Identification and Functional Characterization of a Soybean (Glycine max) Thioesterase that Acts on Intermediates of Fatty Acid Biosynthesis
Abstract
1. Introduction
2. Results
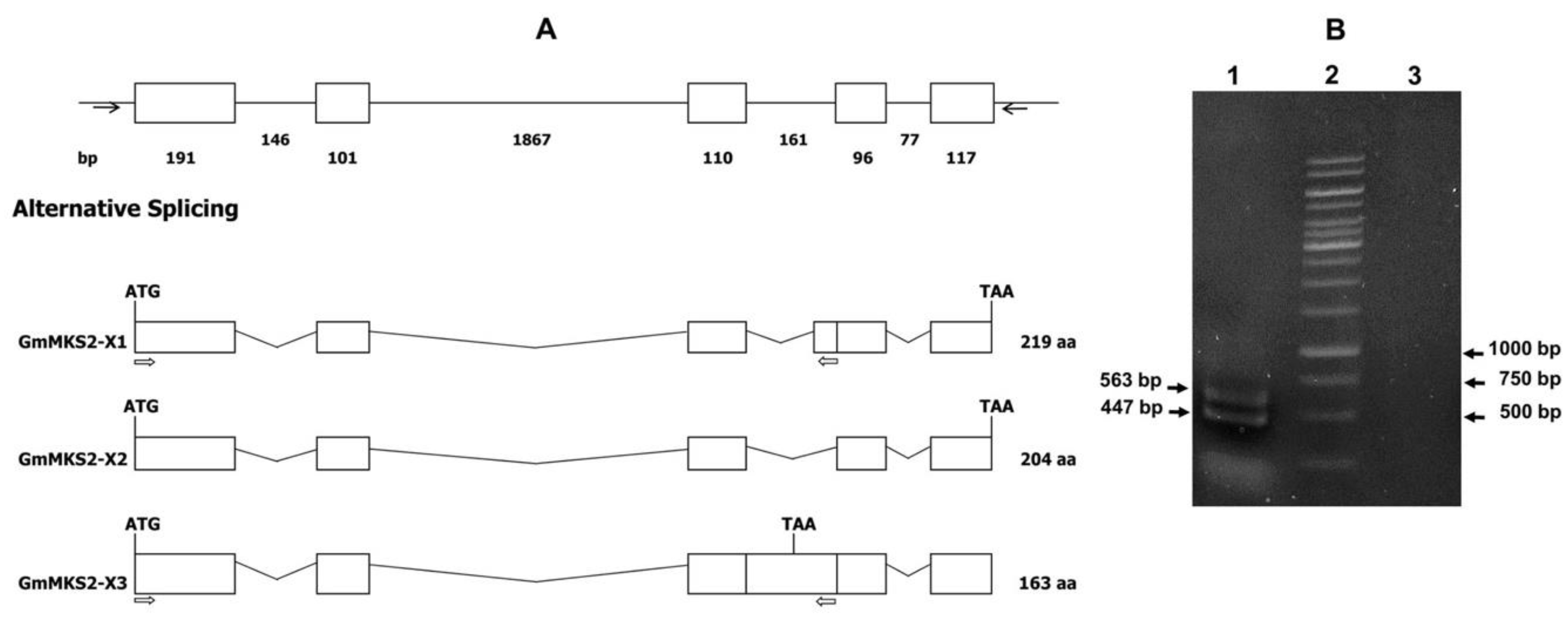
2.1. Identification of Alternatively Spliced MKS2 Isoforms in G. max
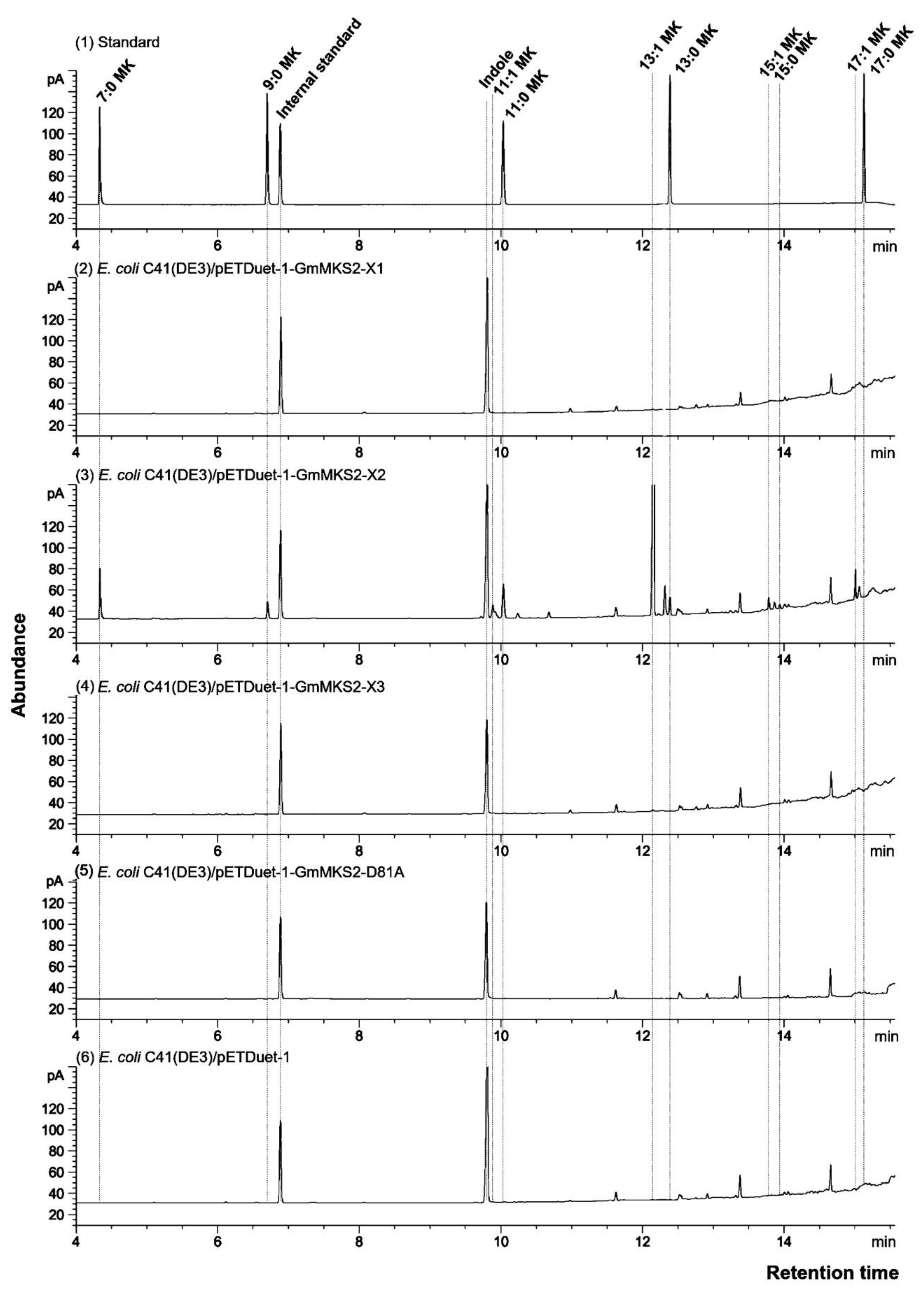
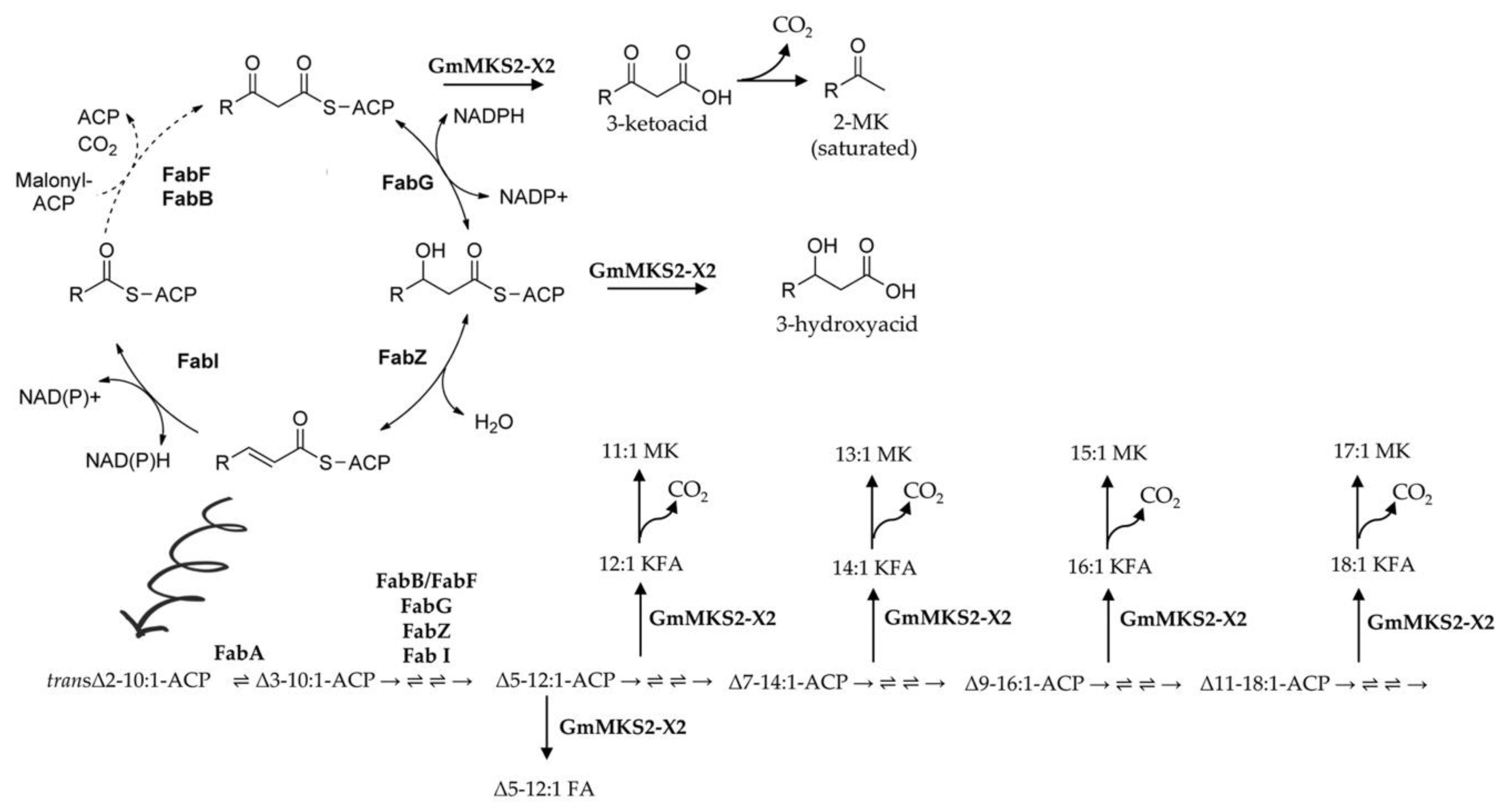
2.2. Expression of GmMSK2-X2, but Not of the Other Two Variants, in E. coli Generated Predominantly Unsaturated 3-Ketoacids
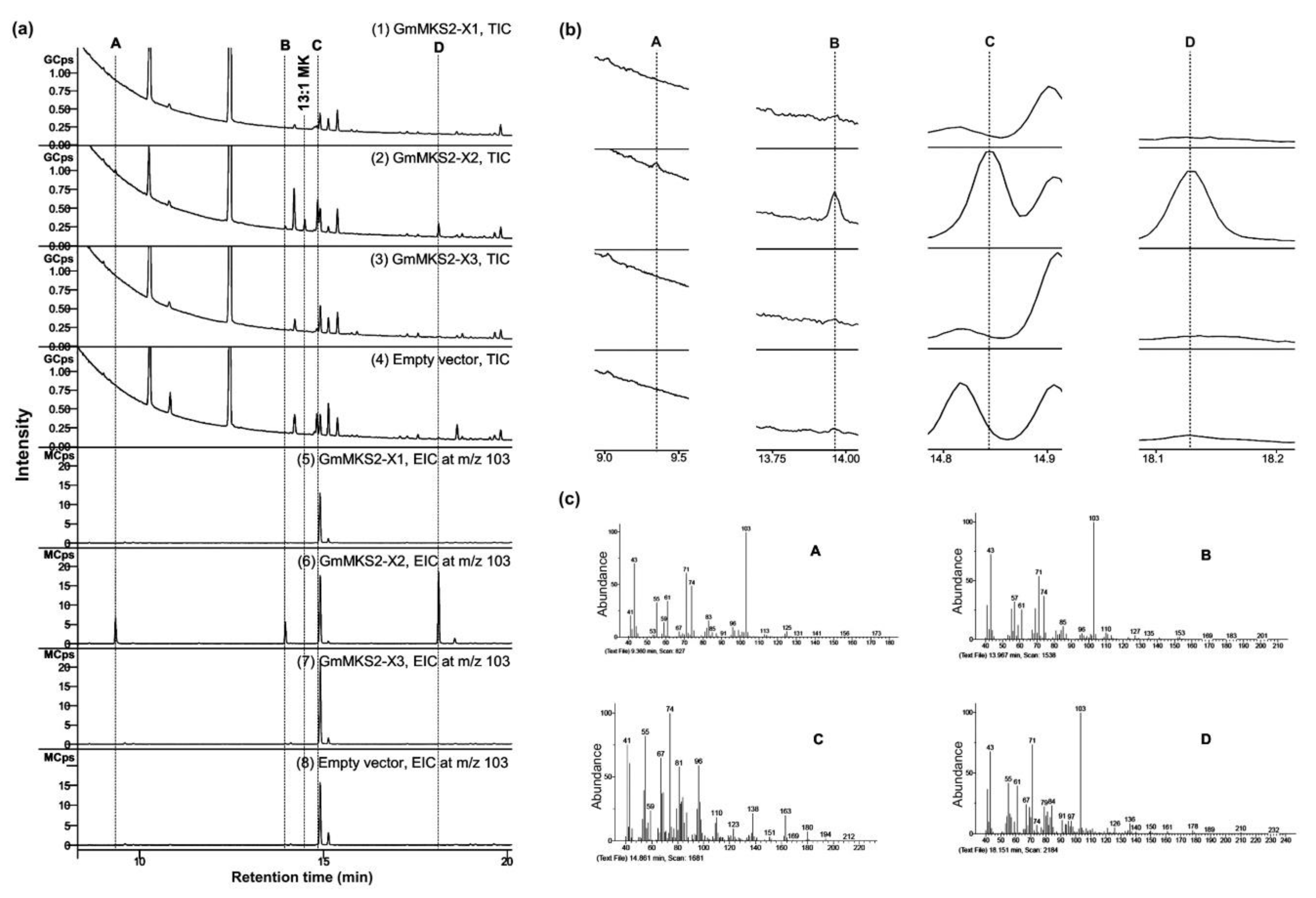
2.3. GmMKS2-X2 Could Also Act on cis-Δ5-dodecenoyl-ACP and 3-hydroxyacyl-ACPs
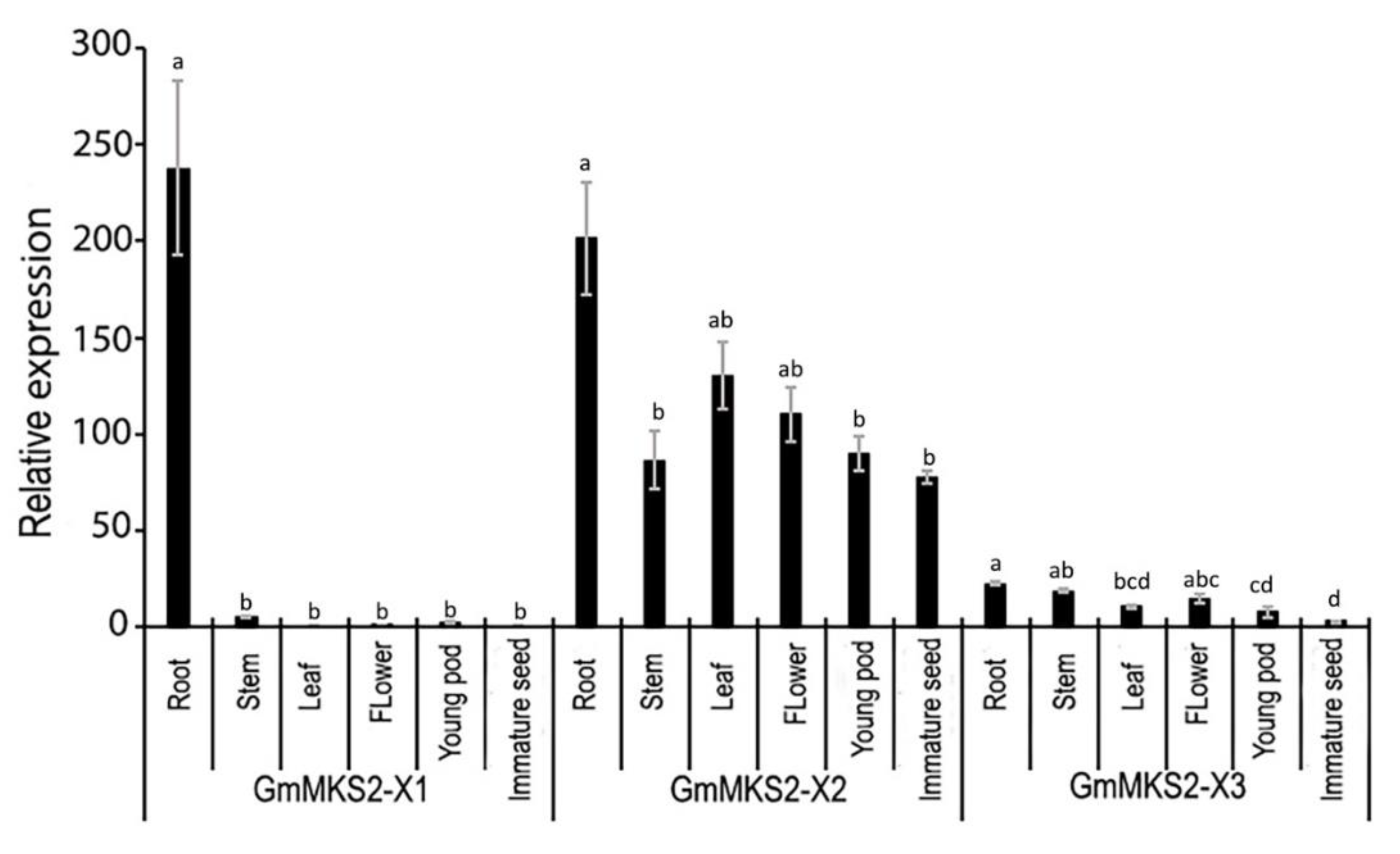
2.4. Expression Analysis of GmMKS2 in Different Tissues of Soybean Plants
2.5. Promoter Prediction and Identification of cis-Regulatory Elements in the GmMKS2 Promoter
3. Discussion
3.1. Soybean Contains Only a Single Functional Ortholog of the Wild Tomato Methylketone Synthase 2 (ShMKS2)
3.2. The Main Isoform of GmMKS2 Has Broad Substrate Specificities toward a Wide Range of Acyl-ACPs That Vary in Terms of Chain Length, Oxidation State, and Saturation Degree
3.3. Possible Biological Roles of GmMKS2 in Soybean
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Bioinformatics
4.3. Identification and Isolation of GmMKS2 Transcripts
4.4. Expression of GmMKS2 Isoforms in E. coli
4.5. Profiling 3-Ketoacids in Bacterial Culture Media and in Soybean Roots via Identification and Quantification of Their Decarboxylated Products
4.6. Determination of Free Fatty Acids in Bacterial Culture Media and in Soybean Roots
4.7. In Silico Analysis of Regulatory Elements in the Promoter Region of GmMKS2 Gene
4.8. Gene Expression Analysis by qRT- PCR
4.9. Accession Number
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ACP | acyl carrier protein |
| ALT | acyl-lipid thioesterase |
| ANOVA | analysis of variance |
| APRT | adenine phosphoribosyl transferase |
| BLAST | basic local alignment search tool |
| FAS | fatty acid synthase |
| HSD | honest significant difference |
| IPTG | isopropylthio-β-galactoside |
| MeJA | methyl jasmonate |
| MeSA | methyl salicylate |
| MKS2 | methylketone synthase 2 |
| MKS1 | methylketone synthase 1 |
| ORF | open reading frame |
| PAGE | polyacrylamide gel electrophoresis |
| qRT-PCR | quantitative real-time polymerase chain reaction |
| SDS | sodium dodecyl sulfate |
| SE | standard error |
| TE | thioesterase |
| TSA | transcriptome shortgun assembly |
References
- Brown, A.P.; Affleck, V.; Fawcett, T.; Slabas, A.R. Tandem affinity purification tagging of fatty acid biosynthetic enzymes in Synechocystis sp. PCC6803 and Arabidopsis thaliana. J. Exp. Bot. 2006, 57, 1563–1571. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Li-Beisson, Y.; Shorrosh, B.; Beisson, F.; Andersson, M.X.; Arondel, V.; Bates, P.D.; Baud, S.; Bird, D.; DeBono, A.; Durrett, T.P.; et al. Acyl-Lipid Metabolism. Arab. Book 2010, 8, e0133. [Google Scholar] [CrossRef] [PubMed]
- Salas, J.J.; Ohlrogge, J.B. Characterization of substrate specificity of plant FatA and FatB acyl-ACP thioesterases. Arch. Biochem. Biophy. 2002, 403, 25–34. [Google Scholar] [CrossRef]
- Jing, F.; Cantu, D.C.; Tvaruzkova, J.; Chipman, J.P.; Nikolau, B.J.; Yandeau-Nelson, M.D.; Reilly, P.J. Phylogenetic and experimental characterization of an acyl-ACP thioesterase family reveals significant diversity in enzymatic specificity and activity. BMC Biochem. 2011, 12, 44. [Google Scholar] [CrossRef] [PubMed]
