Integrative Transcriptomics and Metabolomics Reveal the Key Metabolic Pathways in Endophyte-Infected Rice Seedlings Resistance to Na2CO3 Stress
Abstract
1. Introduction
2. Results
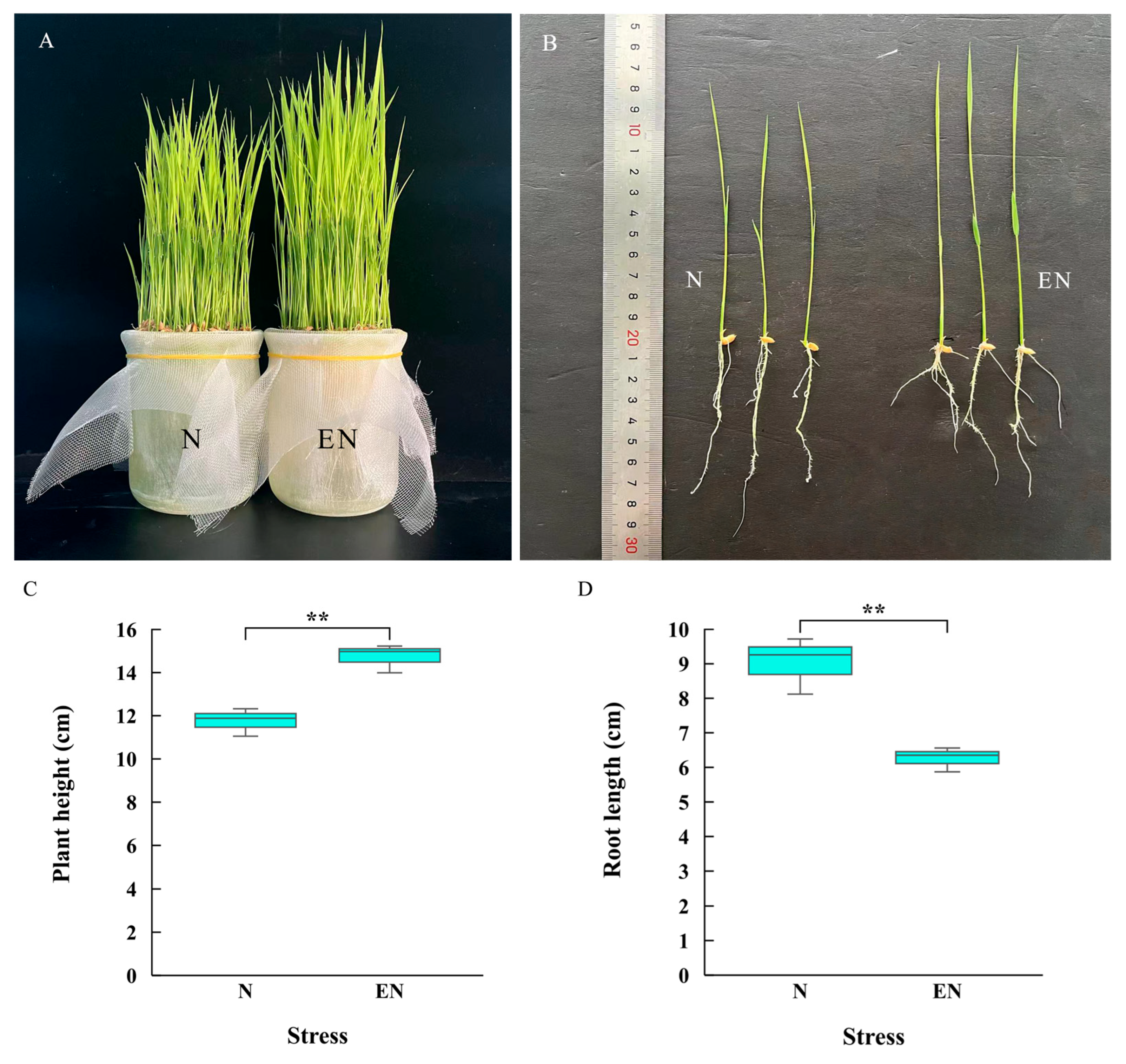
2.1. Growth Indexes of Rice Seedlings
