Comprehensive Genome-Wide Characterization of L-Type Lectin Receptor-like Kinase (L-LecRLK) Genes in Wheat (Triticum aestivum L.) and Their Response to Abiotic Stress
Abstract
1. Introduction
2. Results
2.1. Identification and Characterization of TaL-LecRLK Genes in Wheat
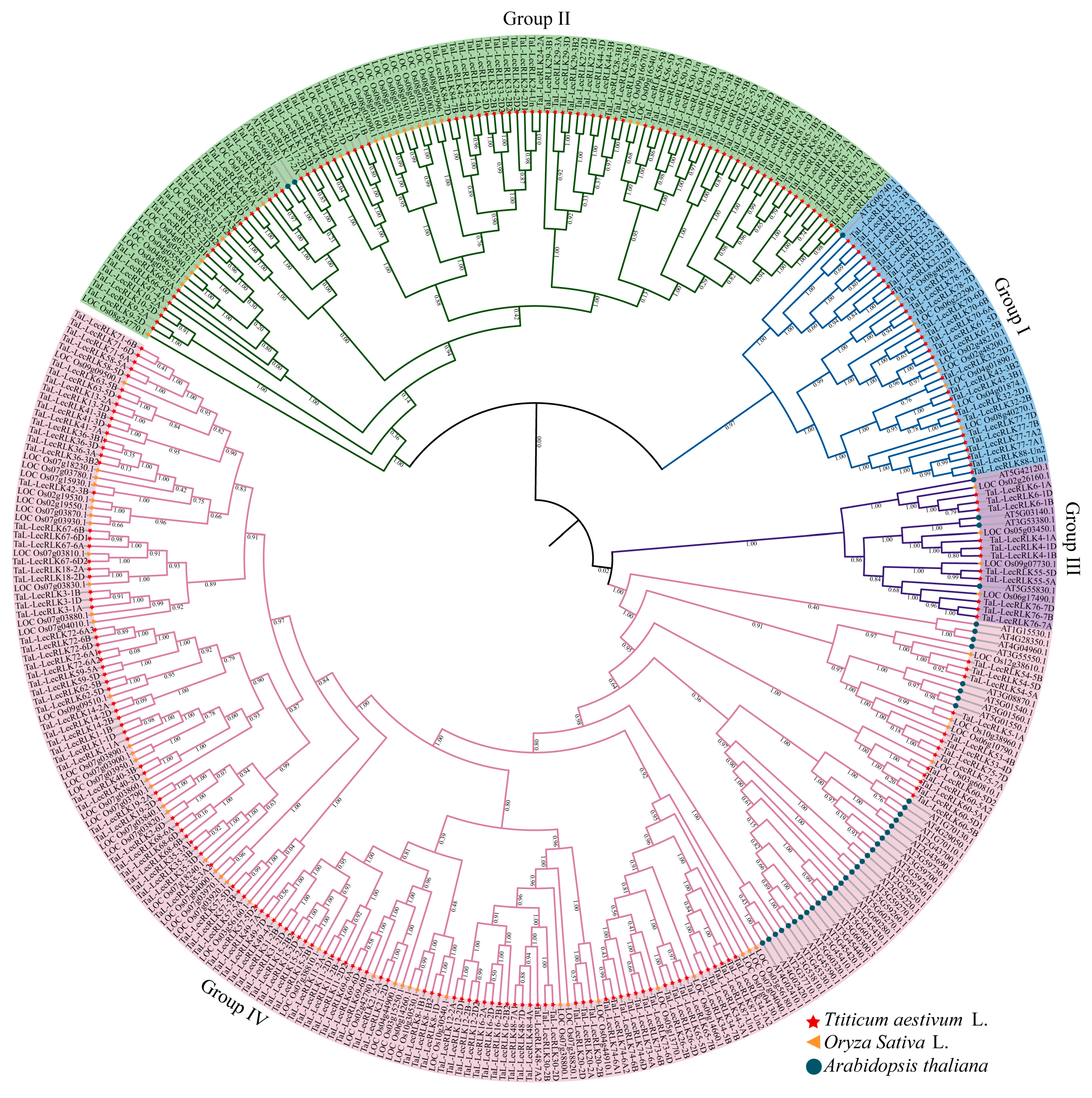
2.2. Phylogenetic Relationships and Classification of TaL-LecRLK Proteins
2.3. Gene Structure and Conserved Motif Analysis of TaL-LecRLKs
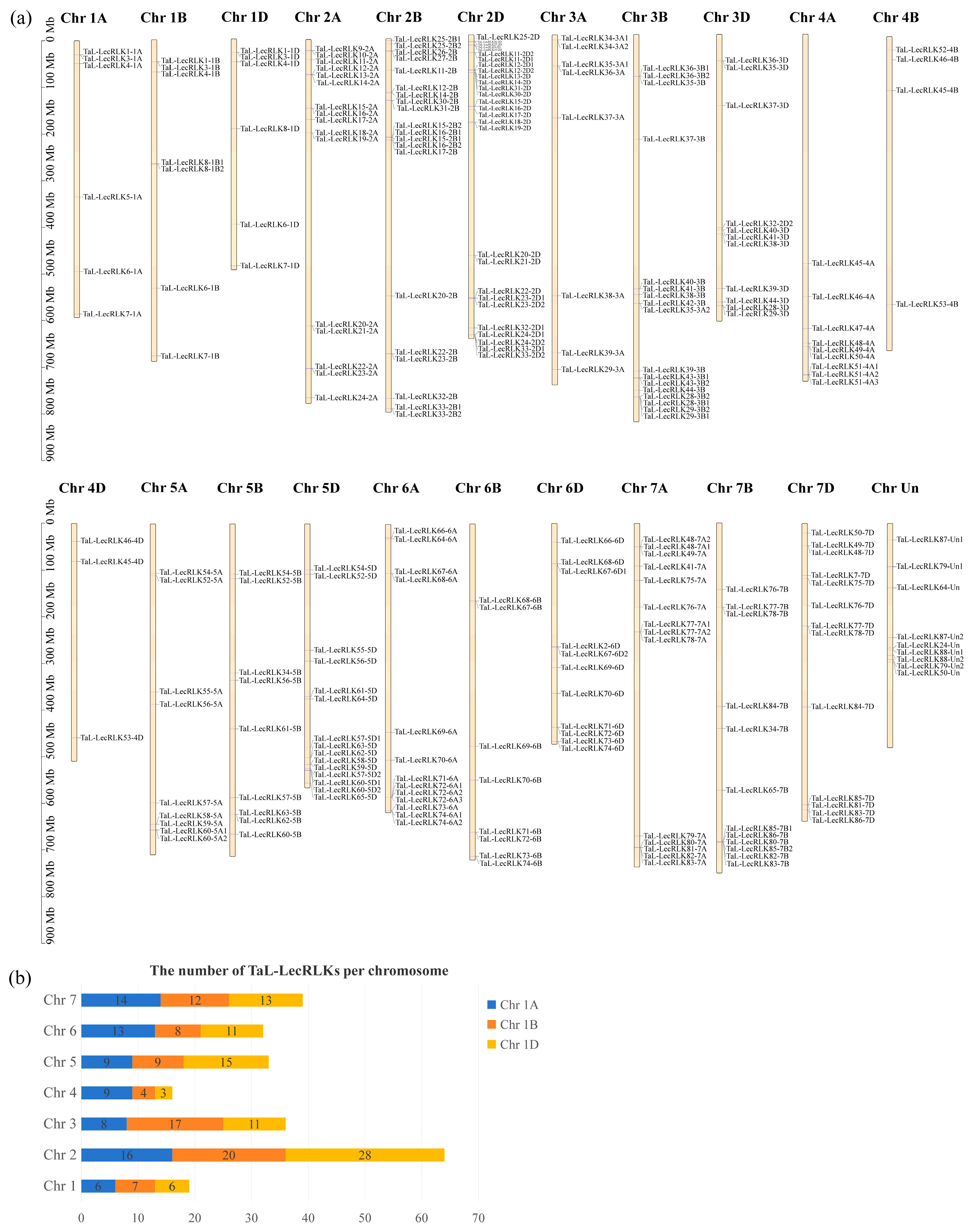
2.4. Chromosomal Localization and Homoeolog Identification of the TaL-LecRLKs
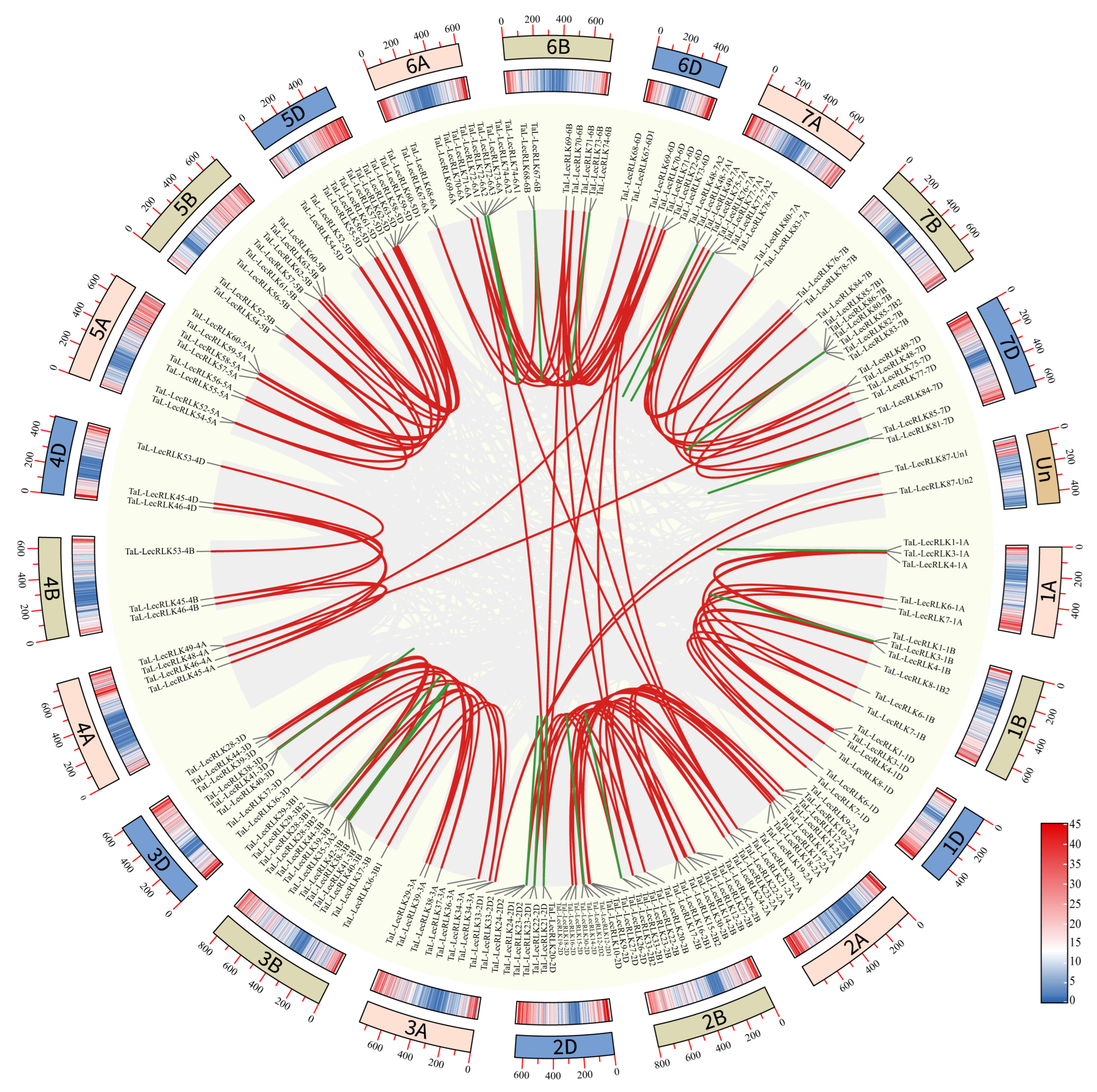
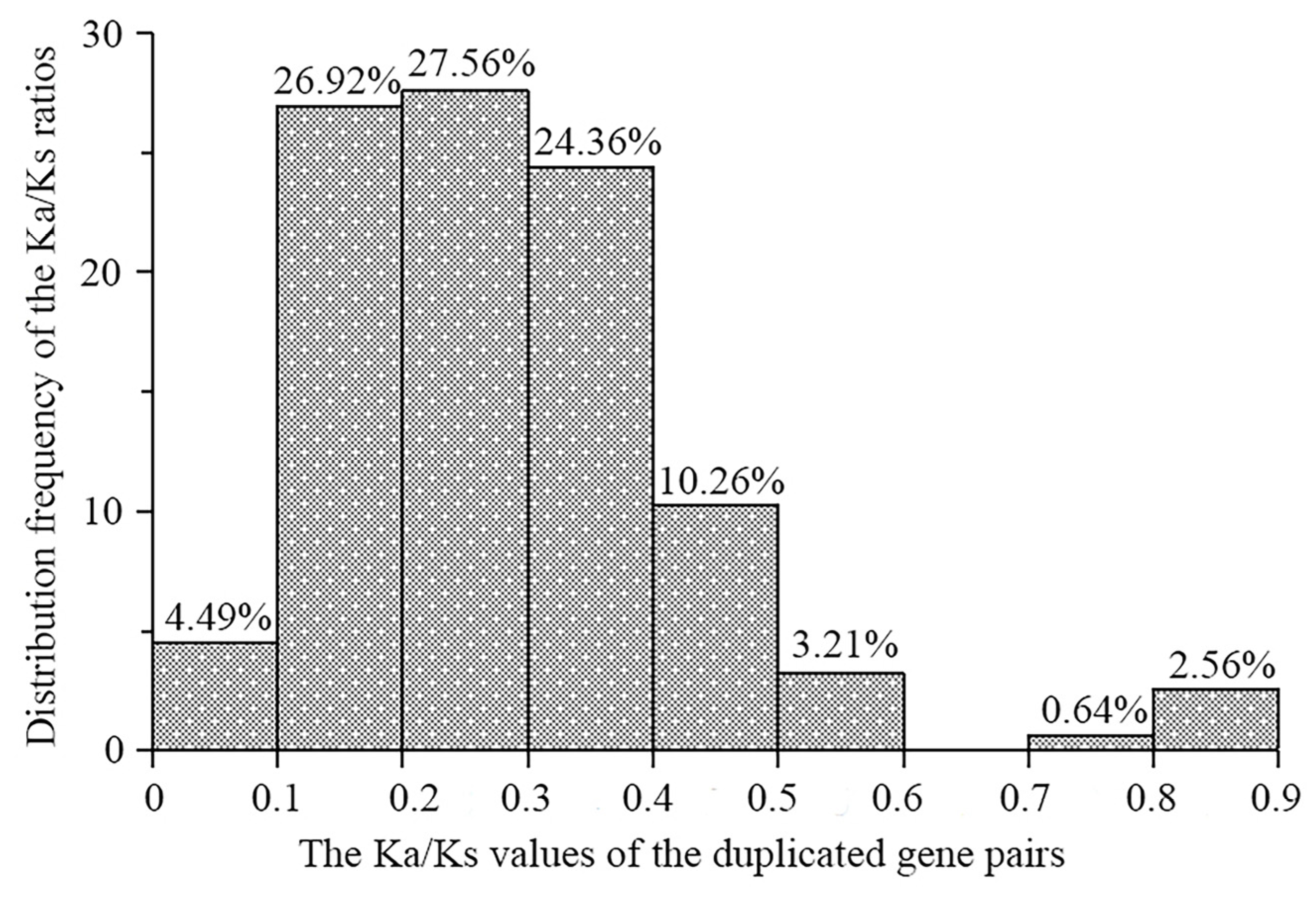
2.5. Duplication and Syntenic Analyses of the L-Type LecRLK Genes
2.6. Analysis of Cis-Acting Elements in the Promoters of the TaL-LecRLK Genes
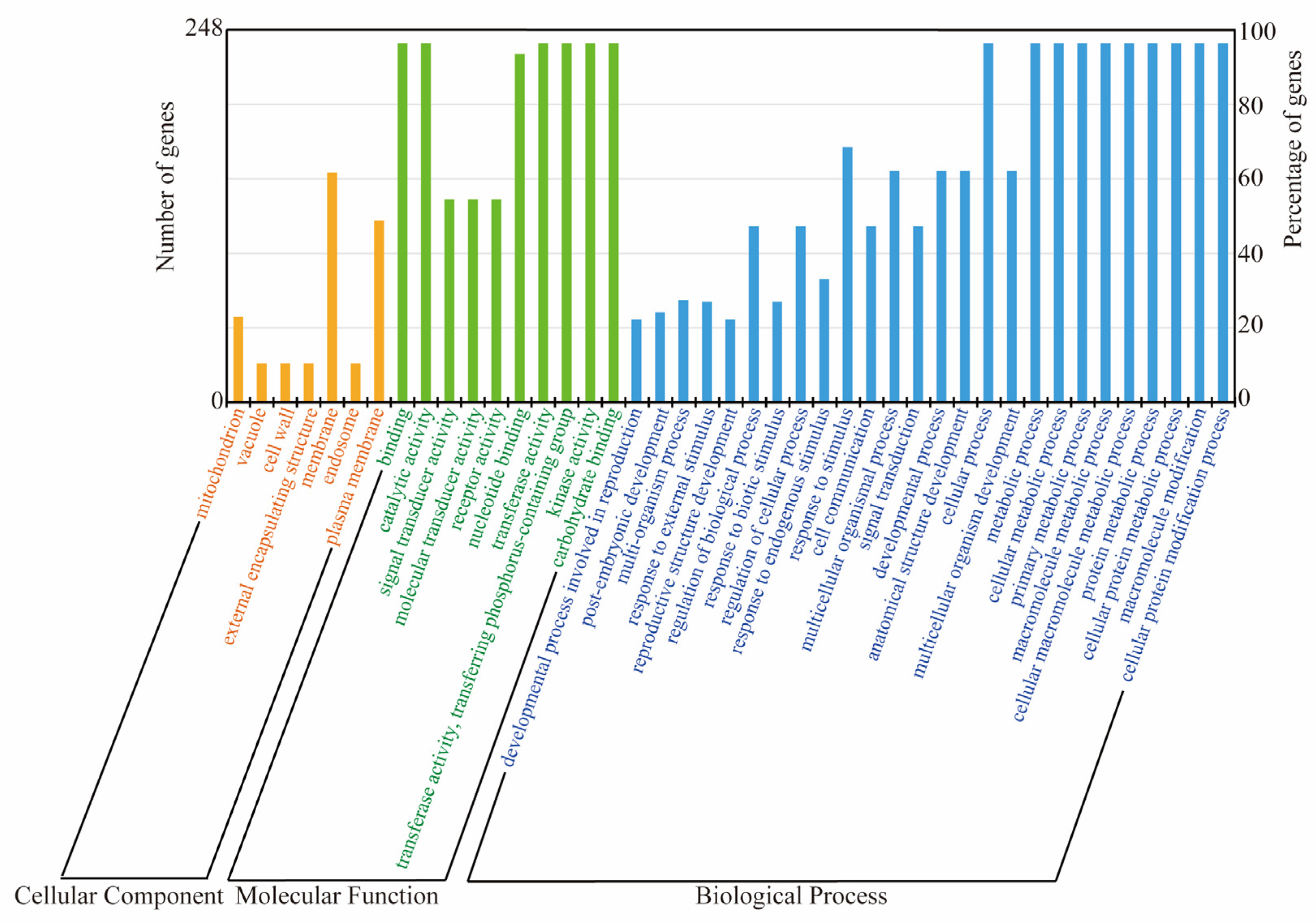
2.7. GO Analysis of the TaL-LecRLK Genes
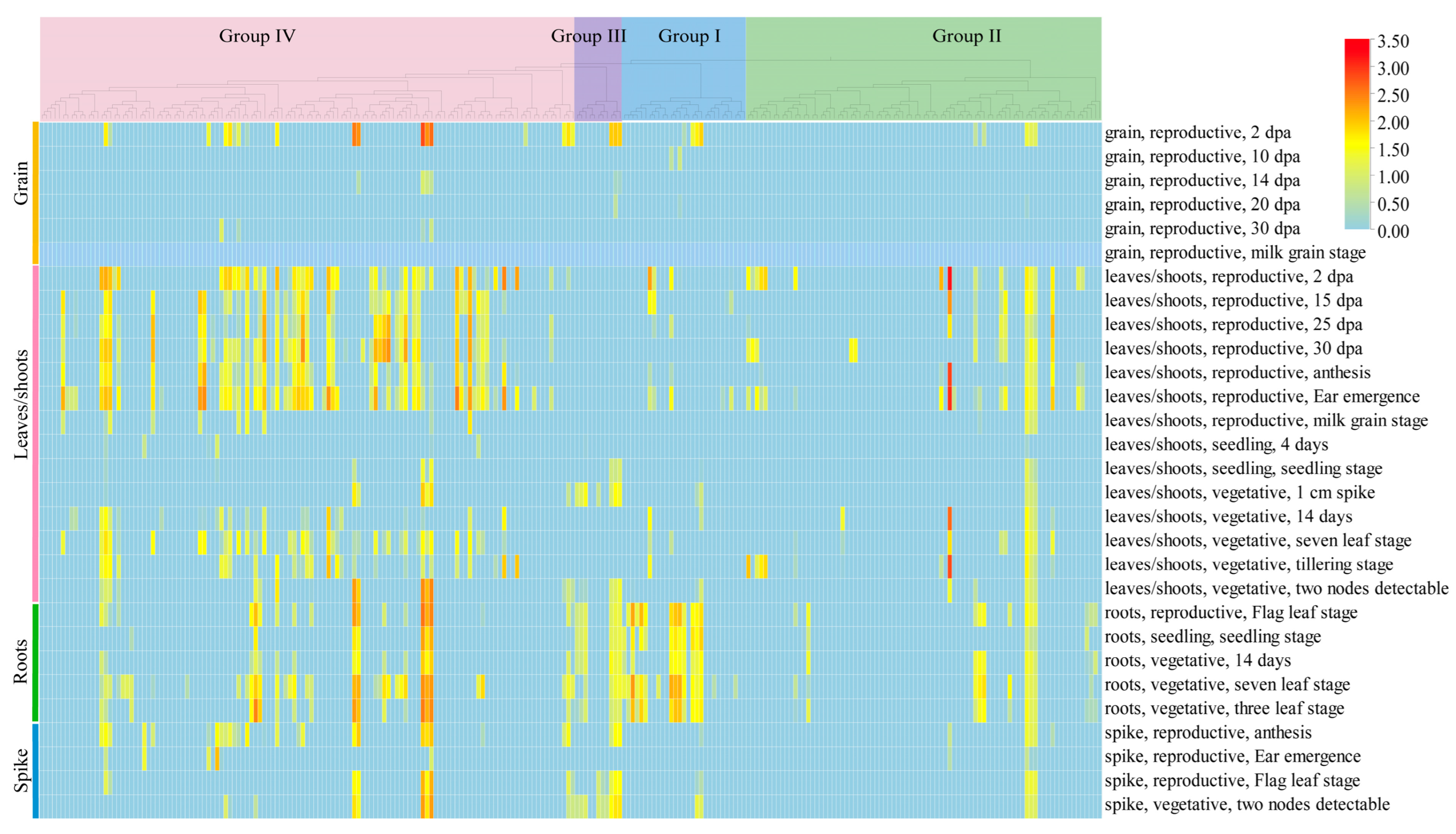
2.8. Spatiotemporal Expression Patterns of the TaL-LecRLK Genes
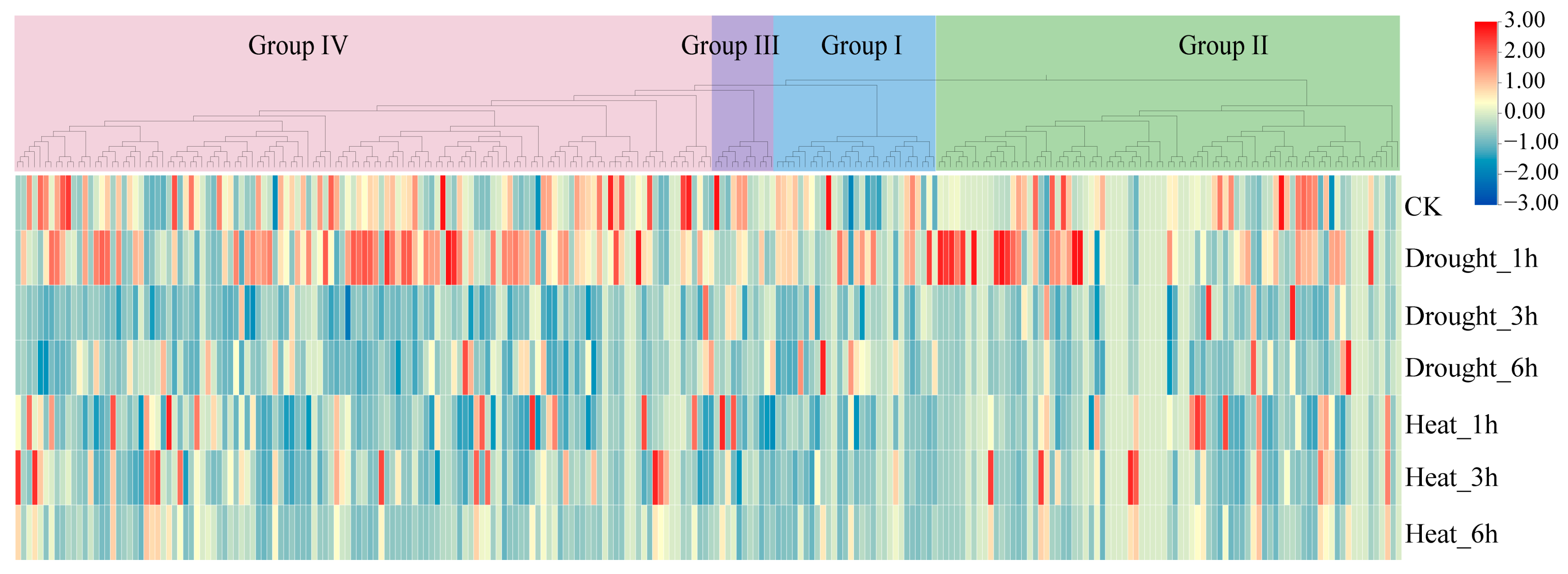
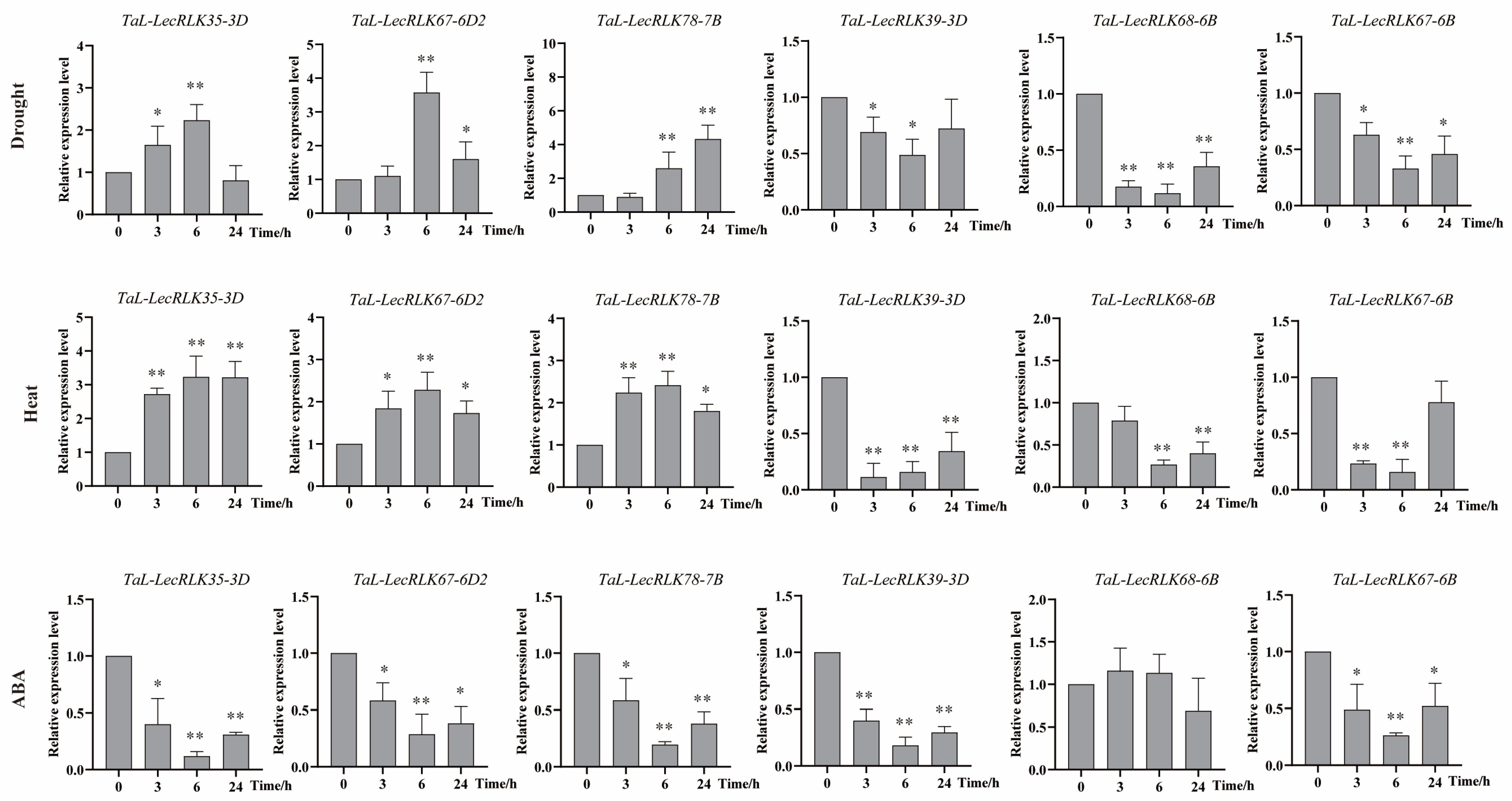
2.9. Abiotic Stress-Responsive Profiling of the TaL-LecRLK Genes
3. Discussion
4. Materials and Methods
4.1. Sequences’ Acquisition and Identification of the L-Type Lectin Receptor-like Kinase (L-LecRLK) Genes in Wheat
4.2. Physicochemical Properties and Subcellular Localization Prediction of TaL-LecRLKs
4.3. Phylogenetic Analysis and Classification of TaL-LecRLK Proteins
4.4. Gene Structure and Conserved Motifs’ Analysis of TaL-LecRLKs
4.5. Chromosomal Distribution and Homoeolog Identification of TaL-LecRLK Genes
4.6. Duplication and Syntenic Analysis of L-Type LecRLK Genes
4.7. Cis-Regulatory Element and Gene Ontology (GO) Analysis of TaL-LecRLKs
4.8. Tissue Expression Profiling of TaL-LecRLK Genes
4.9. Plant Cultivation, Growth Conditions, and Stress Treatments
4.10. RNA Isolation, RNA-Seq Library Preparation, and Illumina HiSeq 2000 Sequencing
4.11. Quantitative Real-Time PCR (qRT-PCR) Analysis and Statistical Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ammar, M.K.; Hanafi, R.S.; Choucry, M.A.; Handoussa, J. Structural, functional, nutritional composition and analytical profiling of Triticum aestivum L. Appl. Biol. Chem. 2023, 66, 48. [Google Scholar] [CrossRef]
- Li, J.; Yang, J.; Li, Y.; Ma, L. Current strategies and advances in wheat biology. Crop J. 2020, 8, 879–891. [Google Scholar] [CrossRef]
- Wang, J.; Luo, M.C.; Chen, Z.; You, F.M.; Wei, Y.; Zheng, Y.; Dvorak, J. Aegilops tauschii single nucleotide polymorphisms shed light on the origins of wheat D-genome genetic diversity and pinpoint the geographic origin of hexaploid wheat. New Phytol. 2013, 19, 925–937. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Guo, Y.; Kang, L.; Yin, C.; Bi, A.; Xu, D.; Zhang, Z.; Zhang, J.; Yang, X.; Xu, J.; et al. Population genomics unravels the Holocene history of bread wheat and its relatives. Nat. Plants 2023, 9, 403–419. [Google Scholar] [CrossRef]
- IWGSC. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361, eaar7191. [Google Scholar] [CrossRef]