- Cantu, D.C.; Chen, Y.; Reilly, P.J. Thioesterases: A new perspective based on their primary and tertiary structures. Protein Sci. 2010, 19, 1281–1295. [Google Scholar] [CrossRef] [PubMed]
- Voelker, T.A.; Worrell, A.C.; Anderson, L.; Bleibaum, J.; Fan, C.; Hawkins, D.J.; Radke, S.E.; Davies, H.M. Fatty acid biosynthesis redirected to medium chains in transgenic oilseed plants. Science 1992, 257, 72–74. [Google Scholar] [CrossRef] [PubMed]
- Jones, A.; Davies, H.M.; Voelker, T.A. Palmitoyl-Acyl Carrier Protein (ACP) Thioesterase and the Evolutionary Origin of Plant Acyl-ACP Thioesterases. Plant. Cell 2007, 7, 359. [Google Scholar]
- Dörmann, P.; Voelker, T.A.; Ohlrogge, J.B. Accumulation of Palmitate in Arabidopsis Mediated by the Acyl-Acyl Carrier Protein Thioesterase FATB1. Plant. Physiol. 2002, 123, 637–644. [Google Scholar] [CrossRef]
- Yu, G.; Nguyen, T.T.H.; Guo, Y.; Schauvinhold, I.; Auldridge, M.E.; Bhuiyan, N.; Ben-Israel, I.; Iijima, Y.; Fridman, E.; Noel, J.P.; et al. Enzymatic Functions of Wild Tomato Methylketone Synthases 1 and 2. Plant. Physiol. 2010, 154, 67–77. [Google Scholar] [CrossRef]
- Pulsifer, I.P.; Lowe, C.; Narayaran, S.A.; Busuttil, A.S.; Vishwanath, S.J.; Domergue, F.; Rowland, O. Acyl-lipid thioesterase1–4 from Arabidopsis thaliana form a novel family of fatty acyl-acyl carrier protein thioesterases with divergent expression patterns and substrate specificities. Plant. Mol. Biol. 2014, 84, 549–563. [Google Scholar] [CrossRef]
- Williams, W.G.; Kennedy, G.G.; Yamamoto, R.T.; Thacker, J.D.; Bordner, J. 2-Tridecanone: A naturally occurring insecticide from the wild tomato Lycopersicon hirsutum f. glabratum. Science 1980, 207, 888–889. [Google Scholar] [CrossRef] [PubMed]
- Kalinger, R.S.; Pulsifer, I.P.; Rowland, O. Elucidating the substrate specificities of acyl-lipid thioesterases from diverse plant taxa. Plant. Physiol. Biochem. 2018, 127, 104–118. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, D.S.; Lević, J.D.; Sredanović, S.A. Fatty acid composition of various soybean products. Food Feed Res. 2011, 37, 65–70. [Google Scholar]
- Davies, H.M. Medium chain acyl-ACP hydrolysis activities of developing oilseeds. Phytochemistry 1993, 33, 1353–1356. [Google Scholar] [CrossRef]
- Dörmann, P.; Spener, F.; Ohlrogge, J.B. Characterization of two acyl-acyl carrier protein thioesterases from developing Cuphea seeds specific for medium-chain- and oleoyl-acyl carrier protein. Planta 1993. [Google Scholar] [CrossRef]
- Goh, E.B.; Baidoo, E.E.K.; Keasling, J.D.; Beller, H.R. Engineering of bacterial methyl ketone synthesis for biofuels. Appl. Environ. Microbiol. 2012, 78, 70–80. [Google Scholar] [CrossRef]
- Antonious, G.F.; Dahlman, D.L.; Hawkins, L.M. Insecticidal and Acaricidal Performance of Methyl Ketones in Wild Tomato Leaves. Bull. Environ. Contam. Toxicol. 2003, 71, 400–407. [Google Scholar] [CrossRef]
- Patton, S. The Methyl Ketones of Blue Cheese and their Relation to its Flavor. J. Dairy Sci. 1950, 33, 680–684. [Google Scholar] [CrossRef]
- López-Lara, I.M.; Nogales, J.; Pech-Canul, Á.; Calatrava-Morales, N.; Bernabéu-Roda, L.M.; Durán, P.; Cuéllar, V.; Olivares, J.; Alvarez, L.; Palenzuela-Bretones, D.; et al. 2-Tridecanone impacts surface-associated bacterial behaviours and hinders plant–bacteria interactions. Environ. Microbiol. 2018, 20, 2049–2065. [Google Scholar] [CrossRef]
