Identification and Expression Profiles of Xyloglucan Endotransglycosylase/Hydrolase Family in Response to Drought Stress in Larix kaempferi
Abstract
1. Introduction
2. Results
2.1. Identification and Characterization of XTHs in L. kaempferi
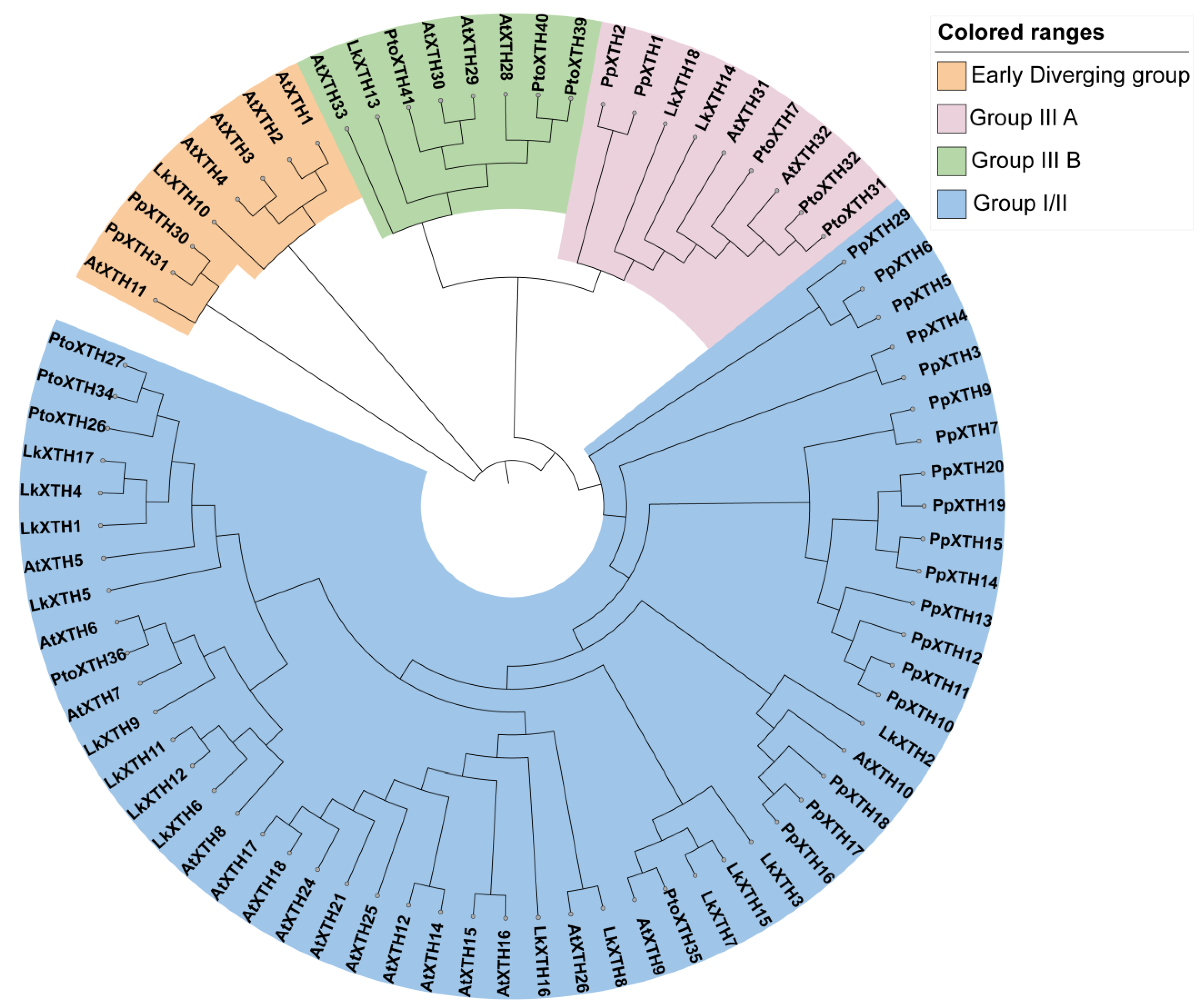
2.2. Phylogenetic Analysis of XTH Proteins
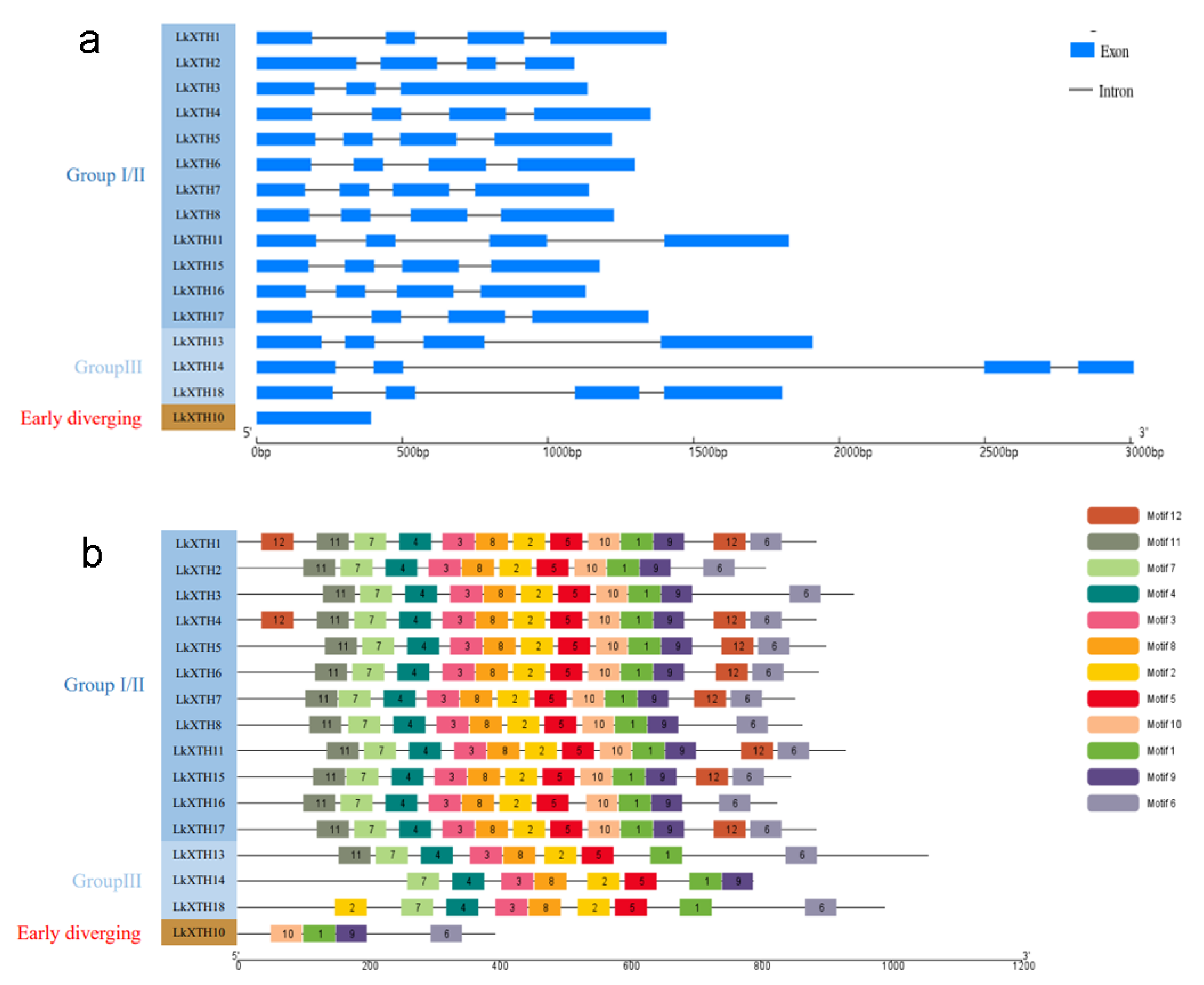
2.3. Gene Structures and Conserved Motif Analyses of LkXTH
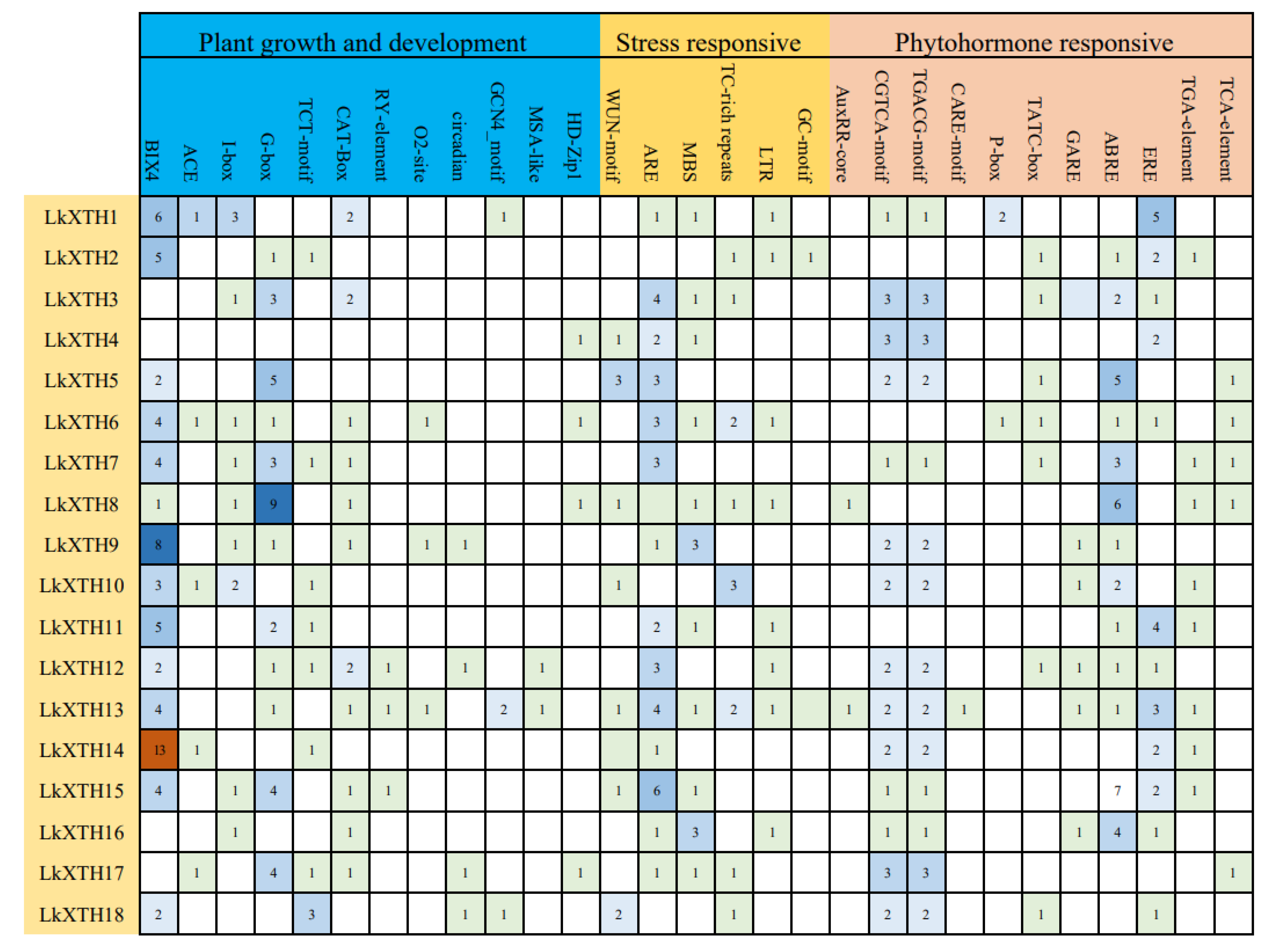
2.4. Cis-Element Analyses of the Promoter Regions of LkXTH Genes
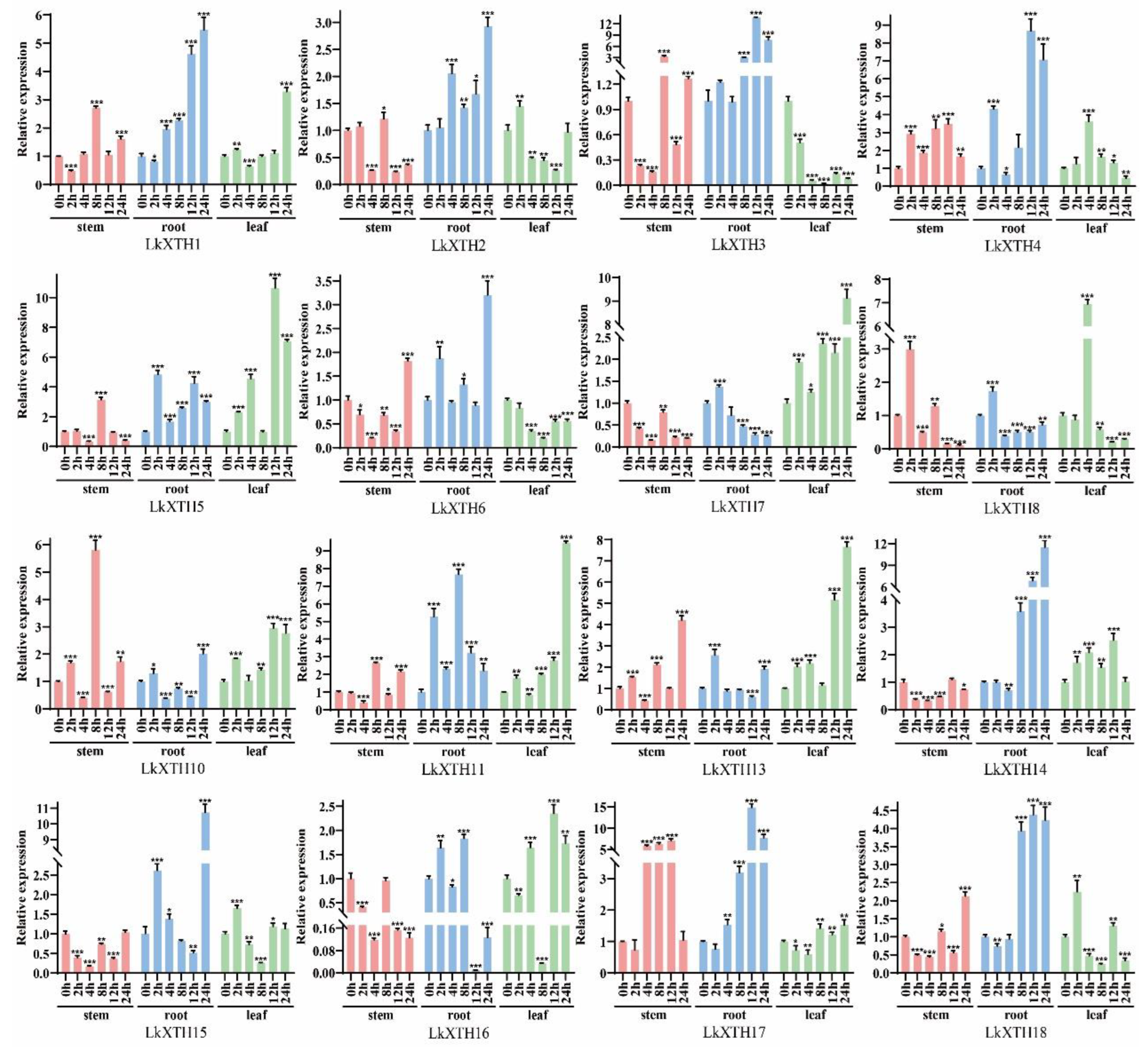
2.5. Expression Pattern Analyses of LkXTH Genes in Different Organs
2.6. LkXTH Genes Respond to Drought Stress in Different Organs
2.7. Osmotolerance of Yeast Transformants
3. Discussion
4. Materials and Methods
4.1. Plant Material and Treatments
4.2. Identification of XTH Genes in L. kaempferi
4.3. Phylogenetic Analysis
4.4. Conserved Motif and Gene Structure Analyses
4.5. Cis-Regulatory Element Analyses
4.6. RNA Extraction and Quantitative Real-Time PCR Expression Analyses
4.7. Osmotolerance of Yeast Transformants
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Cosgrove, D.J. Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 2005, 6, 850–861. [Google Scholar] [CrossRef] [PubMed]
- Shinohara, N.; Sunagawa, N.; Tamura, S.; Yokoyama, R.; Ueda, M.; Igarashi, K.; Nishitani, K. The plant cell-wall enzyme AtXTH3 catalyses covalent cross-linking between cellulose and cello-oligosaccharide. Sci. Rep. 2007, 7, 46099. [Google Scholar] [CrossRef]
- Smithers, E.T.; Luo, J.; Dyson, R.J. A continuum mechanics model of the plant cell wall reveals interplay between enzyme action and cell wall structure. Eur. Phys. J. E 2024, 47, 1. [Google Scholar] [CrossRef] [PubMed]