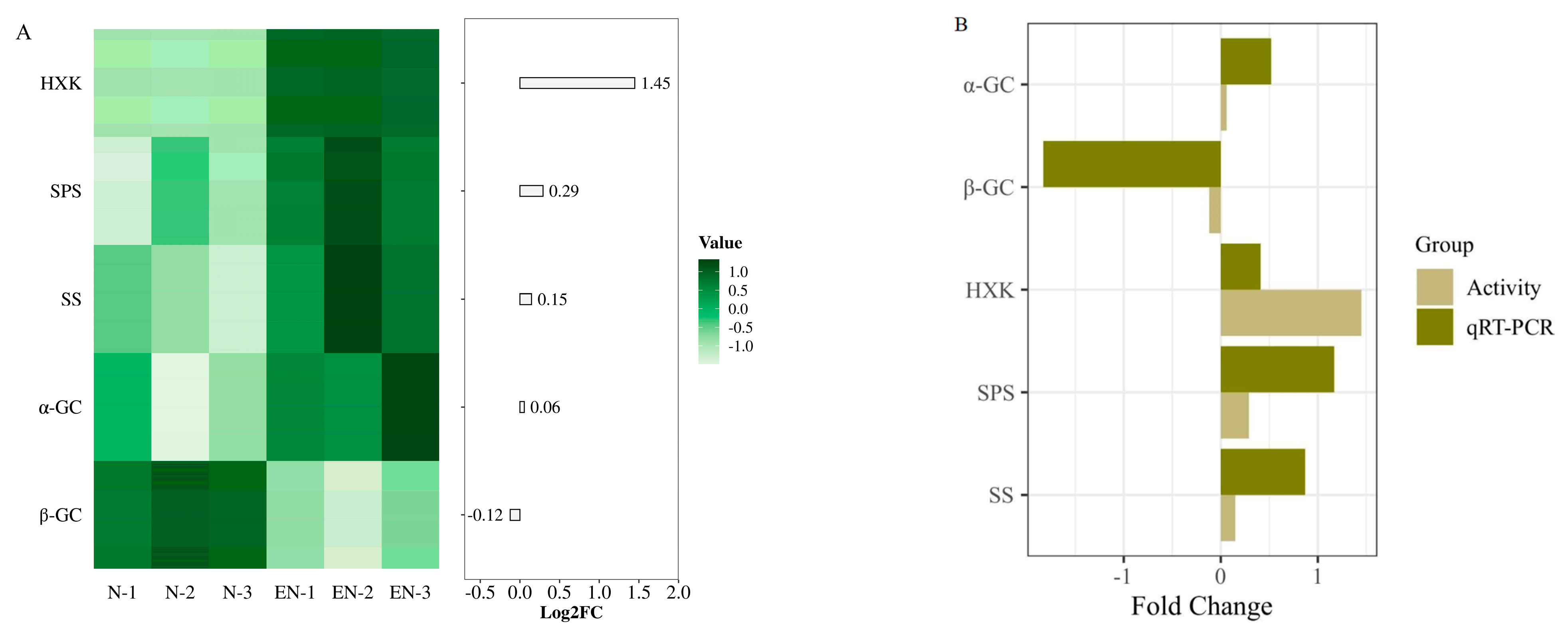
2.2. Changes and Validation of Sucrose Metabolism-Related Enzyme Activities
