Abstract
Wild rice relatives having the same AA genome as domesticated rice (Oryza sativa) comprise the primary gene pool for rice genetic improvement. Among them, O. meridionalis and O. rufipogon are found in the northern part of Australia. Three Australian wild rice strains, Jpn1 (O. rufipogon), Jpn2, and W1297 (O. meridionalis), and one cultivated rice cultivar Taichung 65 (T65) were used in this study. A recurrent backcrossing strategy was adopted to produce chromosomal segment substitution lines (CSSLs) carrying chromosomal segments from wild relatives and used for trait evaluation and genetic analysis. The segregation of the DNA marker RM136 locus on chromosome 6 was found to be highly distorted, and a recessive lethal gene causing abortion at the seed developmental stage was shown to be located between two DNA markers, KGC6_10.09 and KGC6_22.19 on chromosome 6 of W1297. We name this gene as SEED DEVELOPMENT 1 (gene symbol: SDV1). O. sativa is thought to share the functional dominant allele Sdv1-s (s for sativa), and O. meridionalis is thought to share the recessive abortive allele sdv1-m (m for meridionalis). Though carrying the sdv1-m allele, the O. meridionalis accessions can self-fertilize and bear seeds. We speculate that the SDV1 gene may have been duplicated before the divergence between O. meridionalis and the other AA genome Oryza species, and that O. meridionalis has lost the function of the SDV1 gene and has kept the function of another putative gene named SDV2.
1. Introduction
Rice (Oryza sativa) is one of the most important staple crops in the world. It feeds about one-third of the world population. Wild rice relatives having the same AA genome as domesticated rice comprise the primary gene pool for rice genetic improvement and include the following species; O. rufipogon, O. meridionalis, O. glumaepatula, O. barthii, O. longistaminata. Another domesticated Oryza species O. glaberrima (African rice) also has an AA gemome, and contributes to rice improvement. Though there are several reproductive barriers among these species as described below, transfer of useful genes such as disease resistance gene from AA genome Oryza species to rice has been successful via hybridization.
O. meridionalis and O. rufipogon are found in the northern part of Australia [1]. O. rufipogon is inferred to be the direct progenitor of O. sativa [2], and widely distributed not only in Australia but also in South and South East Asia and New Guinea. On the other hand, the distribution of O. meridionalis is confined to the northern parts of Australia and Irian Jaya, Indonesia [1]. Molecular data provides support for the divergence of O. meridionalis from the other AA genome Oryza species [3,4,5,6,7]. This is reflected by low pollen fertility of the hybrids between O. meridionalis and the other AA genome species [8,9], with almost no progeny being produced from the selfing of the hybrids. To utilize the rice breeding potential of wild relatives of rice, a recurrent backcrossing strategy has been adopted to produce chromosomal segment substitution lines (CSSLs) carrying chromosomal segments from wild relatives of rice in the genetic background of cultivated rice [10,11,12,13]. Subsequent backcrossing with O. sativa as pollen parent was successful, because the F1 plants between O. sativa and its wild relatives retained female fertility.
To elucidate the genetic potential for the improvement of cultivated rice using these wild species, we produced three kinds of CSSLs with different Australian wild rice strains in the same genetic background. As a model agronomic trait, we selected late-heading, because the wild rice strains in this study head later than the recurrent parent Taichung 65 by about 50 days, and heading-time is easily scored. We have succeeded in mapping the late-heading time genes from these wild rice strains (see below) and found a new genetic distortion phenomenon in the Oryza. In this study, we report the genetic mechanism of the new distortion phenomenon.
2. Results
2.1. Mapping of Photoperiod Sensitivity Gene
Three wild rice strains, Jpn1, Jpn2, and W1297, and one cultivated rice cultivar Taichung 65 (T65) were used in this study. We bred various CSSLs in a T65 genetic background incorporating the three Australian wild rice strains, W1297, Jpn1, and Jpn2, chromosomal segments by recurrent backcrossing (see Material and Methods). Hereafter, the backcrossing populations using Jpn1, Jpn2, and W1297 as donor parent are described as BCnFm (Jpn1), BCnFm (Jpn2), and BCnFm (W1297), respectively. “n” and “m” represent numbers of backcrossing and selfing, respectively. The frequency distributions of days to heading of the three BC3F2 populations are shown in Figure 1. All populations showed bimodal distributions. A total of 94 DNA markers covering the whole 12 chromosomes and showing polymorphism between T65 and the three wild rice strains were subjected to preliminary linkage analysis using bulked DNA from the three BC3F2 populations. Only one marker RM136, located 568 kbp away from a photoperiod sensitivity gene HD1 [14], showed heterozygosity in all the bulk DNAs. Chi square values for the independence between genotypes of RM136 and days to heading (early and late heading divided by the dotted line in Figure 1) were 26.880, 81.073, and 86.693 for Jpn1, Jpn2, and W1297, respectively, all highly significant (P < 0.0001). These results suggest that the three strains from Australia carry photoperiod sensitive alleles of the HD1 locus, because heterozygotes and homozygotes of these strains at the RM136 locus headed much later than the homozygotes of T65. This cultivar proved to carry a photoperiod insensitive allele at the HD1(= Se1) locus [15], which behaved as an early heading-time allele in a usual cropping season in Japan [16,17,18].

Figure 1.
Frequency distributions of days to heading of the three BC3F2 populations using T65 as the recurrent parent. Jpn1, Jpn2, and W1297 were respectively used as donor parent in subfigure (a), (b), and (c). Three classified genotypes were assessed for RM136 as indicated: white, homozygous for T65, grey, heterozygous, black, homozygous for wild rice strains, Jpn1 (a), Jpn2 (b), and W1297 (c). Dotted lines dividing each population into early heading and late heading were drawn for chi-square analysis (see text).
In the BC3F2 (W1297), the segregation of the RM136 locus was highly distorted: very few homozygotes of W1297 appeared. To check if this phenomenon was specific to the cross with W1297 as donor parent, and to evaluate this phenomenon more clearly under a more uniform genetic background, BC4F2 populations with the same cross combinations were subject to further study. As for W1297, the BC3F1 plants producing the BC3F2 population for the analysis was backcrossed again to produce BC4F1 plants. Among them, late flowering plants were selected to produce BC4F2 populations. As for Jpn1 and Jpn2, different BC3F1 plants from that producing the BC3F2 population for the above experiment were backcrossed to produce BC4F1 plants. Among them, late flowering plants were selected to produce BC4F2 populations.
