Abstract
In this study, we examine a means for developing near-IR fluorescent sensors through streamlined, site-specific coupling with peptide-based receptors. As the penultimate step of solid-phase synthesis of a peptide-based receptor, we show a simple means of labeling the N’ terminus with the near IR fluorophore IR-783 to afford a viable fluorescent sensor after cleavage from the resin. The proof-of-concept probe utilized a biotin mimetic peptide sequence as the receptive moiety. Here we revealed a “turn-on” fluorescence enhancement upon binding of the biotin mimetic probe to its intended streptavidin target. Not all peptide-receptive moieties tested were able to generate such an enhancement upon target binding, and as such, the rationale for the observed fluorescence response properties is discussed.
1. Introduction
A common problem in probe/sensor research is often proper labeling of the recognition element, requiring proper determination of where the receptors are labeled and, in some cases, how many times the receptor has been labeled. To turn a receptor into a standalone probe or sensor, proper labeling with a reporter in a well-defined manner is required. Such reporters may be responsive to the binding event, wherein molecular recognition by the receptor invokes some form of signal transduction via the reporter (i.e., fluorescence). In looking to identify a means for generating a fluorescence-based turn-on probe directly in line with solid-phase synthesis techniques, we explored the use of the near-IR fluorophore IR-783 as a terminal coupling moiety to be incorporated prior to cleavage during solid-phase synthesis of a peptide aptamer. The use of near-infrared (NIR) fluorophores as in vivo probes has increased significantly over recent years [1,2,3,4]. In the 700–900 nm NIR region, light penetrates deep below the surface of tissue [5,6]. When compared to the other light regions, NIR shows better applicability for in vivo imaging because of characteristics such as: deep tissue penetration, high sensitivity, little to no photodamage to samples, and relatively low autofluorescence interference from biological systems [1,2,7,8,9]. Heptamethine cyanine NIR dyes [5,10] such as IR-783 have often been used as reporters for probe development because of their large Stoke’s shifts, high extinction coefficients, and ability to be spotted deep within tissues [1,2,10,11,12]. Due to the ease of modification via the central chloro group, heptamethine cyanine dyes have long been incorporated with synthetic receptors for sensor development, such as their functionalization with crown ethers to generate detectors for metal ions [13,14]. Because IR-783 is non-toxic and water soluble, it is applicable for patient use without causing deleterious effects [3,11,15]. When compared to other NIR imaging materials, IR-783 has shown high sensitivity for imaging with good stability [6,7]. Past applications of IR-783 have seen its use in conjunction with nanobubbles for imaging breast cancer, where it was used for dual-mode imaging, and increased the rate and binding ability to tumors, specifically [16]. In another study, the uptake and retention of IR-783 in tumor tissues and cells were tested, with results showing that the dye was retained predominantly in cancer cells [4]. Specific targeting of IR-783 by incorporating a peptide-based receptor (aptamer) could impart different selectivity and may permit other diseased tissues to be preferentially observed depending on the nature of the receptive moiety. Synthesized peptides are commonly explored for molecular biosensors because of their high affinity to proteins, structure variety, high stability, and their small size is more amenable to deep tissue penetration than larger protein-based receptors like immunoglobulin, which are largely restricted to the vasculature [17,18,19,20,21]. Synthesis of peptide-based receptors in high purity and yield is carried out following standard solid-phase peptide synthesis (SPPS) techniques [20,22]. In this work, we describe a methodology for direct terminal covalent attachment of the NIR dye IR-783 which allows for peptide-based receptors to be labeled with a single fluorophore reporter through a simple coupling reaction step during solid-phase synthesis prior to cleavage from the resin.
As opposed to large fluorescent proteins, direct signaling cannot be achieved by peptides alone, which is why it is important to conjugate the peptides with signaling moieties [20]. In comparison to genetic coupling of fluorescent reporter proteins to receptors for expression of fusion-based probes [23], the use of synthetic bioconjugation or solid-phase synthesis strategies for coupling receptors to small molecule reporters can be carried out in higher yield and purity. Several strategies for cross-linking fluorophores to thiols include the use of alkylating agents such as maleimides, which have preferential reactivity to thiol over other nucleophiles. Haloacetamides, epoxides, and benzylic halides also offer reactivity to thiols, but may react with other components such as amine-bearing groups, among other residues [24]. Effective fluorophore labeling of peptide have previously used thiol reactive succinimidyl ester derivatives of near-infrared cyanine dyes for attachment to cysteine residues on peptide-based receptors [25]. A new strategy of using the 7-oxanorbornadiene framework for coupling dansyl to thiol groups present on peptides has also been used to generate fluorogenic probes [26]. Amino groups on residues or the primary N-terminal amine itself can be targeted by the use of ethylenediamine carbodiimide (EDC) coupling and has been widely used to modify peptides with a variety of functional reporters such as nanoparticles [27] and macrocyclic contrast agents [28]. Amide bond formation between an amine and a carboxyl group can be catalyzed by carbodiimide formation of an activate ester which is often further reacted with N-hydroxysuccinimide (NHS) for conversion to an amine reactive ester. Peptides have long been conjugated to fluorophores using such NHS-ester coupling chemistries or via heterobifunctional cross-linkers. Some of these fluorophores utilized include boron dipyromethene difluoride (BODIPY) and carboxy-seminaphtho-fluorescein (SNAFL), which have been coupled to peptides to observe trafficking within cells. In such works, peptide motifs, such as CKGGAKL, KKKRK, KDEL, and SDYQRL, have been conjugated to these fluorophores in order to observe targeting to peroxisomes, nucleus, the endoplasmic reticulum, and the trans-Golgi network, respectively [29,30]. Aromatic isothiocyanate groups are also commonly used (i.e., FITC) for labeling of target peptides at reactive nucleophilic sites such as amines, thiols, or hydroxyl groups, and, to some extent, DTPA dianhydrides and aldehyde/ketone-based conjugation schemes may also be employed [31]. Because these methods have the drawback of non-specific labeling of the peptide at locations that may inherently alter their binding capabilities, more recently developments have made use of bioorthogonal click chemistry to provide a means for site-specifically labeling peptide receptors. One such demonstration for a CD13 targeting probe utilized a dimeric NGR peptide synthesized to contain an alkyne that allows specific coupling to an azide-terminated Cy5.5 fluorophore (Cy5.5-N3) [32].
