Characterization of the Soybean GPAT Gene Family Identifies GmGPAT1 as a Key Protein in Salt Stress Tolerance
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Treatments
2.2. Soybean GPAT Gene Family Identification
2.3. GmGPAT Evolution, Gene Structure, and Synteny Analyses
2.4. GmGPAT Promoter Analysis
2.5. Subcellular Localization
2.6. qPCR
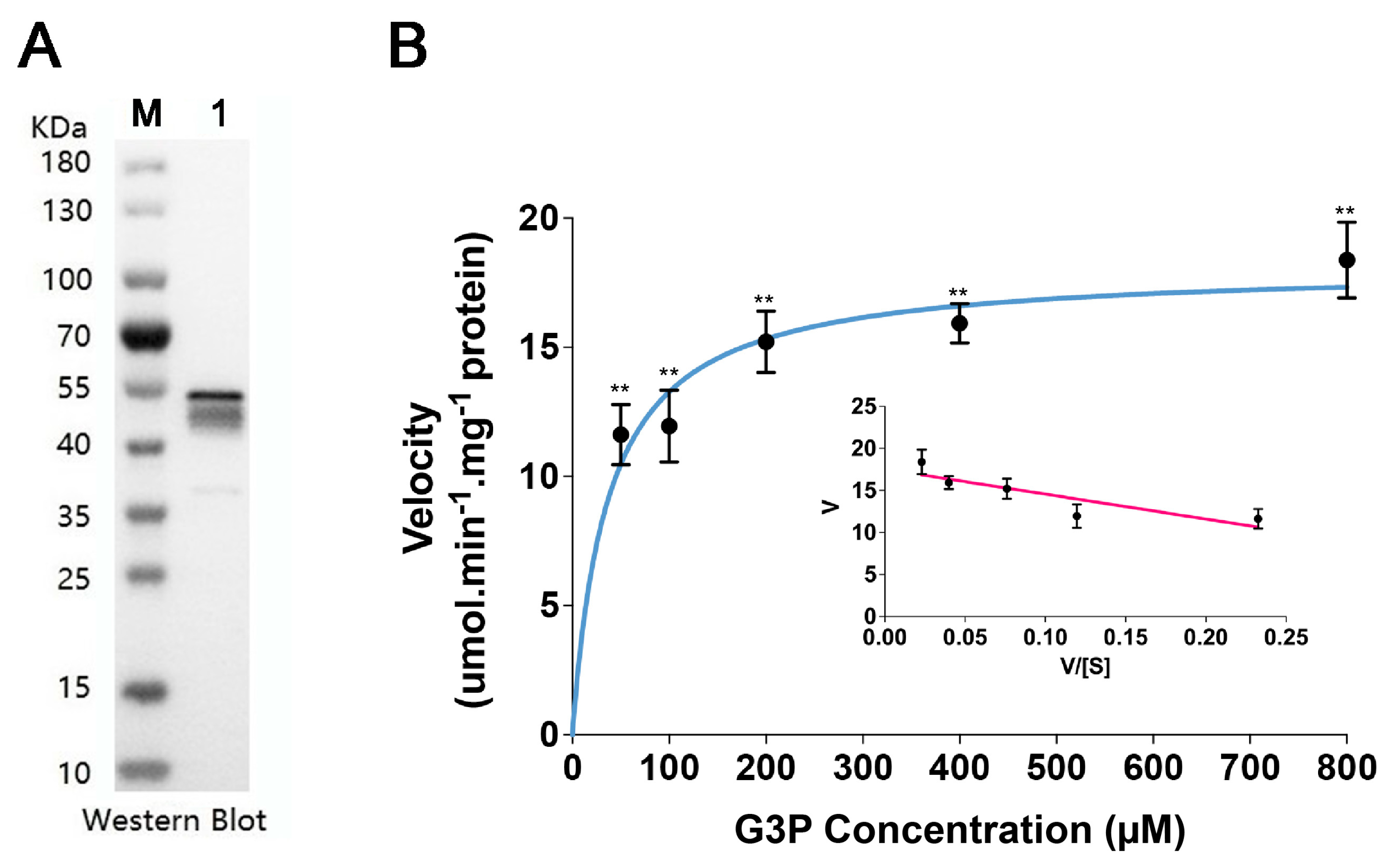
2.7. Recombinant Protein Expression and Enzyme Kinetic Activity Assays
2.8. Agrobacterium-Mediated Overexpression and Knockout of GmGPAT1
2.9. Reactive Oxygen and Antioxidant Enzyme Analyses
2.10. Haplotype Analyses of GmGPAT1
2.11. Statistical Analysis
3. Results
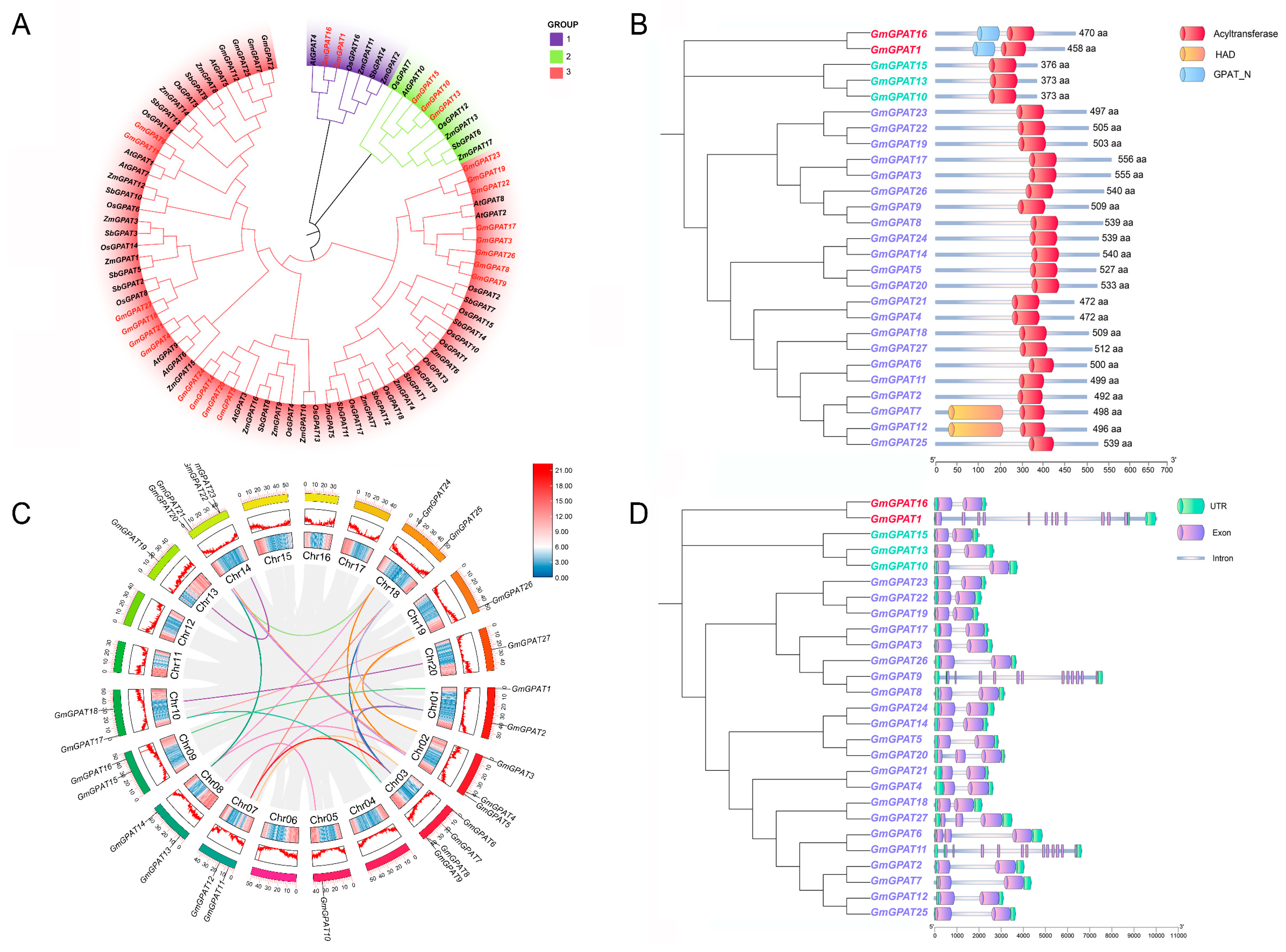
3.1. Soybean GPAT Identification
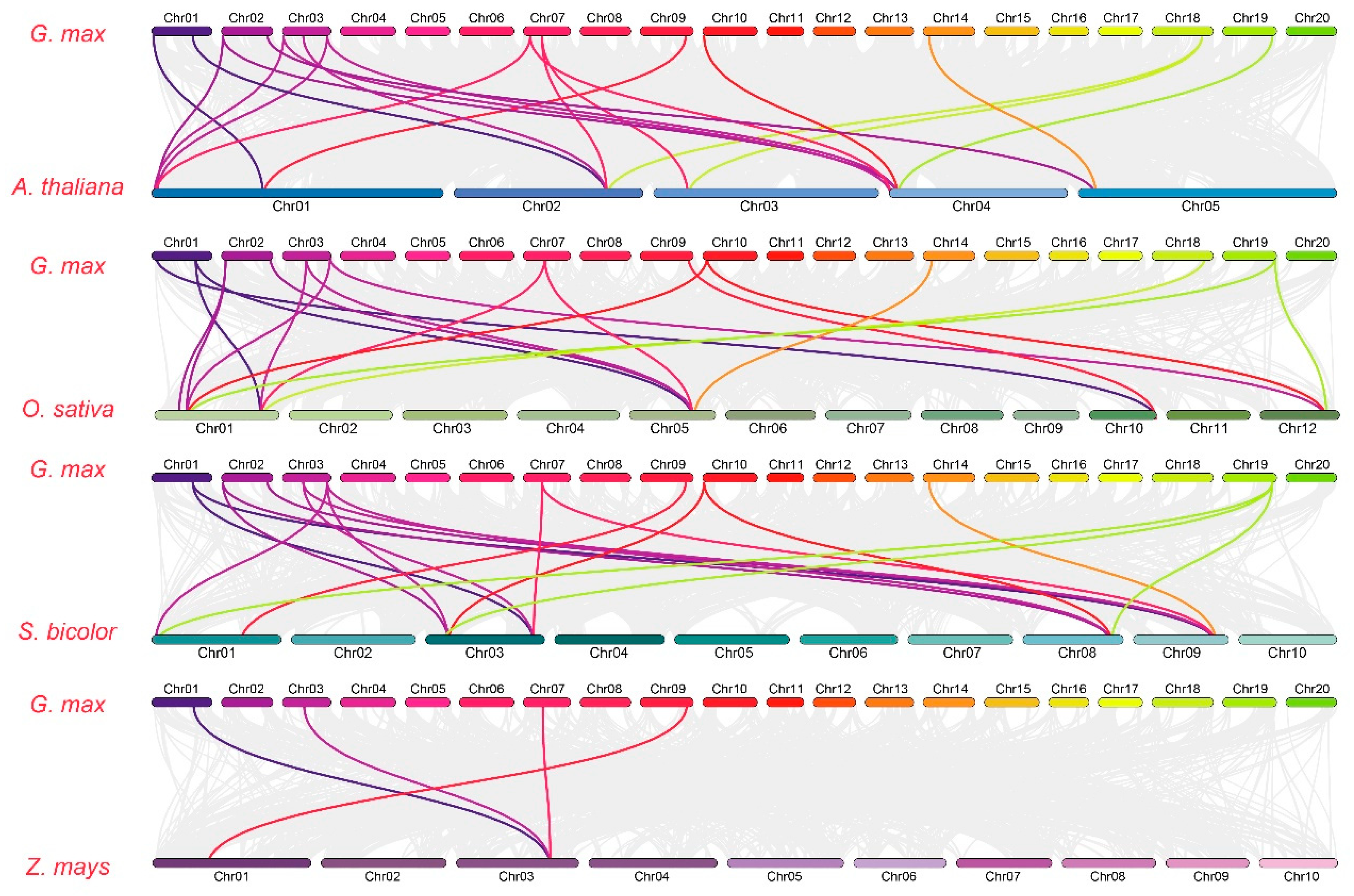
3.2. GmGPAT Synteny and Gene Structure Analyses
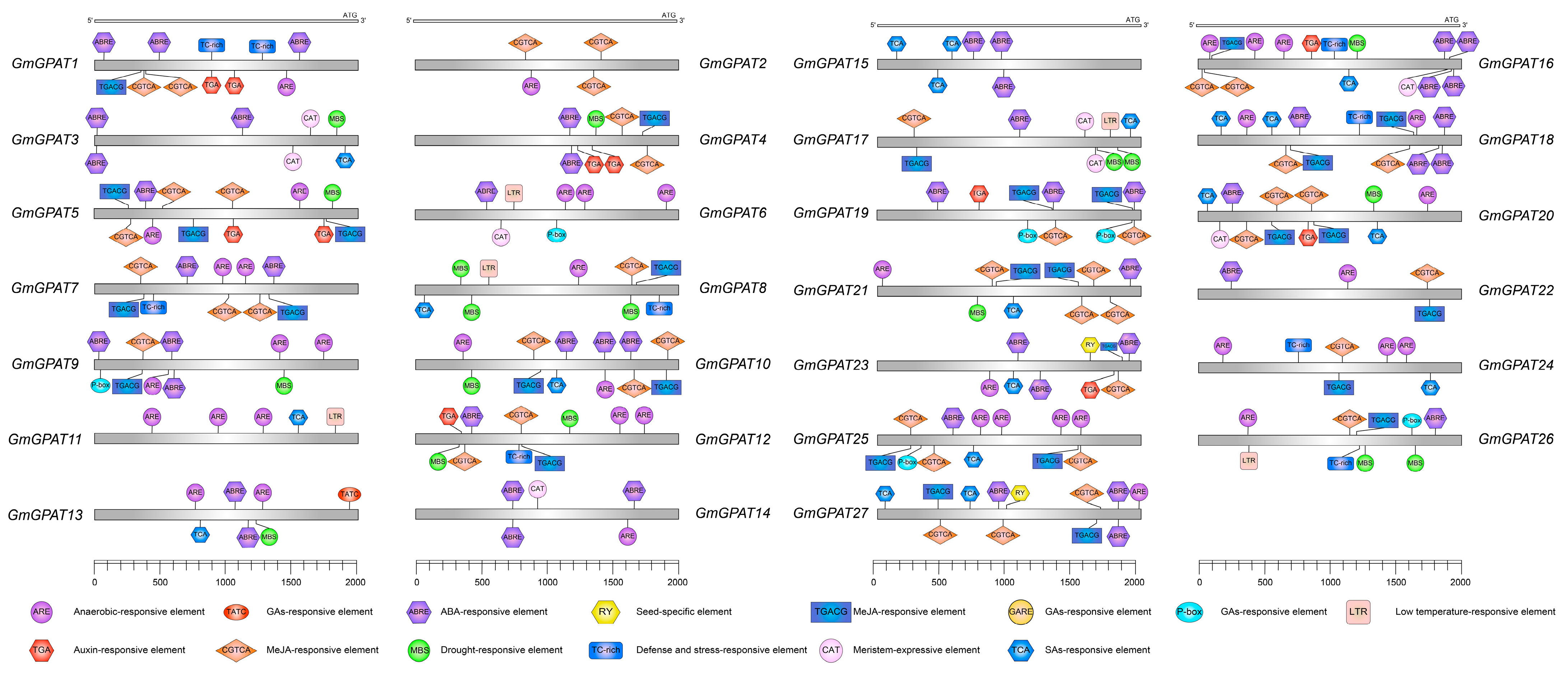
3.3. GmGPAT Promoter Regulatory Element Analysis
3.4. GmGPAT Subcellular Localization Analysis
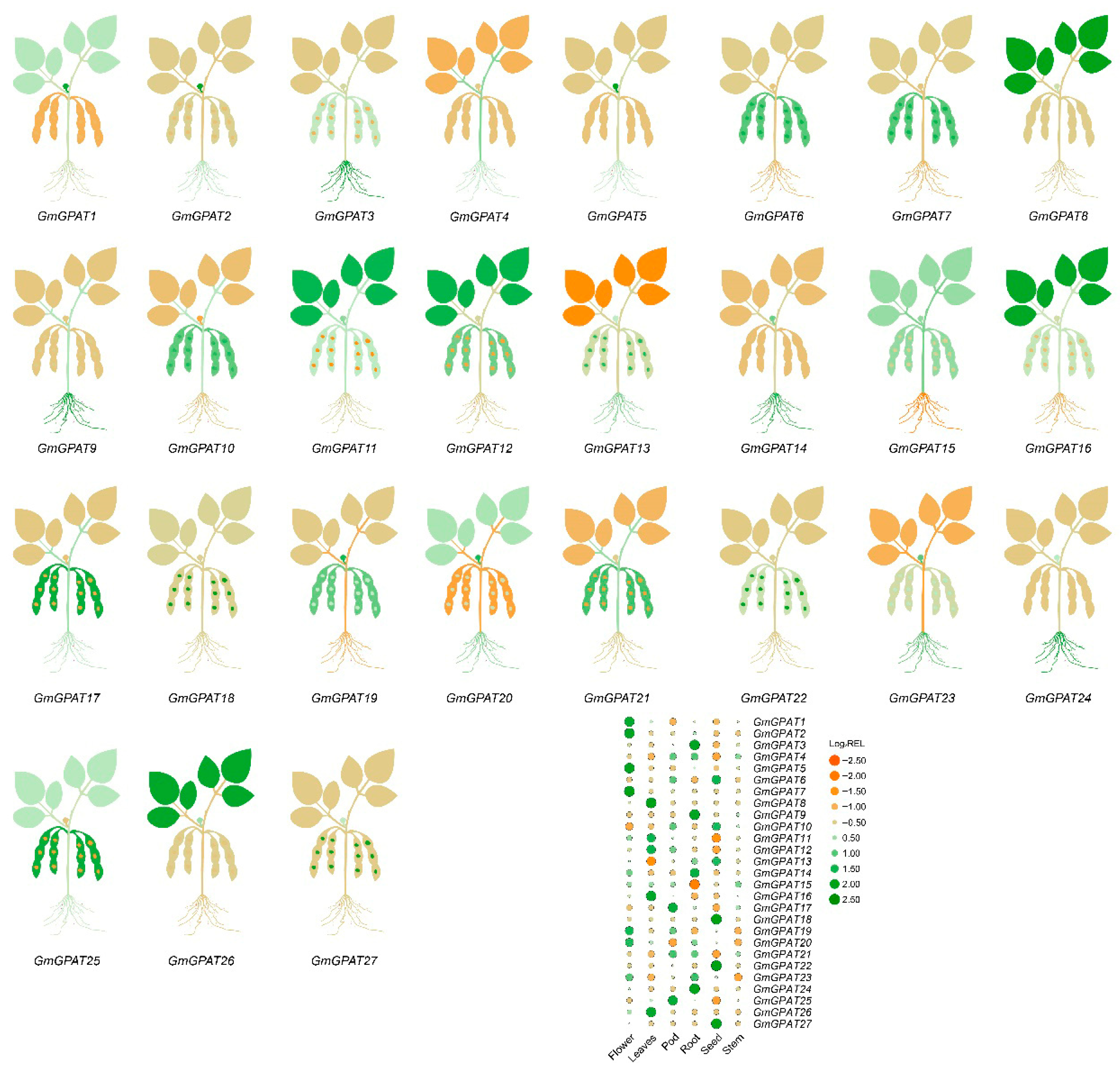
3.5. GmGPAT Tissue-Specific Expression Pattern Analysis
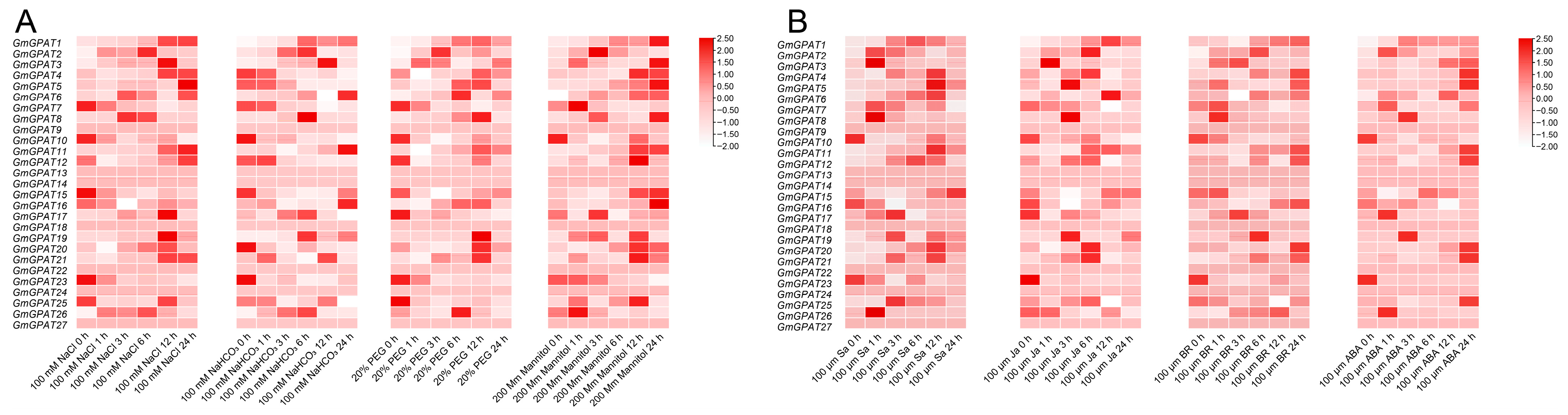
3.6. Analysis of GmGPAT Expression Patterns Under Different Stresses
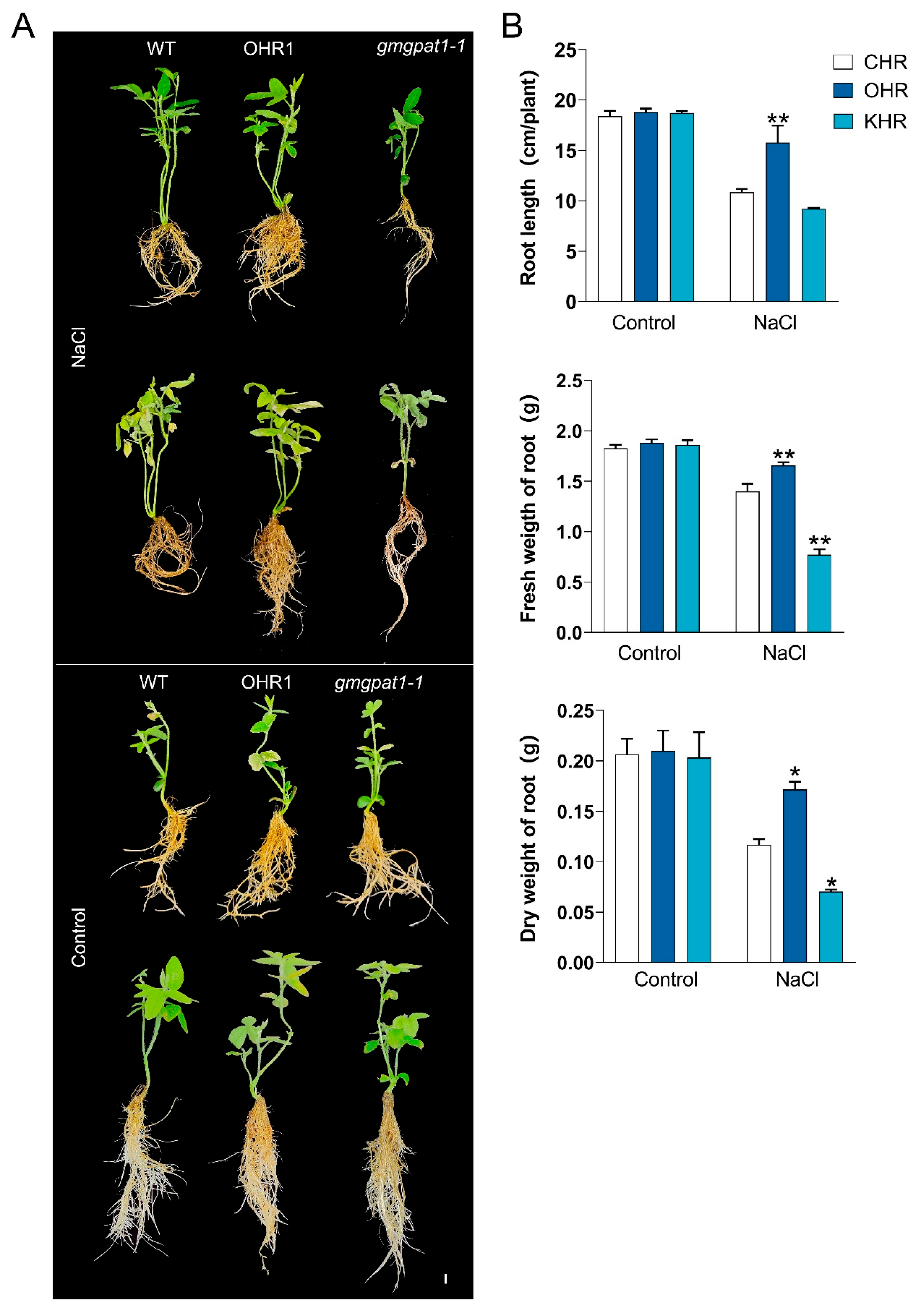
3.7. GmGPAT1 Overexpression Enhances Salt Tolerance
3.8. GmGPAT1 Alleviates Oxidative Stress and Maintains Redox Homeostasis Under Salt Stress
3.9. GmGPAT1 Haplotype Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zou, J.; Wei, Y.; Jako, C.; Kumar, A.; Selvaraj, G.; Taylor, D.C. The Arabidopsis thaliana TAG1 mutant has a mutation in a diacylglycerol acyltransferase gene. Plant J. 1999, 19, 645–653. [Google Scholar] [CrossRef] [PubMed]
- Wendel, A.A.; Lewin, T.M.; Coleman, R.A. Glycerol-3-phosphate acyltransferases: Rate limiting enzymes of triacylglycerol biosynthesis. Biochim. Biophys. Acta 2009, 1791, 501–506. [Google Scholar] [CrossRef]
- Hurlock, A.K.; Roston, R.L.; Wang, K.; Benning, C. Lipid trafficking in plant cells. Traffic 2014, 15, 915–932. [Google Scholar] [CrossRef]
