Environmental Factors, Developmental Genes and Oxidative Stress Determine Inter-Species Variability in Seed Longevity in Salicaceae
Abstract
1. Introduction
2. Results
2.1. Germination Decreased with Seed Dispersal
2.2. Decreased Storage Temperature and Humidity Increased Seed Longevity in Seven Salix Species
2.3. Decreased Storage Temperature and Humidity Increased Seed Longevity in Six Populus Species
2.4. Salix and Populus Seeds Maintain High Viability After Three Years Storage at Both −20 °C and in LN
2.5. Correlation Studies Between Seed Longevity P50 and Meteorological Data
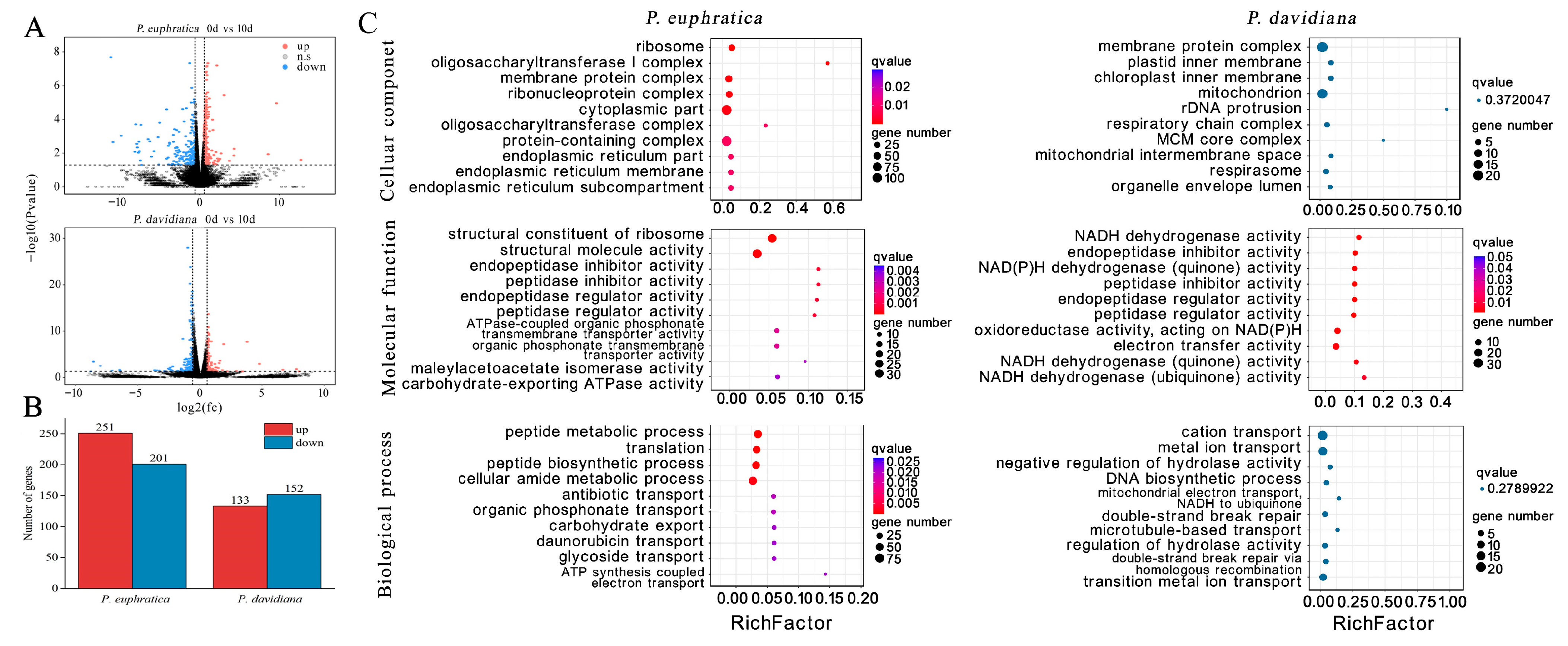
2.6. Transcriptome Analysis of P. euphratica (S2) (Shorter-Lived) and P. davidiana (Longer-Lived)
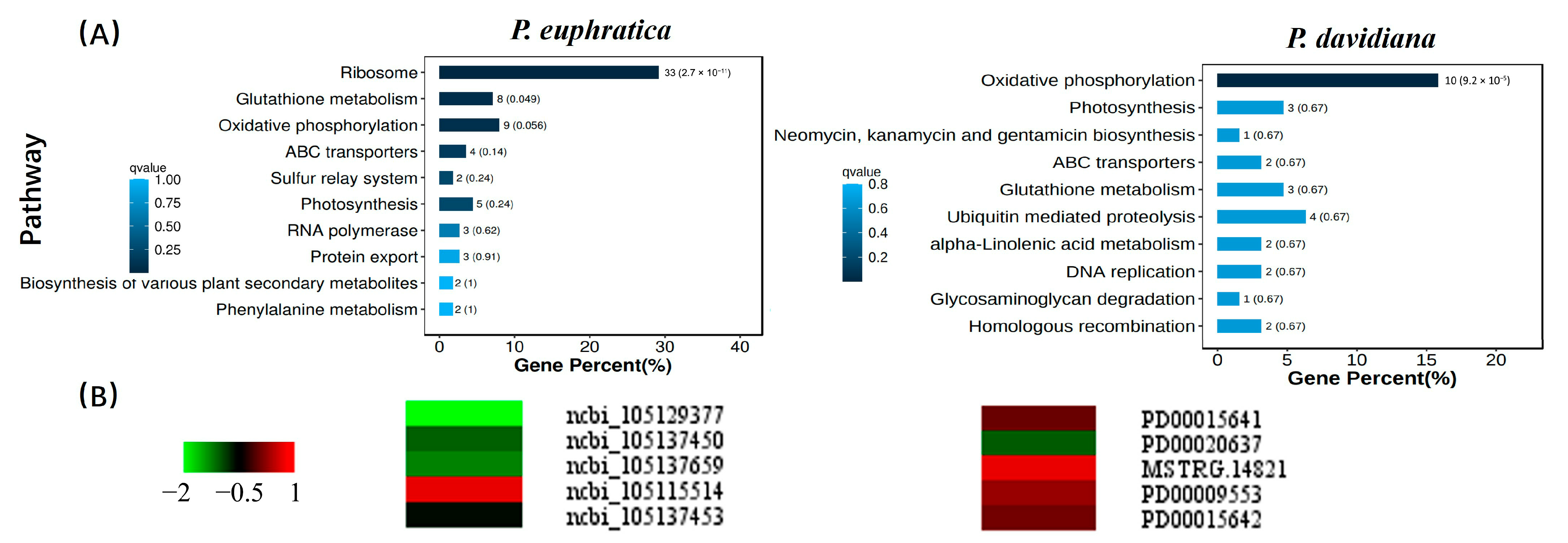
2.7. Late Maturation Genes Showed Dramatic Differences Between Species with Shorter- and Longer-Lived Seeds
2.8. Longer-Lived Seeds (Populus davidiana) Show Stable Levels of Glutathione Metabolism Genes During Ageing
3. Discussion
3.1. Initial Seed Quality and the Glassy State Directly Affects Conservation Outcomes
3.2. Transcriptome Analysis of Populus Dry-Stored Seeds Reveals the Importantce of Late Maturation Genes in Seed Lifespan
3.3. Seed Lifespan of Populus Species Was Strongly Correlated with Environmental Factors
4. Materials and Methods
4.1. Capsule Collection and Seed Cleaning
4.2. Seed Desiccation Treatments, Storage Conditions and Germination Test
4.3. Water Sorption Isotherm Construction
4.4. Meteorological Data of the Sampling Sites
4.5. Seed Ageing and RNA Extraction, Library Construction and Sequencing
4.6. Differentially Expressed Genes (DEGs) and Pathway Enrichment Analysis
4.7. Differential Scanning Calorimetry
4.8. qPCR Test
4.9. Data Analysis and Presentation
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nadarajan, J.; Walters, C.; Pritchard, H.W.; Ballesteros, D.; Colville, L. Seed longevity—The evolution of knowledge and a conceptual framework. Plants 2023, 12, 471. [Google Scholar] [CrossRef]
- Colville, L.; Pritchard, H.W. Seed life span and food security. New Phytol. 2019, 224, 557–562. [Google Scholar] [CrossRef]
- Ballesteros, D.; Pritchard, H.W. The cryobiotechnology of oaks: An integration of approaches for the long-term ex situ conservation of species. Forests 2020, 11, 1281. [Google Scholar] [CrossRef]
- Probert, R.; Daws, M.I.; Hay, F. Ecological correlates of seed longevity: A comparative study on 195 species. Ann. Bot. 2009, 104, 57–69. [Google Scholar] [CrossRef]
- Merritt, D.; Martyn, A.; Ainsley, P.; Young, R.; Seed, L.; Thorpe, M.; Hay, F.; Commander, L.; Shackelford, N.; Offord, C.; et al. A continental-scale study of seed lifespan in experimental storage examining seed, plant, and environmental traits associated with longevity. Biodiver. Conserv. 2014, 23, 1081–1104. [Google Scholar] [CrossRef]