2.2. Mapping of Segregation Distortion Gene
In the BC4F2 (W1297), the genotype of RM136 was distorted again (data not shown). In our preliminary experiment, among the published DNA markers around RM136, RM314 [19] located at 4,845kb, RM276 [19] at 6,231kb, RM7023 [20] at 6972kb, RM3628 [20] at 23,738kb and RM5314 [20] at 24,843kb on the IRGSP 1.0 pseudomolecule for chromosome 6 were fixed for the T65 allele. On the other hand, RM6818 and RM193 (Table 1) were segregating. These results suggest that the cause of segregation distortion is located between RM7023 and RM3628. Because other published DNA markers in our stocks failed in amplification of W1297 or did not distinguish T65 from W1297, we designed new DNA markers (Table 1), and performed linkage analysis. For the five consecutive markers from KGC6_12.02 to KGC6_19.48, only homozygotes of T65 and heterozygotes appeared (Figure 2), and no recombination occurred among the five markers (Table 2). The ratio of 64 homozygotes of T65: 119 heterozygotes fitted very well to 1:2 (χ2(1:2) = 0.221, P = 0.638).

Table 1.
Primer sequences of DNA markers designed or redesigned for linkage analysis of SDV1 gene.
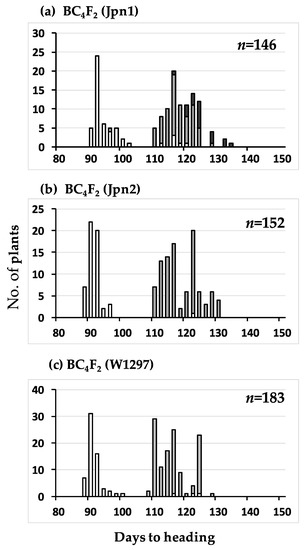
Figure 2.
Frequency distributions of days to heading of the three BC4F2 populations using T65 as recurrent parent. Jpn1, Jpn2, and W1297 were respectively used as donor parent in subfigure (a), (b) and (c). Three classified genotypes were assessed for KGC6_12.02 as indicated: white, homozygous for T65, grey, heterozygous, black, homozygous for wild rice strains, Jpn1 (a), Jpn2 (b), and W1297 (c).

Table 2.
Haplotypes around the segregation distortion region on rice chromosome 6 of BC4F2 (W1297).
The distorted segregation ratio 1:2:0 can be explained by one pair of recessive lethal genes. If the lethality occurred at the seedling stage, about 25% of seedlings would be expected to die. However, our visual observation did not fit with such a phenomenon. We then speculated that segregation distortion occurred during seed development. If so, seed fertility of the heterozygotes should be lower than that of the T65 homozygotes by about 25%. To test this, we examined the seed fertility of each of the BC4F2 plants. In the BC4F2 (W1297) population, the heterozygotes for KGC6_12.02 showed lower seed fertility than the homozygotes of the T65 allele (Figure 3). If lower fertility was caused by one recessive gene, many of the sterile seeds were expected to be aborted after fertilization. Therefore, sterile seeds were dehusked to see if sterility occurred before or after fertilization (Figure 4). The proportion of seeds aborted after fertilization for heterozygotes for KGC6_12.02 was higher than that for homozygotes of the T65 allele (Figure 5). These results suggested that homozygotes of the W1297 allele for KGC6_12.02 die at the seed developmental stage.
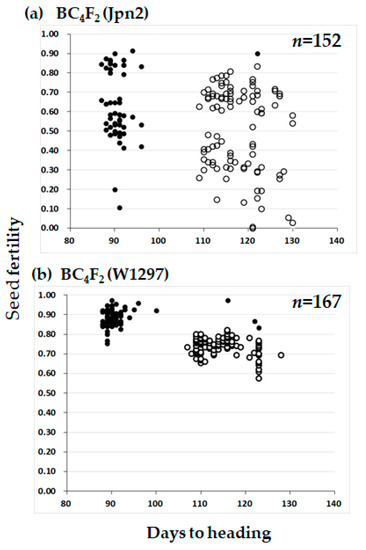
Figure 3.
Scatter diagram of days to heading and seed fertility in the two BC4F2 populations using T65 as the recurrent parent. Jpn2 and W1297 were respectively used as donor parent in subfigure (a) and (b). Two classified genotypes were assessed for KGC6_12.02 as indicated: solid circle, homozygous for T65; open circle, heterozygous. In (b), plants used for testcross (Table 3) or damaged by birds in Figure 2 were removed in this figure.

Figure 4.
Sterile seeds aborted after fertilization (top) and normal fertile seeds (bottom) found in the BC4F2 (W1297). One unit of the rightmost scale indicates 1 mm.
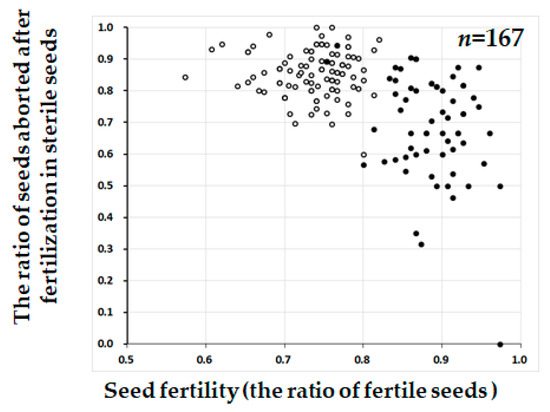
Figure 5.
The scatter diagram between seed fertility (the ratio of fertile seeds) (X-axis) and the ratio of seeds aborted after fertilization in sterile seeds (Y-axis) in the BC4F2 population (W1297). Two classified genotypes were assessed for KGC6_12.02 as indicated: solid circle, homozygous for T65; open circle, heterozygous. Plants used for testcross (Table 3) or damaged by birds in Figure 2 were removed in this figure.