- Zhou, X.R.; Bhandari, S.; Johnson, B.S.; Kotapati, H.K.; Allen, D.K.; Vanhercke, T.; Bates, P.D. Reorganization of Acyl Flux through the Lipid Metabolic Network in Oil-Accumulating Tobacco Leaves. Plant Physiol. 2020, 182, 739–755. [Google Scholar] [CrossRef]
- Zhang, M.; Fan, J.; Taylor, D.C.; Ohlrogge, J.B. DGAT1 and PDAT1 acyltransferases have overlapping functions in Arabidopsis triacylglycerol biosynthesis and are essential for normal pollen and seed development. Plant Cell 2009, 21, 3885–3901. [Google Scholar] [CrossRef]
- Xu, X.; Yan, B.; Zhao, Y.; Wang, F.; Zhao, X.; He, L.; Xu, J.; Zhao, C. Characterization and expression analysis of GPAT gene family in maize. Can. J. Plant Sci. 2019, 99, 577–588. [Google Scholar] [CrossRef]
- Waschburger, E.; Kulcheski, F.R.; Veto, N.M.; Margis, R.; Margis-Pinheiro, M.; Turchetto-Zolet, A.C. Genome-wide analysis of the Glycerol-3-Phosphate Acyltransferase (GPAT) gene family reveals the evolution and diversification of plant GPATs. Genet. Mol. Biol. 2018, 41 (Suppl. 1), 355–370. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.; Wu, S.; Zhang, D.; Li, Z.; Xie, K.; An, X.; Ma, B.; Hou, Q.; Dong, Z.; Tian, Y.; et al. Genome-wide analysis of maize GPAT gene family and cytological characterization and breeding application of ZmMs33/ZmGPAT6 gene. Theor. Appl. Genet. 2019, 132, 2137–2154. [Google Scholar] [CrossRef]
- Men, X.; Shi, J.; Liang, W.; Zhang, Q.; Lian, G.; Quan, S.; Zhu, L.; Luo, Z.; Chen, M.; Zhang, D. Glycerol-3-Phosphate Acyltransferase 3 (OsGPAT3) is required for anther development and male fertility in rice. J. Exp. Bot. 2017, 68, 513–526. [Google Scholar] [CrossRef]
- Zhou, Y.; Huang, X.; Hu, T.; Chen, S.; Wang, Y.; Shi, X.; Yin, M.; Li, R.; Wang, J.; Jia, X. Genome-Wide Analysis of Glycerol-3-Phosphate Acyltransferase (GPAT) Family in Perilla frutescens and Functional Characterization of PfGPAT9 Crucial for Biosynthesis of Storage Oils Rich in High-Value Lipids. Int. J. Mol. Sci. 2023, 24, 15106. [Google Scholar] [CrossRef]