- Walters, C.; Wheeler, L.; Grotenhuis, J. Longevity of seeds stored in a genebank: Species characteristics. Seed Sci. Res. 2005, 15, 1–20. [Google Scholar] [CrossRef]
- Buitink, J.; Leprince, O. Intracellular glasses and seed survival in the dry state. Comptes Rendus Biol. 2008, 331, 788–795. [Google Scholar] [CrossRef]
- Chen, H.; Osuna, D.; Colville, L.; Lorenzo, L.O.; Graeber, K.; Kuster, H.; Leubner-Metzger, G.; Kranner, I. Transcriptome-wide mapping of pea seed ageing reveals a pivotal role for genes related to oxidative stress and programmed cell death. PLoS ONE 2013, 8, e78471. [Google Scholar] [CrossRef] [PubMed]
- Sommerville, K.D.; Newby, Z.J.; Yenson, A.J.M.; Offord, C.A. Are orthodox Australian rainforest seeds short-lived in storage? Aust. J. Bot. 2023, 71, 340–352. [Google Scholar] [CrossRef]
- Buijs, G.; Willems, L.A.J.; Kodde, J.; Groot, S.P.C.; Bentsink, L. Evaluating the EPPO method for seed longevity analyses in Arabidopsis. Plant Sci. 2020, 301, 110644. [Google Scholar] [CrossRef]
- Gerna, D.; Ballesteros, D.; Arc, E.; Stoggl, W.; Seal, C.; Marami-Zonouz, N.; Na, C.S.; Kranner, I.; Roach, T. Does oxygen affect ageing mechanisms of seeds? A matter of cytoplasmic physical state. J. Exp. Bot. 2022, 73, 2631–2649. [Google Scholar] [CrossRef]
- Roach, T.; Nagel, M.; Börner, A.; Eberle, C.; Kranner, I. Changes in tocochromanols and glutathione reveal differences in the mechanisms of seed ageing under seedbank conditions and controlled deterioration in barley. Environ. Exp. Bot. 2018, 156, 8–15. [Google Scholar] [CrossRef]
- Waterworth, W.; Balobaid, A.; West, C. Seed longevity and genome damage. Biosci. Rep. 2024, 44, BSR20230809. [Google Scholar] [CrossRef] [PubMed]
- Roberts, E.H. Predicting the storage life of seeds. Seed Sci. Technol. 1973, 1, 499–514. [Google Scholar]
- Ellis, R.H.; Roberts, E.H. Improved equations for the prediction of seed longevity. Ann. Bot. 1980, 45, 13–30. [Google Scholar] [CrossRef]
- Dickie, J.B.; Ellis, R.H.; Kraak, H.L.; Ryder, K.; Tompsett, P.B. Temperature and seed storage longevity. Ann. Bot. 1990, 65, 197–204. [Google Scholar] [CrossRef]
- Pence, V.C. Cryopreservation of recalcitrant seed. In Biotechnology Agriculture Forest; Bajaj, Y.P.S., Ed.; Springer: Berlin, Germany, 1995; pp. 29–50. [Google Scholar]
- Kim, D.H. Extending Populus seed longevity by controlling seed moisture content and temperature. PLoS ONE 2018, 13, e0203080. [Google Scholar] [CrossRef]
- Chen, K.Y.; Wang, J.D.; Xiang, R.Q.; Xue, D.Y.; Yun, Q.Z.; Huang, Y.; Sun, H.; Chen, J.H. Backbone phylogeny of Salix based on genome skimming data. Plant Divers. 2024, 47, 178–188. [Google Scholar] [CrossRef]
- Ministry of Ecology and Environmental of the Peoples’ Republic of China, 2023. Chinese Red List. Available online: http://www.mee.gov.cn (accessed on 13 April 2023).