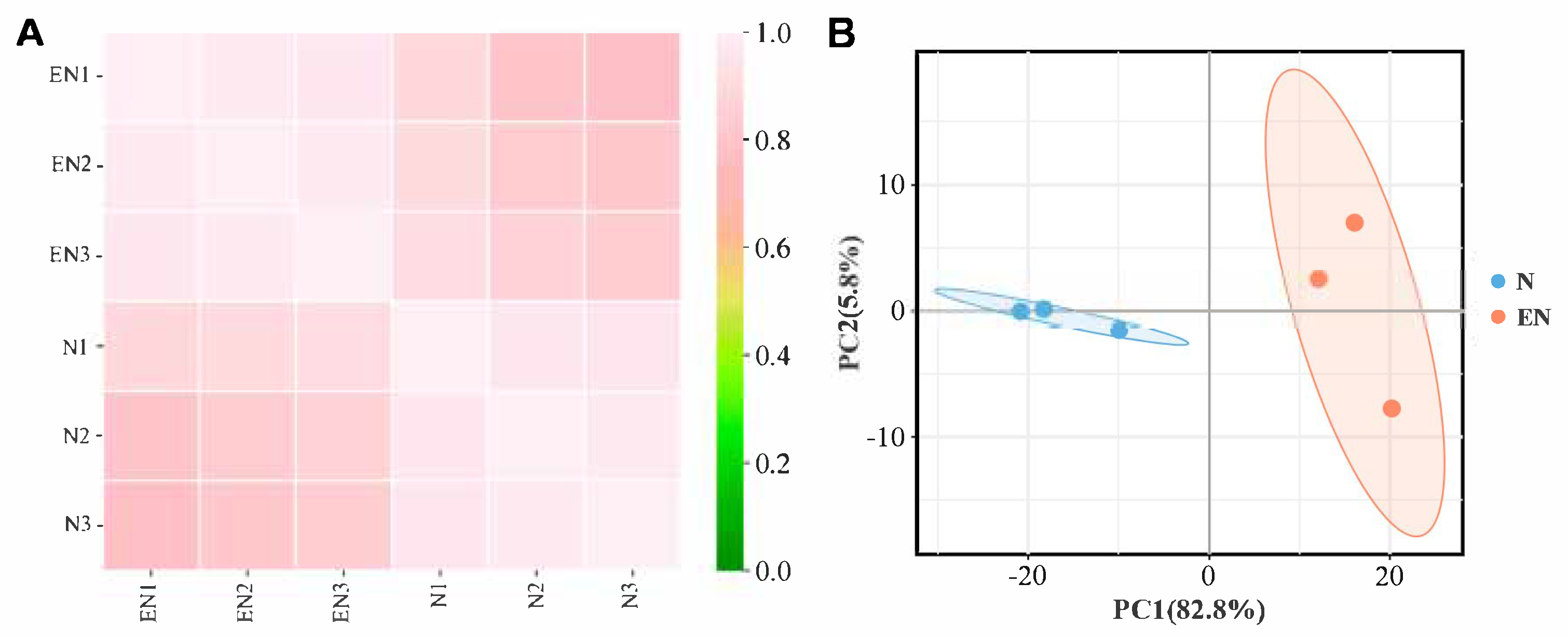
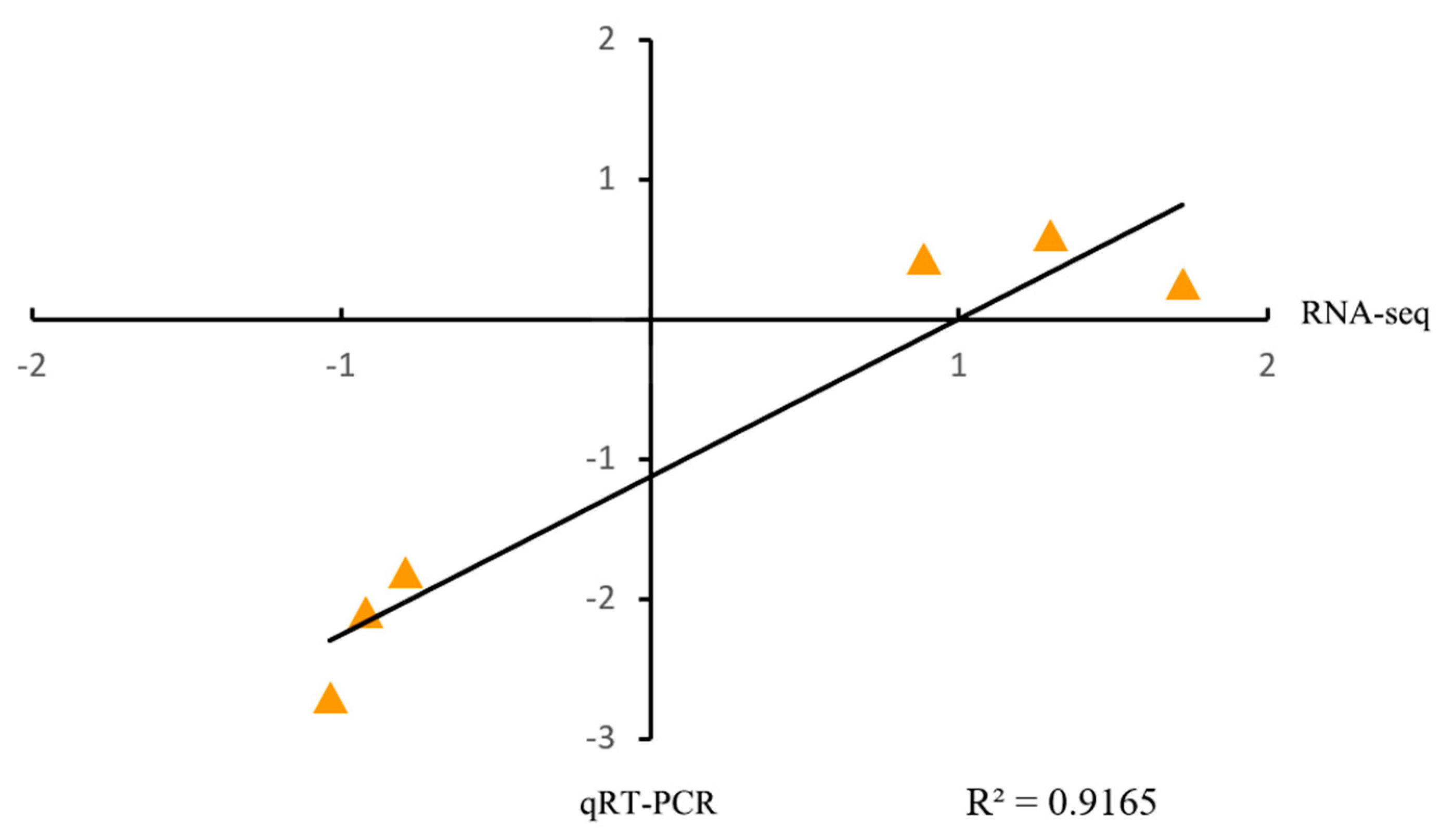
2.3. Analysis and Validation