- Qiao, T.; Zhang, L.; Yu, Y.; Pang, Y.; Tang, X.; Wang, X.; Li, L.; Li, B.; Sun, Q. Identification and expression analysis of xyloglucan endotransglucosylase/hydrolase (XTH) family in grapevine (Vitis vinifera L.). PeerJ 2022, 10, e13546. [Google Scholar] [CrossRef]
- Wu, J.; Zong, Y.; Tu, Z.; Yang, L.; Li, W.; Cui, Z.; Hao, Z.; Li, H. Genome-wide identification of XTH genes in Liriodendron chinense and functional characterization of LcXTH21. Front. Plant Sci. 2022, 13, 1014339. [Google Scholar] [CrossRef]
- Yang, L.; Chen, Y.; Liu, X.J.; Zhang, S.; Han, Q.Q. Genome-wide identification and expression analysis of xyloglucan endotransglucosylase/hydrolase genes family in Salicaceae during grafting. BMC Genom. 2023, 24, 676. [Google Scholar] [CrossRef]
- Li, X.L.; Su, Q.F.; Feng, Y.C.; Gao, X.H.; Wang, B.C.; Tahir, M.M.; Yang, H.J.; Zhao, Z.Y. Identification and analysis of the xyloglucan endotransferase/hydrolase (XTH) family genes in apple. Sci. Hortic. 2023, 315, 111990–112003. [Google Scholar] [CrossRef]
- Nishitani, K. The role of endoxyloglucan transferase in the organization of plant cell walls. Int. Rev. Cytol. 1997, 173, 157–206. [Google Scholar]
- Wang, M.; Xu, Z.C.; Ding, A.M.; Kong, Y.Z. Genome-wide identification and expression profiling analysis of the xyloglucan endotransglucosylase/hydrolase gene family in tobacco (Nicotiana tabacum L.). Genes 2018, 9, 273. [Google Scholar] [CrossRef]
- Nishikubo, N.; Takahashi, J.; Roos, A.A.; Derba-Maceluch, M.; Piens, K.; Brumer, H.; Teeri, T.T.; Mellerowicz, H. Xyloglucan endo-transglycosylase-mediated xyloglucan rearrangements in developing wood of hybrid aspen. Plant Physiol. 2011, 155, 399–413. [Google Scholar] [CrossRef]
