Abstract
Heat stress transcription factor (Hsf) families play important roles in abiotic stress responses. However, previous studies reported that HsfBs genes may play diverse roles in response to heat stress. Here, we conducted functional analysis on a woodland strawberry Class B Hsf gene, FvHsfB1a, to improve thermotolerance. The structure of FvHsfB1a contains a typical Hsf domain for DNA binding at the N-terminus, and FvHsfB1a belongs to the B1 family of Hsfs. The FvHsfB1a protein was localized in the nucleus. The FvHsfB1a gene was expressed in various strawberry tissues and highly induced by heat treatment. Under heat stress conditions, ectopic expression of FvHsfB1a in Arabidopsis improves thermotolerance, with higher germination and survival rates, a longer primary root length, higher proline and chlorophyll contents, lower malonaldehyde (MDA) and O2− contents, better enzyme activities, and greater expression of heat-responsive and stress-related genes compared to WT. FvWRKY75 activates the promoter of the FvHsfB1a gene through recognizing the W-box element. Similarly, FvWRKY75-OE lines also displayed a heat-tolerant phenotype, exhibiting more proline and chlorophyll contents, lower MDA and O2− contents, and higher enzyme activities under heat stress. Taken together, our study indicates that FvHsfB1a is a positive regulator of heat stress.
1. Introduction
The growth and development of plants require suitable environmental conditions, generally including temperature, light, water, and soil nutrients [1]. Adverse environmental factors, especially heat stress, cause several morphological and physiological changes in plants. This includes protein denaturation, plasma membrane disruption, and reactive oxygen species (ROS) accumulation, and can lead to plant death in severe cases [2,3,4,5]. Plants have a series of complex physiological mechanisms and molecular regulatory networks to minimize the damage caused by the external environmental stresses [6,7].
The heat shock transcription factors (Hsfs) family is generally known to play a central role in regulating abiotic stress response [8,9,10]. Based on the conserved Hsf DNA-binding domain (DBD) and oligomerization domain (OD) regions, Hsfs can be divided into three classes in higher plants, namely HsfA, HsfB, and HsfC [11,12]. In a previous study, HsfAs were found to be master regulators of thermotolerance by regulating the expression of downstream genes in plants [13,14,15,16]. In contrast to HsfAs, HsfBs have no transcriptional activity, because they lack a C-terminal activator domain (CTAD) [17]. Previous studies reported that HsfBs are induced by heat stress and involved in the regulation of thermotolerance in many plant species [18,19]. For example, AtHsfB1 and AtHsfB2b act as suppressors of the expression of Hsfs and several heat shock protein genes but are necessary for acquiring thermotolerance [20]; PpHsf5 in peaches and HsfB1 in grapes were reported to play a positive role in thermotolerance [21,22]. In addition, some HsfBs genes, such as OsHsfB2b from rice, CarHsfB2 from chickpeas, GmHsfB2b from soybeans, and ZmHsf08 (Class B gene) from maize, play important roles in drought and salt tolerance responses [23,24,25,26]. For HsfCs members, it is demonstrated that HsfC2a from wheat and HsfC1b from tall fescue regulate the heat-responsive genes to increase heat tolerance, and VaHsfC1 from grapes is involved in multiple abiotic stresses [27,28,29,30].
The strawberry is cultivated widely in the world and is threatened by a range of adverse environmental factors. With global warming, the increasing number of extreme temperature events has severely affected the growth, development, productivity, and quality of strawberry, posing a considerable challenge to modern agricultural farming [31,32]. Seventeen Hsf genes have been identified in the diploid woodland strawberry (Fragaria vesca), and different FvHsfs have distinct responses to various biotic and abiotic stresses at the transcriptional level [33]. However, the molecular mechanisms of the FvHsfs genes involved in abiotic stress remain unclear. Herein, we cloned the FvHsfB1a gene from ‘Ruegen’ strawberry leaves and found that FvHsfB1a was induced by heat stress treatment. We then explored its function in heat stress response. The overexpression of the FvHsfB1a gene enhances heat stress resistance. Compared with WT plants, heat-responsive and stress-related genes in FvHsfB1a-OE lines were rapidly and strongly induced by heat stress treatment. Furthermore, the FvHsfB1a gene was directly regulated by the FvWRKY75 TF. The overexpression of the FvWRKY75 gene in Arabidopsis also improved heat tolerance. Overall, we have provided evidence that FvHsfB1a may play an important role in improving strawberry thermotolerance.
2. Results
2.1. Isolation and Characterization of FvHsfB1a
The FvHsfB1a gene was isolated from ‘Ruegen’ leaves and the CDS sequence of FvHsfB1a was 873 bp, encoding a protein containing 290 amino acids (Figure 1A). The predicted molecular weight and isoelectric point of FvHsfB1a protein were predicted to be 32.01 kDa and 5.96 M. The structure of FvHsfB1a contained a typical HSF domain for DNA binding at the N-terminus (Figure 1A). Phylogenetic analysis showed that the sequence of FvHsfB1a was the closest homolog to RcHSF24 (class B1) (Figure 1B).
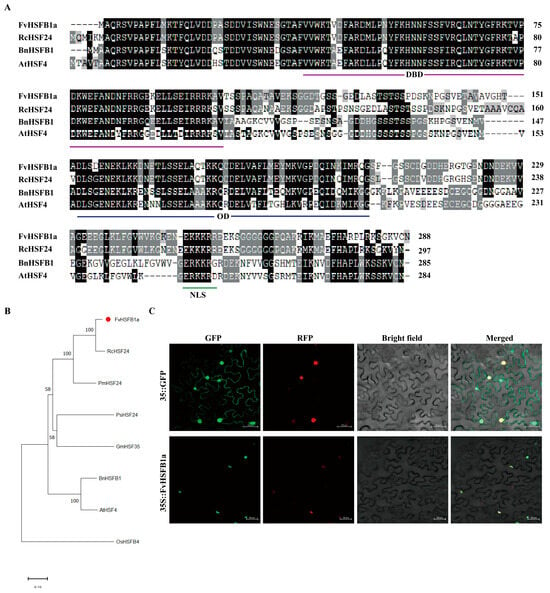
Figure 1.