In contrast to the above approaches, a solid-phase synthesis technique could provide a simple, low-cost, and streamlined means for linking a single reporter moiety to a peptide receptor of interest by direct conjugation of the reporter to the peptide N-terminus during synthesis of the receptor sequence. We have found only one scenario in the literature in which such a strategy has been implemented utilizing dansylated peptide, wherein dansyl chloride was used to cap the end of a tetrapeptide upon completion of solid-phase synthesis [33]. The resulting dansylated peptide served as a probe component for a combinatorial peptide screening process, in which receptors for the tetrapeptide were screened and afforded fluorescence if capable of binding the dansyl probe. In this paper, we demonstrate a related but distinct approach in which we carry out covalent attachment of IR-783 to peptide-based receptors at the penultimate step of peptide synthesis. This proof-of-concept is demonstrated with a characteristic HPQ-bearing peptide (VSHPQAPF) capable of binding to streptavidin [34,35,36]. This biotin-mimicking peptide motif is known to bind to streptavidin at the same location as native biotin but with lower affinity [34,35]. The biotin-streptavidin bond is one of the strongest non-covalent interactions in nature, and as such competition between the peptide mimic and native biotin for the same recognition site on streptavidin will favor the native biotin [34,36]. In our work, we examined the formation of a fluorescent probe using the biotin mimetic peptide to directly interact with the streptavidin target which we found to create a “turn-on” fluorescence response upon binding. The coupling strategy of using free amine present during solid-phase synthesis for covalent attachment amine substitution with the reactive chloro moiety of IR-783 allows a means for capping the end of a synthesized peptides with the fluorescent IR-783 reporter. This has proved useful in affording a fluorescently labeled peptide which exhibits “turn-on” fluorescence enhancement when bound by its target for the case of the biotin mimetic peptide but was found to not be the case in general as other peptide receptors did not produce a “turn-on” response with binding. The binding to streptavidin and controlled release by competitive binding with native biotin allowed this fluorescence signal to be attenuated back to its original state (Figure 1). Here we offer a new strategy to convert an identified peptide-based aptamer sequence into a useful sensing moiety by a straightforward IR-783 labeling technique. Utilizing fluorescence techniques along with microscale thermophoresis [37] we provide an assessment of the response and binding ability of the synthesized probe.
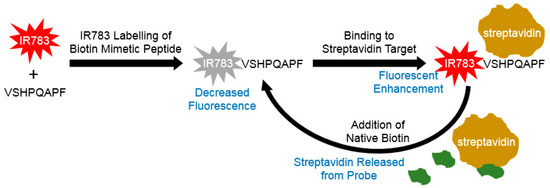
Figure 1.
Schematic of IR-783 labeled peptide sequence VSHPQAPF serving as biotin mimic for recognition by streptavidin. Binding results in induction of turn-on fluorescence enhancement that is reversible upon addition of native biotin to displace the probe returning it to weak fluorescence.