- Rose, J.K.C.; Braam, J.; Fry, S.C.; Nishitani, K. The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: Current perspectives and a new unifying nomenclature. Plant Cell Physiol. 2002, 43, 1421–1435. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Liu, A.; Qu, X.; Liang, J.; Song, M. Genome-wide identification, and phylogenetic and expression profiling analyses of XTH gene families in Brassica rapa L. and Brassica oleracea L. BMC Genom. 2020, 21, 782. [Google Scholar] [CrossRef]
- Fu, M.; Liu, C.; Wu, F. Genome-wide identification, characterization, and expression analysis of xyloglucan endotransglucosylase/hydrolase genes family in barley (Hordeum vulgare). Molecules 2019, 24, 1935. [Google Scholar] [CrossRef]
- Yokoyama, R.; Nishitani, K. A comprehensive expression analysis of all members of a gene family encoding cell-wall enzymes allowed us to predict cis-regulatory regions involved in cell-wall construction in specific organs of Arabidopsis. Plant Cell Physiol. 2001, 42, 1025–1033. [Google Scholar] [CrossRef]
- Baumann, M.J.; Eklof, J.M.; Michel, G.; Kallas, A.M.; Teeri, T.T.; Czjzek, M.; Brumer, H.I. Structural evidence for the evolution of xyloglucanase activity from xyloglucan endo-transglycosylases: Biological implications for cell wall metabolism. Plant Cell 2007, 19, 1947–1963. [Google Scholar] [CrossRef] [PubMed]
- Campbell, P.; Braam, J. Xyloglucan endotransglycosylases: Diversity of genes, enzymes, and potential wall-modifying functions. Trends Plant Sci. 1999, 4, 361–366. [Google Scholar] [CrossRef]
- Jiang, Y.; Wang, Y.; Qin, R.; Jiang, X.; Man, R.; Gai, Y. Site-directed mutagenesis and molecular docking simulations identified the crucial substrate binding site asparagine 101 of xyloglucan endotransglycosylase/hydrolase (XTH) in woody plants. Ind. Crops Prod. 2024, 222, 119478. [Google Scholar] [CrossRef]