- Ramírez-González, R.; Borrill, P.; Lang, D.; Harrington, S.; Brinton, J.; Venturini, L.; Davey, D.; Jacobs, J.; van Ex, F.; Pashaet, A.; et al. The transcriptional landscape of polyploid wheat. Science 2018, 361, eaar6089. [Google Scholar] [CrossRef]
- Sun, Y.; Qiao, Z.; Muchero, W.; Chen, J.G. Lectin receptor-like kinases: The sensor and mediator at the plant cell surface. Front. Plant Sci. 2020, 11, 596301. [Google Scholar] [CrossRef]
- Vaid, N.; Macovei, A.; Tuteja, N. Knights in action: Lectin receptor-like kinases in plant development and stress responses. Mol. Plant 2013, 6, 1405–1418. [Google Scholar] [CrossRef]
- De Coninck, T.; Van Damme, E.J.M. Plant lectins: Handymen at the cell surface. Cell Surf. 2022, 8, 100091. [Google Scholar] [CrossRef]
- Bellande, K.; Bono, J.J.; Savelli, B.; Jamet, E.; Canut, H. Plant lectins and lectin receptor-like kinases: How do they sense the outside? Int. J. Mol. Sci. 2017, 18, 1164. [Google Scholar] [CrossRef]
- Reidling, J.C.; Miller, M.A.; Steele, R.E. Sweet Tooth, a novel receptor protein-tyrosine kinase with C-type lectin-like extracellular domains. J. Biol. Chem. 2000, 275, 10323–10330. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.L.; Huang, Y.; Shi, P.H.; Yu, M.; Xie, J.B.; Xie, L. Duplication and diversification of lectin receptor-like kinases (LecRLK) genes in soybean. Sci. Rep. 2018, 8, 5861. [Google Scholar] [CrossRef] [PubMed]
- Vaid, N.; Pandey, P.K.; Tuteja, N. Genome-wide analysis of lectin receptor-like kinase family from Arabidopsis and rice. Plant Mol. Biol. 2012, 80, 365–388. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Liu, Y.W.; Zhou, J.M.; Zhao, S.P.; Zhang, X.H.; Min, D.H. Genome-wide analysis of the lectin receptor-like kinase family in foxtail millet (Setaria italica L.). Plant Cell Tiss. Organ Cult. 2016, 127, 335–346. [Google Scholar] [CrossRef]
- Ahmed, F.F.; Dola, F.S.; Islam, M.S.U.; Zohra, F.T.; Akter, N.; Rahman, S.M.; Sarkar, M.A.R. Genome-wide comprehensive identification and in silico characterization of lectin receptor-like kinase gene family in barley (Hordeum vulgare L.). Genet. Res. 2024, 2024, 2924953. [Google Scholar] [CrossRef]
- Shiu, S.H.; Bleecker, A.B. Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl. Acad. Sci. USA 2001, 98, 10763–10768. [Google Scholar] [CrossRef]
- Tang, D.; Wang, G.; Zhou, J.M. Receptor kinases in plant-pathogen interactions: More than pattern recognition. Plant Cell 2007, 29, 618–637. [Google Scholar] [CrossRef]
- Naithani, S.; Chookajorn, T.; Ripoll, D.R.; Nasrallah, J.B. Structural modules for receptor dimerization in the S-locus receptor kinase extracellular domain. Proc. Natl. Acad. Sci. USA 2007, 104, 12211–12216. [Google Scholar] [CrossRef]
- Wang, Y.; Bouwmeester, K. L-type lectin receptor kinases: New forces in plant immunity. PLoS Pathog. 2017, 13, e1006433. [Google Scholar] [CrossRef]
- Osterne, V.J.S.; Sloover, G.D.; Van Damme, E.J.M. Revisiting legume lectins: Structural organization and carbohydrate-binding properties. Carbohydr. Res. 2024, 544, 109241. [Google Scholar] [CrossRef]