Analysis of FvHsfB1a transcription factor. (A) Amino acid sequence alignment of FvHsfB1a; DBD: DNA-binding domain; OD: oligomerization domain; NLS: nuclear localization signal of Fragaria vesca, Rosa chinensis, Brassica napus, and Arabidopsis thaliana. (B) Phylogenetic tree analysis of FvHsfB1a. (C) Subcellular localization of FvHsfB1a in tobacco leaves and RFP as a nuclear location marker.
The subcellular localization of FvHsfB1a protein was detected via green fluorescent protein (GFP) and red fluorescent protein (RFP) as a nuclear location marker. As shown in Figure 1C, the recombinant pCAMBIA1302-FvHsfB1a-GFP was localized only in the nucleus.
2.2. Expression Profiles of FvHsfB1a in Diploid Strawberry
The tissue-specific expression patterns of FvHsfB1a were measured in the diploid strawberry ‘Ruegen’. The FvHsfB1a gene was differentially expressed in different tissues, with the lowest expression in leaves and the highest expression in the stem (Figure 2A). Then, we investigated the expression profiles of the FvHsfB1a gene by using mature strawberry leaves under abiotic stress conditions. Under heat stress treatment, FvHsfB1a gene expression was significantly induced and reached a peak with a 43.7-fold increase after 24 h of treatment (Figure 2B).
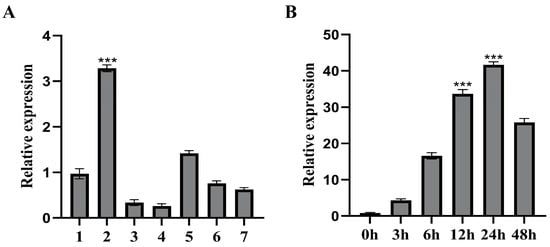
Figure 2.
The expression profile of FvHsfB1a in diploid strawberry. (A) The tissue-specific expression of the FvHsfB1a gene in diploid strawberry. 1: Root; 2: stem; 3: young leaves; 4: mature leaves; 5: flowers; 6: young fruits; 7: mature fruits. (B) Relative expression levels of the FvHsfB1a gene under heat stress (42 °C). Three independent biological replicates were used for the experiment. *** p < 0.001.
2.3. FvHsfB1a Improves Thermotolerance in Arabidopsis
To examine the function of FvHsfB1a under heat stress, we generated FvHsfB1a transgenic Arabidopsis plants, and two FvHsfB1a-OE T3 homozygous lines were selected by using PCR and qRT-PCR for future tests (Figure S1). As shown in Figure 3, no significant differences in seed germination and seedling growth were observed between the WT and OE lines under normal conditions. After treatment with heat stress for 7 d, the WT seeds exhibited the lowest germination rates, especially under the 42 °C treatment (Figure 3A,B). In contrast, the FvHsfB1a-OE seeds had higher germination rates in 37 °C and 42 °C treatments for 7 d compared to WT (Figure 3A,B). Meanwhile, the performance of FvHsfB1a-OE and WT Arabidopsis seedlings was evaluated after 45 °C treatment for 3 h. Compared with WT, there were better performances, higher survival rates, and longer root lengths in FvHsfB1a-OE seedlings observed (Figure 3C,D). Additionally, the heat tolerance of WT and FvHsfB1a-OE mature plants was also detected. Compared with WT, FvHsfB1a-OE lines showed a lower degree of leaf wilting, higher chlorophyll and proline contents, lower MDA and O2− contents, and better superoxide (SOD) and peroxidase (POD) activities after heat stress treatment (Figure 4A–G). These results suggest that the FvHsfB1a gene could improve thermotolerance in Arabidopsis.
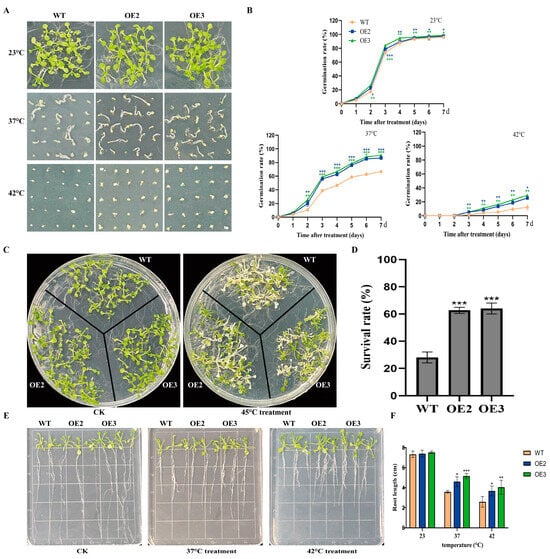
Figure 3.
Heat tolerance analysis of WT and FvHsfB1a-OE lines. (A,B) Germination phenotypes and germination rates of FvHsfB1a-OEs and WT seeds under heat stress conditions. (C,D) Phenotypes and survival rates of FvHsfB1a-OEs and WT seedlings under heat stress. A sample of 7 d old seedlings grown on 1/2 MS medium was treated at 45 °C for 4 h, and then underwent recovery for 7 d. (E,F) The phenotype and root length of WT and FvHsfB1a-OE seedlings before and after 37 °C and 42 °C treatments, respectively. Three independent biological replicates were used for the experiment. *** p < 0.001, ** p < 0.01, * p < 0.05.