- Michalak, M.; Plitta, B.; Tadeusz, T.C.; Chmielarz, P.; Suszka, J. Desiccation tolerance and cryopreservation of seeds of black poplar (Populus nigra L.), a disappearing tree species in Europe. Europ. J. For. Res. 2015, 134, 53–60. [Google Scholar] [CrossRef]
- Hay, F.R.; Probert, R.J. Advances in seed conservation of wild plant species: A review of recent research. Conser. Physiol. 2013, 1. [Google Scholar] [CrossRef] [PubMed]
- Ma, T.; Wang, J.; Zhou, G.; Yue, Z.; Hu, Q.; Chen, Y.; Liu, B.; Qiu, Q.; Wang, Z.; Zhang, J.; et al. Genomic insights into salt adaptation in a desert poplar. Nat. Commun. 2013, 4, 97. [Google Scholar] [CrossRef]
- Fang, C.F.; Zhao, S.D.; Skvortsov, A.K. SALICACEAE. In Flora of China; Science Press: Beijing, China; Missouri Botanical Garden Press: St. Louis, MO, USA, 1999; p. 139. [Google Scholar]
- Hou, J.; Sun, Y.; Wang, L.; Jiang, Y.Z.; Chen, N.; Tong, S. Genome-wide analysis of the homeobox gene family and identification of drought-responsive members in Populus trichocarpa. Plants 2021, 10, 2284. [Google Scholar] [CrossRef]
- Ingvarsson, P.K.; Bernhardsson, C. Genome-wide signatures of environmental adaptation in European aspen (Populus tremula) under current and future climate conditions. Evol. Appl. 2020, 13, 132–142. [Google Scholar] [CrossRef]
- Leprince, O.; Pellizzaro, A.; Berriri, S.; Buitink, J. Late seed maturation: Drying without dying. J. Exp. Bot. 2017, 68, 827–841. [Google Scholar] [CrossRef]
- Nakajima, S.; Ito, H.; Tanaka, R. Chlorophyll b reductase plays an essential role in maturation and storability of Arabidopsis seeds. Plant Physiol. 2012, 160, 261–273. [Google Scholar] [CrossRef]
- Andrási, N.; Pettkó-Szandtner, A.; Szabados, L. Diversity of plant heat shock factors: Regulation, interactions, and functions. J. Exp. Bot. 2021, 72, 1558–1575. [Google Scholar] [CrossRef] [PubMed]
- Righetti, K.; Vu, J.L.; Pelletier, S.; Vu, B.; Glaab, E.; Lalanne, D.; Pasha, A.; Patel, R.; Provart, N.; Verdier, J.; et al. Inference of longevity-related genes from a robust coexpression network of seed maturation identifies regulators linking seed storability to biotic defense-related pathways. Plant Cell 2015, 27, 2692–2708. [Google Scholar] [CrossRef] [PubMed]