- Morillo, S.A.; Tax, F.E. Functional analysis of receptor-like kinases in monocots and dicots. Curr. Opin. Plant Biol. 2006, 9, 460–469. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Weide, R.; Govers, F.; Bouwmeester, K. L-type lectin receptor kinases in Nicotiana benthamiana and tomato and their role in Phytophthora resistance. J. Exp. Bot. 2015, 66, 6731–6743. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Shang, J.; Chen, D.; Lei, C.; Zou, Y.; Zhai, W.; Liu, G.; Xu, J.; Ling, Z.; Cao, G.; et al. A B-lectin receptor kinase gene conferring rice blast resistance. Plant J. 2006, 46, 794–804. [Google Scholar] [CrossRef] [PubMed]
- Barre, A.; Hervé, C.; Lescure, B.; Rougé, P. Lectin receptor kinases in plants. Crit. Rev. Plant Sci. 2002, 21, 379–399. [Google Scholar] [CrossRef]
- Wang, R.; Li, C.; Jia, Z.; Su, Y.; Ai, Y.; Li, Q.; Guo, X.; Tao, Z.; Lin, F.; Liang, Y. Reversible phosphorylation of a lectin-receptor-like kinase controls xylem immunity. Cell Host Microbe 2023, 31, 2051–2066.e7. [Google Scholar] [CrossRef]
- Chen, K.; Wang, Q.; Yu, X.; Wang, C.; Gao, J.; Zhang, S.; Cheng, S.; You, S.; Zheng, H.; Lu, J.; et al. OsSRF8 interacts with OsINP1 and OsDAF1 to regulate pollen aperture formation in rice. Nat. Commun. 2024, 15, 4512. [Google Scholar] [CrossRef]
- Xiao, W.; Hu, S.; Zou, X.; Cai, R.; Liao, R.; Lin, X.; Yao, R.; Guo, X. Lectin receptor-like kinase LecRK-VIII.2 is a missing link in MAPK signaling-mediated yield control. Plant Physiol. 2021, 186, 445–458. [Google Scholar] [CrossRef]
- Mehla, S.; Singh, Y.; Kumar, U.; Balyan, P.; Singh, K.P.; Dhankher, O.P. Overexpression of rice lectin receptor-like kinase, OsLec-RLK, confers salinity stress tolerance and increases seed yield in pigeon pea (Cajanus cajan (L.) Millsp.). Plant Cell Rep. 2024, 43, 230. [Google Scholar] [CrossRef]
- Wu, F.; Qu, D.; Zhang, X.; Sun, Y.; Wang, J.; Zhu, D.; Yang, L.; Liu, X.; Tian, W.; Wang, L.; et al. PaLectinL7 enhances salt tolerance of sweet cherry by regulating lignin deposition in connection with PaCAD1. Tree Physiol. 2023, 43, 1986–2000. [Google Scholar] [CrossRef]
- Ma, N.; Liu, C.; Li, H.; Wang, J.; Zhang, B.; Lin, J.; Chang, Y. Genome-wide identification of lectin receptor kinases in pear: Functional characterization of the L-type LecRLK gene PbLRK138. Gene 2018, 661, 11–21. [Google Scholar] [CrossRef]
- Yang, X.; Chen, Z.; Lu, J.; Wei, X.; Yao, Y.; Lv, W.; Han, J.; Fei, J. Identification and characterization of the LecRLKs gene family in maize, and its role under biotic and abiotic stress. Biology 2024, 14, 20. [Google Scholar] [CrossRef] [PubMed]
- Xiong, G.; Cui, D.; Tian, Y.; Schwarzacher, T.; Heslop-Harrison, J.S.; Liu, Q. Genome-wide identification of the lectin receptor-like kinase gene family in Avena sativa and its role in salt stress tolerance. Int. J. Mol. Sci. 2024, 25, 12754. [Google Scholar] [CrossRef] [PubMed]
- Biswas, S.; Mondal, R.; Srivastava, A.; Trivedi, M.; Singh, S.K.; Mishra, Y. In silico characterization, molecular phylogeny, and expression profiling of genes encoding legume lectin-like proteins under various abiotic stresses in Arabidopsis thaliana. BMC Genom. 2022, 23, 480. [Google Scholar] [CrossRef] [PubMed]
- Shumayla; Sharma, S.; Pandey, A.K.; Singh, K.; Upadhyay, S.K. Molecular characterization and global expression analysis of lectin receptor kinases in bread wheat (Triticum aestivum). PLoS ONE 2016, 11, e0153925. [Google Scholar] [CrossRef]