- Liu, F.; Xia, Y.; Wu, L.; Fu, D.; Hayward, A.; Luo, J.; Yan, X.; Xiong, X.; Fu, P.; Wu, G.; et al. Enhanced seed oil content by overexpressing genes related to triacylglyceride synthesis. Gene 2015, 557, 163–171. [Google Scholar] [CrossRef]
- Payá-Milans, M.; Venegas-Calerón, M.; Salas, J.J.; Garcés, R.; Martínez-Force, E. Cloning, heterologous expression and biochemical characterization of plastidial sn-glycerol-3-phosphate acyltransferase from Helianthus annuus. Phytochemistry 2015, 111, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Sui, N.; Li, M.; Zhao, S.J.; Li, F.; Liang, H.; Meng, Q.W. Overexpression of glycerol-3-phosphate acyltransferase gene improves chilling tolerance in tomato. Planta 2007, 226, 1097–1108. [Google Scholar] [CrossRef]
- Lv, Y.; Zhang, X.; Luo, L.; Yang, H.; Li, P.; Zhang, K.; Liu, F.; Wan, Y. Characterization of glycerol-3-phosphate acyltransferase 9 (AhGPAT9) genes, their allelic polymorphism and association with oil content in peanut (Arachis hypogaea L.). Sci. Rep. 2020, 10, 14648. [Google Scholar] [CrossRef]
- Cui, Y.; Ma, J.; Liu, G.; Wang, N.; Pei, W.; Wu, M.; Li, X.; Zhang, J.; Yu, J. Genome-Wide Identification, Sequence Variation, and Expression of the Glycerol-3-Phosphate Acyltransferase (GPAT) Gene Family in Gossypium. Front. Genet. 2019, 10, 116. [Google Scholar] [CrossRef]
- Liu, H.; Wei, L.; Zhu, J.; Zhang, B.; Gan, Y.; Zheng, Y. Identification of GmGPATs and their effect on glycerolipid biosynthesis through seed-specific expression in soybean. Mol. Biol. Rep. 2022, 49, 9585–9592. [Google Scholar] [CrossRef]
- Muñoz, C.F.; Weusthuis, R.A.; D’Adamo, S.; Wijffels, R.H. Effect of Single and Combined Expression of Lysophosphatidic Acid Acyltransferase, Glycerol-3-Phosphate Acyltransferase, and Diacylglycerol Acyltransferase on Lipid Accumulation and Composition in Neochloris oleoabundans. Front. Plant Sci. 2019, 10, 1573. [Google Scholar] [CrossRef] [PubMed]