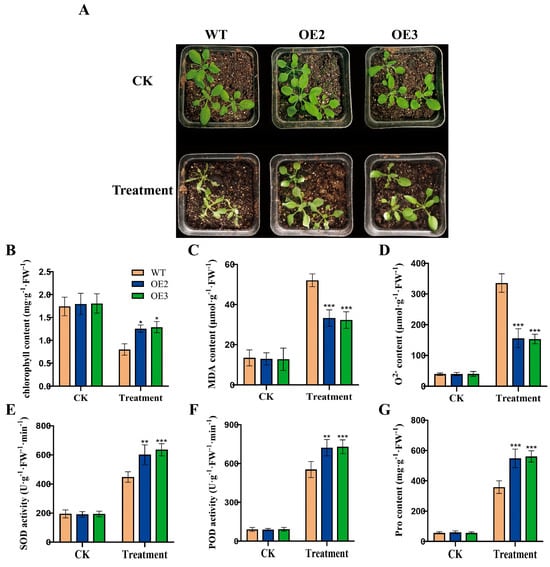
Figure 4.
Thermotolerance analysis of WT and FvHsfB1a-OE lines. (A) Phenotypes of FvHsfB1a-OEs and WT plants before and after heat stress treatment for 7 d. (B–G) The chlorophyll, MDA, O2−, SOD activity, POD activity, and proline content of WT and FvHsfB1a-OE plants under heat stress, respectively. Three independent biological replicates were used for the experiment. *** p < 0.001, ** p < 0.01, * p < 0.05.
2.4. Overexpression of FvHsfB1a Regulates the Expression of Stress-Related Genes
To further investigate the regulation of FvHsfB1a in response to heat stress, the expression profiles of several heat-responsive and stress-related genes were detected by using qRT-PCR in the WT and FvHsfB1a-OE lines. After treatment with heat stress, the expression levels of heat-responsive genes (AtHSP70b and AtHSP101) and stress-related genes (AtP5CS1, AtSOD, and AtPOD) were significantly upregulated in FvHsfB1a-OE lines as compared with the WT (Figure 5).
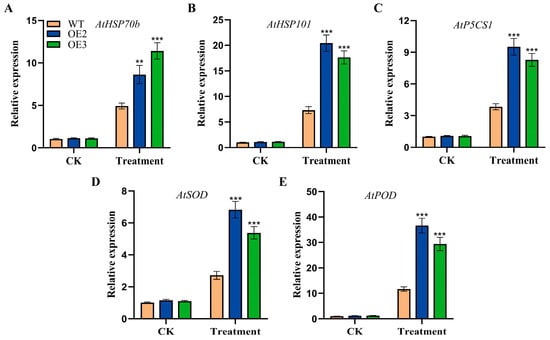
Figure 5.
The expression of stress-related genes in WT and FvHsfB1a-OE lines under cold stress. The gene expression levels of 2-week-old Arabidopsis seedlings treated at 42 °C for 2 d were detected by using qRT-PCR. (A–E) The transcript level of AtHSP70b, AtHSP101, AtP5CS1, AtSOD and AtPOD gene in mature plants under heat stress, respectively. Three independent biological replicates were used for the experiment. *** p < 0.001, ** p < 0.01.
2.5. FvWRKY75 Can Active the Promoter of FvHsfB1a Gene
Bioinformatics analysis showed that the promoter of the FvHsfB1a gene contains several WRKY binding elements (Figure 6A). Based on previous findings, FvWRKY70 and FvWRKY75 were selected for a yeast one-hybrid assay to determine their relationship with FvHsfB1a. As shown in Figure 6B,C, the FvWRKY75 directly bound to the FvHsfB1a promoter and influenced its expression in the yeast one-hybrid and LUC assays.
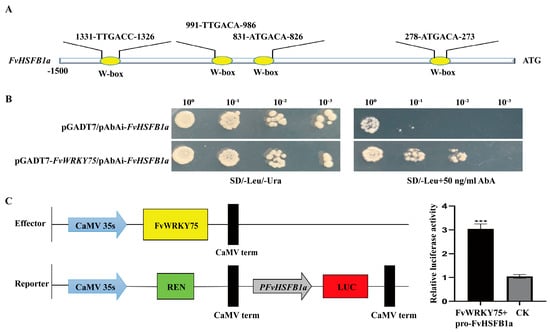
Figure 6.
FvWRKY75 activated FvHsfB1a expression. (A) Bioinformatics analysis showed that the promoter of FvHsfB1a contained WRKY binding elements. (B) Analysis of the activation of FvHsfB1a transcription by FvWRKY75 using a yeast one-hybrid assay. (C) The luciferase activity assay showed that FvWRKY75 binds to the FvHsfB1a promoter. *** p < 0.001.
2.6. FvWRKY75 Enhances Thermotolerance in Arabidopsis
Previously, we cloned the FvWRKY75 gene and generated two FvWRKY75-OE transgenic Arabidopsis plants [34]. In this study, we examined the role of the FvWRKY75 gene in the response to high-temperature stress. During heat treatment, the FvWRKY75-OE transgenic plants exhibited greater insensitivity to heat stress with a better phenotype compared to the WT (Figure 7A). Compared with the WT, lower MDA and O2− contents, higher chlorophyll and proline contents, and better peroxidase activity were observed in the FvWRKY75-OE plants (Figure 7B–G), which suggested that FvWRKY75 is also a positive regulator of heat tolerance.
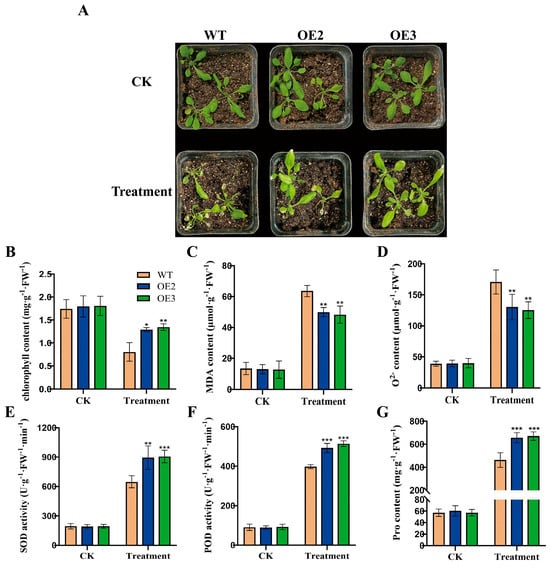
Figure 7.