- Lai, Z.; Vinod, K.; Zheng, Z.; Fan, B.; Chen, Z. Roles of Arabidopsis WRKY3 and WRKY4 transcription factors in plant responses to pathogens. BMC Plant Biol. 2008, 8, 68. [Google Scholar] [CrossRef] [PubMed]
- Sano, N.; Rajjou, L.; North, H. Staying alive: Molecular aspects of seed longevity. Plant Cell Physiol. 2016, 57, 660–674. [Google Scholar] [CrossRef]
- Pritchard, H.W.; Dickie, J.B. Predicting seed longevity: The use and abuse of seed viability equations. In Seed Conservation: Turning Science into Practice, 1st ed.; Smith, R.D., Dickie, J.B., Linington, S.H., Prtichard, H.W., Probert, R.J., Eds.; Royal Botanic Gardens, Kew: London, UK, 2003; pp. 653–722. [Google Scholar]
- Francisqueti, A.M.; Marin, R.R.; Hengling, M.M.; Hosomi, S.T.; Pritchard, H.W.; Custodio, C.C.; Machado-Neto, N.B. Orchid seeds are not always short lived in a conventional seed bank! Ann. Bot. 2024, 133, 941–951. [Google Scholar] [CrossRef]
- Maroder, H.L.; Prego, I.A.; Facciuto, G.R.; Maldonado, S.B. Storage behavior of Salix alba and Salix matsudana seeds. Ann. Bot. 2000, 86, 1017–1021. [Google Scholar] [CrossRef]
- Ballesteros, D.; Pence, V. Survival and Death of Seeds during Liquid Nitrogen Storage: A case study on seeds with short lifespans. Cryoletters 2017, 38, 8–289. [Google Scholar]
- Simpson, J.D.; Daigle, B. Five years’ storage of seeds from three willow species. Nativ. Plant J. 2009, 10, 63–67. [Google Scholar] [CrossRef]
- Pence, V.C.; Meyer, A.; Linsky, J.; Gratzfeld, J.; Pritchard, H.W.; Westwood, M.; Bruns, E.B. Defining exceptional species-A conceptual framework to expand and advance ex situ conservation of plant diversity beyond conventional seed banking. Biol. Conserv. 2022, 266, 109440. [Google Scholar] [CrossRef]
- Wani, S.H.; Anand, S.; Singh, B.; Bohra, A.; Joshi, R. WRKY transcription factors and plant defense responses: Latest discoveries and future prospects. Plant Cell Rep. 2021, 40, 1071–1085. [Google Scholar] [CrossRef]
- Wan, Y.; Mao, M.; Wan, D.; Yang, Q.; Yang, F.; Mandlaa; Li, G.; Wang, R. Identification of the WRKY gene family and functional analysis of two genes in Caragana intermedia. BMC Plant Biol. 2018, 18, 31. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.; Zentgraf, U. A HECT E3 ubiquitin ligase negatively regulates Arabidopsis leaf senescence through degradation of the transcription factor WRKY53. Plant J. 2010, 63, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Gao, Y.; Li, H.; Yang, S.; Liu, Y. Over-expression of SlWRKY39 leads to enhanced resistance to multiple stress factors in tomato. J. Plant Biol. 2015, 58, 52–60. [Google Scholar] [CrossRef]