2. Materials and Methods
For this proof of concept, a biotin-mimetic peptide aptamer capable of binding to streptavidin was utilized as the receptor moiety, having the sequence (Val-Ser-His-Pro-Gln-Ala-Pro-Phe). Standard solid-phase synthesis was carried out to produce the peptide. In brief, 1 mmole scale synthesis (using 1 g of rink amide resin) was conducted in a filtered polypropylene reaction vessel. Before synthesis, the resin was swelled in methylene chloride, the solvent was filtered, and then any initial Fmoc group was deprotected by shaking in a mixture of 4 mL of piperidine and 16 mL of NMP for 30 min followed by washing with NMP, methanol, and methylene chloride. The first amino acid coupling was carried out by introducing to 1 g of resin a solution of pre-mixed 4 mmole of Fmoc protected amino acid phenylalanine, 4mmole 1-hydroxybenzotriazole, and 4mmole diisopropylcarbodiimide in 20 mL of N-methylpyrrolidone (NMP). After 3 h of coupling under shaking conditions at room temperature, the resin was washed with NMP, methanol, and methylene chloride. A sample of the resin was examined using a common ninhydrin-based Kaiser test to determine the presence of any unreacted primary amines which would be indicative of incomplete completing thereby necessitating further coupling with the same amino acid mixture. After confirmation of the completed coupling, the Fmoc group was deprotected by shaking in a mixture of 4 mL of piperidine and 16 mL of NMP for 30 min followed by washing of the resin as mentioned above. This process was repeated for each amino acid in the sequence C’term-F-P-A-Q-P-H-S-V-N’term from the C terminus to the N terminus using 4 mmole of each of the following protected amino acids respectively (Fmoc-Phe-OH, Fmoc-Pro-OH, Fmoc-Ala-OH, Fmoc-Gln-(Trt)-OH, Fmoc-Pro-OH, Fmoc-His-(Trt)-OH, Fmoc-Ser-(tBu)-OH, Fmoc-Val-OH). For each of these step, the same 4 mmole of Fmoc protected amino acid, 4 mmole 1-hydroxybenzotriazole, and 4 mmole diisopropylcarbodiimide in 20 mL of N-methylpyrrolidone (NMP) were used for coupling for 3 h or longer until the Kaiser test confirmed complete coupling. After the final protected valine was finally coupled to the resin, the Fmoc group was deprotected with 20% piperidine in 20 mL of NMP for 30 min (Figure S4). To covalently link the NIR dye to the exposed terminal primary amino group of valine at the end of the peptide immobilized on the resin, the resin was first washed with methylene chloride and dried under flowing air for 30 min followed by vacuum desiccator for 2 h. The dried resin was then added to a mixture of 1 mmole of IR-783 and 2 mmole of DIPEA in 20 mL of dimethylformamide followed by shaking for at least 48 h at room temperature. After this coupling of IR-783, the resin was washed to remove unreacted IR-783. The labeled peptide was finally cleaved from the resin by addition of 20 mL of 88% trifluoroacetic acid (TFA), 5% phenol, 5% water, and 2% triisopropylsilane with shaking for 2 h followed by collection of the filtrate into a round bottom flask. Alternating washes of methylene chloride and TFA were added to the resin and filtered into the round bottom flask. Rotary evaporation at 80 °C was used to remove TFA and methylene chloride. The remaining liquid was added dropwise to 40 mL of cold diethyl ether. The dark blue precipitate was centrifuged and re-suspended in fresh diethyl ether (40 mL). This washing step was performed four times to remove trace impurities. The dark blue sample was finally collected in water, frozen, and lyophilized. Lyophilized samples were stored at −20 °C until use. The same procedure was carried out for the IR783 labeled CD47 peptide mimic having the sequence Glu-Val-Thr-Glu-Leu-Thr-Arg-Glu-Gly Glu.
UV visible spectra were recorded on a Biotek Epoch 2 Microplate Spectrophotometer and fluorescence spectra were recorded on a TECAN Infinite 200 PRO. Lyophilized streptavidin stock (New England Biolabs) was diluted to 5 mg/mL in phosphate buffered saline (PBS), and the lyophilized IR-783 coupled VSHPQAPF peptide was diluted to 19 mg/mL in PBS. For microscale thermophoresis, a Nanotemper Monolith NT.115 blue/red microscale thermophoresis system was employed with the RED dectector using excitation wavelengths of 600–650 nm. Using microscale thermophoresis, the initial confirmation of binding of the IR-783 VSHPQAPF to streptavidin was carried out using standard capillaries with 45 μM of IR-783 VSHPQAPF and 45 μM streptavidin concentration as compared to a sample of 45 μM of IR-783 VSHPQAPF without streptavidin. Further assays using microscale thermophoresis with premium capillaries used a constant amount of 120 μM of IR-783 VSHPQAPF and varying the concentration of streptavidin from 22.5 μM to 0.69 nM to determine the binding ability of the probe. The initial red fluorescence was also recorded using the Nanotemper monolith. To determine if the binding of streptavidin to the biotin mimicking peptide probe was reversible in the presence of biotin, a series of binding tests examined by microscale thermophoresis using premium capillaries was implemented over several dilutions of biotin (New England Biolabs). Specifically, 40 μM streptavidin was mixed with 40 μM of peptide probe (IR-783 VSHPQAPF) along with a varying concentrations of biotin (1 mM–0.32 nM) after 10 min the samples were loaded into capillaries and microscale thermophoresis was performed for 3 sets of experiments. After finding an appropriate biotin concentration for inhibiting the biotin-mimetic peptide probe interaction with streptavidin, a series of samples were examined for their red fluorescence, as well as their thermophoretic signal using the Nanotemper system with either 40 μM of IR-783 VSHPQAPF, 40 μM of IR-783 VSHPQAPF with 40 μM streptavidin, or 40 μM IR-783 VSHPQAPF with 40 μM streptavidin and 200 μM biotin. Fluorescence spectra of the 40 μM of IR-783 VSHPQAPF with 40 μM streptavidin sample, as well as the 40 μM IR-783 VSHPQAPF with 40 μM streptavidin and 200 μM biotin were more closely examined using a TECAN Infinite 200 PRO fluorescence microplate reader for conducting emission scans operating at a 618 nm excitation wavelength. Similarly, the Nanotemper system was used to examine 150 μM of IR783 labeled CD47 peptide mimic in terms of its fluorescence and MST signal in the presence and absence of 50 μM human SIRPalpha.
3. Results
3.1. Spectra of IR-783 Conjugated VSHPQAPF
After conjugation of the IR-783 to the VSHPQAPF peptide, we observed a blue shift in the fluorescence spectra with an emission maxima of ~770 nm after coupling, as compared to ~800 nm for the standalone IR-783 prior to coupling (Figure 2). Such a blue shift is a common observation for heptamethine cyanine dyes functionalized at their chlorocyclohexenyl ring, and other reports of modification of the chloro moiety in IR783 specifically indicated such a blue shift in the spectrum [38,39]. A blue shift in the absorption spectra could also be observed after covalent coupling (Figure S5).