2.3. Segregation Distortion Caused by Abortion During Seed Development
To confirm this hypothesis, the following experiments were performed. First, pollen fertility was examined for all BC4F2 (W1297) plants, with the result that all plants showed more than 90% pollen fertility (data not shown), suggesting that pollen sterility was not the cause of the distorted segregation ratio. Second, reciprocal backcrossing of heterozygotes for KGC6_12.02 to T65 to produce a BC5F1 generation was undertaken. The BC5F1 from both cross combinations showed the segregation ratio fitted a 1 heterozygote:1 homozygote ratio for the T65 allele (Table 3), indicating that normal gene segregation occurred at both the egg and pollen developmental stage. The BC4F2 plants used for backcrossing were also selfed to produce a BC4F3 generation. DNA was extracted from the embryo of the fertile seeds. The segregation ratio was largely distorted from 1:2:1 at the KGC6_12.02 locus, and no homozygotes for the W1297 allele for KGC6_12.02 appeared, indicating that segregation distortion occurred during seed development, and was not caused by ungerminated fertile seeds, though the segregation ratio did not fit to a 1:2:0 ratio. (Table 3). The BC4F3 plants deriving from selfed seeds of the BC4F2 plants heterozygous for the KGC6_12.02 locus also showed distorted segregation, and the ratio fitted a 1: 2: 0 ratio, confirming the other experimental results (Table 3). Taken together, all the experimental results indicated that a recessive lethal gene causing abortion at the seed developmental stage was located between KGC6_10.09 and KGC6_22.19 on chromosome 6 of W1297 (Table 2).

Table 3.
Segregation of progeny of BC4F2 (W1297) heterozygous for KGC6_12.02 genotype.
The same segregation distortion was also found in the BC4F2 (Jpn2) population (Figure 2). The ratio of 59 homozygotes of T65 allele: 97 heterozygotes at the KGC6_12.02 locus fitted very well to 1:2 (χ2(1: 2) = 0.556, P = 0.456), and no homozygotes of Jpn2 allele appeared. The seed fertility of heterozygotes of KGC6_12 was lower than that of the homozygote of T65 allele, supporting the view that a recessive lethal gene causing abortion at seed developmental stage was located close to KGC6_12 of Jpn2 (Figure 3). The seed fertility of BC4F2 (Jpn2) was highly variable for both homozygotes of the T65 allele and heterozygote at the KGC6_12.02 locus, suggesting that other genetic factor(s) were involved in the large variance of seed fertility. Our preliminary results showed low pollen fertility might be responsible for low seed fertility of some plants (unpublished data). Therefore, the cause of the seed sterility was not investigated further. For Jpn1, both BC3F2 (Jpn1) and BC4F2(Jpn1) showed that normal gene segregation occurred around the HD1 locus (Figure 1 and Figure 2).
These results indicated that the two Australian O. meridionalis strains, W1297 and Jpn2, carry a recessive lethal gene causing abortion at the seed developmental stage, which was located between the two DNA markers, KGC6_10.09 and KGC6_22.19, spanning 12 Mb on chromosome 6, and that the Australian O. rufipogon strain Jpn1 does not carry such a gene.
3. Discussion
There have been many genes conferring hybrid seed sterility, hybrid pollen sterility, and segregation distortion found on Oryza chromosome 6 in inter-and intra-specific crosses, most of which O. sativa is involved with [21,22,23,24,25,26,27]. However, to our knowledge, the segregation distortion caused by seed abortion after fertilization has not been reported in the genus Oryza. We name this gene SEED DEVELOPMENT 1 (gene symbol: SDV1), according to the gene nomenclature system for rice [28], because this gene is involved in the early seed developmental stage. In the intraspecific crosses among O. sativa, there have been no reports of gene distortion or partial seed sterility phenomena as described above on chromosome 6, though other phenomena have been reported [21,22,23,24,25,26,27]. Therefore, all O. sativa is thought to share the same functional dominant allele found in T65. This allele was called Sdv1-s (s for sativa). The homozygotes of the W1297 allele and the Jpn2 allele of this locus do not exist in the T65 genetic background probably because they die at the early seed development stage. W1297 and Jpn2 have originated from different places in Australia: W1297 is from Northern Territory, and Jpn2 is from Queensland. According to Juliano et al. [29], most crosses between Northern Territory and Queensland accessions produced sterile hybrids. Our preliminary results showed the hybrids from the reciprocal crosses between W1297 and Jpn2 were highly sterile (unpublished data). DNA marker-based analyses showed O. meridionalis genetic differentiation corresponding to geographic origin [29]. Further, in the CSSL lines of an O. meridionalis accession, W1625, chromosomal segments in a T65 genetic background [12], no lines were fixed for the W1625 chromosomal segment on which SDV1 locus is located (https://shigen.nig.ac.jp/rice/Oryzabase/locale/change?lang=en). The results described above on the whole suggest that all O. meridionalis share the recessive abortive allele. This allele was called sdv1-m (m for meridionalis).
Though carrying the recessive abortive allele in homozygous form at the SDV1 locus, the O. meridionalis accessions can self-fertilize and bear seeds. We speculate that the SDV1 gene may have been duplicated before the divergence between O. meridionalis and the other AA genome Oryza species, and that O. meridionalis has lost the function of the SDV1 gene and has kept the function of the other gene while O. sativa kept the function of the SDV1 gene and has lost the function of the other gene (Figure 6). Such duplication and loss of reproductive barrier-related genes has been reported; Yamagata et al. [30] found that the reciprocal loss of duplicated genes encoding mitochondrial ribosomal protein L27, essential for the later stage of pollen development, causes hybrid pollen sterility in F1 hybrid between O. sativa and O. glumaepatula. Nguyen et al. [31] reported that the duplication and loss of function of genes encoding RNA polymerase III subunit C4 hybrid causes pollen sterility in F1 hybrid between O. sativa and O. nivara (annual form of O. rufipogon). Ichitani et al. [32] performed linkage analysis of hybrid chlorosis genes in rice, and found that the causal recessive genes hca1-1 and hca2-1 are located on the distal region of the short arm of chromosome 12 and 11, respectively, known to be highly conserved as a duplicated chromosomal segment.
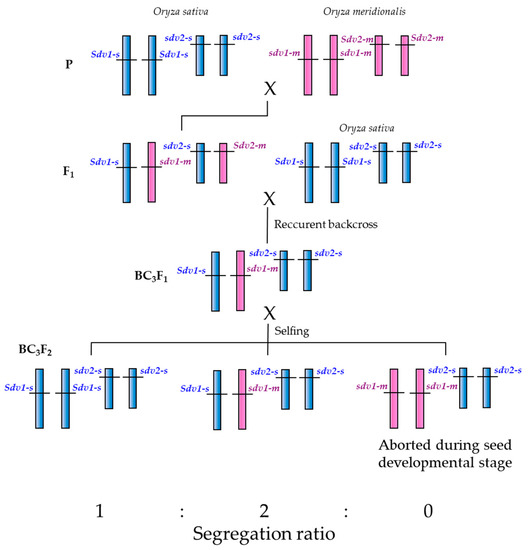
Figure 6.