- Li, X.C.; Zhu, J.; Yang, J.; Zhang, G.R.; Xing, W.F.; Zhang, S.; Yang, Z.N. Glycerol-3-phosphate acyltransferase 6 (GPAT6) is important for tapetum development in Arabidopsis and plays multiple roles in plant fertility. Mol. Plant 2012, 5, 131–142. [Google Scholar] [CrossRef]
- Chen, X.; Chen, G.; Truksa, M.; Snyder, C.L.; Shah, S.; Weselake, R.J. Glycerol-3-phosphate acyltransferase 4 is essential for the normal development of reproductive organs and the embryo in Brassica napus. J. Exp. Bot. 2014, 65, 4201–4215. [Google Scholar] [CrossRef]
- Ouyang, L.L.; Li, H.; Yan, X.J.; Xu, J.L.; Zhou, Z.G. Site-Directed Mutagenesis from Arg195 to His of a Microalgal Putatively Chloroplastidial Glycerol-3-Phosphate Acyltransferase Causes an Increase in Phospholipid Levels in Yeast. Front. Plant Sci. 2016, 7, 286. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Yang, J.; Yang, X.; Cao, Z.; Cai, S.; Wang, B.; Ye, J.; Fu, M.; Zhang, W.; Rao, S. Transcriptome and miRNA sequencing analyses reveal the regulatory mechanism of α-linolenic acid biosynthesis in Paeonia rockii. Food Res. Int. 2022, 155, 111094. [Google Scholar] [CrossRef]
- Sui, N.; Tian, S.; Wang, W.; Wang, M.; Fan, H. Overexpression of Glycerol-3-Phosphate Acyltransferase from Suaeda salsa Improves Salt Tolerance in Arabidopsis. Front. Plant Sci. 2017, 8, 1337. [Google Scholar] [CrossRef]
- Sui, N.; Li, M.; Shu, D.F.; Zhao, S.J.; Meng, Q.W. Antisense-mediated depletion of tomato chloroplast glycerol-3-phosphate acyltransferase affects male fertility and increases thermal tolerance. Physiol. Plant. 2007, 130, 301–314. [Google Scholar] [CrossRef]
- Sun, S.K.; Yang, N.N.; Chen, L.J.; Irfan, M.; Zhao, X.H.; Li, T.L. Characterization of LpGPAT gene in Lilium pensylvanicum and response to cold stress. Biomed Res. Int. 2015, 2015, 792819. [Google Scholar] [CrossRef]
- Wu, F.; Chen, Z.; Zhang, F.; Zheng, H.; Li, S.; Gao, Y.; Yang, J.; Sui, N. Identification and Transcriptome Analysis of Genes Related to Membrane Lipid Regulation in Sweet Sorghum under Salt Stress. Int. J. Mol. Sci. 2022, 23, 5465. [Google Scholar] [CrossRef]