Figure 2.
Fluorescence spectra of IR783 (solid line) and IR783 covalently linked to VSHPQAPF (dashed line) showing blue shift in the emission peak.
3.2. Properties of IR-783 Conjugated VSHPQAPF
3.2.1. Target Binding Ability
The previously discovered VSHPQAPF sequence [35] used in this proof of concept study was selected for its ability to mimic biotin in regard to its binding of streptavidin as a well-known target/receptor pair. To confirm that covalent coupling of IR-783 to the N terminus of the VSHPQAPF did not cause inactivation of the peptides ability to bind to streptavidin, we performed a quantitative binding assay using microscale thermophoresis. We observed a concentration-dependent increase in the MST signal for mixtures of IR-783 VSHPQAPF with increasing concentrations of streptavidin when more than 1 μM of streptavidin was added (Figure 3). Below approximately 1 μM of streptavidin, there was no significant change in the MST trace, indicating that no binding could be observed at streptavidin concentrations below this level. Additional experiments revealed the affinity of the IR783-VSHPQAPF binding to streptavidin to be 20 ± 5 μM (Figure S6), which is in good agreement with the literature values for the VSHPQAPF peptide binding to streptavidin previously reported from 4.1 μM to 79.5 μM [35].
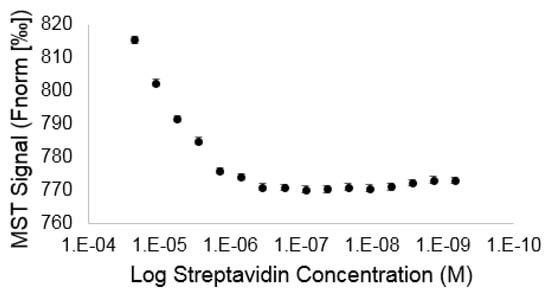
Figure 3.
Microscale thermophoresis signals resulting from binding induced changes in hydration shell corresponding to distinct thermophoretic signals for the probe (IR783-VSHPQAPF) with increasing concentrations of streptavidin target.
3.2.2. Fluorescence Enhancement
We observed an interesting “turn on” enhancement in fluorescence for the IR-783 VSHPQAPF probe upon binding to streptavidin, which occurred at concentrations greater than approximately 1 μM, which is near the same cut-off point for detectable binding as quantified by MST. Analysis of the fluorescence enhancement revealed a linear response with respect to target streptavidin concentration (Figure 4 and Figure S3). We suspect that turn-on enhancement upon binding is based on blocking of a weak photoinduced electron transfer mechanism seen among other heptamethine cyanine dye-based sensors that possess receptive moieties coupled via amination of the chlorocyclohexenyl ring [40,41]. The close proximity of the histidine to the IR-783 could potentially serve as the PET acceptor to weakly quench fluorescence, whereupon binding of the HPQ motif to streptavidin may block this transfer resulting in turn-on fluorescence enhancement. This does not suggest however that all IR-783 couple peptide receptors produce “turn-on” fluorescence upon binding, as we have produced additional IR-783 probes capable of binding to hSIRPalpha that reveal binding but do not elicit a significant fluorescence enhancement (Figure S1).
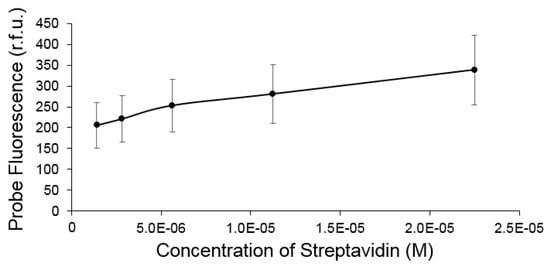
Figure 4.
The fluorescence intensity of the IR783-VSHPQAPF was observed to increase linearly with increasing concentrations of streptavidin.
3.2.3. Reversibility of Probe Enhancement
To examine if this turn-on fluorescence enhancement was reversible, native biotin was added as a means for competitive elution of the probe. We found that by increasing the amount of biotin from 40 μM to 200 μM we could observe a decrease in fluorescence intensity (Figure 5a). This demonstrates the binding and fluorescence enhancement to streptavidin was indeed reversible by the decrease in signal of the IR783 VSHPQAPF probe when released from streptavidin due to competitive binding by biotin at the same streptavidin target site. The fluorescence spectra of the probe in the bound and unbound state reflected only a change in intensity and no significant peak shifts were observed (Figure 5b).
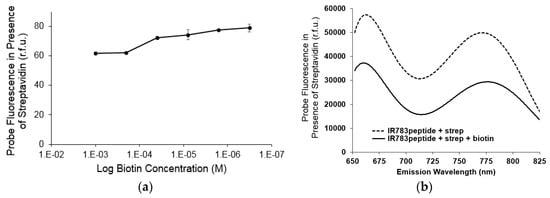
Figure 5.
The biotin mimetic probe IR-783 VSHPQAPF, when bound to streptavidin, has enhanced fluorescence, which is reversible when native biotin is subsequently added to competitively elute the probe from the streptavidin target: (a) Addition of increasing amount of biotin shows a significant decrease in fluorescence when more than one equivalent of biotin was combined with respect to the streptavidin concentration; (b) Fluorescence spectra show that the addition of 3 equivalents of biotin results in a decrease in intensity of the signal but no significant peak shift.