A genetic model that explains segregation distortion and seed sterility assuming gene duplication and loss of gene function for seed development.
There are other models explaining the hybrid incompatibility (abortion, lethality, or weakness) known as the Bateson–Dobzhansky–Muller (BDM) model (for a review, Bomblies et al. [33]). In the incompatibility caused by the two nonallelic dominant genes, if the one locus is heterozygote or fixed for the incompatibility-causing allele, both heterozygotes and homozygotes of the incompatibility-causing allele of the other locus should show incompatibility. If the one locus is fixed for the normal allele, incompatibility does not occur. In the incompatibility caused by the heterozygote on one locus, only heterozygotes should show incompatibility. Therefore, these models cannot explain the segregation distortion in this study. In the hybrid breakdown model proposed by Oka [34], the combination of the heterozygotes or the homozygotes of recessive alleles at one locus and the homozygotes of recessive alleles at the other locus show incompatibility. This model cannot explain the segregation in this study either. Therefore, the gene duplication model as described above fits the phenomenon in this study best.
As the counterpart of SDV1, “the other” putative gene is named SEED DEVELOPMENT 2 (gene symbol: SDV2). SDV1 and SDV2 are thought to be derived from duplication. O. meridionalis accessions and O. sativa accessions should carry the functional allele and the unfunctional allele at the SDV2 locus, respectively. We name these respective alleles Sdv2-m and sdv2-s. The presence and chromosomal location of SDV2 have not been elucidated. We are undertaking the genetic analysis of SDV2, tracing back to earlier backcrossing populations.
If useful genes of O. meridionalis for rice genetic improvement are located close to sdv1-m, the introgression of these genes into O. sativa genetic background should be combined with Sdv2-m. Therefore, the chromosomal location of SDV2 and tightly linked DNA markers to it are urgently needed.
In the frequently cited high-density rice genetic linkage map by Harushima et al. [35], the centromeric region of chromosome six is located between 64.7 cM and 65.7 cM. Some of the DNA marker sequences located on the centromeric region are available in NCBI (https://www.ncbi.nlm.nih.gov). C574 (accession name: D15395) is located at 13,685 kb, and G294 (accession name: D14774) is located at 17,056 kb in Nipponbare genome (Os-Nipponbare-Reference-IRGSP-1.0). Therefore the physical size of the centromeric region is at least 3371 kb. The candidate chromosomal region of SDV1 encompasses this region (Table 1 and Table 2). Recombination events were, in general, highly suppressed around the centromere. Our result is consistent with that. The combination of high resolution linkage analysis with gene expression analysis, gene disruption, and association study will be necessary to identify the SDV1 gene.
Seed development is dissected into embryogenesis and endosperm development. We are undertaking microscopic observation of seed development of Sdv1-s sdv1-m heterozygotes in the T65 background to define the cause of the seed abortion. Several genes required for embryogenesis and endosperm development have been reported [36,37,38,39]. Identification of the SDV1 and SDV2 genes will contribute to the molecular genetics of seed development.
Direct evidence supporting the gene model was that the DNA from the embryo of aborted seeds deriving from the heterozygote of KGC6_1202 was homozygous for the W1297 allele. In our preliminary experiments, we tried to extract DNA from them, modifying the method below so that the DNA concentration would be higher. A few embryos were homozygous for the W1297 allele. However, PCR failed in most cases. This suggests that embryogenesis stops at an early stage in the homozygotes of sdv1-m. One alternative approach might be to extract DNA from developing seeds, not from mature seeds. Combination of microscopic observation of developing embryo and DNA genotyping will contribute to understanding the abortion mechanism caused by the sdv1-m gene.
According to the chloroplast genome analyses by Wambugu et al. [40], Yin et al. [41] and Sotowa et al. [42], O. rufipogon in Australia carries a chloroplast genome similar to that of O. meridionalis rather than that of O. rufipogon in Asia and O. sativa, probably because of chloroplast capture (introgression). During the process of chloroplast capture, some nuclear genome genes could be shared by O. rufipogon in Australia and O. meridionalis. However, they might carry distinct alleles on SDV1 and SDV2 loci. When the DNA sequences of these alleles of the two loci are uncovered, this information can be applied for the analysis of plants growing in the wild, and possible ongoing hybridization between O. rufipogon and O. meridionalis can be monitored in the Northern part of Australia, in which the two species are sympatric. Hybrids have been found in the wild and confirmed by molecular analysis [43], but the low frequency of these hybrids and the continued existence of the two distinct AA genome taxa in the northern Australian environment may be explained by these genes that create a reproductive barrier.
4. Materials and Methods
4.1. Plant Material
Three wild rice strains, Jpn1, Jpn2, and W1297, and one cultivated rice cultivar Taichung 65 (T65) were used in this study. W1297 is a strain of O. meridionalis collected in Darwin, Northern Territory, Australia, and provided by National Institute of Genetics, Mishima, Japan. Jpn1 and Jpn2 were collected in Australia with the permission from the Queensland government, under the EcoAccess program [42]. Judging from its perennial life history, typical of Australian O. rufipogon, and Indel marker genotypes, Jpn1 was classified as O. rufipogon. The Australian O. rufipogon population at the Jpn1 site has been shown to have a chloroplast similar to that of O. meridionalis and a nuclear genome closer to O. rufipogon [44] suggesting it may need to be considered as a distinct taxon. Jpn2 was distinct with a short anther, typical of O. meridionalis, and perennial life history in its habitat in Queensland, Australia [42]. O. meridionalis is now described as including both annual and perennial types [45]. It had Indel marker genotypes that were the same as 18 O. meridionalis Core collection accessions. It was treated as a type of O. meridionalis based on five Indel DNA markers that reflect varietal differentiation in comparisons, such as Indica–Japonica, temperate Japonica–tropical Japonica with high accuracy [46,47]. Our visual observations indicated that the three wild rice strains each showed a uniform phenotype in the first growing year, suggesting that they had been fixed for at least the loci controlling agronomic traits. Before anthesis, the panicles of the wild rice strains were covered with bags made of glassine paper to force self-fertilization in every generation. The selfed progeny also showed uniform phenotypes. The preliminary analysis of DNA markers covering the whole 12 chromosomes indicated that they were homozygous at all the DNA marker loci. T65 is a Japonica cultivar used frequently in the study of rice genetics, as a recurrent parent of CSSLs, isogenic lines, and the study of induced mutation [12,21,48].