- Wang, N.; Xia, E.; Gao, L. Genome-wide analysis of WRKY family of transcription factors in common bean, Phaseolus vulgaris: Chromosomal localization, structure, evolution and expression divergence. Plant Gene 2016, 5, 22–30. [Google Scholar] [CrossRef]
- Khan, M.I.; Zhang, Y.; Liu, Z.; Hu, J.; Liu, C.; Yang, S.; Hussain, A.; Ashraf, M.F.; Noman, A.; She, L.; et al. CaWRKY40b in pepper acts as a negative regulator in response to Ralstonia solanacearum by directly modulating defense genes including CaWRKY40. Int. J. Mol. Sci. 2018, 19, 1403. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Chen, W.; Xu, Z.; Chen, M.; Yu, D. Functions of WRKYs in plant growth and development. Trends Plant Sci. 2023, 28, 630–645. [Google Scholar] [CrossRef]
- Sun, S.; Ma, W.; Mao, P. Genomic identification and expression profiling of WRKY genes in alfalfa (Medicago sativa) elucidate their responsiveness to seed vigor. BMC Plant Biol. 2023, 23, 568. [Google Scholar] [CrossRef]
- Hundertmark, M.; Buitink, J.; Leprince, O.; Hincha, D.K. The reduction of seed-specific dehydrins reduces seed longevity in Arabidopsis thaliana. Seed Sci. Res. 2011, 21, 165–173. [Google Scholar] [CrossRef]
- Eriksson, S.; Eremina, N.; Barth, A.; Danielsson, J.; Harryson, P. Membrane-induced folding of the plant stress dehydrin Lti30. Plant Physiol. 2016, 171, 932–943. [Google Scholar]
- Ballesteros, D.; Walters, C. Detailed characterization of mechanical properties and molecular mobility within dry seed glasses: Relevance to the physiology of dry biological systems. Plant J. 2011, 68, 607–619. [Google Scholar] [CrossRef] [PubMed]
- Chatelain, E.; Hundertmark, M.; Leprince, O.; Gall, S.L.; Satour, P.; Deligny-penninck, S.; Rogniaux, H.; Buitink, J. Temporal profiling of the heat-stable proteome during late maturation of Medicago truncatula seeds identifies a restricted subset of late embryogenesis abundant proteins associated with longevity. Plant Cell Environ. 2012, 35, 1440–1455. [Google Scholar] [CrossRef] [PubMed]