Competitive elution by native biotin was further validated by the MST signal. In looking at a pre-mixture of streptavidin and the biotin mimetic IR-783 VSHPQAPF probe, it was found that the probe was displaced from streptavidin’s biotin recognition site when native biotin was added. This provided further confirmation that the probe was bound specifically at the biotin recognition site on the streptavidin target (Figure 6).

Figure 6.
Microscale thermophoresis showing distinct signals for the IR783-VSHPQAPF probe before and after binding to streptavidin revealing a clear binding event and again returning to its original signal upon addition of native biotin confirming the release of the probe from streptavidin.
4. Discussion
The modular nature of the fluorophore coupling strategy presented here allows for peptide-based receptors to be converted into fluorescent probes as the penultimate step during solid phase peptide synthesis. Specifically, we demonstrated covalent labeling via the reactive chloro-group of IR-783 for substitution at the central position with the VSHPQAPF peptide that exhibits the characteristic biotin mimetic sequence (HPQ). The resulting probe yielded a blue-shifted fluorescence emission spectrum, which upon binding to the biotin recognition site on streptavidin was able to yield a turn-on enhancement of the intensity of the fluorescence emission spectra. We suspect that the location of particular residues (such as histidine) on the peptide-based receptor moiety in close proximity to the IR-783 may fulfil photo-induced electron transfer-based quenching. Recent work has shown that nearby tyrosine, tryptophan, methionine, or histidine residues could quench fluorescence dyes by serving as acceptors for photo-induced electron transfer [42]. Fluorescein dyes for instance have shown reduced fluorescence when in close proximity to Trp and Tyr residues, where the quenching via electron transfer is known to be very sensitive to the interaction between the dye and amino acid which is easily influenced by changes in the local environment and structure [43]. Similarly, oxazine and rhodamine dye derivatives could be driven to a non-fluorescent state depending on the quenching distance with nearby Trp [44]. In recent works, photo-induced electron transfer as a quenching mechanism has been designed into heptamethine cyanine dyes though strategic placement of aromatic amines to show the feasibility of designing “turn-on” near-IR fluorescence sensors for protons and metal cations [45]. As with these sensors, our introduction of peptides at the central position of the heptamethine cyanine dye may follow a similar mechanism to describe the recovery in fluorescence by suppression of photo-induced electron transfer. To reveal whether all peptide receptors would achieve the same turn-on fluorescence enhancement upon target binding, we also examined a receptor sequence that lacked the residues Tyr, Met, Trp, and His that are known to take part in photo-induced electron transfer. Specifically, the peptide sequence (Glu-Val-Thr-Glu-Leu-Thr-Arg-Glu-Gly Glu) was previously found [46] to bind to hSIRPalpha and was synthesized with a terminal IR-783 group, as was performed for the biotin-mimetic peptide. With this CD47 mimetic probe, we found it did not exhibit fluorescence enhancement upon target binding to hSIRPalpha in contrast to the biotin mimetic probe binding to streptavidin. Through microscale thermophoresis, we revealed that the CD47 mimetic probe could bind to hSIRPalpha, but while the MST signals confirmed binding there was no noticeable change in fluorescence upon binding.
Researchers have previously explored enhancement in fluorescence for chemosensors, often making use of photo-induced electron transfer in which fluorophores are linked to a targeted quenching moiety that, upon binding to a receptor, lowers the quencher’s HOMO level or otherwise increases the fluorophore to quencher distance in some cases by cleaving the fluorophore linked quencher. As such, turn-on fluorescence activation by photo-induced electron transfer-based approaches have long been applied for modular aptamer-based chemosensors [47]. We identified the potential of our methodology to afford a simple strategy for generating fluorescent probes by direct coupling of the IR-783 at the end of the peptide synthesis. From analysis of the fluorescent enhancement observed for the biotin mimetic probe upon streptavidin binding, we find a linear increase in fluorescence of the IR-783 VSHPQAPF upon addition of streptavidin above approximately 1 μM. This linearity could be very desirable from the perspective of biochemical sensing, where it is generally preferred for fluorescence values to closely fit an expected response.
For accurate confirmation that the close proximity of the IR-783 moiety did not prevent the peptide portion from entering the binding site of streptavidin, we examined competitive binding in the presence of native biotin. The return of the probe signal to its original fluorescence upon addition of biotin indicated that the biotin mimetic IR783-VSHPQAPF probe was indeed originally occupying the same streptavidin recognition site as that of the native biotin. The reversibility of the fluorescence enhancement of the probe was confirmed by this elution of the probe from the target by addition of the native biotin to displace the biotin mimetic probe. There may be several reasons for the release of the probe resulting in the observed return to the lower fluorescence intensity state as that seen for the standalone probe. We suspect the histidine present in the probe may participate in photo-induced electron transfer with IR-783 when it has become freed from the streptavidin binding site upon the addition of native biotin. The binding interactions of the histidine on biotin-mimetic peptides are known to include direct and indirect hydrogen bonds with streptavidin as well as hydrophobic interactions [48] which may have the potential to disrupt photo-induced electron transfer-based quenching. The complexity is further increased by the possibility of conformational changes in the probe leading to greater distances between the histidine and fluorophore or alternatively changes in the electron delocalization as extended peptides are known to exhibit larger HOMO-LUMO gaps than helical structures [49]. The biotin-mimetic peptides are one such sequence known to take on a helical character when bound to streptavidin [48]. In looking at the competition with native biotin, we see that at greater than 1 equivalent of biotin added per molecule of streptavidin, there is a significant decrease in the observed probe fluorescence, indicating the strong biotin interaction with streptavidin is displacing the probe from the multiple recognition sites present on streptavidin. This approach to fluorophore labeling of peptide-based receptors in a streamlined manner during solid-phase peptide synthesis has indeed proved to be versatile in affording probes with site-specific IR-783 functionalization. The resulting probes can even possess turn-on fluorescence enhancement depending on the sequence of the receptive moiety. Future studies will warrant a closer examination of the changes in fluorescent properties for such probes as determined by the role of individual amino acid residues, their proximity to the N’ terminal labelled IR-783 fluorophore, and the extent of target binding induced rearrangement in the receptor conformation. From this, we may begin to more deeply identify probable design criteria for future iterations of this probe generation approach. In looking forward, applications of this approach may provide a simple strategy for producing NIR fluorescent probes with coupled peptides which may soon find utility for targeted in vivo imaging depending on the specificity of the peptide sequence for a target site of interest.