We bred CSSLs in a T65 genetic background incorporating the three Australian wild rice strains, W1297, Jpn1, and Jpn2, chromosomal segments by recurrent backcrossing. First T65 was crossed with W1297, Jpn1, and Jpn2 as pollen parents. One plant per each wild rice strain was used for producing the F1 generation. Then the F1 was backcrossed with T65 as a pollen parent in all subsequent backcross generations with some exception described above. A total of 39, 43, and 33 BC1F1 plants were obtained using W1297, Jpn1, and Jpn2 as donor parent, respectively. All the BC1F1 plants were backcrossed with T65. One BC2F1 plant originating from each BC1F1 plant was backcrossed with T65 to produce BC3F1. One BC3F1 plant originating from each BC2F1 plant was backcrossed with T65 to produce BC4F1. W1297, Jpn1, and Jpn2 have many characters different from T65, such as late heading, red pericarp, long awn, and easy shattering. Some BC3F1 plants had such characteristics in T65 genetic background, which was suitable for genetic dissection of these characters. As a model character, late heading was selected. We selected the latest BC3F1 plants and collected seeds from these plants to produce a BC3F2 generation. As shown above, because the segregation of genes conferring days to heading did not fit the expected Mendelian single gene segregation, we focused on the analysis of the distorted segregation. The BC4F2 generations deriving from the late heading BC4F1 plants were also examined.
Plant cultivation followed Ichitani et al. [48]. Germinated seeds were sown in nursery beds in a greenhouse. About two weeks after sowing, seedlings were transferred out of the greenhouse. About 30 days after the sowing date, seedlings were planted in a paddy field at the Experimental Farm of Kagoshima University, Kagoshima, Japan. The fertilizers applied were 4, 6, and 5 g/m2, respectively, for N, K2O, and P2O5. Plant spacing was 15 × 30 cm. Sowing and transplanting were done respectively on May 31 and June 24 in 2015, on May 27and June 28 in 2016, May 25 and July 4 in 2017, respectively. Hybridization was performed as follows: For emasculation, panicles of the egg donor were soaked in hot water at 43 ℃ for 7 min. For pollination, the upper half of the open spikelets were cut about 30 min after emasculation. All the closed spikelets were cut off. Then pollen of the pollen donor was scattered on them. After pollination, panicles were covered with bags made of glassine paper. At least one panicle was left without pollination to check whether emasculation was complete.
4.2. Trait Evaluation
Heading date was recorded for each plant when the first developing panicle emerged from the leaf sheath of the flag leaf. Heading date was converted into days to heading. Seed fertility was evaluated by collecting 50 seeds from the upper side of each of the three panicles, using a modification of the method of Wan and Ikehashi [49], counting fertile and sterile spikelets on the upper half of 3–4 panicles for each plant. Seeds were scored as fertile or sterile. In the W1297 cross, sterile seeds were dehusked to see if sterility occurred before or after fertilization. The BC4F2 plants that produced the BC5F1 generation were dug up, and transferred from the paddy field to a glass house a day before pollination. We empirically know that rice plants undergoing such a treatment show lower seed fertility, probably because of root damage. Therefore, we did not evaluate seed fertility of these plants. Panicles of some plants were damaged by birds after heading. This is the reason for the inconsistency in BC4F2 plant number among tables and figures.
Pollen fertility of the BC4F2 (W1297) population was evaluated using iodine-potassium iodide solution. Panicles were collected about three days after emerging from the leaf sheath of the flag leaf, and dried in paper bags at room temperature. All the anthers in a spikelet collected one day before anthesis were cleaved to gather pollen on a glass slide. Pollen were stained with iodine-potassium iodide solution. More than 200 pollen grains were scored for each individual. Densely stained pollen with a normal size were scored as fertile. The other pollen were scored as sterile.
4.3. DNA Analysis
DNA from leaves and embryo from fertile seeds was extracted according to Ichitani et al. [48] with some modifications: Each leaf tip, 2.5 cm long from a single plant, or embryo from dehusked seeds was put in a well of a 96-deep-well plate. Then 100 μL of extraction buffer (100 mM Tris–HCl (pH 8.0), 1 M KCl, and 10 mM EDTA) was added with a 5-mm-diameter stainless steel ball to the well. After being covered with a hard lid, the plate was shaken hard (ShakeMaster ver. 1.2; BioMedical Science Inc., Tokyo, Japan) for 1 min to grind the leaves or embryos. After centrifuging, the plate was incubated at 70 °C for half an hour, then at room temperature for half an hour. Then 10 μL of the supernatant was recovered and 8 μL of 2-propanol was added. After centrifuging, the supernatant was discarded and the DNA pellet was rinsed with 50 μL of 70% ethanol. The DNA pellet was dried and dissolved in 50 μL of sterilized distilled water. It was very difficult to separate the embryo from the other part of seed completely. However, our preliminary experiment indicated that even if DNA was extracted from the whole dehusked seeds produced by a heterozygote for a DNA marker such as KGC6_12.02 (Table 1), DNA marker segregation was observed, suggesting that DNA from the parts of the dehusked seed other than the embryo was negligible. PCR mixture, cycle, electrophoresis, DNA staining, gel image documentation also followed Ichitani et al. [48].
4.4. DNA Markers
Most published PCR-based DNA markers for Oryza are based on an O. sativa genome sequence such as Nipponbare (Os-Nipponbare-Reference-IRGSP-1.0, [50]) and 9311 (GCA_0000046551, [51]). However, a preliminary survey comparing the genome of Nipponbare (IRGSP 1.0) and that of O. meridionalis accession (GCA_000338895.2. [7]) showed that there were many discrepancies between them, leading to expected failure in amplification from the O. meridionalis genome when using O. sativa genome-based DNA markers. Our strategy of designing co-dominant DNA markers was that insertion/deletion (indel) polymorphisms ranging from 5 to 100 base pairs were searched for between the Nipponbare and the O. meridionalis genomes. Then, the indels found only between O. meridionalis and Nipponbare, not between meridionalis and two Indica cultivars, 93-11 and HR-12 (GCA_000725085), were selected. The event causing such indels were thought to have occurred in Japonica rice after Japonica-Indica differentiation. T65 is a typical Japonica cultivar. Our preliminary survey showed that T65 shared the banding patterns of Nipponbare in most of the DNA markers examined [52]. Therefore, the indels as described above were expected to show polymorphism between T65 and O. meridionalis. The selected indels were screened based on sequence similarity surrounding indels between Nipponbare and the O. meridionalis genomes. The primer design followed Busung et al. [53].