- Yu, C.S.; Lin, C.J.; Hwang, J.K. Predicting subcellular localization of proteins for Gram-negative bacteria by support vector machines based on n-peptide compositions. Protein Sci. 2004, 13, 1402–1406. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.H.; Tang, H.; Wang, X.; Paterson, A.H. PGDD: A database of gene and genome duplication in plants. Nucleic Acids Res. 2013, 41, D1152–D1158. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Liu, W.; Xie, Y.; Ma, J.; Luo, X.; Nie, P.; Zuo, Z.; Lahrmann, U.; Zhao, Q.; Zheng, Y.; Zhao, Y. IBS: An illustrator for the presentation and visualization of biological sequences. Bioinformatics 2015, 31, 3359–3361. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.D.; Cho, Y.H.; Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Zhao, Y.; Cao, P.; Cui, Y.; Liu, D.; Li, J.; Zhao, Y.; Yang, S.; Zhang, B.; Zhou, R.; Sun, M. Enhanced production of seed oil with improved fatty acid composition by overexpressing NAD(+) -dependent glycerol-3-phosphate dehydrogenase in soybean. J. Integr. Plant Biol. 2021, 63, 1036–1053. [Google Scholar] [CrossRef]
- Tóth, K.; Batek, J.; Stacey, G. Generation of soybean (Glycine max) transient transgenic roots. Curr. Protoc. Plant Biol. 2016, 1, 1–13. [Google Scholar] [CrossRef]
- Fryer, M.J.; Oxborough, K.; Mullineaux, P.M.; Baker, N.R. Imaging of photo-oxidative stress responses in leaves. J. Exp. Bot. 2002, 53, 1249–1254. [Google Scholar] [CrossRef]
- Velikova, V.; Yordanov, I.; Edreva, A. Oxidative stress and some antioxidant systems in acid rain-treated bean plants: Protective role of exogenous polyamines. Plant Sci. 2000, 151, 59–66. [Google Scholar] [CrossRef]
- Giannopolitis, C.N.; Ries, S.K. Superoxide dismutase. I. occurrence in higher plants. J. Plant Physiol. 1977, 59, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Nakano, Y.; Asada, K. Hydrogen peroxide is scavenged by ascorbate-specific peroxidase in spinach chloroplasts. Plant Cell Physiol. 1981, 22, 867–880. [Google Scholar] [CrossRef]