- Kornberg, A.; Ochoa, S.; Mehler, A.H. Spectrophotometric studies on the decarboxylation of β-keto acids. J. Biol. Chem. 1948, 174, 159–172. [Google Scholar]
- Benning, M.M.; Wesenberg, G.; Liu, R.; Taylor, K.L.; Dunaway-Mariano, D.; Holden, H.M. The three-dimensional structure of 4-hydroxybenzoyl-CoA thioesterase from Pseudomonas sp. strain CBS-3. J. Biol. Chem. 1998, 273, 33572–33579. [Google Scholar] [CrossRef] [PubMed]
- Thoden, J.B.; Holden, H.M.; Zhuang, Z.; Dunaway-Mariano, D. X-ray crystallographic analyses of inhibitor and substrate complexes of wild-type and mutant 4-hydroxybenzoyl-CoA thioesterase. J. Biol. Chem. 2002, 277, 27468–27476. [Google Scholar] [CrossRef]
- Shahmuradov, I.A.; Umarov, R.K.; Solovyev, V.V. TSSPlant: A new tool for prediction of plant Pol II promoters. Nucl. Acids Res. 2017, 45, gkw1353. [Google Scholar] [CrossRef] [PubMed]
- Zuo, Y.C.; Li, Q.Z. Identification of TATA and TATA-less promoters in plant genomes by integrating diversity measure, GC-Skew and DNA geometric flexibility. Genomics 2011, 97, 112–120. [Google Scholar] [CrossRef] [PubMed]
- Higo, K.; Ugawa, Y.; Iwamoto, M.; Korenaga, T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucl. Acids Res. 1999, 27, 297–300. [Google Scholar] [CrossRef] [PubMed]
- Zolotarov, Y.; Strömvik, M. De Novo Regulatory Motif Discovery Identifies Significant Motifs in Promoters of Five Classes of Plant Dehydrin Genes. PLoS ONE 2015, 10, e0129016. [Google Scholar] [CrossRef] [PubMed]
- Ciolkowski, I.; Wanke, D.; Birkenbihl, R.P.; Somssich, I.E. Studies on DNA-binding selectivity of WRKY transcription factors lend structural clues into WRKY-domain function. Plant. Mol. Biol. 2008, 68, 81–92. [Google Scholar] [CrossRef]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. WRKY transcription factors. Trends Plant. Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef]
- Khuat, V.L.U.; Bui, V.T.T.; Tran, H.T.D.; Truong, N.X.; Nguyen, T.C.; Mai, P.H.H.; Dang, T.L.A.; Dinh, H.M.; Pham, H.T.A.; Nguyen, T.T.H. Characterization of Solanum melongena Thioesterases Related to Tomato Methylketone Synthase 2. Genes 2019, 10, 549. [Google Scholar] [CrossRef]
- Jing, F.; Yandeau-Nelson, M.D.; Nikolau, B.J. Identification of active site residues implies a two-step catalytic mechanism for acyl-ACP thioesterase. Biochem. J. 2018, 475, 3861–3873. [Google Scholar] [CrossRef]
- Fridman, E.; Wang, J.; Iijima, Y.; Froehlich, J.E.; Gang, D.R.; Ohlrogge, J.; Pichersky, E. Metabolic, genomic, and biochemical analyses of glandular trichomes from the wild tomato species Lycopersicon hirsutum identify a key enzyme in the biosynthesis of methylketones. Plant. Cell 2005, 17, 1252–1267. [Google Scholar] [CrossRef]
- Cronan, J.E.; Birge, C.H.; Vagelos, P.R. Evidence for two genes specifically involved in unsaturated fatty acid biosynthesis in Escherichia coli. J. Bacteriol. 1969, 100, 601–604. [Google Scholar] [PubMed]
- Cronan, J.E. A bacterium that has three pathways to regulate membrane lipid fluidity. Mol. Microbiol. 2006, 60, 256–259. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Cronan, J.E. Escherichia coli Unsaturated Fatty Acid Synthesis. J. Biol. Chem. 2009, 284, 29526–29535. [Google Scholar] [CrossRef] [PubMed]