Author Contributions
Conceptualization, K.I.; methodology, K.I.; validation, D.T. and M.U.; formal analysis, D.T., M.U., and K.I.; investigation, D.T. and M.U.; resources, S.T., T.S., R.H., R.I., K.I.; data curation, D.T., M.U., K.I.; writing—original draft preparation, D.T. and K.I.; writing—review and editing, D.T., M.U., S.T., T.S., R.H., R.I. and K.I.; visualization, D.T., M.U., and K.I.; supervision, K.I.; project administration, R.I.; funding acquisition, R.I.
Funding
This research was funded by JSPS KAKENHI Grant Number JP16H05777 from the Japan Society for the Promotion of Science.
Acknowledgments
We are grateful to the National Institute of Genetics for their kind provision of W1297 seeds. We thank Mr. Masaaki Ikenoue, Mr. Nishiobino Tsubasa, Ms. Yoko Nakashima and Ms. Asako Kobai for their technical assistance.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Henry, R.J.; Rice, N.; Waters, D.L.E.; Kasem, S.; Ishikawa, R.; Hao, Y.; Dillon, S.; Crayn, D.; Wing, R.; Vaughan, D. Australian Oryza: Utility and conservation. Rice 2010, 3, 235–241. [Google Scholar] [CrossRef]
- Huang, X.; Kurata, N.; Wei, X.; Wang, Z.-X.; Wang, A.; Zhao, Q.; Zhao, Y.; Liu, K.; Lu, H.; Li, W.; et al. A map of rice genome variation reveals the origin of cultivated rice. Nature 2012, 490, 497–501. [Google Scholar] [CrossRef] [PubMed]
- Ohtsubo, H.; Cheng, C.; Ohsawa, I.; Tsuchimoto, S.; Ohtsubo, E. Rice retroposon p-SINE1 and origin of cultivated rice. Breed. Sci. 2004, 54, 1–11. [Google Scholar] [CrossRef]
- Cheng, C.; Tsuchimoto, S.; Ohtsubo, H.; Ohtsubo, E. Evolutionary relationships among rice species with AA genome based on SINE insertion analysis. Genes Genet. Syst. 2002, 77, 323–334. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.-H.; Kurata, N.; Akimoto, M.; Ohtsubo, H.; Ohtsubo, E. Identification and characterization of Australian wild rice strains of Oryza meridionalis and Oryza rufipogon by SINE insertion polymorphism. Genes Genet. Syst. 2005, 80, 129–134. [Google Scholar] [CrossRef][Green Version]
- Zhang, Q.-J.; Zhu, T.; Xia, E.-H.; Shi, C.; Liu, Y.-L.; Zhang, Y.; Liu, Y.; Jiang, W.-K.; Zhao, Y.-J.; Mao, S.-Y.; et al. Rapid diversification of five Oryza AA genomes associated with rice adaptation. Proc. Natl. Acad. Sci. USA 2014, 111, E4954–E4962. [Google Scholar] [CrossRef] [PubMed]
- Stein, J.C.; Yu, Y.; Copetti, D.; Zwickl, D.J.; Zhang, L.; Zhang, C.; Chougule, K.; Gao, D.; Iwata, A.; Goicoechea, J.L.; et al. Genomes of 13 domesticated and wild rice relatives highlight genetic conservation, turnover and innovation across the genus Oryza. Nat. Genet. 2018, 50, 285–296. [Google Scholar] [CrossRef] [PubMed]
- Naredo, M.E.B.; Juliano, A.B.; Lu, B.-R.; Jackson, M.T. Hybridization of AA genome rice species from Asia and Australia I. Crosses and development of hybrids. Genet. Resour. Crop Evol. 1997, 44, 17–23. [Google Scholar] [CrossRef]
- Naredo, M.E.B.; Juliano, A.B.; Lu, B.-R.; Jackson, M.T. Taxonomic status of Oryza glumaepatula Steud. II. Hybridization between New World diploids and AA genome species from Asia and Australia. Genet. Resour. Crop Evol. 1998, 45, 205–214. [Google Scholar] [CrossRef]
- Shim, R.A.; Angeles, E.R.; Ashikari, M.; Takashi, T. Development and evaluation of Oryza glaberrima Steud. chromosome segment substitution lines (CSSLs) in the background of O. sativa L. cv. Koshihikari. Breed. Sci. 2010, 60, 613–619. [Google Scholar] [CrossRef]
- Hirabayashi, H.; Sato, H.; Nonoue, Y.; Kuno-Takemoto, Y.; Takeuchi, Y.; Kato, H.; Nemoto, H.; Ogawa, T.; Yano, M.; Imbe, T.; et al. Development of introgression lines derived from Oryza rufipogon and O. glumaepatula in the genetic background of japonica cultivated rice (O. sativa L.) and evaluation of resistance to rice blast. Breed. Sci. 2010, 60, 604–612. [Google Scholar] [CrossRef]
- Yoshimura, A.; Nagayama, H.; Sobrizal; Kurakazu, T.; Sanchez, P.L.; Doi, K.; Yamagata, Y.; Yasui, H. Introgression lines of rice (Oryza sativa L.) carrying a donor genome from the wild species, O. glumaepatula Steud. and O. meridionalis Ng. Breed. Sci. 2010, 60, 597–603. [Google Scholar] [CrossRef]
- Arbelaez, J.D.; Moreno, L.T.; Singh, N.; Tung, C.-W.; Maron, L.G.; Ospina, Y.; Martinez, C.P.; Grenier, C.; Lorieux, M.; McCouch, S. Development and GBS-genotyping of introgression lines (ILs) using two wild species of rice, O. meridionalis and O. rufipogon, in a common recurrent parent, O. sativa cv. Curinga. Mol. Breed. 2015, 35, 81. [Google Scholar] [CrossRef] [PubMed]
- Yano, M.; Katayose, Y.; Ashikari, M.; Yamanouchi, U.; Monna, L.; Fuse, T.; Baba, T.; Yamamoto, K.; Umehara, Y.; Nagamura, Y.; et al. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the arabidopsis flowering time gene CONSTANS. Plant Cell 2000, 12, 2473–2483. [Google Scholar] [CrossRef]