- Kaur, H.; Petla, B.P.; Kamble, N.U.; Singh, A.; Rao, V.; Salvi, P.; Ghosh, S.; Majee, M. Differentially expressed seed aging responsive heat shock protein OsHSP18.2 implicates in seed vigor, longevity and improves germination and seedling establishment under abiotic stress. Front Plant Sci. 2015, 6, 160286. [Google Scholar] [CrossRef] [PubMed]
- Jungunz, I.; Link, K.; Vogel, F.; Voll, L.M.; Sonnewald, S.; Sonnewald, U. AtHsp70-15-deficient Arabidopsis plants are characterized by reduced growth, a constitutive cytosolic protein response and enhanced resistance to TuMV. Plant J. 2011, 66, 983–995. [Google Scholar] [CrossRef]
- Wang, R.; Zhang, Y.; Kieffer, M.; Yu, H.; Kepinski, S.; Estelle, M. HSP90 regulates temperature-dependent seedling growth in Arabidopsis by stabilizing the auxin co-receptor F-box protein TIR1. Nat. Commun. 2016, 7, 10269. [Google Scholar] [CrossRef]
- Giesguth, M.; Sahm, A.; Simon, S.; Dietz, K. Redox-dependent translocation of the heat shock transcription factor AtHSFA8 from the cytosol to the nucleus in Arabidopsis thaliana. FEBS Lett. 2015, 589, 718–725. [Google Scholar] [CrossRef]
- Wang, N.; Liu, W.; Yu, L.; Guo, Z.; Chen, Z.; Jiang, S.; Xu, H.; Fang, H.; Wang, Y.; Zhang, Z.; et al. HEAT SHOCK FACTOR A8a modulates flavonoid synthesis and drought tolerance. Plant Physiol. 2020, 184, 13. [Google Scholar] [CrossRef] [PubMed]
- Hernández Estévez, I.; Rodríguez Hernández, M. Plant glutathione S-transferases: An overview. Plant Gene 2020, 23, 100233. [Google Scholar] [CrossRef]
- Liu, D.; Liu, Y.; Rao, J.; Wang, G.; Li, H.; Ge, F.; Chen, C. Overexpression of the glutathione S-transferase gene from Pyrus pyrifolia fruit improves tolerance to abiotic stress in transgenic tobacco plants. Mol. Biol. 2013, 47, 591–601. [Google Scholar] [CrossRef]
- Hu, T.; Yang, J.; Wu, X.; Huang, X.; Chen, Z.; Wu, Y. Overexpression of a specific OsGSTL2 isoenzyme improves glyphosate and chlorsulfuron tolerance of transgenic rice plants. Afr. J. Agric. Res. 2013, 8, 1520–1527. [Google Scholar] [CrossRef]
- Dreesen, D.R. Propagation protocol for container willows in the southwestern US using seeds. Nativ. Plant J. 2003, 4, 118–124. [Google Scholar] [CrossRef]