Heat tolerance analyses of WT and FvWRKY75-OE lines. (A) Phenotypes of FvWRKY75-OEs and WT plants with or without heat treatment for 7 d. (B–G) The chlorophyll, MDA, O2−, SOD activity, POD activity, and proline content of WT and FvWRKY75-OE plants under heat stress, respectively. Three independent biological replicates were used for the experiment. *** p < 0.001, ** p < 0.01, * p < 0.05.
3. Discussion
Heat stress severely affects the growth, development, production, and yields of most agricultural crops [35,36,37,38]. The Hsf family has been well-identified in many plant species and functions as a key regulatory component involved in various stress responses [10,12,39]. In strawberries, 17 Hsfs were identified based on the wild diploid woodland strawberry (Fragaria vesca) [33]. Here, we cloned an Hsf TF gene, FvHsfB1a, from ‘Ruegen’ leaves, and explored its characterization. The amino acid sequence of FvHsfB1a showed all the characteristics of Class B HSFs, and the sequence of FvHsfB1a showed high homology with that of RcHsf24, which belongs to Class B1 Hsfs (Figure 1A,B). The FvHsfB1a protein was localized only in the nucleus (Figure 1C,D). Some Hsf B TF genes, including OsHSF2b, CarHSFB2, and ZmHsf08, were upregulated under abiotic stresses and proved to play a role in heat stress response [23,24,26]. In the current study, we found that the FvHsfB1a gene was induced by heat stress (Figure 2), which suggested that the FvHsfB1a gene may be involved in the heat stress response in strawberry.
Previous studies reported that HsfBs genes may play diverse roles in abiotic stress responses. For example, the overexpression of VdHSFB1 and VvHSFB1 in grapes improved heat tolerance, and the overexpression of CarHSFB2 and GmHsfB2b improved resistance to drought and salt stress in transgenic plants [22,23,25]. However, OsHsfB2b and ZmHsf08 (a member of HSFB1) negatively regulated drought and salt tolerance in rice and maize [24,26]. To better understand the function of the FvHsfB1a gene in strawberry, two FvHsfB1a-OE Arabidopsis lines were generated and used for future tests (Figure S1). After heat stress treatment, FvHsfB1a-OE lines showed increased insensitivity to high temperature, longer root length, higher germination rates, and higher survival rates than the WT (Figure 3E,F), which suggested that FvHsfB1a may regulate the growth of roots in response to heat stress. Under stress conditions, chlorophyll degradation and membrane lipid peroxidation occur in plants, resulting in a decrease in chlorophyll content and an increase in ROS (O2− and H2O2) and MDA content. Therefore, chlorophyll, MDA, O2−, and H2O2 contents are commonly used to assess the severity of stress damage in plants [40,41,42]. In addition, antioxidant enzymes, such as POD, CAT, and APX, play important roles in scavenging oxidative damage caused by ROS accumulation [43,44]. In this study, it was noticed that FvHsfB1a-OE lines had a lower MDA content and higher chlorophyll and proline contents than WT after stress treatment; meanwhile, a lower O2− content and higher SOD and POD activities were presented for the former rather than the latter (Figure 4), which suggested that FvHsfB1a could attenuate heat stress damage to plants by modulating ROS scavenging and increasing antioxidant enzyme activity.
Heat shock proteins (HSPs) were known as the target genes of HSFs and play important roles in heat stress regulation [45,46]. HSP70 and HSP101 are induced by heat stress and confer improved heat tolerance in Arabidopsis [47,48]. In the current study, AtHSP70b and AtHSP101 were significantly induced in FvHsfB1a-OE lines compared to in WT plants under heat stress (Figure 5A,B). This suggests that FvHsfB1a could influence the expression of these HSP genes to improve thermotolerance in transgenic plants. The P5CS gene participated in the synthesis of proline, which contributes to the positive regulation of plant dehydration and osmotic stress [49]. The upregulation of the AtP5CS1 gene in the FvHsfB1a-OE lines under heat treatment may contribute to the accumulation of proline, which can reduce the injury caused by stress (Figure 5C). Many studies reported the connection between antioxidant enzyme genes and Hsfs in scavenging ROS [50,51]. In the current study, the stress-related genes (AtSOD and AtPOD) showed higher expression levels in transgenic plants under heat stress conditions (Figure 5D,E). All of the results suggest that the overexpression of the FvHsfB1a gene improves thermotolerance by upregulating the expression of heat-responsive genes (AtHSP70b and AtHSP101) and stress-related genes (AtP5CS1, AtSOD, and AtPOD).
In this study, the Y1 H and LUC assays showed that the FvWRKY75 transcription factor directly binds to the promoter region of the FvHsfB1a gene (Figure 6). It has been reported that the WRKY TFs play important roles in abiotic stress responses, especially WRKY75 [52,53,54]. For example, the overexpression of PtrWRKY75 in poplar and AhWRKY75 in peanuts improves heat, drought, and salt tolerance; the overexpression of MdWRKY75 in apple regulates the expression of Hsf genes to enhance thermotolerance [55,56,57]. In a previous study, we cloned the FvWRKY75 gene and found that the FvWRKY75 gene plays an active role in response to salt stress [34]. Herein, we explored the role of the FvWRKY75 gene in the response to high-temperature stress. It was noticed that the FvWRKY75-OE transgenic Arabidopsis plants exhibited a better phenotype compared to the WT under heat treatment, and lower MDA and O2− contents, higher chlorophyll and proline contents, and better SOD and POD activities were observed in the former than in the latter (Figure 7). This suggests that FvWRKY75 is also a positive regulator of thermotolerance in strawberry.
In conclusion, we suggest a model of the WRKY-HSF transcriptional regulatory cascade for regulating heat stress in strawberry. In response to heat stress, FvHsfB1a acts as a positive regulator to improve thermotolerance by modulating ROS scavenging, enhancing antioxidant enzyme activity, and being directly regulated by the FvWRKY75 TF. These results provide physiological and molecular evidence highlighting the significance of FvHsfB1a in plant responses to heat stress.