of RNA-Seq Data
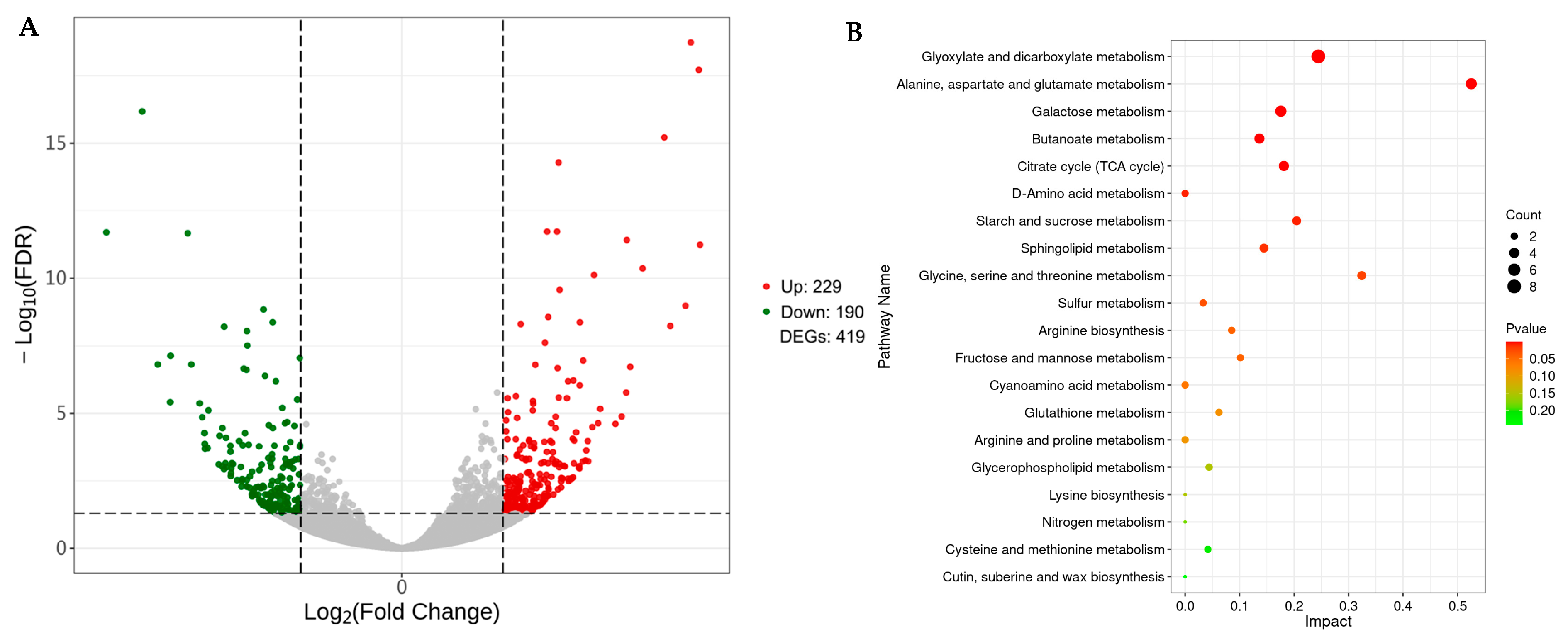
2.4. Screening and Functional Analysis of DEGs