- Cheng, Z.; Zhang, X.; Yao, W.; Gao, Y.; Zhao, K.; Guo, Q.; Zhou, B.; Jiang, T. Genome-wide identification and expression analysis of the xyloglucan endotransglucosylase/hydrolase gene family in poplar. BMC Genom. 2021, 22, 804. [Google Scholar] [CrossRef]
- Gao, Y.; Wang, L.; Li, D.; Qi, D.; Fang, F.; Luo, Y.; Zhang, H.; Zhang, S. Genome-wide characterization of the xyloglucan endotransglucosylase/hydrolase family genes and their response to plant hormone in sugar beet. Plant Physiol. Biochem. 2024, 206, 108239. [Google Scholar] [CrossRef]
- Li, Q.; Li, H.; Yin, C.; Wang, X.; Jiang, Q.; Zhang, R.; Ge, F.; Chen, Y.; Yang, L. Genome-wide identification and characterization of xyloglucan endotransglycosylase/hydrolase in Ananas comosus during development. Genes 2019, 10, 537. [Google Scholar] [CrossRef]
- Rai, K.M.; Thu, S.W.; Balasubramanian, V.K.; Cobos, C.J.; Disasa, T.; Mendu, V. Identification, characterization, and expression analysis of cell wall related genes in Sorghum bicolor (L.) Moench, a food, fodder, and biofuel crop. Front. Plant Sci. 2016, 7, 1287. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, R.; Rose, J.; Nishitani, K. A surprising diversity and abundance of xyloglucan endotransglucosylase/hydrolases in rice classification and expression analysis. Plant Physiol. 2004, 134, 1088–1099. [Google Scholar] [CrossRef]
- Yokoyama, R.; Uwagaki, Y.; Sasaki, H.; Harada, T.; Hiwatashi, Y.; Hasebe, M.; Nishitani, K. Biological implications of the occurrence of 32 members of the XTH (xyloglucan endotransglucosylase/hydrolase) family of proteins in the bryophyte Physcomitrella patens. Plant J. 2010, 64, 645–656. [Google Scholar] [CrossRef] [PubMed]
- Morales-Quintana, L.; Carrasco-Orellana, C.; Beltran, D.; Alejandra Moya-Leon, M.; Herrera, R. Molecular insights of a xyloglucan endo-transglycosylase/hydrolase of radiata pine (PrXTH1) expressed in response to inclination: Kinetics and computational study. Plant Physiol. Biochem. 2019, 136, 155–161. [Google Scholar] [CrossRef]