5. Conclusions
In summary, we offer a straightforward approach to generating fluorescent probes from peptide-based receptors. While loss of binding ability is often a concern in probe development when using less-specific labeling strategies, these site-specifically labelled receptors were found to be capable of retaining binding to their intended targets. Interestingly, the sequence of the peptide itself was found to impact the fluorescence intensity of the reporter while the reporter in the case of the peptides tested did not prevent target binding.
Supplementary Materials
The following are available online at http://www.mdpi.com/2227-9040/6/4/47/s1, Supplementary Figures S1–S6.
Author Contributions
Conceptualization, E.K.; Methodology, E.K., L.K., and B.L.; Formal Analysis, J.J.; Investigation, E.K., L.K., and B.L.; Writing-Original Draft Preparation, L.K.; Writing-Review & Editing, J.J.; Supervision, J.J.; Project Administration, J.J.
Funding
The authors acknowledge the financial support from the National Heart, Lung and Blood Institute. LeNaiya Kydd is supported by the National Institutes of Health (NIH) training award, NIH T32 HL134613. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
References
- Zhang, E.; Luo, S.; Tan, X.; Shi, C. Mechanistic study of IR-780 dye as a potential tumor targeting and drug delivery agent. Biomaterials 2014, 35, 771–778. [Google Scholar] [CrossRef] [PubMed]
- Kiyose, K.; Aizawa, S.; Sasaki, E.; Kojima, H.; Hanaoka, K.; Terai, T.; Urano, Y.; Nagano, T. Molecular design strategies for near-infrared ratiometric fluorescent probes based on the unique spectral properties of aminocyanines. Chem. A Eur. J. 2009, 15, 9191–9200. [Google Scholar] [CrossRef] [PubMed]
- Hilderbrand, S.A.; Kelly, K.A.; Weissleder, R.; Tung, C.-H. Monofunctional near-infrared fluorochromes for imaging applications. Bioconj. Chem. 2005, 16, 1275–1281. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Shi, C.; Tong, R.; Qian, W.; Zhau, H.E.; Wang, R.; Zhu, G.; Cheng, J.; Yang, V.W.; Cheng, T.; et al. Near IR heptamethine cyanine dye–mediated cancer imaging. Clin. Cancer Res. 2010, 16, 2833–2844. [Google Scholar] [CrossRef] [PubMed]
- Licha, K.; Riefke, B.; Ebert, B.; Grötzinger, C. Cyanine dyes as contrast agents in biomedical optical imaging. Acad. Radiol. 2002, 9, 320S–322S. [Google Scholar] [CrossRef]
- Li, C.; Greenwood, T.R.; Bhujwalla, Z.M.; Glunde, K. Synthesis and characterization of glucosamine-bound near-infrared probes for optical imaging. Org. Lett. 2006, 8, 3623–3626. [Google Scholar] [CrossRef] [PubMed]
- Kuang, Y.; Zhang, K.; Cao, Y.; Chen, X.; Wang, K.; Liu, M.; Pei, R. Hydrophobic IR-780 dye encapsulated in cRGD-conjugated solid lipid nanoparticles for NIR imaging-guided photothermal therapy. ACS Appl. Mater. Interfaces 2017, 9, 12217–12226. [Google Scholar] [CrossRef] [PubMed]
- Cheng, G.; Fan, J.; Sun, W.; Cao, J.; Hu, C.; Peng, X. A near-infrared fluorescent probe for selective detection of HClO based on Se-sensitized aggregation of heptamethine cyanine dye. Chem. Commun. 2014, 50, 1018–1020. [Google Scholar] [CrossRef] [PubMed]
- Butte, P.V.; Mamelak, A.; Parrish-Novak, J.; Drazin, D.; Shweikeh, F.; Gangalum, P.R.; Chesnokova, A.; Ljubimova, J.Y.; Black, K. Near-infrared imaging of brain tumors using the Tumor Paint BLZ-100 to achieve near-complete resection of brain tumors. Neurosurg. Focus 2014, 36, E1. [Google Scholar] [CrossRef] [PubMed]
- Kiyose, K.; Kojima, H.; Urano, Y.; Nagano, T. Development of a ratiometric fluorescent zinc ion probe in near-infrared region, based on tricarbocyanine chromophore. J. Am. Chem. Soc. 2006, 128, 6548–6549. [Google Scholar] [CrossRef] [PubMed]
- Mujumdar, R.B.; Ernst, L.A.; Mujumdar, S.R.; Waggoner, A.S. Cyanine dye labeling reagents containing isothiocyanate groups. Cytom. J. Int. Soc. Anal. Cytol. 1989, 10, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Song, F.; Lu, E.; Wang, Y.; Zhou, W.; Fan, J.; Gao, Y. Heptamethine cyanine dyes with a large stokes shift and strong fluorescence: A paradigm for excited-state intramolecular charge transfer. J. Am. Chem. Soc. 2005, 127, 4170–4171. [Google Scholar] [CrossRef] [PubMed]