4. Materials and Methods
4.1. Plant Materials, Growth Conditions, and Stress Treatment
Diploid strawberry cultivar ‘Ruegen’ (Fragia vesca), Nc89 tobacco (Nicotiana tabacum), and Arabidopsis thaliana (Columbia) plants were grown in a growth chamber at 23 °C, a 12 h light (4000 lux)/12 h dark photoperiod, and 60% relative humidity in Anhui Agricultural University (Hefei, China). The leaf, stem, root, flower, green fruit, and mature fruit of ‘Ruegen’ plants were harvested and used to detect the tissue specificity of the FvHsfB1a gene. The two-month-old seedings were subjected to 42 °C treatment in an artificial climate incubator, and mature leaves were collected at a series of time points (0 h, 3 h, 6 h, 12 h, 24 h, and 48 h).
4.2. Structure and Sequence Analysis of FvHsfB1a
The CDS sequence of FvHsfB1a (XM_004288037.1) was collected from NCBI (www.ncbi.nlm.nih.gov, accessed on 10 April 2022) and cloned from ‘Ruegen’ cDNA using the specific primers (Supplemental Table S1). DANMAN (version 6.0) software was used to analyze the homologs of FvHsfB1a and other species (accessed on 20 May 2022). MEGA (version 7.0) software was used to construct the phylogenetic tree by the maximum-likelihood (ML) method (accessed on 20 May 2022).
4.3. Subcellular Localization of FvHsfB1a Protein
The CDS region without the stop codon of FvHsfB1a was cloned and used to construct the fusion vector 35S::FvHsfB1a-eGFPm and then introduced into the A. tumefaciens GV3101 strain to infiltrate the tobacco leaves. Leica fluorescence confocal microscopy (Leica TCS SP8) was used to observe the GFP signal. The subcellular localization assays were used according to the methods of a previous study [58]. The primers used are listed in Table S1.
4.4. Genetic Transformation of Arabidopsis
The CDS sequence of FvHsfB1a was cloned and inserted into the pRI101-AN vector to construct the pRI101-FvHsfB1a fusion vector. The fusion vector was transferred into the A. tumefaciens GV3101 strain and then transferred into the Arabidopsis Columbia-0 (Col-0, abbreviated WT) plants at flower budding by the floral dip method [59]. T3 homozygous transgenic lines were measured by PCR and qRT-PCR with the specific primers, and used for further tests. All of the primers used are listed in Table S1.
4.5. RNA Extraction and RT-qPCR
The CTAB methods were used to isolate the total RNA of strawberry and Arabidopsis plants. Each cDNA sample was reverse-transcribed and then used for quantitative real-time PCR (qRT-PCR) according to the established protocols. Three technical replicates of each sample were analyzed for qRT-PCR. The primers used are listed in Table S1.
4.6. Heat Stress Assay of Transgenic Arabidopsis Plants
To test the function of FvHsfB1a and FvWRKY75 genes, WT, FvHsfB1a-OE, and FvWRKY75-OE lines were treated with heat stress. At first, seeds of WT and FvHsfB1a-OE lines were treated in a chamber at 37 °C and 42 °C in darkness for 7 days for heat stress. After heat treatment, all Arabidopsis seeds were transferred into a growth chamber for 4 days under normal growing conditions (23/18 °C day/night), and then the germination rates were measured. Meanwhile, seeds of FvHsfB1a-OE lines and WT were sterilized and seeded on an MS medium plate. A sample of 7-day-old Arabidopsis seedlings was subjected to heat stress at 45 °C for 3 h, and then underwent recovery for 5 days under normal conditions to record phenotypes and count survival rates. Furthermore, 3-week-old WT, FvHsfB1a-OE, and FvWRKY75-OE lines were subjected to heat stress at 42/37 °C (day/night) for 7 days, and samples were collected to measure the physiological parameters. A sample of 3-week-old plants was subjected to heat stress at 42 °C for 3 h, and the leaves were harvested to analyze the expression profiles of related genes.
4.7. Yeast One-Hybrid Assay
The CDS region of FvWRKY75 (XM_004310052.2) was cloned and inserted into the plasmid vector pGADT7 with the GAL4 activation domain. PlantCARE online software (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/, accessed on 20 May 2022) was used to identify the cis-acting elements in the promoter region. The promoter region (1500 bp) of FvHsfB1a was cloned and inserted into the pAbAi vector. Yeast one-hybrid assays were conducted according to the previously reported methods [58]. The primers used for the yeast one-hybrid assays are listed in Table S1.
4.8. Dual Luciferase Activity Assay
The CDS region of FvWRKY75 was cloned and inserted into the pGreenII-62SK vector, named pGreenII-62SK-FvWRKY75, and defined as an effector. The promoter region (1500 bp) of FvHsfB1a was inserted into the pGreenII-0800-LUC vector and named pGreenII-0800-FvHsfB1a, which acts as a reporter. Dual luciferase activity assays were conducted according to the previously reported methods [58]. All of the primers used are listed in Table S1.
4.9. Data Analysis
The data are the average of three repeated experiments and are expressed as the mean ± SD. The statistical analysis was determined by using SPSS system (version 20) software. Duncan’s multiple range test was used to identify significant differences among treatment means (* p < 0.05, ** p < 0.01, and *** p < 0.001).
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/plants14152392/s1, Figure S1: The identification of FvHsfB1a transgenic Arabidopsis thaliana lines. A: PCR assay of FvHsfB1a transgenic lines. B: Relative expression of FvHsfB1a gene in different transgenic lines. Note: 1: Wild-type Arabidopsis thaliana; 2: negative; 3: line 1; 4: line 2; 5: line 3; 6: positive; Table S1: The primers used in this study.