- Lv, D.; Wang, G.; Xiong, L.R.; Sun, J.X.; Chen, Y.; Guo, C.L.; Yu, Y.; He, H.L.; Cai, R.; Pan, J.S. Genome-wide identification and characterization of lectin receptor-like kinase gene family in cucumber and expression profiling analysis under different treatments. Genes 2020, 11, 1032. [Google Scholar] [CrossRef]
- Haider, M.S.; De Britto, S.; Nagaraj, G.; Gurulingaiah, B.; Shekhar, R.; Ito, S.I.; Jogaiah, S. Genome-wide identification, diversification, and expression analysis of lectin receptor-like kinase (LecRLK) gene family in cucumber under biotic stress. Int. J. Mol. Sci. 2021, 22, 6585. [Google Scholar] [CrossRef]
- Birchler, J.A.; Yang, H. The multiple fates of gene duplications: Deletion, hypofunctionalization, subfunctionalization, neofunctionalization, dosage balance constraints, and neutral variation. Plant Cell 2022, 34, 2466–2474. [Google Scholar] [CrossRef]
- Cui, L.; Cheng, H.; Yang, Z.; Xia, C.; Zhang, L.; Kong, X. Comparative analysis reveals different evolutionary fates and biological functions in wheat duplicated genes (Triticum aestivum L.). Plants 2023, 12, 3021. [Google Scholar] [CrossRef]
- Li, Y.; Xiang, R.; Liu, K.; Ahmad, B.; Zhang, X.; Yang, L.; Tian, Y.; Shi, X.; Du, G.; Wang, L. Genomic-organization and expression profiling of lectin receptor kinases genes suggest their involvement in multiple biological processes. Sci. Hortic. 2024, 329, 113042. [Google Scholar] [CrossRef]
- Wang, T.; Duan, S.; Xu, C.; Wang, Y.; Zhang, X.; Xu, X.; Chen, L.; Han, Z.; Wu, T. Pan-genome analysis of 13 Malus accessions reveals structural and sequence variations associated with fruit traits. Nat. Commun. 2023, 14, 7377. [Google Scholar] [CrossRef]
- Jeffares, D.C.; Penkett, C.J.; Bähler, J. Rapidly regulated genes are intron poor. Trends Genet. 2008, 24, 375–378. [Google Scholar] [CrossRef] [PubMed]
- Chung, B.Y.; Simons, C.; Firth, A.E.; Brown, C.M.; Hellens, R.P. Effect of 5’UTR introns on gene expression in Arabidopsis thaliana. BMC Genom. 2006, 7, 120. [Google Scholar] [CrossRef] [PubMed]
- Aviña-Padilla, K.; Ramírez-Rafael, J.A.; Herrera-Oropeza, G.E.; Muley, V.Y.; Valdivia, D.I.; Díaz-Valenzuela, E.; García-García, A.; Varela-Echavarría, A.; Hernández-Rosales, M. Evolutionary perspective and expression analysis of intronless genes highlight the conservation of their regulatory role. Front. Genet. 2021, 12, 654256. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Zhou, L.; Jiang, P.; Lu, R.; Halford, N.G.; Liu, C. Genome-wide identification of sucrose nonfermenting-1-related protein kinase (SnRK) genes in barley and RNA-seq analyses of their expression in response to abscisic acid treatment. BMC Genom. 2021, 22, 300. [Google Scholar] [CrossRef]
- Marcussen, T.; Sandve, S.R.; Heier, L.; Spannagl, M.; Pfeifer, M.; International Wheat Genome Sequencing Consortium; Jakobsen, K.S.; Wulff, B.B.; Steuernagel, B.; Mayer, K.F.; et al. Ancient hybridizations among the ancestral genomes of bread wheat. Science 2014, 345, 1250092. [Google Scholar] [CrossRef]