- Doi, K.; Izawa, T.; Fuse, T.; Yamanouchi, U.; Kubo, T.; Shimatani, Z.; Yano, M.; Yoshimura, A. Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev. 2004, 18, 926–936. [Google Scholar] [CrossRef] [PubMed]
- Ichitani, K.; Okumoto, Y.; Tanisaka, T. photoperiod sensitivity gene of Se-1 locus found in photoperiod insensitive rice cultivars of the northern limit region of rice cultivation. Jpn. J. Breed. 1997, 47, 145–152. [Google Scholar] [CrossRef][Green Version]
- Ichitani, K.; Okumoto, Y.; Tanisaka, T. Genetic analysis of the rice cultivar Kasalath with special reference to two photoperiod sensitivity loci, E1 and Se-1. Jpn. J. Breed. 1998, 48, 51–57. [Google Scholar] [CrossRef][Green Version]
- Inoue, H.; Nishida, H.; Okumoto, Y.; Tanisaka, T. Identification of an early heading time gene found in the taiwanese rice cultivar Taichung 65. Jpn. J. Breed. 1998, 48, 103–108. [Google Scholar] [CrossRef]
- Temnykh, S.; Park, W.D.; Ayres, N.; Cartinhour, S.; Hauck, N.; Lipovich, L.; Cho, Y.G.; Ishii, T.; McCouch, S.R. Mapping and genome organization of microsatellite sequences in rice (Oryza sativa L.). Theor. Appl. Genet. 2000, 100, 697–712. [Google Scholar] [CrossRef]
- McCouch, S.R.; Teytelman, L.; Xu, Y.; Lobos, K.B.; Clare, K.; Walton, M.; Fu, B.; Maghirang, R.; Li, Z.; Xing, Y.; et al. Development and Mapping of 2240 New SSR Markers for Rice ( Oryza sativa L.). DNA Res. 2002, 9, 199–207. [Google Scholar] [CrossRef]
- Sano, Y. Genetic comparisons of chromosome 6 between wild and cultivated rice. Jpn. J. Breed. 1992, 42, 561–572. [Google Scholar] [CrossRef]
- Matsubara, K.; Khin-Thidar; Sano, Y. A gene block causing cross-incompatibility hidden in wild and cultivated rice. Genetics 2003, 165, 343–352. [Google Scholar] [PubMed]
- Koide, Y.; Ogino, A.; Yoshikawa, T.; Kitashima, Y.; Saito, N.; Kanaoka, Y.; Onishi, K.; Yoshitake, Y.; Tsukiyama, T.; Saito, H.; et al. Lineage-specific gene acquisition or loss is involved in interspecific hybrid sterility in rice. Proc. Natl. Acad. Sci. USA 2018, 115, E1955–E1962. [Google Scholar] [CrossRef] [PubMed]
- Yanagihara, S.; McCouch, S.R.; Ishikawa, K.; Ogi, Y.; Maruyama, K.; Ikehashi, H. Molecular analysis of the inheritance of the S-5 locus, conferring wide compatibility in Indica/Japonica hybrids of rice (O. sativa L.). Theoret. Appl. Genet. 1995, 90, 182–188. [Google Scholar] [CrossRef]
- Liu, Y.S.; Zhu, L.H.; Sun, J.S.; Chen, Y. Mapping QTLs for defective female gametophyte development in an inter-subspecific cross in Oryza Sativa L. Theor. Appl. Genet. 2001, 102, 1243–1251. [Google Scholar] [CrossRef]
- Yang, J.; Zhao, X.; Cheng, K.; Du, H.; Ouyang, Y.; Chen, J.; Qiu, S.; Huang, J.; Jiang, Y.; Jiang, L.; et al. A killer-protector system regulates both hybrid sterility and segregation distortion in rice. Science 2012, 337, 1336–1340. [Google Scholar] [CrossRef]
- Long, Y.; Zhao, L.; Niu, B.; Su, J.; Wu, H.; Chen, Y.; Zhang, Q.; Guo, J.; Zhuang, C.; Mei, M.; et al. Hybrid male sterility in rice controlled by interaction between divergent alleles of two adjacent genes. Proc. Natl. Acad. Sci. USA 2008, 105, 18871–18876. [Google Scholar] [CrossRef]
- McCouch, S.R.; CGSNL (Committee on Gene Symbolization, N. and L., Rice Genetics Cooperative). Gene Nomenclature System for Rice. Rice 2008, 1, 72–84. [Google Scholar] [CrossRef]
- Juliano, A.B.; Naredo, M.E.B.; Lu, B.-R.; Jackson, M.T. Genetic differentiation in Oryza meridionalis Ng based on molecular and crossability analyses. Genet. Resour. Crop Evol. 2005, 52, 435–445. [Google Scholar] [CrossRef]
- Yamagata, Y.; Yamamoto, E.; Aya, K.; Win, K.T.; Doi, K.; Sobrizal; Ito, T.; Kanamori, H.; Wu, J.; Matsumoto, T.; et al. Mitochondrial gene in the nuclear genome induces reproductive barrier in rice. Proc. Natl. Acad. Sci. USA 2010, 107, 1494–1499. [Google Scholar] [CrossRef]
- Nguyen, G.N.; Yamagata, Y.; Shigematsu, Y.; Watanabe, M.; Miyazaki, Y.; Doi, K.; Tashiro, K.; Kuhara, S.; Kanamori, H.; Wu, J.; et al. Duplication and loss of function of genes encoding RNA polymerase III subunit C4 causes hybrid incompatibility in rice. G3 Genes Genom. Genet. 2017, 7, 2565–2575. [Google Scholar] [CrossRef] [PubMed]
- Ichitani, K.; Takemoto, Y.; Iiyama, K.; Taura, S.; Sato, M. Chromosomal location of HCA1 and HCA2, hybrid chlorosis genes in rice. Int. J. Plant Genom. 2012, 2012. [Google Scholar] [CrossRef] [PubMed]
- Bomblies, K.; Weigel, D. Hybrid necrosis: Autoimmunity as a potential gene-flow barrier in plant species. Nat. Rev. Genet. 2007, 8, 382–393. [Google Scholar] [CrossRef] [PubMed]
- Oka, H.I. Phylogenetic differentiation of cultivated rice. XV. Complementary lethal genes in rice. Jpn. J. Genet. 1957, 32, 83–87. [Google Scholar] [CrossRef]