- Kordali, S.; Kotan, R.; Mavi, A.; Cakir, A.; Ala, A.; Yildirim, A. Determination of the Chemical Composition and Antioxidant Activity of the Essential Oil of Artemisia dracunculus and of the Antifungal and Antibacterial Activities of Turkish Artemisia absinthium, A. dracunculus, Artemisia santonicum, and Artemisia spi. essential oils. J. Agric. Food Chem. 2005, 53, 9452–9458. [Google Scholar] [PubMed]
- Van Poecke, R.M.P.; Posthumus, M.A.; Dicke, M. Herbivore-induced volatile production by Arabidopsis thaliana leads to attraction of the parasitoid Cotesia rubecula: Chemical, behavioral, and gene-expression analysis. J. Chem. Ecol. 2001, 27, 1911–1928. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Chen, C.; Chen, Z. Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant. Mol. Biol. 2003, 51, 21–37. [Google Scholar] [CrossRef]
- Kalde, M.; Barth, M.; Somssich, I.E.; Lippok, B. Members of the Arabidopsis WRKY Group III Transcription Factors Are Part of Different Plant Defense Signaling Pathways. Mol. Plant-Microbe Interact. 2003, 16, 295–305. [Google Scholar] [CrossRef]
- Eulgem, T.; Somssich, I.E. Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant. Biol. 2007, 10, 366–371. [Google Scholar] [CrossRef]
- Dellagi, A.; Heilbronn, J.; Avrova, A.O.; Montesano, M.; Palva, E.T.; Stewart, H.E.; Toth, I.K.; Cooke, D.E.L.; Lyon, G.D.; Birch, P.R.J. A Potato Gene Encoding a WRKY-like Transcription Factor Is Induced in Interactions with Erwinia carotovora subsp. atroseptica and Phytophthora infestans and Is Coregulated with Class I Endochitinase Expression. Mol. Plant-Microbe Interact. 2000, 13, 1092–1101. [Google Scholar] [CrossRef]
- Laloi, C.; Mestres-Ortega, D.; Marco, Y.; Meyer, Y.; Reichheld, J.-P. The Arabidopsis Cytosolic Thioredoxin h 5 Gene Induction by Oxidative Stress and Its W-Box-Mediated Response to Pathogen Elicitor. Plant. Physiol. 2004, 134, 1006–1016. [Google Scholar] [CrossRef] [PubMed]
- Foster, R.; Izawa, T.; Chua, N.H. Plant bZIP proteins gather at ACGT elements. FASEB J. 1994, 8, 192–200. [Google Scholar] [CrossRef] [PubMed]
- Toledo-Ortiz, G.; Huq, E.; Quail, P.H. The Arabidopsis Basic/Helix-Loop-Helix Transcription Factor Family. Plant. Cell 2003, 15, 1749–1770. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Zou, H.F.; Wei, W.; Hao, Y.J.; Tian, A.G.; Huang, J.; Liu, Y.F.; Zhang, J.S.; Chen, S.Y. Soybean GmbZIP44, GmbZIP62 and GmbZIP78 genes function as negative regulator of ABA signaling and confer salt and freezing tolerance in transgenic Arabidopsis. Planta 2008, 228, 225–240. [Google Scholar] [CrossRef] [PubMed]
- Uno, Y.; Furihata, T.; Abe, H.; Yoshida, R.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc. Natl. Acad. Sci. USA 2000, 97, 11632–11637. [Google Scholar] [CrossRef] [PubMed]
- Alves, M.; Dadalto, S.; Gonçalves, A.; De Souza, G.; Barros, V.; Fietto, L. Plant bZIP Transcription Factors Responsive to Pathogens: A Review. Int. J. Mol. Sci. 2013, 14, 7815–7828. [Google Scholar] [CrossRef] [PubMed]
- Feller, A.; Machemer, K.; Braun, E.L.; Grotewold, E. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant. J. 2011, 66, 94–116. [Google Scholar] [CrossRef]
- Sun, X.; Wang, Y.; Sui, N. Transcriptional regulation of bHLH during plant response to stress. Biochem. Biophys. Res. Commun. 2018, 503, 397–401. [Google Scholar] [CrossRef]