- Feng, X.; Chen, Q.; Wu, W.; Wang, J.; Li, G.; Xu, S.; Shao, S.; Liu, M.; Zhong, C.; Wu, C.I.; et al. Genomic evidence for rediploidization and adaptive evolution following the whole-genome triplication. Nat. Commun. 2024, 15, 1635. [Google Scholar] [CrossRef]
- Ren, R.; Wang, H.; Guo, C.; Zhang, N.; Zeng, L.; Chen, Y.; Ma, H. Widespread whole genome duplications contribute to genome complexity and species diversity in angiosperms. Mol. Plant 2018, 11, 414–428. [Google Scholar] [CrossRef]
- Li, J.; Cui, C.; Han, F.; Liu, J. Genome-wide identification and analysis of the UBA2 gene family in wheat (Triticum aestivum L.). BMC Genom. 2025, 26, 180. [Google Scholar] [CrossRef]
- Das, M.; Haberer, G.; Panda, A.; Das Laha, S.; Ghosh, T.C.; Schäffner, A.R. Expression pattern similarities support the prediction of orthologs retaining common functions after gene duplication events. Plant Physiol. 2016, 171, 2343–2357. [Google Scholar] [CrossRef]
- Yang, Q.; Han, X.M.; Gu, J.K.; Liu, Y.J.; Yang, M.J.; Zeng, Q.Y. Functional and structural profiles of GST gene family from three Populus species reveal the sequence-function decoupling of orthologous genes. New Phytol. 2019, 221, 1060–1073. [Google Scholar] [CrossRef]
- Schmitz, R.J.; Grotewold, E.; Stam, M. Cis-regulatory sequences in plants: Their importance, discovery, and future challenges. Plant Cell 2022, 34, 718–741. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Wang, C.; Ding, Z.; Zhao, Y.; Dai, J.; Li, J.; Huang, H.; Wang, T.; Zhu, M.; Feng, M.; et al. A plant NLR receptor employs ABA central regulator PP2C-SnRK2 to activate antiviral immunity. Nat. Commun. 2024, 15, 3205. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Wang, Z.; Dai, X.; Gao, J.; Zhao, J.; Ma, R.; Chen, Y.; Sun, Y.; Ma, H.; Li, S.; et al. A COMPASS histone H3K4 trimethyltransferase pentamer transactivates drought tolerance and growth/biomass production in Populus trichocarpa. New Phytol. 2024, 241, 1950–1972. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Zhang, F.; Du, X.; Zhang, X.; Huang, X.; Li, Z.; Zhang, Y.; Gan, P.; Li, H.; Li, M.; et al. TaANK-TPR1 enhances wheat resistance against stripe rust via controlling gene expression and protein activity of NLR protein TaRPP13L1. Dev. Cell 2025, 60, 1–17. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, J.; Li, J.; Li, H.; Zhang, G. The functional and regulatory mechanisms of the Thellungiella salsuginea Ascorbate Peroxidase 6 (TsAPX6) in response to salinity and water deficit stresses. PLoS ONE 2016, 11, e0154042. [Google Scholar] [CrossRef]
- Sato, H.; Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Complex plant responses to drought and heat stress under climate change. Plant J. 2024, 117, 1873–1892. [Google Scholar] [CrossRef]
- Yekondi, S.; Liang, F.C.; Okuma, E.; Radziejwoski, A.; Mai, H.W.; Swain, S.; Singh, P.; Gauthier, M.; Chien, H.C.; Murata, Y.; et al. Nonredundant functions of Arabidopsis LecRKV.2 and LecRK-Vll.1 in controlling stomatal immunity andjasmonate-mediated stomatal closurey. New Phytol. 2018, 218, 253–268. [Google Scholar] [CrossRef]