- Osato, Y.; Yokoyama, R.; Nishitani, K. A principal role for AtXTH18 in Arabidopsis thaliana root growth: A functional analysis using RNAi plants. J. Plant Res. 2006, 119, 153–162. [Google Scholar] [CrossRef]
- Yang, Z.; Zhang, R.; Zhou, Z. The XTH gene family in Schima superba: Genome-wide identification, expression profiles, and functional interaction network analysis. Front. Plant Sci. 2022, 13, 911761. [Google Scholar] [CrossRef]
- Braam, J. Regulated expression of the calmodulin-related TCH genes in cultured Arabidopsis cells: Induction by calcium and heat shock. Proc. Natl. Acad. Sci. USA 1992, 89, 3213–3216. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Zhang, A.; Yang, L.; Wei, J.; Bei, J.; Xu, Z.; Wang, X.; Chen, B. Identification of XTH family genes and expression analysis of endosperm weakening in lettuce (Lactuca sativa L.). Agronomy 2024, 14, 324. [Google Scholar] [CrossRef]
- Tiika, R.J.; Wei, J.; Cui, G.; Ma, Y.; Yang, H.; Duan, H. Transcriptome-wide characterization and functional analysis of xyloglucan endo-transglycosylase/hydrolase (XTH) gene family of Salicornia europaea L. under salinity and drought stress. BMC Plant Biol. 2021, 21, 491. [Google Scholar] [CrossRef]
- Cho, S.K.; Kim, J.E.; Park, J.; Eom, T.J.; Kim, W.T. Constitutive expression of abiotic stress-inducible hot pepper CaXTH3, which encodes a xyloglucan endotransglucosylase/hydrolase homolog, improves drought and salt tolerance in transgenic Arabidopsis plants. FEBS Lett. 2006, 580, 3136–3144. [Google Scholar] [CrossRef]