- Harushima, Y.; Yano, M.; Shomura, A.; Sato, M.; Shimano, T.; Kuboki, Y.; Yamamoto, T.; Lin, S.Y.; Antonio, B.A.; Parco, A.; et al. A high-density rice genetic linkage map with 2275 markers using a single F2 population. Genetics 1998, 148, 479–494. [Google Scholar] [PubMed]
- Huang, X.; Peng, X.; Sun, M.-X. OsGCD1 is essential for rice fertility and required for embryo dorsal-ventral pattern formation and endosperm development. New Phytol. 2017, 215, 1039–1058. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Lu, Z.; Wang, X.; Ouyang, Y.; Chen, W.; Xie, K.; Wang, D.; Luo, M.; Luo, J.; Yao, J. Imprinted gene OsFIE1 modulates rice seed development by influencing nutrient metabolism and modifying genome H3K27me3. Plant J. 2016, 87, 305–317. [Google Scholar] [CrossRef] [PubMed]
- Hara, T.; Katoh, H.; Ogawa, D.; Kagaya, Y.; Sato, Y.; Kitano, H.; Nagato, Y.; Ishikawa, R.; Ono, A.; Kinoshita, T.; et al. Rice SNF2 family helicase ENL1 is essential for syncytial endosperm development. Plant J. 2015, 81, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.K.; Kitano, H.; Satoh, H.; Nagato, Y. How is embryo size genetically regulated in rice? Development 1996, 122, 2051–2058. [Google Scholar]
- Wambugu, P.W.; Brozynska, M.; Furtado, A.; Waters, D.L.; Henry, R.J. Relationships of wild and domesticated rices (Oryza AA genome species) based upon whole chloroplast genome sequences. Sci. Rep. 2015, 5, 13957. [Google Scholar] [CrossRef]
- Yin, H.; Akimoto, M.; Kaewcheenchai, R.; Sotowa, M.; Ishii, T.; Ishikawa, R. Inconsistent diversities between nuclear and plastid genomes of aa genome species in the genus Oryza. Genes Genet. Syst. 2015, 90, 269–281. [Google Scholar] [CrossRef] [PubMed]
- Sotowa, M.; Ootsuka, K.; Kobayashi, Y.; Hao, Y.; Tanaka, K.; Ichitani, K.; Flowers, J.M.; Purugganan, M.D.; Nakamura, I.; Sato, Y.-I.; et al. Molecular relationships between Australian annual wild rice, Oryza meridionalis, and two related perennial forms. Rice 2013, 6, 26. [Google Scholar] [CrossRef] [PubMed]
- Moner, A.M.; Henry, R.J. Oryza meridionalis Ng. In The Wild Oryza Genomes; Mondal, T.K., Henry, R., Eds.; Springer-Verlag: Heidelberg, Germany, 2018; pp. 177–182. [Google Scholar]
- Brozynska, M.; Copetti, D.; Furtado, A.; Wing, R.A.; Crayn, D.; Fox, G.; Ishikawa, R.; Henry, R.J. Sequencing of australian wild rice genomes reveals ancestral relationships with domesticated rice. Plant Biotechnol. J. 2017, 15, 765–774. [Google Scholar] [CrossRef] [PubMed]
- Moner, A.M.; Furtado, A.; Chivers, I.; Fox, G.; Crayn, D.; Henry, R.J. Diversity and evolution of rice progenitors in australia. Ecol. Evol. 2018, 8, 4360–4366. [Google Scholar] [CrossRef] [PubMed]
- Ichitani, K.; Taura, S.; Sato, M.; Kuboyama, T. Distribution of Hwc2-1, a causal gene of a hybrid weakness, in the World Rice Core collection and the Japanese Rice mini Core collection: Its implications for varietal differentiation and artificial selection. Breed. Sci. 2016, 66, 776–789. [Google Scholar] [CrossRef] [PubMed]
- Muto, C.; Ishikawa, R.; Olsen, K.M.; Kawano, K.; Bounphanousay, C.; Matoh, T.; Sato, Y.-I. Genetic diversity of the wx flanking region in rice landraces in northern Laos. Breed. Sci. 2016, 66, 580–590. [Google Scholar] [CrossRef][Green Version]
- Ichitani, K.; Yamaguchi, D.; Taura, S.; Fukutoku, Y.; Onoue, M.; Shimizu, K.; Hashimoto, F.; Sakata, Y.; Sato, M. Genetic analysis of ion-beam induced extremely late heading mutants in rice. Breed. Sci. 2014, 64, 222–230. [Google Scholar] [CrossRef]
- Wan, J.; Ikehashi, H. Identification of two types of differentiation in cultivated rice (Oryza sativa L.) detected by polymorphism of isozymes and hybrid sterility. Euphytica 1997, 94, 151–161. [Google Scholar] [CrossRef]
- Kawahara, Y.; de la Bastide, M.; Hamilton, J.P.; Kanamori, H.; McCombie, W.R.; Ouyang, S.; Schwartz, D.C.; Tanaka, T.; Wu, J.; Zhou, S.; et al. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 2013, 6, 4. [Google Scholar] [CrossRef]
- Yu, J.; Hu, S.; Wang, J.; Wong, G.K.-S.; Li, S.; Liu, B.; Deng, Y.; Dai, L.; Zhou, Y.; Zhang, X.; et al. A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 2002, 296, 79–92. [Google Scholar] [CrossRef]
- Ichitani, K.; Taura, S.; Tezuka, T.; Okiyama, Y.; Kuboyama, T. Chromosomal location of HWA1and HWA2, complementary hybrid weakness genes in rice. Rice 2011, 4, 29–38. [Google Scholar] [CrossRef]
- Busungu, C.; Taura, S.; Sakagami, J.-I.; Ichitani, K. Identification and linkage analysis of a new rice bacterial blight resistance gene from XM14, a mutant line from IR24. Breed. Sci. 2016, 66, 636–645. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).