- Simpson, S.D.; Nakashima, K.; Narusaka, Y.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Two different novel cis-acting elements of erd1, a clpA homologous Arabidopsis gene function in induction by dehydration stress and dark-induced senescence. Plant. J. 2003, 33, 259–270. [Google Scholar] [CrossRef]
- Abe, H.; Urao, T.; Ito, T.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant. Cell 2003, 15, 63–78. [Google Scholar] [CrossRef]
- Nesi, N.; Debeaujon, I.; Jond, C.; Pelletier, G.; Caboche, M.; Lepiniec, L. The TT8 Gene Encodes a Basic Helix-Loop-Helix Domain Protein Required for Expression of DFR and BAN Genes in Arabidopsis Siliques. Plant. Cell 2000, 12, 1863. [Google Scholar] [CrossRef] [PubMed]
- Baudry, A.; Heim, M.A.; Dubreucq, B.; Caboche, M.; Weisshaar, B.; Lepiniec, L. TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant. J. 2004, 39, 366–380. [Google Scholar] [CrossRef] [PubMed]
- Hichri, I.; Heppel, S.C.; Pillet, J.; Léon, C.; Czemmel, S.; Delrot, S.; Lauvergeat, V.; Bogs, J. The Basic Helix-Loop-Helix Transcription Factor MYC1 Is Involved in the Regulation of the Flavonoid Biosynthesis Pathway in Grapevine. Mol. Plant. 2010, 3, 509–523. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.-Y.; Tian, A.-G.; Zou, H.-F.; Xie, Z.-M.; Lei, G.; Huang, J.; Wang, C.-M.; Wang, H.-W.; Zhang, J.-S.; Chen, S.-Y. Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant. Biotech. J. 2008, 6, 486–503. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Wang, P.; Hou, L.; Zhao, S.; Zhao, C.; Xia, H.; Li, P.; Zhang, Y.; Bian, X.; Wang, X. Global Analysis of WRKY Genes and Their Response to Dehydration and Salt Stress in Soybean. Front. Plant. Sci. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Kargiotidou, A.; Deli, D.; Galanopoulou, D.; Tsaftaris, A.; Farmaki, T. Low temperature and light regulate delta 12 fatty acid desaturases (FAD2) at a transcriptional level in cotton (Gossypium hirsutum). J. Exp. Bot. 2008, 59, 2043–2056. [Google Scholar] [CrossRef] [PubMed]
- Ishizaki-Nishizawa, O.; Fujii, T.; Ohtani, T.; Toguri, T. Low-Temperature Resistance of Higher Plants is Significantly Enhanced by a Non-Specific Δ9 Desaturase from Cyanobacteria. In Physiology, Biochemistry and Molecular Biology of Plant Lipids; Springer Netherlands: Dordrecht, The Netherlands, 1997; pp. 212–214. [Google Scholar]
- Gibson, S.; Falcone, D.L.; Browse, J.; Somerville, C. Use of transgenic plants and mutants to study the regulation and function of lipid composition. Plant. Cell Environ. 1994, 17, 627–637. [Google Scholar] [CrossRef]
- Kim, E.; Hwang, S.; Lee, I. SoyNet: A database of co-functional networks for soybean Glycine max. Nucl. Acids Res. 2017, 45, D1082–D1089. [Google Scholar] [CrossRef]
- Laloum, T.; Martín, G.; Duque, P. Alternative Splicing Control of Abiotic Stress Responses. Trends Plant. Sci. 2018, 23, 140–150. [Google Scholar] [CrossRef]
- Chojnacki, S.; Cowley, A.; Lee, J.; Foix, A.; Lopez, R. Programmatic access to bioinformatics tools from EMBL-EBI update: 2017. Nucl. Acids Res. 2017, 45, W550–W553. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evolution. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Ichihara, K.; Fukubayashi, Y. Preparation of fatty acid methyl esters for gas-liquid chromatography. J. Lipid Res. 2010, 51, 635–640. [Google Scholar] [CrossRef] [PubMed]