- Balagué, C.; Gouget, A.; Bouchez, O.; Souriac, C.; Haget, N.; Boutet-Mercey, S.; Govers, F.; Roby, D.; Canut, H. The Arabidopsis thaliana lectin receptor kinase LecRK-I.9 is required for full resistance to Pseudomonas syringae and affects jasmonate signalling. Mol. Plant Pathol. 2017, 18, 937–948. [Google Scholar] [CrossRef]
- Sun, M.; Qian, X.; Chen, C.; Cheng, S.; Jia, B.; Zhu, Y.; Sun, X. Ectopic expression of GsSRK in Medicago sativa reveals its involvement in plant architecture and salt stress responses. Front. Plant Sci. 2018, 9, 226. [Google Scholar] [CrossRef]
- Osman, M.E.M.; Osman, R.S.H.; Elmubarak, S.A.; Ibrahim, M.A.; Abakar, H.B.M.; Dirar, A.I.; Konozy, E.H.E. In silico analysis of L- and G-type lectin receptor kinases in tomato: Evolution, diversity, and abiotic responses. BMC Genom. 2024, 25, 1143. [Google Scholar] [CrossRef]
- Bolser, D.M.; Staines, D.M.; Perry, E.; Kersey, P.J. Ensembl plants: Integrating tools for visualizing, mining, and analyzing plant genomic data. Methods Mol. Biol. 2017, 1533, 1–31. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, Y.; de la Bastide, M.; Hamilton, J.P.; Kanamori, H.; McCombie, W.R.; Ouyang, S.; Schwartz, D.C.; Tanaka, T.; Wu, J.; Zhou, S.; et al. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 2013, 6, 4. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef]
- Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Gonzales, N.R.; Gwadz, M.; Lu, S.; Marchler, G.H.; Song, J.S.; Thanki, N.; Yamashita, R.A.; et al. The conserved domain database in 2023. Nucleic Acids Res. 2023, 51, D384–D388. [Google Scholar] [CrossRef]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; de Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E.; et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012, 40, W597–W603. [Google Scholar] [CrossRef]
- Chou, K.C.; Shen, H.B. Plant-mPLoc: A top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS ONE 2010, 5, e11335. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Edgar, R.C. Muscle5: High-accuracy alignment ensembles enable unbiased assessments of sequence homology and phylogeny. Nat. Commun. 2022, 13, 6968. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Schilling, S.; Kennedy, A.; Pan, S.; Jermiin, L.S.; Melzer, R. Genome-wide analysis of MIKC-type MADS-box genes in wheat: Pervasive duplications, functional conservation and putative neofunctionalization. New Phytol. 2020, 225, 511–529. [Google Scholar] [CrossRef]
- Xu, X.; Zhang, L.; Zhao, W.; Fu, L.; Han, Y.; Wang, K.; Yan, L.; Li, Y.; Zhang, X.H.; Min, D.H. Genome-wide analysis of the serine carboxypeptidase-like protein family in Triticum aestivum reveals TaSCPL184-6D is involved in abiotic stress response. BMC Genom. 2021, 22, 350. [Google Scholar] [CrossRef]
- Zhang, Z. KaKs_calculator 3.0: Calculating selective pressure on coding and non-coding sequences. Genom. Proteom. Bioinf. 2022, 20, 536–540. [Google Scholar] [CrossRef]
- Han, Y.; Zhang, L.; Yan, L.; Xiong, X.; Wang, W.; Zhang, X.H.; Min, D.H. Genome-wide analysis of TALE superfamily in Triticum aestivum reveals TaKNOX11-A is involved in abiotic stress response. BMC Genom. 2022, 23, 89. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Tian, T.; Liu, Y.; Yan, H.; You, Q.; Yi, X.; Du, Z.; Xu, W.; Su, Z. agriGO v2.0: A GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 2017, 45, W122–W129. [Google Scholar] [CrossRef] [PubMed]