- Choi, J.Y.; Seo, Y.S.; Kim, S.J.; Kim, W.T.; Shin, J.S. Constitutive expression of CaXTH3, a hot pepper xyloglucan endotransglucosylase/hydrolase, enhanced tolerance to salt and drought stresses without phenotypic defects in tomato plants (Solanum lycopersicum cv. Dotaerang). Plant Cell Rep. 2011, 30, 867–877. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Wang, W.; Sun, J.; Ding, M.; Zhao, R.; Deng, S.; Wang, F.; Hu, Y.; Wang, Y.; Lu, Y.; et al. Populus euphratica XTH overexpression enhances salinity tolerance by the development of leaf succulence in transgenic tobacco plants. J. Exp. Bot. 2013, 64, 4225–4238. [Google Scholar] [CrossRef]
- Jiang, Y.; Li, Y.; Lu, C.; Tang, Y.; Jiang, X.; Gai, Y. Isolation and characterization of Populus xyloglucan endotransglycosylase/hydrolase (XTH) involved in osmotic stress responses. Int. J. Biol. Macromol. 2020, 155, 1277–1287. [Google Scholar] [CrossRef] [PubMed]
- De Caroli, M.; Manno, E.; Piro, G.; Lenucci, M.S. Ride to cell wall: Arabidopsis XTH11, XTH29 and XTH33 exhibit different secretion pathways and responses to heat and drought stress. Plant J. 2021, 107, 448–466. [Google Scholar] [CrossRef]
- Iurlaro, A.; De Caroli, M.; Sabella, E.; De Pascali, M.; Rampino, P.; De Bellis, L.; Perrotta, C.; Dalessandro, G.; Piro, G.; Fry, S.C.; et al. Drought and heat differentially affect XTH expression and XET activity and action in 3-day-old seedlings of Durum Wheat cultivars with different stress susceptibility. Front. Plant Sci. 2016, 7, 1686. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, H.; Fu, J.; Du, Y.; Qu, J.; Song, Y.; Wang, P. The GmXTH1 gene improves the drought stress resistance of soybean seedlings. Mol. Breed. 2022, 42, 3. [Google Scholar] [CrossRef]