- Matsuba, Y.; Nguyen, T.T.H.; Wiegert, K.; Falara, V.; Gonzales-Vigil, E.; Leong, B.; Schafer, P.; Kudrna, D.; Wing, R.A.; Bolger, A.M.; et al. Evolution of a Complex Locus for Terpene Biosynthesis in Solanum. Plant. Cell 2013, 25, 2022–2036. [Google Scholar] [CrossRef] [PubMed]
- Yanagisawa, S.; Schmidt, R.J. Diversity and similarity among recognition sequences of Dof transcription factors. Plant. J. 1999, 17, 209–214. [Google Scholar] [CrossRef] [PubMed]
- Yanagisawa, S. Dof1 and Dof2 transcription factors are associated with expression of multiple genes involved in carbon metabolism in maize. Plant. J. 2000, 21, 281–288. [Google Scholar] [CrossRef] [PubMed]
- Vom Endt, D.; Soares e Silva, M.; Kijne, J.W.; Pasquali, G.; Memelink, J. Identification of a Bipartite Jasmonate-Responsive Promoter Element in the Catharanthus roseus ORCA3 Transcription Factor Gene That Interacts Specifically with AT-Hook DNA-Binding Proteins. Plant. Physiol. 2007, 144, 1680–1689. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Chung, H.-J.; Thomas, T.L. Isolation of a novel class of bZIP transcription factors that interact with ABA-responsive and embryo-specification elements in the Dc3 promoter using a modified yeast one-hybrid system. Plant. J. 1997, 11, 1237–1251. [Google Scholar] [CrossRef]
- Hartmann, U.; Sagasser, M.; Mehrtens, F.; Stracke, R.; Weisshaar, B. Differential combinatorial interactions of cis-acting elements recognized by R2R3-MYB, BZIP, and BHLH factors control light-responsive and tissue-specific activation of phenylpropanoid biosynthesis genes. Plant. Mol. Biol. 2005, 57, 155–171. [Google Scholar] [CrossRef]
- Chinnusamy, V. ICE1: A regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 2003, 17, 1043–1054. [Google Scholar] [CrossRef]
- Ross, E.J.H. Activation of the Oryza sativa non-symbiotic haemoglobin-2 promoter by the cytokinin-regulated transcription factor, ARR1. J. Exp. Bot. 2004, 55, 1721–1731. [Google Scholar] [CrossRef]
- Eulgem, T. Early nuclear events in plant defence signalling: Rapid gene activation by WRKY transcription factors. EMBO J. 1999, 18, 4689–4699. [Google Scholar] [CrossRef] [PubMed]
- Mohanty, B.; Krishnan, S.P.T.; Swarup, S.; Bajic, V. Detection and Preliminary Analysis of Motifs in Promoters of Anaerobically Induced Genes of Different Plant Species. Ann. Bot. 2005, 96, 669–681. [Google Scholar] [CrossRef] [PubMed]
- Piechulla, B.; Merforth, N.; Rudolph, B. Identification of tomato Lhc promoter regions necessary for circadian expression. Plant. Mol. Biol. 1998, 38, 655–662. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, M.; Hanada, A.; Yamauchi, Y.; Kuwahara, A.; Kamiya, Y.; Yamaguchi, S. Gibberellin Biosynthesis and Response during Arabidopsis Seed Germination. Plant. Cell 2003, 15, 1591–1604. [Google Scholar] [CrossRef] [PubMed]
- Menkens, A.E.; Schindler, U.; Cashmore, A.R. The G-box: A ubiquitous regulatory DNA element in plants bound by the GBF family of bZIP proteins. Trends Biochem. Sci. 1995, 20, 506–510. [Google Scholar] [CrossRef]
- Hudson, M.E.; Quail, P.H. Identification of Promoter Motifs Involved in the Network of Phytochrome A-Regulated Gene Expression by Combined Analysis of Genomic Sequence and Microarray Data. Plant. Physiol. 2003, 133, 1605–1616. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, K.; Fujita, Y.; Katsura, K.; Maruyama, K.; Narusaka, Y.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Transcriptional Regulation of ABI3- and ABA-responsive Genes Including RD29B and RD29A in Seeds, Germinating Embryos, and Seedlings of Arabidopsis. Plant. Mol. Biol. 2006, 60, 51–68. [Google Scholar] [CrossRef] [PubMed]