Author Contributions
Conceptualization, H.X. (Hao Xue) and Q.C.; validation and formal analysis, Q.C., T.M., S.L. and H.X. (Hanxiu Xie); data curation, Q.C. and T.M.; writing—original draft preparation, T.M. and Q.C.; writing—review and editing, H.X. (Hao Xue) and K.Y.; funding acquisition, H.X. (Hao Xue). All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Natural Science Foundation of Anhui Province (No. 2108085QC120).
Data Availability Statement
Data are contained within the article and can be made available on request.
Conflicts of Interest
The authors have no conflict of interest to declare.
References
- Choudhury, F.K.; Rivero, R.M.; Blumwald, E.; Mittler, R. Reactive oxygen species, abiotic stress and stress combination. Plant J. 2017, 90, 856–867. [Google Scholar] [CrossRef]
- Wahid, A.; Gelani, S.; Ashraf, M.; Foolad, M.R. Heat tolerance in plants: An overview. Environ. Exp. 2007, 61, 199–223. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Nahar, K.; Alam, M.M.; Roychowdhury, R.; Fujita, M. Physiological, biochemical, and molecular mechanisms of heat stress tolerance in plants. Int. J. Mol. Sci. 2013, 14, 9643–9684. [Google Scholar] [CrossRef]
- Gechev, T.; Petrov, V. Reactive oxygen species and abiotic stress in plants. Int. J. Mol. Sci. 2020, 21, 7433. [Google Scholar] [CrossRef] [PubMed]
- Zandalinas, S.I.; Fritschi, F.B.; Mittler, R. Global warming, climate change, and environmental pollution: Recipe for a multifactorial stress combination disaster. Trends Plant Sci. 2021, 26, 588–599. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Erpen, L.; Devi, H.S.; Grosser, J.W.; Dutt, M. Potential use of the DREB/ERF, MYB, NAC and WRKY transcription factors to improve abiotic and biotic stress in transgenic plants. Plant Cell Tissue Organ Cult. 2017, 132, 1–25. [Google Scholar] [CrossRef]
- Wang, W.; Vinocur, B.; Shoseyov, O.; Altman, A. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 2004, 9, 244–252. [Google Scholar] [CrossRef]
- Xue, G.P.; Sadat, S.; Drenth, J.; Mcintyre, C.L. The heat shock factor family from Triticum aestivum in response to heat and other major abiotic stresses and their role in regulation of heat shock protein genes. J. Exp. Bot. 2014, 65, 539–557. [Google Scholar] [CrossRef]
- Guo, M.; Liu, J.H.; Ma, X.; Luo, D.X.; Gong, Z.H.; Lu, M.H. The plant heat stress transcription factors (HSFs): Structure, regulation, and function in response to abiotic stresses. Front. Plant Sci. 2016, 7, 114. [Google Scholar] [CrossRef]
- Doring, P.; Treuter, E.; Kistner, C.; Lyck, R.; Chen, A.; Nover, L. The role of AHA motifs in the activator function of tomato heat stress transcription factors HsfA1 and HsfA2. Plant Cell 2000, 12, 265–278. [Google Scholar] [CrossRef]
- Jacob, P.; Hirt, H.; Bendahmane, A. The heat-shock protein/chaperone network and multiple stress resistance. Plant Biotechnol. J. 2017, 15, 405–414. [Google Scholar] [CrossRef]
- Liu, H.C.; Charng, Y.Y. Common and distinct functions of arabidopsis class A1 and A2 heat shock factors in diverse abiotic stress responses and development. Plant Physiol. 2013, 163, 276–290. [Google Scholar] [CrossRef]
- Chen, S.; Yu, M.; Li, H.; Wang, Y.; Lu, Z.; Zhang, Y.; Liu, M.; Qiao, G.; Wu, L.; Han, X.; et al. SaHsfA4c from Sedum alfredii hance enhances cadmium tolerance by regulating ROS-scavenger activities and heat shock proteins expression. Front. Plant Sci. 2020, 11, 142. [Google Scholar] [CrossRef]
- Gai, W.X.; Ma, X.; Li, Y.; Xiao, J.J.; Khan, A.; Li, Q.H.; Gong, Z.H. CaHsfA1d improves plant thermotolerance via regulating the expression of stress- and antioxidant-related genes. Int. J. Mol. Sci. 2020, 21, 8374. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Mao, C.J.; Jiang, C.H.; Zhang, L.; Peng, S.Y.; Zhang, Y. One heat shock transcription factor confers high thermal tolerance in clematis plants. Int. J. Mol. Sci. 2021, 22, 2900. [Google Scholar] [CrossRef]
- Bharti, K.; Koskull--Döring, P.V.; Bharti, S.; Kumar, P.; Tintschl-Körbitzer, A.; Treuter, E.; Nover, L. Tomato heat stress transcription factor HsfB1 represents a novel type of general transcription coactivator with a histone-like motif interacting with the plant CREB binding protein ortholog HAC1. Plant Cell 2004, 16, 1521–1535. [Google Scholar] [CrossRef] [PubMed]
- Giorno, F.; Guerriero, G.; Baric, S.; Mariani, C. Heat shock transcriptional factors in Malus domestica: Identification, classification and expression analysis. BMC Genom. 2012, 13, 639. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Han, Y.; Zhang, K.; Zhao, F.L.; Li, Y.J.; Zheng, Y.; Wang, Y.J.; Wen, Y.Q. Identification and expression analysis of heat shock transcription factors in the wild Chinese grapevine (Vitis pseudoreticulata). Plant Physiol. Biochem. 2016, 99, 1–10. [Google Scholar] [CrossRef]