- Yuan, W.; Yao, F.; Liu, Y.; Xiao, H.; Sun, S.; Cheng, J.; An, Y.; Chen, N.; Huang, L.; Lu, M.; et al. Identification of the xyloglucan endotransglycosylase/hydrolase genes and the role of PagXTH12 in drought resistance in poplar. J. For. Res. 2024, 4, e039. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Xie, Y.; Li, Z.; Liu, Y.; Sun, X.; Li, J.; Quan, W.; Zeng, Q.; van de Peer, Y.; Zhang, S. The Larix kaempferi genome reveals new insights into wood properties. J. Integr. Plant Biol. 2022, 64, 1364–1373. [Google Scholar] [CrossRef]
- Li, W.; Lee, J.; Yu, S.; Wang, F.; Lv, W.; Zhang, X.; Li, C.; Yang, J. Characterization and analysis of the transcriptome response to drought in Larix kaempferi using PacBio full-length cDNA sequencing integrated with de novo RNA-seq reads. Planta 2021, 253, 28. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Chou, K.; Shen, H. Plant-mPLoc: A top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS ONE 2010, 5, e11335. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; Mcgettigan, P.A.; Mcwilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Shao, L.; Li, L.; Huang, X.; Fu, Y.; Yang, D.; Li, C.; Yang, J. Identification of C2H2 zinc finger genes through genome-wide association study and functional analyses of LkZFPs in response to stresses in Larix kaempferi. BMC Plant Biol. 2023, 23, 298. [Google Scholar] [CrossRef]


| Symbol | Genes ID | Peptide Length (aa) | Molecular Weight (kDa) | Isoelectric Point (pI) | GRAVY | Subcellular Localization |
|---|---|---|---|---|---|---|
| LkXTH1 | OP235906 | 294 | 33.8114 | 8.71 | −0.470 | Cell wall and cytoplasm |
| LkXTH2 | OP235907 | 268 | 30.2061 | 5.72 | −0.199 | Cell wall |
| LkXTH3 | OP235908 | 313 | 35.4489 | 8.58 | −0.342 | Cell wall |
| LkXTH4 | OQ746059 | 294 | 33.7183 | 8.88 | −0.451 | Cell wall and cytoplasm |
| LkXTH5 | OP235909 | 299 | 34.7603 | 8.50 | −0.520 | Cell wall and cytoplasm |
| LkXTH6 | OP235910 | 295 | 34.2441 | 4.79 | −0.395 | Cell wall |