- Rushton, P.J.; Torres, J.T.; Parniske, M.; Wernert, P.; Hahlbrock, K.; Somssich, I.E. Interaction of elicitor-induced DNA-binding proteins with elicitor response elements in the promoters of parsley PR1 genes. EMBO J. 1996, 15, 5690–5700. [Google Scholar] [CrossRef] [PubMed]
- Sutoh, K.; Yamauchi, D. Two cis- acting elements necessary and sufficient for gibberellin-upregulated proteinase expression in rice seeds. Plant. J. 2003, 34, 635–645. [Google Scholar] [CrossRef] [PubMed]
- Shirsat, A.; Wilford, N.; Croy, R.; Boulter, D. Sequences responsible for the tissue specific promoter activity of a pea legumin gene in tobacco. MGG Mol. Gen. Genet. 1989, 215, 326–331. [Google Scholar] [CrossRef]
- Tatematsu, K.; Ward, S.; Leyser, O.; Kamiya, Y.; Nambara, E. Identification of cis-Elements That Regulate Gene Expression during Initiation of Axillary Bud Outgrowth in Arabidopsis. Plant. Physiol. 2005, 138, 757–766. [Google Scholar] [CrossRef] [PubMed]
- Teakle, G.R.; Manfield, I.W.; Graham, J.F.; Gilmartin, P.M. Arabidopsis thaliana GATA factors: Organisation, expression and DNA-binding characteristics. Plant. Mol. Biol. 2002, 50, 43–57. [Google Scholar] [CrossRef] [PubMed]
- Xue, G.-P. Characterisation of the DNA-binding profile of barley HvCBF1 using an enzymatic method for rapid, quantitative and high-throughput analysis of the DNA-binding activity. Nucleic Acids Res. 2002, 30, e77. [Google Scholar] [CrossRef] [PubMed]
- Kagaya, Y.; Ohmiya, K.; Hattori, T. RAV1, a novel DNA-binding protein, binds to bipartite recognition sequence through two distinct DNA-binding domains uniquely found in higher plants. Nucleic Acids Res. 1999, 27, 470–478. [Google Scholar] [CrossRef] [PubMed]
- Maeda, K.; Kimura, S.; Demura, T.; Takeda, J.; Ozeki, Y. DcMYB1 Acts as a Transcriptional Activator of the Carrot Phenylalanine Ammonia-lyase Gene (DcPAL1) in Response to Elicitor Treatment, UV-B Irradiation and the Dilution Effect. Plant. Mol. Biol. 2005, 59, 739–752. [Google Scholar] [CrossRef] [PubMed]
- Ellerström, M.; Stålberg, K.; Ezcurra, I.; Rask, L. Functional dissection of a napin gene promoter: Identification of promoter elements required for embryo and endosperm-specific transcription. Plant. Mol. Biol. 1996, 40, 699–709. [Google Scholar]
- Huang, H.; Mizukami, Y.; Hu, Y.; Ma, H. Isolation and characterization of the binding sequences for the product of the Arabidopsis floral homeotic gene AGAMOUS. Nucleic Acids Res. 1993, 21, 4769–4776. [Google Scholar] [CrossRef]
- Ezcurra, I.; Wycliffe, P.; Nehlin, L.; Ellerstrom, M.; Rask, L. Transactivation of the Brassica napus napin promoter by ABI3 requires interaction of the conserved B2 and B3 domains of ABI3 with different cis-elements: B2 mediates activation through an ABRE, whereas B3 interacts with an RY/G-box. Plant. J. 2000, 24, 57–66. [Google Scholar] [CrossRef]
- Haralampidis, K.; Milioni, D.; Rigas, S.; Hatzopoulos, P. Combinatorial Interaction of Cis Elements Specifies the Expression of the Arabidopsis AtHsp90-1 Gene. Plant. Physiol. 2002, 129, 1138–1149. [Google Scholar] [CrossRef]
- Allen, R.D.; Bernier, F.; Lessard, P.A.; Beachy, R.N. Nuclear factors interact with a soybean beta-conglycinin enhancer. Plant. Cell 1989, 1, 623–631. [Google Scholar] [CrossRef]