- Chen, H.; Visscher, A.M.; Ai, Q.; Yang, L.; Pritchard, H.W.; Li, W.Q. Intra-specific variation in desiccation tolerance of Citrus sinensis “bingtangcheng” (L.) seeds under different environmental conditions in China. Int. J. Mol. Sci. 2023, 24, 7393. [Google Scholar] [CrossRef]
- Griffith, M.; Walker, J.R.; Spies, N.C.; Ainscough, B.J.; Griffith, O.L. Informatics for RNA sequencing: A web resource for analysis on the cloud. PLOS Comput. Biol. 2015, 11, e1004393. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]

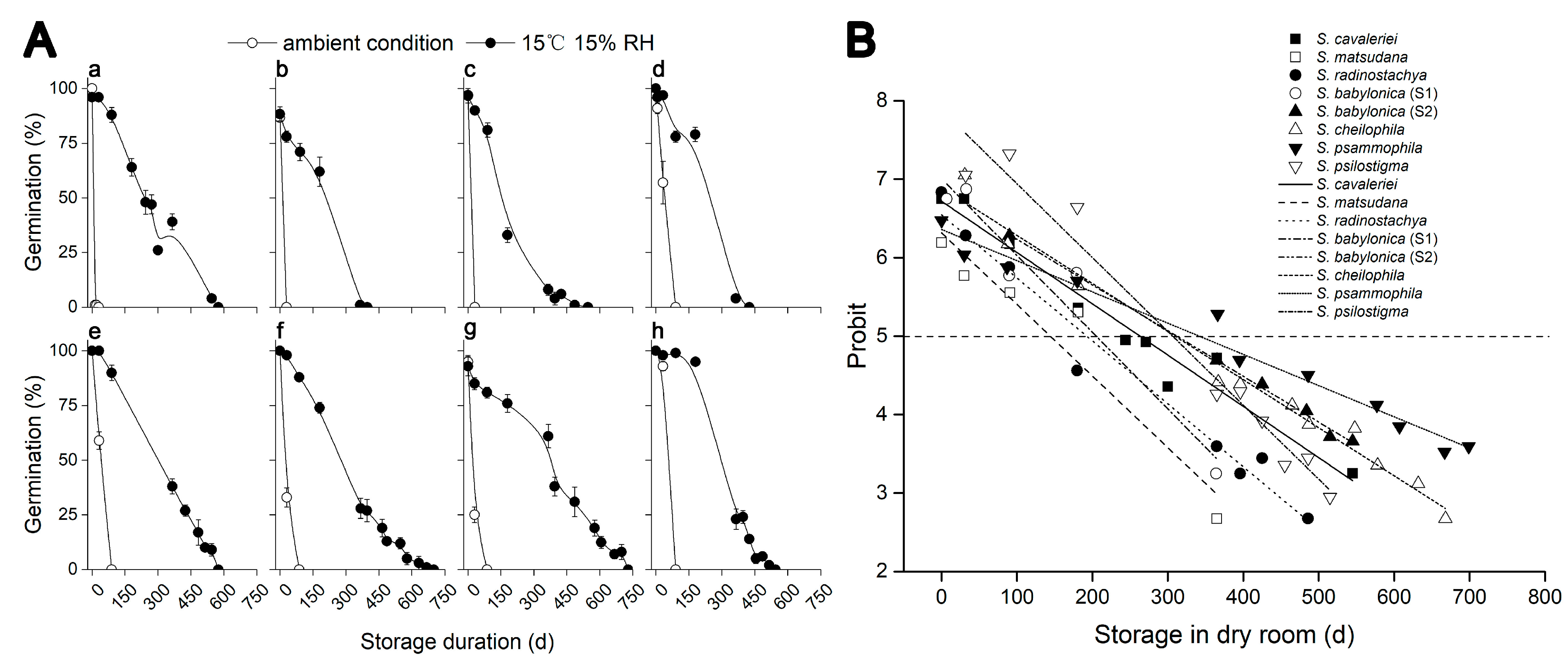
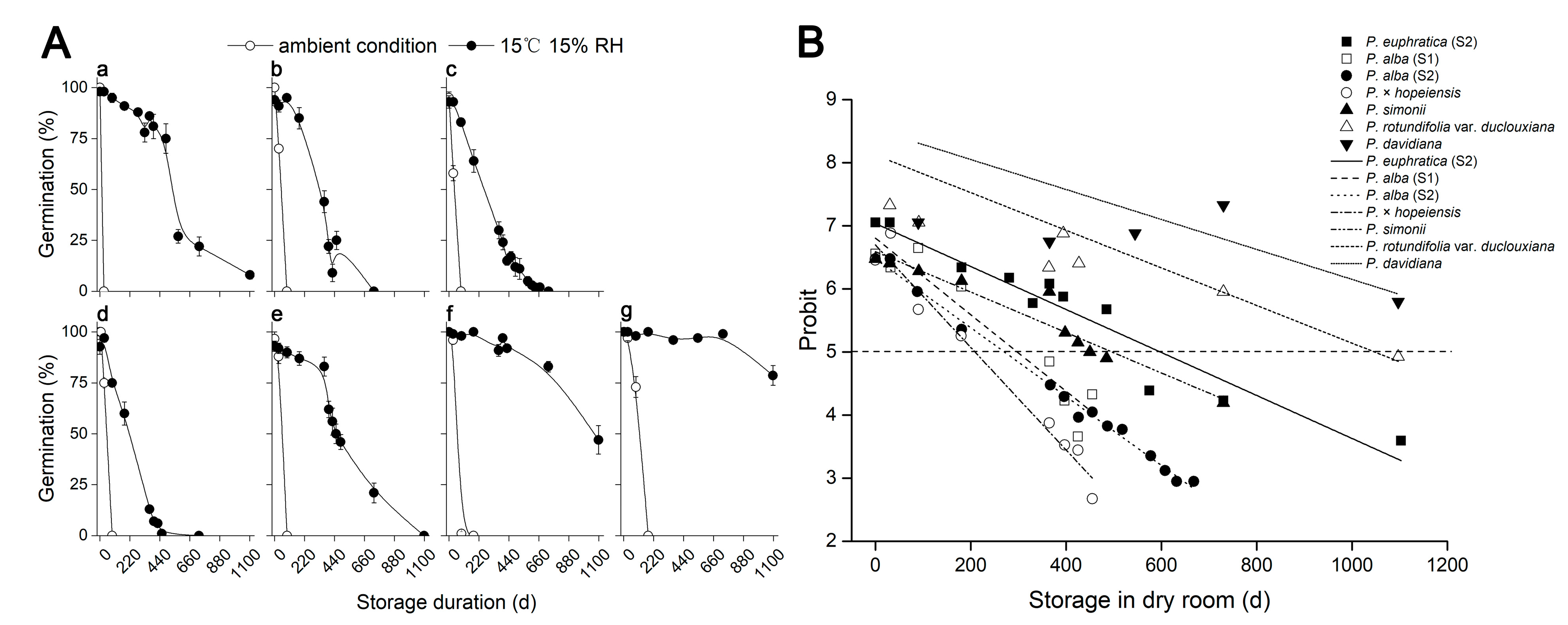
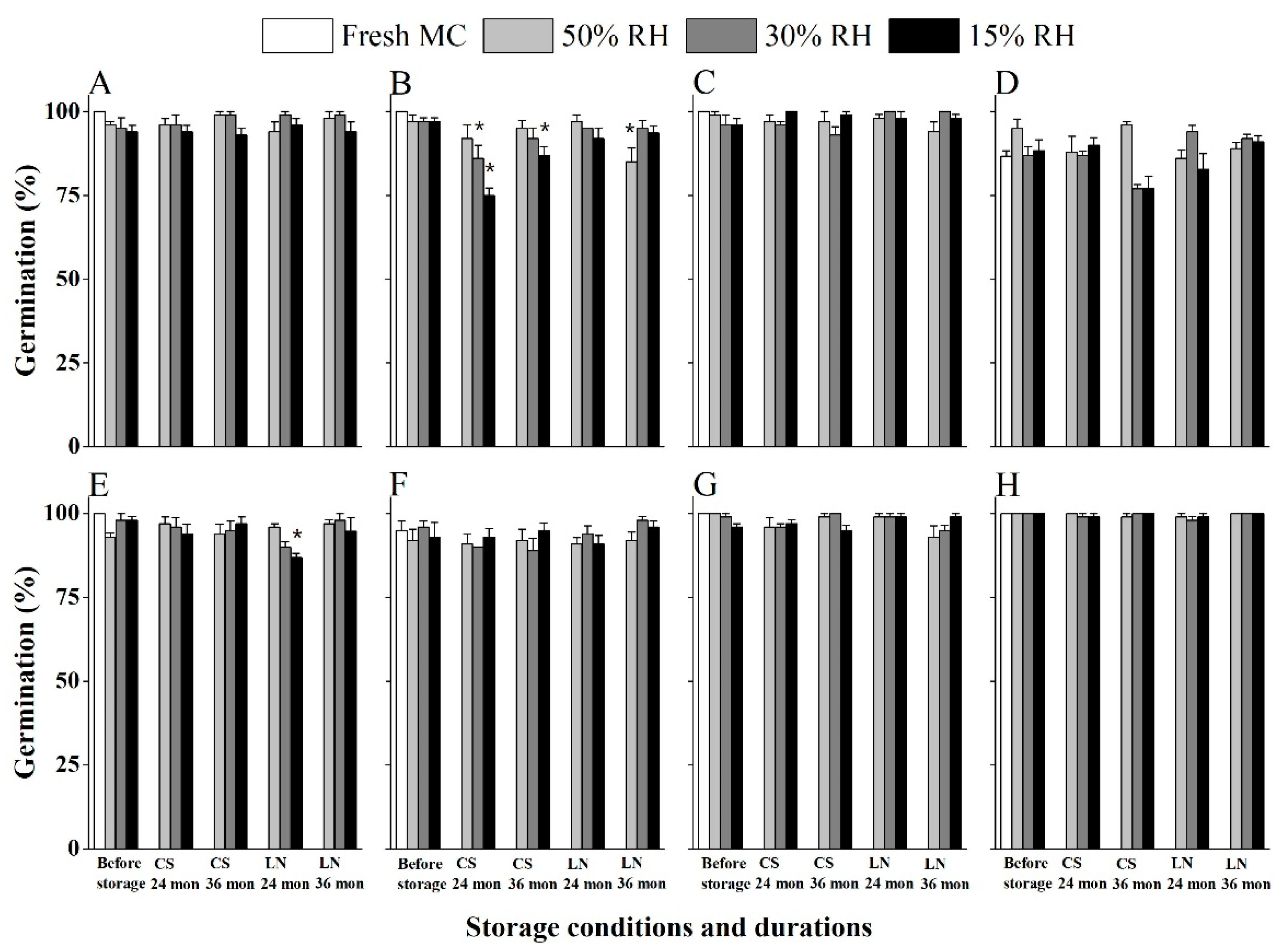

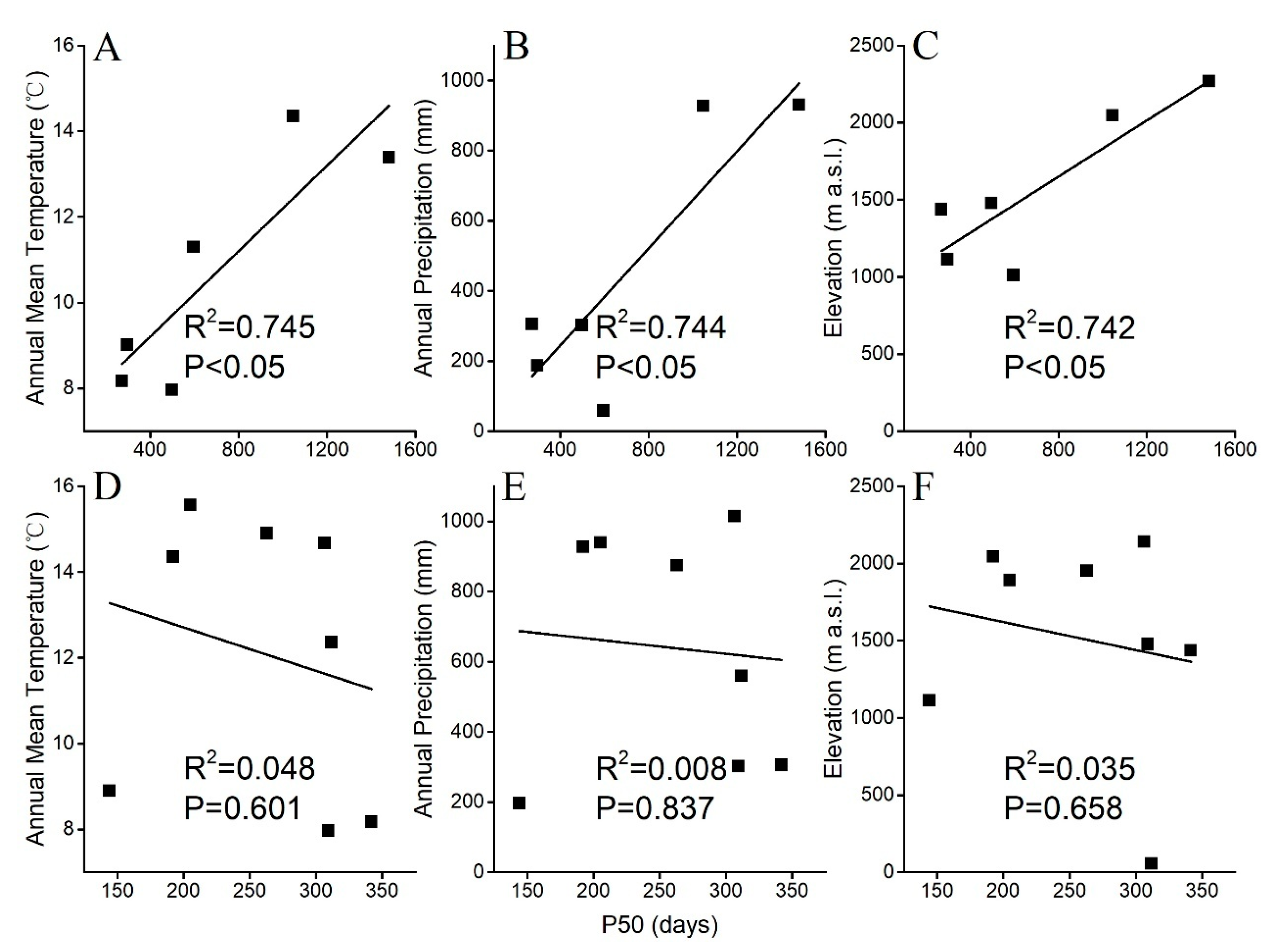


| Species | Maturity Stages | Green Capsules | Yellow Capsules | Start of Dehiscence | Fully Dehiscing | Dispersed | |
|---|---|---|---|---|---|---|---|
| Water Status | |||||||
| Populus euphratica Olivier (S1) | fresh seeds | 85 ± 5.77 a | 100 ± 0 a | 98.33 ± 1.67 a | 81.67 ± 8.33 a | 1.67 ± 1.67 b | |
| dry seeds | 42.5 ± 4.33 c * | 100 ± 0 a | 96.67 ± 1.67 a | 78.33 ± 2.89 b | 0 ± 0 d | ||
| Populus davidiana Dode | fresh seeds | 86 ± 8.57 a | 100 ± 0 a | 99 ± 1 a | 90 ± 2.24 a | 77 ± 7 a | |
| dry seeds | 67 ± 4.64 c | 100 ± 0 a | 100 ± 0 a | 86 ± 1.87 b | 71 ± 3.67 c | ||
| Salix babylonica L. (S1) | fresh seeds | 80 ± 5.70 a | 100 ± 0 a | 98.67 ± 1.33 a | 60 ± 7.64 b | 26.67 ± 5.70 c | |
| dry seeds | 86 ± 4.58 b | 100 ± 0 a | 95.33 ± 0.67 a, b | 60 ± 2.89 c | 18 ± 1.15 d | ||
| Salix psilostigma Anderss. | fresh seeds | 98.33 ± 1.67 a | 100 ± 0 a | 100 ± 0 a | 96.67 ± 1.67 a | NA | |
| dry seeds | 7 ± 2.54 c * | 100 ± 0 a | 100 ± 0 a | 90 ± 2.74 b | NA | ||
| Species | Collection Date (y/m/d) | Longitude | Altitude (m) | 1000 Seeds Weight (mg) |
|---|---|---|---|---|
| Populus alba L. (S1) | 2020/04/23 | 38°25′ N; 106°10′ E | 1077 | 379.6 |
| Populus alba L. (S2) | 2020/04/13 | 37°44′ N; 107°19′ E | 1378 | 312.0 |
| Populus euphratica Oliver (S1) | 2019/09/20 | 41°9′ N; 86°6′ E | 820 | NA |
| Populus euphratica Oliver (S2) | 2020/09/18 | 40°32′ N; 81°17′ E | 960 | 61.2 |
| Populus × hopeiensis Hu & H.F. Chow | 2020/04/13 | 37°44′ N; 107°19′ E | 1377 | 197.2 |
| Populus simonii Carr. | 2020/04/15 | 37°44′ N; 107°04′ E | 1467 | 509.6 |
| Populus davidiana Dode | 2020/03/29 | 25°14′ N; 102°44′ E | 1915 | 110.0 |
| Populus rotundifolia Griff | 2020/05/15 | 25°08′ N; 102°44′ E | 1882 | 136.4 |