- Zhang, R.; Hussain, S.; Wang, Y.; Liu, Y.; Li, Q.; Chen, Y.; Wei, H.; Gao, P.; Dai, Q. Comprehensive Evaluation of Salt Tolerance in Rice (Oryza sativa L.) Germplasm at the Germination Stage. Agronomy 2021, 11, 1569. [Google Scholar] [CrossRef]
- Han, R.; Lu, X.; Gao, G. Analysis of the Principal Components and the Subordinate Function of Alfalfa Drought Resistance. Acta Agrestia Sin. 2006, 14, 142–146. [Google Scholar]
- Dai, H.; Wu, H.; Amanguli, M.; Wang, L.; Maimaiti, A.; Zhang, J. Analysis of Salt-Tolerance and Determination of Salt-Tolerant Evaluation Indicators in Cotton Seedlings of Different Genotypes. Sci. Agric. Sin. 2014, 47, 1290–1300. [Google Scholar]
- Librado, P.; Rozas, J. DnaSP v5: A Software for Comprehensive Analysis of DNA Polymorphism Data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Berkemeyer, M.; Scheibe, R.; Ocheretina, O. A novel, non-redox-regulated NAD-dependent malate dehydrogenase from chloroplasts of Arabidopsis thaliana L. J. Biol. Chem. 1998, 273, 27927–27933. [Google Scholar] [CrossRef]
- Selinski, J.; König, N.; Wellmeyer, B.; Hanke, G.T.; Linke, V.; Neuhaus, H.E.; Scheibe, R. The plastid-localized NAD-dependent malate dehydrogenase is crucial for energy homeostasis in developing Arabidopsis thaliana seeds. Mol. Plant 2014, 7, 170–186. [Google Scholar] [CrossRef]
- Sew, Y.S.; Ströher, E.; Fenske, R.; Millar, A.H. Loss of mitochondrial malate dehydrogenase activity alters seed metabolism lmpairing seed maturation and post-germination growth in Arabidopsis. Plant Physiol. 2016, 171, 849–863. [Google Scholar]
- Wang, Q.J.; Sun, H.; Dong, Q.L.; Sun, T.Y.; Jin, Z.X.; Hao, Y.J.; Yao, Y.X. The enhancement of tolerance to salt and cold stresses by modifying the redox state and salicylic acid content via the cytosolic malate dehydrogenase gene in transgenic apple plants. Plant Biotechnol. J. 2016, 14, 1986–1997. [Google Scholar] [CrossRef]
- Qi, J.; Song, C.P.; Wang, B.; Zhou, J.; Kangasjärvi, J.; Zhu, J.K.; Hao, Y.J.; Yao, Y.X. Reactive oxygen species signaling and stomatal movement in plant responses to drought stress and pathogen attack. J. Integr. Plant Biol. 2018, 60, 805–826. [Google Scholar] [CrossRef]