- Ellis, A.L.; Mason, J.C.; Lee, H.-W.; Strekowski, L.; Patonay, G.; Choi, H.; Yang, J.J. Design, synthesis, and characterization of a calcium-sensitive near infrared dye. Talanta 2002, 56, 1099–1107. [Google Scholar] [CrossRef]
- Górecki, T.; Patonay, G.; Strekowski, L.; Chin, R.; Salazar, N. Synthesis of novel near-infrared cyanine dyes for metal ion determination. J. Heterocycl. Chem. 1996, 33, 1871–1876. [Google Scholar] [CrossRef]
- Harrison, V.S.; Carney, C.E.; MacRenaris, K.W.; Waters, E.A.; Meade, T.J. Multimeric near IR–MR contrast agent for multimodal in vivo imaging. J. Am. Chem. Soc. 2015, 137, 9108–9116. [Google Scholar] [CrossRef] [PubMed]
- Lv, W.; Shen, Y.; Yang, H.; Yang, R.; Cai, W.; Zhang, J.; Yuan, L.; Duan, Y.; Zhang, L. A Novel Bimodal Imaging Agent Targeting HER2 Molecule of Breast Cancer. J. Immunol. Res. 2018, 2018. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.; Yu, F.; Lv, C.; Choo, J.; Chen, L. Fluorescent chemical probes for accurate tumor diagnosis and targeting therapy. Chem. Soc. Rev. 2017, 46, 2237–2271. [Google Scholar] [CrossRef] [PubMed]
- Tung, C.H. Fluorescent peptide probes for in vivo diagnostic imaging. Peptide Sci. Orig. Res. Biomol. 2004, 76, 391–403. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Wu, J.; Di, C.; Zhou, R.; Zhang, H.; Su, P.; Xu, C.; Zhou, P.; Ge, Y.; Liu, D.; et al. A novel peptide-based fluorescence chemosensor for selective imaging of hydrogen sulfide both in living cells and zebrafish. Biosens. Bioelectron. 2017, 92, 602–609. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Wang, J.; Boyd, B.J. Peptide-based biosensors. Talanta 2015, 136, 114–127. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Liu, L.; Zhou, P.; Wu, W.; Wu, J.; Liu, W.; Tang, Y. A peptide-based fluorescent chemosensor for multianalyte detection. Biosens. Bioelectron. 2015, 72, 80–86. [Google Scholar] [CrossRef] [PubMed]
- Pazos, E.; Vazquez, O.; Mascarenas, J.L.; Vazquez, M.E. Peptide-based fluorescent biosensors. Chem. Soc. Rev. 2009, 38, 3348–3359. [Google Scholar] [CrossRef] [PubMed]
- Rajarao, G.K.; Nekhotiaeva, N.; Good, L. Peptide-mediated delivery of green fluorescent protein into yeasts and bacteria. FEMS Microbiol. Lett. 2002, 215, 267–272. [Google Scholar] [CrossRef] [PubMed]
- Hermanson, G.T. Bioconjugate Techniques; Academic Press: Cambridge, MA, USA, 2013. [Google Scholar]
- Bouteiller, C.; Clavé, G.; Bernardin, A.; Chipon, B.; Massonneau, M.; Renard, P.-Y.; Romieu, A. Novel water-soluble near-infrared cyanine dyes: Synthesis, spectral properties, and use in the preparation of internally quenched fluorescent probes. Bioconj. Chem. 2007, 18, 1303–1317. [Google Scholar] [CrossRef] [PubMed]
- Hong, V.; Kislukhin, A.A.; Finn, M. Thiol-selective fluorogenic probes for labeling and release. J. Am. Chem. Soc. 2009, 131, 9986–9994. [Google Scholar] [CrossRef] [PubMed]
- Bartczak, D.; Kanaras, A.G. Preparation of peptide-functionalized gold nanoparticles using one pot EDC/sulfo-NHS coupling. Langmuir 2011, 27, 10119–10123. [Google Scholar] [CrossRef] [PubMed]
- De León-Rodríguez, L.M.; Kovacs, Z. The synthesis and chelation chemistry of DOTA—Peptide conjugates. Bioconj. Chem. 2007, 19, 391–402. [Google Scholar] [CrossRef] [PubMed]
- Pap, E.H.; Dansen, T.B.; van Summeren, R.; Wirtz, K.W. Peptide-based targeting of fluorophores to organelles in living cells. Exp. Cell Res. 2001, 265, 288–293. [Google Scholar] [CrossRef] [PubMed]
- Pap, E.H.; Dansen, T.B.; Wirtz, K.W. Peptide-based targeting of fluorophores to peroxisomes in living cells. Trends Cell Biol. 2001, 11, 10–12. [Google Scholar] [CrossRef]
- Liu, S.; Edwards, D.S. Bifunctional chelators for therapeutic lanthanide radiopharmaceuticals. Bioconj. Chem. 2001, 12, 7–34. [Google Scholar] [CrossRef]
- Li, G.; Xing, Y.; Wang, J.; Conti, P.S.; Chen, K. Near-infrared fluorescence imaging of CD13 receptor expression using a novel Cy5. 5-labeled dimeric NGR peptide. Amino Acids 2014, 46, 1547–1556. [Google Scholar] [CrossRef] [PubMed]