- Ikeda, M.; Mitsuda, N.; Ohme-Takagi, M. Arabidopsis HsfB1 and HsfB2b act as repressors of the expression of heat-inducible Hsfs but positively regulate the acquired thermotolerance. Plant Physiol. 2011, 157, 1243–1254. [Google Scholar] [CrossRef]
- Tan, B.; Yan, L.; Li, H.N.; Lian, X.D.; Cheng, J.; Wang, W. Genome-wide identification of HSF family in peach and functional analysis of PpHSF5 involvement in root and aerial organ development. PeerJ 2021, 9, e10961. [Google Scholar] [CrossRef]
- Chen, H.Y.; Liu, X.N.; Li, S.C.; Yuan, L.; Mu, H.Y.; Wang, Y.; Li, Y.; Duan, W.; Fan, P.G.; Liang, Z.C.; et al. The class B heat shock factor HSFB1 regulates heat tolerance in grapevine. Hortic. Res. 2023, 10, uhad001. [Google Scholar] [CrossRef]
- Ma, H.; Wang, C.T.; Yang, B.; Cheng, H.Y.; Wang, Z.; Mijiti, A.; Ren, C.; Qu, G.H.; Zhang, H.; Ma, L. CarHSFB2, a class B heat shock transcription factor, is involved in different developmental processes and various stress responses in chickpea (Cicer arietinum L.). Plant Mol. Biol. Rep. 2016, 34, 1–14. [Google Scholar] [CrossRef]
- Xiang, J.H.; Ran, J.; Zou, J.; Zhou, X.Y.; Liu, A.L.; Zhang, X.W.; Peng, Y.; Tang, N.; Luo, G.Y.; Chen, X.B. Heat shock factor OsHsfB2b negatively regulates drought and salt tolerance in rice. Plant Cell Rep. 2013, 32, 1795–1806. [Google Scholar] [CrossRef]
- Bian, X.H.; Li, W.; Niu, C.F.; Wei, W.; Hu, Y.; Han, J.Q.; Lu, X.; Tao, J.J.; Jin, M.; Qin, H.; et al. A class B heat shock factor selected for during soybean domestication contributes to salt tolerance by promoting flavonoid biosynthesis. New Phytol. 2020, 225, 268–283. [Google Scholar] [CrossRef]
- Wang, J.; Chen, L.; Long, Y.; Si, W.; Cheng, B.; Jiang, H. A novel heat shock transcription factor (ZmHsf08) negatively regulates salt and drought stress responses in maize. Int. J. Mol. Sci. 2021, 22, 11922. [Google Scholar] [CrossRef]
- Schmidt, R.; Schippers, J.H.; Welker, A.; Mieulet, D.; Guiderdoni, E.; Mueller-Roeber, B. Transcription factor OsHsfC1b regulates salt tolerance and development in Oryza sativa ssp. japonica. AoB Plants 2012, 1, pls011. [Google Scholar] [CrossRef]
- Zhuang, L.L.; Cao, W.; Wang, J.; Yu, J.J.; Yang, Z.M.; Huang, B.R. Characterization and functional analysis of FaHsfC1b from festuca arundinacea conferring heat tolerance in Arabidopsis. Int. J. Mol. Sci. 2018, 19, e2702. [Google Scholar] [CrossRef]
- Jiao, S.Z.; Guo, C.; Yao, W.K.; Zhang, N.B.; Zhang, J.Y.; Xu, W.R. An Amur grape VaHsfC1 is involved in multiple abiotic stresses. Sci. Horti. 2022, 295, 110785. [Google Scholar] [CrossRef]
- Hu, X.J.; Chen, D.D.; Lynne, M.C.; Fernanda, D.M.; Zhang, Z.B.; Drenth, J.; Kalaipandian, S.; Chang, H.P.; Xue, G.P. Heat shock factor C2a serves as a proactive mechanism for heat protection in developing grains in wheat via an ABA-mediated regulatory pathway. Plant Cell Environ. 2017, 41, 79–98. [Google Scholar] [CrossRef] [PubMed]
- Long, S.P.; Ort, D.R. More than taking the heat: Crops and global change. Curr. Opin. Plant Biol. 2010, 13, 241–248. [Google Scholar] [CrossRef]
- Teixeira, E.I.; Fischer, G.; Van Velthuizen, H.; Walter, C.; Ewert, F. Global hot-spots of heat stress on agricultural crops due to climate change. Agric. For. Meteorol. 2013, 170, 206–215. [Google Scholar] [CrossRef]
- Hu, Y.; Han, Y.; Wei, W.; Li, Y.J.; Zhang, K.; Gao, Y.R.; Zhao, F.L.; Feng, J.Y. Identification, isolation, and expression analysis of heat shock transcription factors in the diploid woodland strawberry Fragaria vesca. Front. Plant Sci. 2015, 6, 736. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Jiang, Y.; Xie, H.X.; Wang, K.W.; Yang, K.B.; Cao, Q.; Xue, H. FvWRKY75 positively regulates FvCRK5 to enhance salt stress tolerance. Plants 2025, 14, 1804. [Google Scholar] [CrossRef] [PubMed]
- Nasser, S.; Kemal, K.; Schenk, P.M. Global plant stress signaling: Reactive oxygen species at the cross-road. Front. Plant Sci. 2016, 7, 187. [Google Scholar] [CrossRef]
- Ohama, N.; Sato, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Transcriptional regulatory network of plant heat stress response. Trends Plant Sci. 2017, 22, 53–65. [Google Scholar] [CrossRef]
- Raza, A.; Razzaq, A.; Mehmood, S.S.; Zou, X.; Zhang, X.; Lv, Y.; Xu, J. Impact of climate change on crops adaptation and strategies to tackle its outcome: A review. Plants. 2019, 8, 34. [Google Scholar] [CrossRef]
- Hu, Y.M.; Zhao, T.L.; Guo, Y.F.; Wang, M.; Brachhold, K.; Chu, C.C.; Hanson, A.; Kumar, S.; Lin, R.C.; Long, W.J.; et al. 100 essential questions for the future of agriculture. MODA 2023, 1, 4–12. [Google Scholar] [CrossRef]
- Liu, F.; Xi, M.W.; Liu, T.; Wu, X.Y.; Ju, L.Y.; Wang, D.J. The central role of transcription factors in bridging biotic and abiotic stress responses for plants’ resilience. New Crops. 2024, 1, 100005. [Google Scholar] [CrossRef]