- Bournonville, C.F.; Díaz-Ricci, J.C. Quantitative determination of superoxide in plant leaves using a modified NBT staining method. Phytochem. Anal. PCA 2011, 22, 268–271. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.Y.; Zhang, H.J.; Zheng, Y.Q.; Cong, Y.Q.; Liu, C.T.; Fan, F.; Zheng, C.; Yuan, G.L.; Pan, G.; Yuan, D.Y. Transcription factor OsMADS25 improves rice tolerance to cold stress. Yi Chuan 2021, 43, 1078–1087. [Google Scholar]
- Vijayaraghavareddy, P.; Adhinarayanreddy, V.; Vemanna, R.S.; Sreeman, S.; Makarla, U. Quantification of Membrane Damage/Cell Death Using Evan’s Blue Staining Technique. Bio-Protoc. 2017, 7, e2519. [Google Scholar]
- Cao, D.; Li, Y.; Liu, B.; Kong, F.; Tran, L.S.P. Adaptive mechanisms of soybean grown on salt-affected soils. Land Degrad. Dev. 2018, 29, 1054–1064. [Google Scholar] [CrossRef]
- Jia, Q.; Bai, Y.; Xu, H.; Liu, Q.; Li, W.; Li, T.; Lin, F.; Shen, L.; Xuan, W.; Zhang, W. Mitochondrial GPAT-derived LPA controls auxin-dependent embryonic and postembryonic development. Proc. Natl. Acad Sci. USA 2022, 119, e2212881119. [Google Scholar] [CrossRef] [PubMed]
- Payá-Milans, M.; Aznar-Moreno, J.A.; Balbuena, T.S.; Haslam, R.P.; Gidda, S.K.; Pérez-Hormaeche, J.; Mullen, R.T.; Thelen, J.J.; Napier, J.A.; Salas, J.J. Sunflower HaGPAT9-1 is the predominant GPAT during seed development. Plant Sci. 2016, 252, 42–52. [Google Scholar] [CrossRef]
- Yang, C.; Ma, J.; Qi, C.; Ma, Y.; Xiong, H.; Duan, R. Genome-Wide Identification, Characterization, Evolutionary Analysis, and Expression Pattern of the GPAT Gene Family in Barley and Functional Analysis of HvGPAT18 under Abiotic Stress. Int. J. Mol. Sci. 2024, 25, 6101. [Google Scholar] [CrossRef]
- Zhang, X.; Gao, H.; Liu, Y.; Zhao, H.; Lü, S. Function identification of Arabidopsis GPAT4 and GPAT8 in the biosynthesis of suberin and cuticular wax. Plant Sci. 2024, 339, 111933. [Google Scholar] [CrossRef]