- Kan, W.; Gao, Y.; Zhu, Y.; Wang, Z.; Yang, Z.; Cheng, Y.; Guo, J.; Wang, D.; Tang, C.; Wu, L. Genome-wide identification and expression analysis of TaFDL gene family responded to vernalization in wheat (Triticum aestivum L.). BMC Genom. 2025, 26, 255. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Xing, Q.; Bu, J.; Han, W.; Shen, Z. Integrated metabolomic and transcriptomic analysis reveals the regulatory mechanisms of flavonoid and alkaloid biosynthesis in the new and old leaves of Murraya tetramera Huang. BMC Plant Biol. 2024, 24, 499. [Google Scholar] [CrossRef] [PubMed]
- Burchardt, S.; Czernicka, M.; Kućko, A.; Pokora, W.; Kapusta, M.; Domagalski, K.; Jasieniecka-Gazarkiewicz, K.; Karwaszewski, J.; Wilmowicz, E. Exploring the response of yellow lupine (Lupinus luteus L.) root to drought mediated by pathways related to phytohormones, lipid, and redox homeostasis. BMC Plant Biol. 2024, 24, 1049. [Google Scholar] [CrossRef]
- Udvardi, M.K.; Czechowski, T.; Scheible, W.R. Eleven golden rules of quantitative RT-PCR. Plant Cell 2008, 20, 1736–1737. [Google Scholar] [CrossRef]
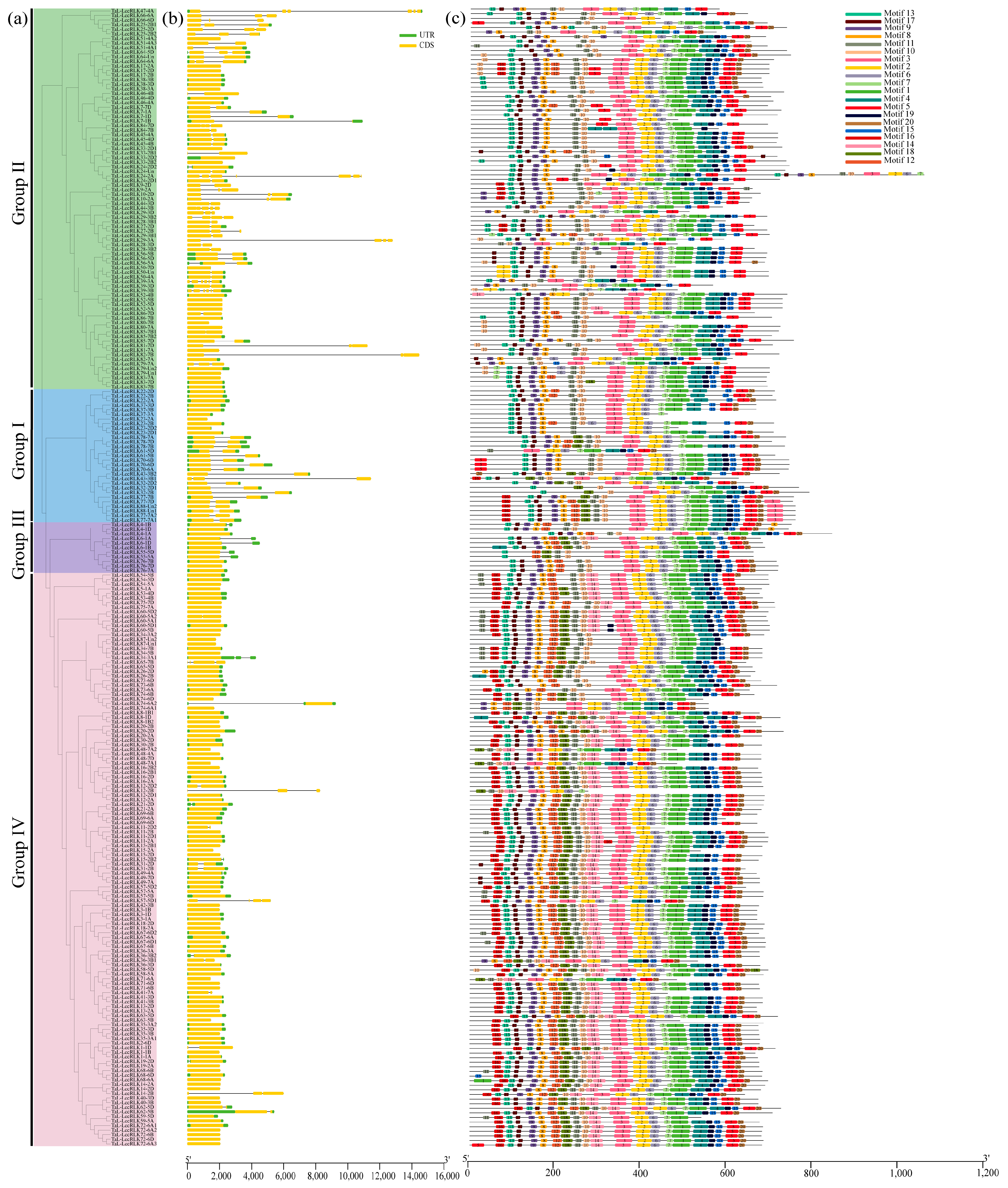
| Subfamily | 0 Intron | 1 Intron | 2 Introns | 3 Introns | 4 Introns | 5 Introns | 6 Introns | Total |
|---|---|---|---|---|---|---|---|---|
| Group I | 10 | 18 | 1 | 0 | 0 | 0 | 0 | 29 |
| Group II | 28 | 31 | 15 | 3 | 2 | 3 | 1 | 83 |
| Group III | 6 | 5 | 0 | 0 | 0 | 0 | 0 | 11 |
| Group IV | 104 | 16 | 3 | 1 | 1 | 0 | 0 | 125 |
| Total | 148 | 70 | 19 | 4 | 3 | 3 | 1 | 248 |
| MOTIF | ID | WIDTH |
|---|---|---|
| 1 | YLHEEWEQVVIHRDIKASNVLLDSSMNGRLGDFGLARLYDH | 41 |
| 2 | KRVSHDSRQGMKEFVAEVVSIGRLRHRNLVQLL | 33 |
| 3 | WEVEFGPHRFSYKDLFRATKGFSEKNLLGRGGFGSVYK | 38 |
| 4 | HVVGTMGYJAPELVRTGKATPETDVFAFGVFLLE | 34 |
| 5 | YDADEAELVLKLGLLCSHPDPSARPSMRQ | 29 |
| 6 | GYCRRKGELLLVYEYMPNGSL | 21 |
| 7 | WPQRYKIIKGVASAL | 15 |
| 8 | INDNHVGIDVNSLVS | 15 |
| 9 | NGNGSNRIVAVEFDT | 15 |
| 10 | HYVLGWSFSSDGPAP | 15 |
| 11 | VLPETVYVGFSAATG | 15 |
| 12 | GAFQNLSLISGKAMQVWVDYD | 21 |
| 13 | TGEVASFSTSFVFAI | 15 |
| 14 | JDISKLPKLPRLGPKPRSKVLEIVLPIAT | 29 |
| 15 | LVDWVWELYGRGAJL | 15 |
| 16 | GLLELTNGTSQLKGHAFHPTP | 21 |
| 17 | GDGMAFFLAPS | 11 |
| 18 | ATQINVTLAPLGVAKPARPLLSA | 23 |
| 19 | VACGRRPIEQNAEDN | 15 |
| 20 | VMQYLDGDAPLPELP | 15 |
| Homoeologous Groups (A:B:D) | All Wheat Genes 1 | All wheat TaL-LecRLK Genes | ||
|---|---|---|---|---|
| Number of Groups | Number of Genes | % of Genes 2 | ||
| 1:1:1 | 35.80% | 24 | 72 | 29.00% |
| n:1:1/1:n:1/1:1:n 3 | 5.70% | 27 | 54 | 21.80% |
| 1:1:0/1:0:1/0:1:1 | 13.20% | 15 | 61 | 24.60% |
| Other ratios | 8.00% | 18 | 57 | 23.00% |
| Orphans/Singletons | 37.10% | - | 4 | 1.60% |
| Total | 99.80% | - | 248 | 100% |
| Category | Function | Site Name | Number | Percentage for Each Category (%) | Percentage of the Total Number (%) |
|---|---|---|---|---|---|
| Hormonal responsiveness (2812, 49.1%) | abscisic acid responsiveness | ABRE | 810 | 28.81 | 13.96 |
