characterization Methods, Variability, and the Need for Standardized Protocols—A Review. Biomedicines. 2025; 13(12):2970. https://doi.org/10.3390/biomedicines13122970
Chicago/Turabian StyleSánchez-Vizcaíno Mengual, Elena, Laura Cordero, and Hernán Pinto. 2025. "Human Blood Exosomes: Isolation and Characterization Methods, Variability, and the Need for Standardized Protocols—A Review" Biomedicines 13, no. 12: 2970. https://doi.org/10.3390/biomedicines13122970
APA StyleSánchez-Vizcaíno Mengual, E., Cordero, L., & Pinto, H. (2025). Human Blood Exosomes: Isolation and Characterization Methods, Variability, and the Need for Standardized Protocols—A Review. Biomedicines, 13(12), 2970. https://doi.org/10.3390/biomedicines13122970