- Tamada, T.; Feese, M.D.; Ferri, S.R.; Kato, Y.; Yajima, R.; Toguri, T.; Kuroki, R. Substrate recognition and selectivity of plant glycerol-3-phosphate acyltransferases (GPATs) from Cucurbita moscata and Spinacea oleracea. Acta Crystallogr. D Biol. Crystallogr. 2004, 60 Pt 2, 13–21. [Google Scholar] [CrossRef]
- Zhu, G.; Gao, W.; Song, X.; Sun, F.; Hou, S.; Liu, N.; Huang, Y.; Zhang, D.; Ni, Z.; Chen, Q. Genome-wide association reveals genetic variation of lint yield components under salty field conditions in cotton (Gossypium hirsutum L.). BMC Plant Biol. 2020, 20, 23. [Google Scholar] [CrossRef]
- Chu, S.; Zhang, X.; Yu, K.; Lv, L.; Sun, C.; Liu, X.; Zhang, J.; Jiao, Y.; Zhang, D. Genome-Wide Analysis Reveals Dynamic Epigenomic Differences in Soybean Response to Low-Phosphorus Stress. Int. J. Mol. Sci. 2020, 21, 6817. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Li, Y.; Sun, Y.; Li, S.; Cai, Q.; Li, S.; Sun, M.; Yu, T.; Meng, X.; Zhang, J. Characterization of the Soybean GPAT Gene Family Identifies GmGPAT1 as a Key Protein in Salt Stress Tolerance. Plants 2025, 14, 2862. https://doi.org/10.3390/plants14182862
Li X, Li Y, Sun Y, Li S, Cai Q, Li S, Sun M, Yu T, Meng X, Zhang J. Characterization of the Soybean GPAT Gene Family Identifies GmGPAT1 as a Key Protein in Salt Stress Tolerance. Plants. 2025; 14(18):2862. https://doi.org/10.3390/plants14182862
Chicago/Turabian StyleLi, Xin, Yunlong Li, Yan Sun, Sinan Li, Quan Cai, Shujun Li, Minghao Sun, Tao Yu, Xianglong Meng, and Jianguo Zhang. 2025. "Characterization of the Soybean GPAT Gene Family Identifies GmGPAT1 as a Key Protein in Salt Stress Tolerance" Plants 14, no. 18: 2862. https://doi.org/10.3390/plants14182862
APA StyleLi, X., Li, Y., Sun, Y., Li, S., Cai, Q., Li, S., Sun, M., Yu, T., Meng, X., & Zhang, J. (2025). Characterization of the Soybean GPAT Gene Family Identifies GmGPAT1 as a Key Protein in Salt Stress Tolerance. Plants, 14(18), 2862. https://doi.org/10.3390/plants14182862