| MeJA-responsiveness | CGTCA-motif | 739 | 26.28 | 12.73 | |
| TGACG-motif | 735 | 26.14 | 12.66 | ||
| gibberellin-responsiveness | P-box | 103 | 3.66 | 1.77 | |
| TATC-box | 44 | 1.56 | 0.76 | ||
| GARE-motif | 28 | 1.00 | 0.48 | ||
| auxin-responsive element | TGA-element | 166 | 5.90 | 2.86 | |
| AuxRR-core | 40 | 1.42 | 0.69 | ||
| salicylic acid responsiveness | TCA-element | 146 | 5.19 | 2.52 | |
| SARE | 1 | 0.04 | 0.02 | ||
| Environmental adaptation (2418, 41.7%) | light responsive element | G-Box | 868 | 35.90 | 14.96 |
| Sp1 | 212 | 8.77 | 3.65 | ||
| GT1-motif | 152 | 6.29 | 2.62 | ||
| ACE | 48 | 1.99 | 0.83 | ||
| MRE | 48 | 1.99 | 0.83 | ||
| 3-AF1 binding site | 13 | 0.54 | 0.22 | ||
| AAAC-motif | 5 | 0.21 | 0.09 | ||
| C-box | 3 | 0.12 | 0.05 | ||
| 4cl-CMA2b | 1 | 0.04 | 0.02 | ||
| anaerobic induction | ARE | 413 | 17.08 | 7.12 | |
| drought-inducibility | MBS | 261 | 10.79 | 4.50 | |
| low-temperature responsiveness | LTR | 171 | 7.07 | 2.95 | |
| anoxic specific inducibility | GC-motif | 127 | 5.25 | 2.19 | |
| defense and stress responsiveness | TC-rich repeats | 86 | 3.56 | 1.48 | |
| wound-responsive element | WUN-motif | 8 | 0.33 | 0.14 | |
| dehydration, low-temperature, salt stresses | DRE | 2 | 0.08 | 0.03 | |
| Plant growth, development, and metabolism (534, 9.1%) | meristem expression | CAT-box | 235 | 44.01 | 4.05 |
| zein metabolism regulation | O2-site | 143 | 26.78 | 2.46 | |
| endosperm expression | GCN4_motif | 52 | 9.74 | 0.90 | |
| seed-specific regulation | RY-element | 36 | 6.74 | 0.62 | |
| circadian control | circadian | 34 | 6.37 | 0.59 | |
| cell cycle regulation | MSA-like | 27 | 5.06 | 0.47 | |
| differentiation of the palisade mesophyll cells | HD-Zip 1 | 7 | 1.31 | 0.12 | |
| Total | 5804 | 100.00 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, W.; Zhang, F.; Wang, J.; Fang, S.; Cheng, Z.; Ma, X.; Fan, J.; Xu, Z.; Chen, X. Comprehensive Genome-Wide Characterization of L-Type Lectin Receptor-like Kinase (L-LecRLK) Genes in Wheat (Triticum aestivum L.) and Their Response to Abiotic Stress. Plants 2025, 14, 1884. https://doi.org/10.3390/plants14121884
Zhao W, Zhang F, Wang J, Fang S, Cheng Z, Ma X, Fan J, Xu Z, Chen X. Comprehensive Genome-Wide Characterization of L-Type Lectin Receptor-like Kinase (L-LecRLK) Genes in Wheat (Triticum aestivum L.) and Their Response to Abiotic Stress. Plants. 2025; 14(12):1884. https://doi.org/10.3390/plants14121884
Chicago/Turabian StyleZhao, Wan, Fuyan Zhang, Jiahuan Wang, Shuai Fang, Zhongjie Cheng, Xuhui Ma, Jialin Fan, Zhaoshi Xu, and Xiaojie Chen. 2025. "Comprehensive Genome-Wide Characterization of L-Type Lectin Receptor-like Kinase (L-LecRLK) Genes in Wheat (Triticum aestivum L.) and Their Response to Abiotic Stress" Plants 14, no. 12: 1884. https://doi.org/10.3390/plants14121884
APA StyleZhao, W., Zhang, F., Wang, J., Fang, S., Cheng, Z., Ma, X., Fan, J., Xu, Z., & Chen, X. (2025). Comprehensive Genome-Wide Characterization of L-Type Lectin Receptor-like Kinase (L-LecRLK) Genes in Wheat (Triticum aestivum L.) and Their Response to Abiotic Stress. Plants, 14(12), 1884. https://doi.org/10.3390/plants14121884