- Schmuck, C.; Heil, M.; Scheiber, J.; Baumann, K. Charge interactions do the job: A combined statistical and combinatorial approach to finding artificial receptors for binding tetrapeptides in water. Angew. Chem. Int. Ed. 2005, 44, 7208–7212. [Google Scholar] [CrossRef] [PubMed]
- Skerra, A.; Schmidt, T.G. Applications of a peptide ligand for streptavidin: The Strep-tag. Biomol. Eng. 1999, 16, 79–86. [Google Scholar] [CrossRef]
- Hundsberger, H.; Önder, K.; Schuller-Götzburg, P.; Virok, D.P.; Herzog, J.; Rid, R. Assembly and use of high-density recombinant peptide chips for large-scale ligand screening is a practical alternative to synthetic peptide libraries. BMC Genom. 2017, 18, 450. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, T.G.; Koepke, J.; Frank, R.; Skerra, A. Molecular interaction between the Strep-tag affinity peptide and its cognate target, streptavidin. J. Mol. Biol. 1996, 255, 753–766. [Google Scholar] [CrossRef] [PubMed]
- Jerabek-Willemsen, M.; Wienken, C.J.; Braun, D.; Baaske, P.; Duhr, S. Molecular interaction studies using microscale thermophoresis. Assay Drug Dev. Technol. 2011, 9, 342–353. [Google Scholar] [CrossRef] [PubMed]
- Awasthi, K.; Nishimura, G. Modification of near-infrared cyanine dyes by serum albumin protein. Photochem. Photobiol. Sci. 2011, 10, 461–463. [Google Scholar] [CrossRef] [PubMed]
- Lou, Z.; Li, P.; Song, P.; Han, K. Ratiometric fluorescence imaging of cellular hypochlorous acid based on heptamethine cyanine dyes. Analyst 2013, 138, 6291–6295. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Yan, M.; Fan, X.-X.; Sun, D.; Yang, S.-Y.; Yang, L.-J.; Li, J.-D.; Jiang, Y.-B. A heptamethine cyanine-based colorimetric and ratiometric fluorescent chemosensor for the selective detection of Ag+ in an aqueous medium. Chem. Commun. 2012, 48, 2243–2245. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Duan, X.; Chen, Z.; Liu, Y.; Xie, T.; Fang, L.; Li, X.; Yin, M.; Tang, B. A near-infrared fluorescent probe for detecting copper (II) with high selectivity and sensitivity and its biological imaging applications. Chem. Commun. 2011, 47, 7755–7757. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Ahsan, S.S.; Santiago-Berrios, M.E.B.; Abruña, H.D.; Webb, W.W. Mechanisms of quenching of Alexa fluorophores by natural amino acids. J. Am. Chem. Soc. 2010, 132, 7244–7245. [Google Scholar] [CrossRef] [PubMed]
- Götz, M.; Hess, S.; Beste, G.; Skerra, A.; Michel-Beyerle, M. Ultrafast electron transfer in the complex between fluorescein and a cognate engineered lipocalin protein, a so-called anticalin. Biochemistry 2002, 41, 4156–4164. [Google Scholar] [CrossRef] [PubMed]
- Vaiana, A.C.; Neuweiler, H.; Schulz, A.; Wolfrum, J.; Sauer, M.; Smith, J.C. Fluorescence quenching of dyes by tryptophan: Interactions at atomic detail from combination of experiment and computer simulation. J. Am. Chem. Soc. 2003, 125, 14564–14572. [Google Scholar] [CrossRef] [PubMed]
- Song, F.; Peng, X.; Lu, E.; Wang, Y.; Zhou, W.; Fan, J. Tuning the photoinduced electron transfer in near-infrared heptamethine cyanine dyes. Tetrahedron Lett. 2005, 46, 4817–4820. [Google Scholar] [CrossRef]
- Rodriguez, P.L.; Harada, T.; Christian, D.A.; Pantano, D.A.; Tsai, R.K.; Discher, D.E. Minimal “Self” peptides that inhibit phagocytic clearance and enhance delivery of nanoparticles. Science 2013, 339, 971–975. [Google Scholar] [CrossRef] [PubMed]
- Sparano, B.A.; Koide, K. A strategy for the development of small-molecule-based sensors that strongly fluoresce when bound to a specific RNA. J. Am. Chem. Soc. 2005, 127, 14954–14955. [Google Scholar] [CrossRef] [PubMed]
- Katz, B.A. Binding to protein targets of peptidic leads discovered by phage display: Crystal structures of streptavidin-bound linear and cyclic peptide ligands containing the HPQ sequence. Biochemistry 1995, 34, 15421–15429. [Google Scholar] [CrossRef] [PubMed]
- Sepunaru, L.; Refaely-Abramson, S.; Lovrinčić, R.; Gavrilov, Y.; Agrawal, P.; Levy, Y.; Kronik, L.; Pecht, I.; Sheves, M.; Cahen, D. Electronic transport via homopeptides: The role of side chains and secondary structure. J. Am. Chem. Soc. 2015, 137, 9617–9626. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).