- Urao, T.; Yamaguchi-Shinozaki, K.; Urao, S.; Shinozaki, K. An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. Plant. Cell 1993, 5, 1529–1539. [Google Scholar] [PubMed]
- Grace, M.L.; Chandrasekharan, M.B.; Hall, T.C.; Crowe, A.J. Sequence and Spacing of TATA Box Elements Are Critical for Accurate Initiation from the β-Phaseolin Promoter. J. Biol. Chem. 2004, 279, 8102–8110. [Google Scholar] [CrossRef] [PubMed]
- Park, H.C.; Kim, M.L.; Kang, Y.H.; Jeon, J.M.; Yoo, J.H.; Kim, M.C.; Park, C.Y.; Jeong, J.C.; Moon, B.C.; Lee, J.H.; et al. Pathogen- and NaCl-Induced Expression of the SCaM-4 Promoter Is Mediated in Part by a GT-1 Box That Interacts with a GT-1-Like Transcription Factor. Plant. Physiol. 2004, 135, 2150–2161. [Google Scholar] [CrossRef] [PubMed]
- Cercos, M.; Gomez-Cadenas, A.; Ho, T.-H.D. Hormonal regulation of a cysteine proteinase gene, EPB-1, in barley aleurone layers: Cis- and trans-acting elements involved in the co-ordinated gene expression regulated by gibberellins and abscisic acid. Plant. J. 1999, 19, 107–118. [Google Scholar] [CrossRef] [PubMed]

| Retention Time (min) | 3-Ketoacids Secreted into Media of E. coli Cells Expressing GmMKS2-X2 | 2-MKs Detected by GC | 2-Methylketone Accumulation (Ng/OD Unit) | |||
|---|---|---|---|---|---|---|
| R1 | R2 | R3 | Mean ± SE | |||
| 4.340 | 3-oxooctanoic acid | 7:0 MK | 619.02 | 547.27 | 576.02 | 580.77 ± 20.85 b |
| 6.707 | 3-oxodecanoic acid | 9:0 MK | 210.98 | 203.95 | 195.00 | 203.31 ± 4.62 c |
| 9.905 | 3-oxododecenoic acid | 11:1 MK | 206.85 | 227.07 | 104.41 | 179.44 ± 37.97 c |
| 10.039 | 3-oxododecanoic acid | 11:0 MK | 521.58 | 529.29 | 467.29 | 506.06 ± 19.51 b |
| 12.159 | 3-oxotetradecenoic acid | 13:1 MK | 5815.96 | 6091.41 | 6161.55 | 6022.97 ± 105.14 a |
| 12.390 | 3-oxotetradecanoic acid | 13:0 MK | 199.25 | 222.36 | 173.19 | 198.27 ± 14.20 c |
| 13.867 | 3-oxohexadecenoic acid | 15:1 MK | 141.92 | 151.64 | 118.91 | 137.49 ± 9.71 c |
| 13.786 | 3-oxohexadecanoic acid | 15:0 MK | 98.18 | 72.29 | 34.47 | 68.31 ± 18.50 c |
| 15.066 | 3-oxooctadecenoic acid | 17:1 MK | 172.27 | 172.08 | 151.65 | 165.33 ± 6.96 c |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tran, H.T.D.; Le, N.T.; Khuat, V.L.U.; Nguyen, T.T.H. Identification and Functional Characterization of a Soybean (Glycine max) Thioesterase that Acts on Intermediates of Fatty Acid Biosynthesis. Plants 2019, 8, 397. https://doi.org/10.3390/plants8100397
Tran HTD, Le NT, Khuat VLU, Nguyen TTH. Identification and Functional Characterization of a Soybean (Glycine max) Thioesterase that Acts on Intermediates of Fatty Acid Biosynthesis. Plants. 2019; 8(10):397. https://doi.org/10.3390/plants8100397
Chicago/Turabian StyleTran, Huong Thi Diem, Nhan Trong Le, Vy Le Uyen Khuat, and Thuong Thi Hong Nguyen. 2019. "Identification and Functional Characterization of a Soybean (Glycine max) Thioesterase that Acts on Intermediates of Fatty Acid Biosynthesis" Plants 8, no. 10: 397. https://doi.org/10.3390/plants8100397
APA StyleTran, H. T. D., Le, N. T., Khuat, V. L. U., & Nguyen, T. T. H. (2019). Identification and Functional Characterization of a Soybean (Glycine max) Thioesterase that Acts on Intermediates of Fatty Acid Biosynthesis. Plants, 8(10), 397. https://doi.org/10.3390/plants8100397