- Hu, L.; Li, H.; Pang, H.; Fu, J. Responses of antioxidant gene, protein and enzymes to salinity stress in two genotypes of Perennial ryegrass (Lolium perenne) diffffering in salt tolerance. J. Plant Physiol. 2012, 169, 146–156. [Google Scholar] [CrossRef]
- Tomar, R.S.; Kataria, S.; Jajoo, A. Behind the scene: Critical role of reactive oxygen species and reactive nitrogen species in salt stress tolerance. J. Agron. Crop Sci. 2021, 207, 577–588. [Google Scholar] [CrossRef]
- Sharma, P.; Jha, A.B.; Dubey, R.S.; Pessarakli, M. Reactive oxygen species, oxidative damage, and antioxidative defense mechanism in plants under stressful conditions. J. Bot. 2012, 4, 217037. [Google Scholar] [CrossRef]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef]
- Sun, T.T.; Wang, C.; Liu, R.; Zhang, Y.; Wang, Y.C.; Wang, L.Q. ThHSFA1 confers salt stress tolerance through modulation of reactive oxygen species scavenging by directly regulating ThWRKY4. Int. J. Mol. Sci. 2021, 22, 5048. [Google Scholar] [CrossRef]
- Hu, W.H.; Hu, G.C.; Han, B. Genome-wide survey and expression profiling of heat shock proteins and heat shock factors revealed overlapped and stress specific response under abiotic stresses in rice. Plant Sci. 2009, 176, 583–590. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.Y.; Huang, W.L.; Yang, Z.M.; Liu, J.; Huang, B.R. Transcriptional regulation of heat shock proteins and ascorbate peroxidase by CtHsfA2b from African bermudagrass conferring heat tolerance in Arabidopsis. Sci. Rep. 2016, 6, 28021. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.Y.; Juan, Y.T.; Hsu, Y.H.; Wu, S.H.; Liao, H.T.; Fung, R.W.; Charng, Y.Y. Interplay between heat shock proteins HSP101 and HSA32 prolongs heat acclimation memory posttranscriptionally in Arabidopsis. Plant Physiol. 2013, 161, 2075–2084. [Google Scholar] [CrossRef]
- Guo, M.; Liu, J.H.; Ma, X.; Zhai, Y.F.; Gong, Z.H.; Lu, M.H. Genome-wide analysis of the Hsp70 family genes in pepper (Capsicum annuum L.) and functional identification of CaHsp70-2 involvement in heat stress. Plant Sci. 2016, 252, 246–256. [Google Scholar] [CrossRef]
- Butt, H.I.; Yang, Z.E.; Gong, Q.; Chen, E.Y.; Wang, X.Q.; Zhao, G.; Ge, X.Y.; Zhang, X.Y.; Li, F.G. GaMYB85, an R2R3 MYB gene, in transgenic Arabidopsis plays an important role in drought tolerance. BMC Plant Biol. 2017, 17, 142. [Google Scholar] [CrossRef]
- Li, F.; Zhang, H.; Zhao, H.; Gao, T.; Song, A.; Jiang, J.; Chen, F.; Chen, S. Chrysanthemum CmHSFA4 gene positively regulates salt stress tolerance in transgenic chrysanthemum. Plant Biotechnol. J. 2018, 16, 1311–1321. [Google Scholar] [CrossRef]
- Panchuk, I.I.; Volkov, R.A.; Schoffl, F. Heat stress- and heat shock transcription factor-dependent expression and activity of ascorbate peroxidase in Arabidopsis. Plant Physiol. 2002, 129, 838–853. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Zhang, D.W.; Lv, K.W.; Zhang, X.M.; Cheng, Z.H.; Li, R.H.; Zhou, B.; Jiang, T.B. Functional characterization of poplar WRKY75 in salt and osmotic tolerance. Plant Sci. 2019, 289, 110259. [Google Scholar] [CrossRef]
- Li, W.X.; Pang, S.Y.; Lu, Z.G.; Jin, B. Function and mechanism of WRKY transcription factors in abiotic stress responses of plants. Plants 2020, 9, 1515. [Google Scholar] [CrossRef]
- Jiang, J.J.; Ma, S.H.; Ye, N.H.; Jiang, M.; Cao, J.S.; Zhang, J.H. WRKY transcription factors in plant responses to stresses. J. Integr. Plant Biol. 2017, 59, 86–101. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhou, Y.; Zhang, D.; Tang, X.; Li, Z.; Shen, C.; Han, X.; Deng, W.; Yin, W.; Xia, X. PtrWRKY75 overexpression reduces stomatal aperture and improves drought tolerance by salicylic acid-induced reactive oxygen species accumulation in poplar. Environ. Exp. Bot. 2020, 176, 104117. [Google Scholar] [CrossRef]
- Zhu, H.; Jiang, Y.N.; Guo, Y.; Huang, J.B.; Zhou, M.H.; Tang, Y.Y.; Sui, J.M.; Wang, J.S.; Qiao, L.X. A novel salt inducible WRKY transcription factor gene, AhWRKY75, confers salt tolerance in transgenic peanut. Plant Physiol. Bioch. 2021, 160, 175–183. [Google Scholar] [CrossRef]
- Zhang, Z.; Yang, C.; Xi, J.; Wang, Y.; Guo, J.; Liu, Q.; Liu, Y.; Ma, Y.; Zhang, J.; Ma, F.; et al. The MdHSC70-MdWRKY75 module mediates basal apple thermotolerance by regulating the expression of heat shock factor genes. Plant Cell 2024, 36, 3631–3653. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Xue, H.; Zhang, F.; Jiang, Q.; Yang, S.; Yue, P.T.; Wang, F.; Zhang, Y.Y.; Li, L.G.; He, P.; et al. The miR156/SPL module regulates apple salt stress tolerance by activating MdWRKY100 expression. Plant Biotech. J. 2020, 19, 311–323. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).