| Populus tomentosa Carrière | 2020/04/22 | 38°28′ N; 106°12′ E | 1105 | 311.2 |
| Populus cathayana Rehd. | 2020/07/13 | 41°22′ N; 111°44′ E | 619 | 581.2 |
| Species | Collection Date (y/m/d) | Longitude | Altitude (m) | 1000 Seeds Weight (mg) |
|---|---|---|---|---|
| Salix babylonica L. (S1) | 2020/03/29 | 25°01′ N; 102°40′ E | 1856 | 131.2 |
| Salix babylonica L. (S2) | 2020/04/19 | 40°00′ N; 116°19′ E | 35 | 132.0 |
| Salix cavaleriei H. Lév | 2020/09/25 | 25°04′ N; 102°22′ E | 1768 | 219.6 |
| Salix matsudana Koidz. | 2020/04/ | 38°30′ N; 106°15′ E | 1104 | 100.8 |
| Salix radinostachya C.K. Schneid. | 2020/05/25 | 25°08′ N; 102°44′ E | 1931 | 68.8 |
| Salix cheilophila C.K. Schneid. | 2020/04/15 | 37°44′ N; 107°04′ E | 1468 | 76.4 |
| Salix psammophila C. Wang & Chang Y. Yang | 2020/04/13 | 37°44′ N; 107°19′ E | 1396 | 76.4 |
| Salix wilhelmsiana M. Bieb. | 2020/05/11 | 38°25′ N; 106°10′ E | 1115 | 62.0 |
| Salix psilostigma Andersson | 2020/05/23 | 25°13′ N; 99°17′ E | 2299 | 96.8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Ai, Q.; Hu, X.; Lin, L.; Yang, X.; Pritchard, H.W.; Cai, J.; He, H.; Chen, H. Environmental Factors, Developmental Genes and Oxidative Stress Determine Inter-Species Variability in Seed Longevity in Salicaceae. Plants 2025, 14, 2861. https://doi.org/10.3390/plants14182861
Zhang X, Ai Q, Hu X, Lin L, Yang X, Pritchard HW, Cai J, He H, Chen H. Environmental Factors, Developmental Genes and Oxidative Stress Determine Inter-Species Variability in Seed Longevity in Salicaceae. Plants. 2025; 14(18):2861. https://doi.org/10.3390/plants14182861
Chicago/Turabian StyleZhang, Xiaoyin, Qin Ai, Xiaojian Hu, Liang Lin, Xiangyun Yang, Hugh W. Pritchard, Jie Cai, Huajie He, and Hongying Chen. 2025. "Environmental Factors, Developmental Genes and Oxidative Stress Determine Inter-Species Variability in Seed Longevity in Salicaceae" Plants 14, no. 18: 2861. https://doi.org/10.3390/plants14182861
APA StyleZhang, X., Ai, Q., Hu, X., Lin, L., Yang, X., Pritchard, H. W., Cai, J., He, H., & Chen, H. (2025). Environmental Factors, Developmental Genes and Oxidative Stress Determine Inter-Species Variability in Seed Longevity in Salicaceae. Plants, 14(18), 2861. https://doi.org/10.3390/plants14182861