| LkXTH7 | OP235911 | 283 | 32.4874 | 5.51 | −0.280 | Cell wall |
| LkXTH8 | OQ746060 | 287 | 32.7222 | 4.47 | −0.362 | Cell wall |
| LkXTH10 | OQ746061 | 130 | 14.6555 | 8.96 | −0.624 | Cell wall |
| LkXTH11 | OQ746062 | 309 | 35.5432 | 8.15 | −0.449 | Cell wall |
| LkXTH13 | OQ746063 | 351 | 39.3741 | 6.07 | −0.405 | Cell wall |
| LkXTH14 | OQ746064 | 262 | 30.2513 | 9.52 | −0.388 | Cell wall |
| LkXTH15 | OQ746065 | 281 | 31.6536 | 4.91 | −0.219 | Cell wall |
| LkXTH16 | OP235912 | 274 | 31.0488 | 5.00 | −0.332 | Cell wall and cytoplasm |
| LkXTH17 | OP235913 | 294 | 33.6462 | 8.21 | −0.416 | Cell wall and cytoplasm |
| LkXTH18 | OP235914 | 329 | 37.3069 | 7.65 | −0.316 | Cell wall |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, Y.; Qin, R.; Wang, Y.; Liu, C.; Gai, Y. Identification and Expression Profiles of Xyloglucan Endotransglycosylase/Hydrolase Family in Response to Drought Stress in Larix kaempferi. Plants 2025, 14, 1882. https://doi.org/10.3390/plants14121882
Jiang Y, Qin R, Wang Y, Liu C, Gai Y. Identification and Expression Profiles of Xyloglucan Endotransglycosylase/Hydrolase Family in Response to Drought Stress in Larix kaempferi. Plants. 2025; 14(12):1882. https://doi.org/10.3390/plants14121882
Chicago/Turabian StyleJiang, Yan, Ruodong Qin, Yuqian Wang, Cuishuang Liu, and Ying Gai. 2025. "Identification and Expression Profiles of Xyloglucan Endotransglycosylase/Hydrolase Family in Response to Drought Stress in Larix kaempferi" Plants 14, no. 12: 1882. https://doi.org/10.3390/plants14121882
APA StyleJiang, Y., Qin, R., Wang, Y., Liu, C., & Gai, Y. (2025). Identification and Expression Profiles of Xyloglucan Endotransglycosylase/Hydrolase Family in Response to Drought Stress in Larix kaempferi. Plants, 14(12), 1882. https://doi.org/10.3390/plants14121882