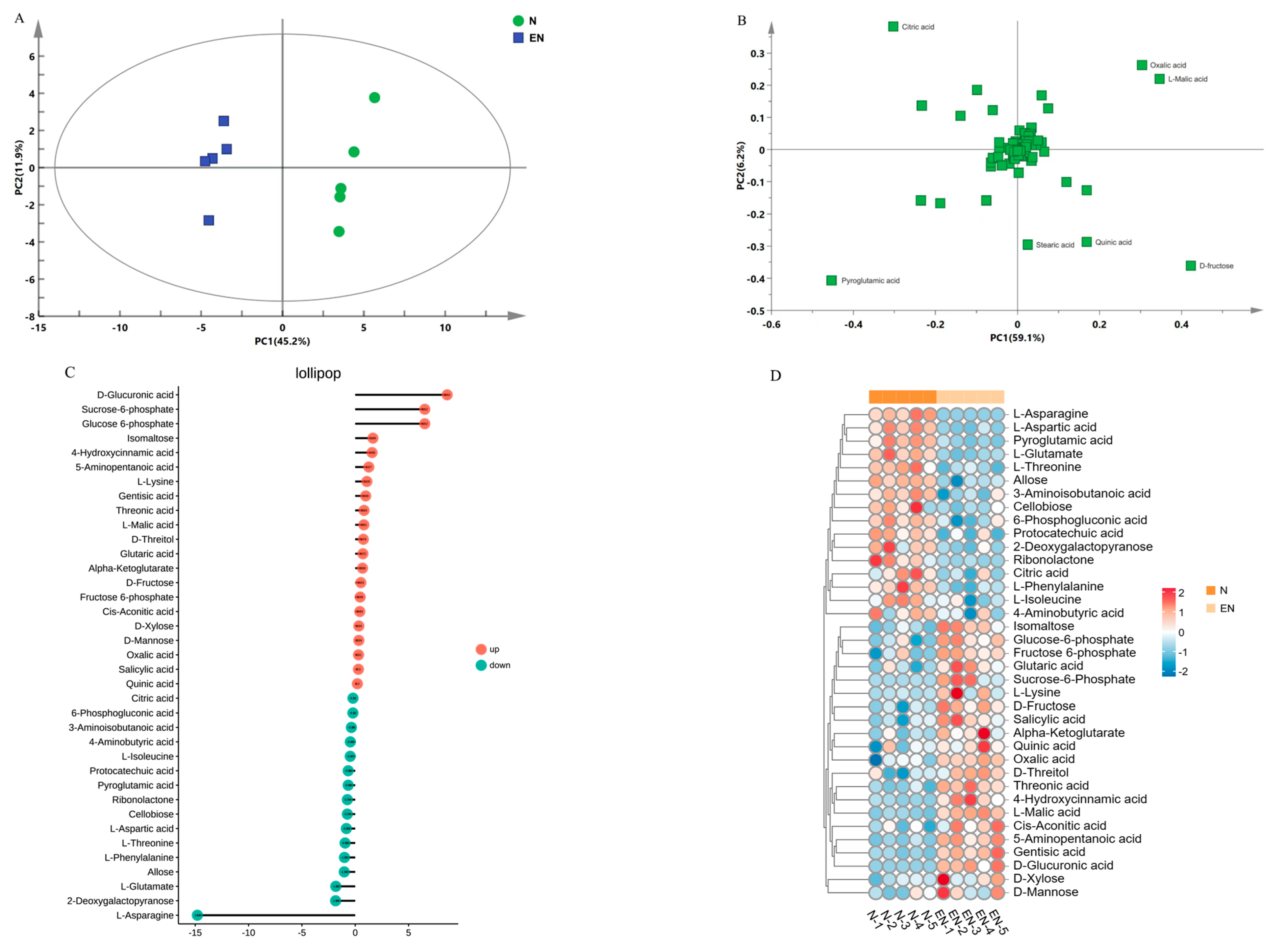
2.5. Assessment and Bioinformatics Analysis of DAMs
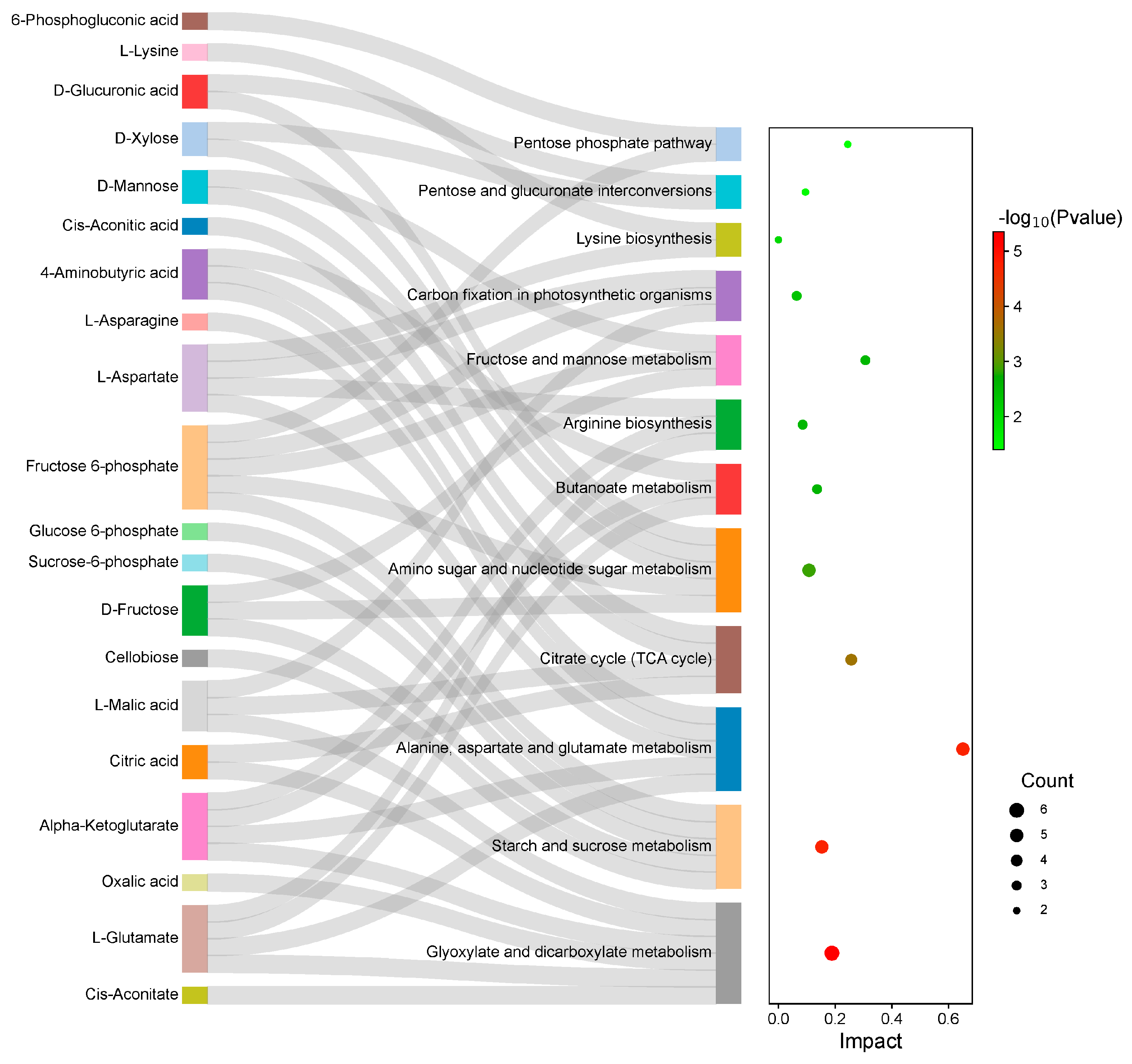
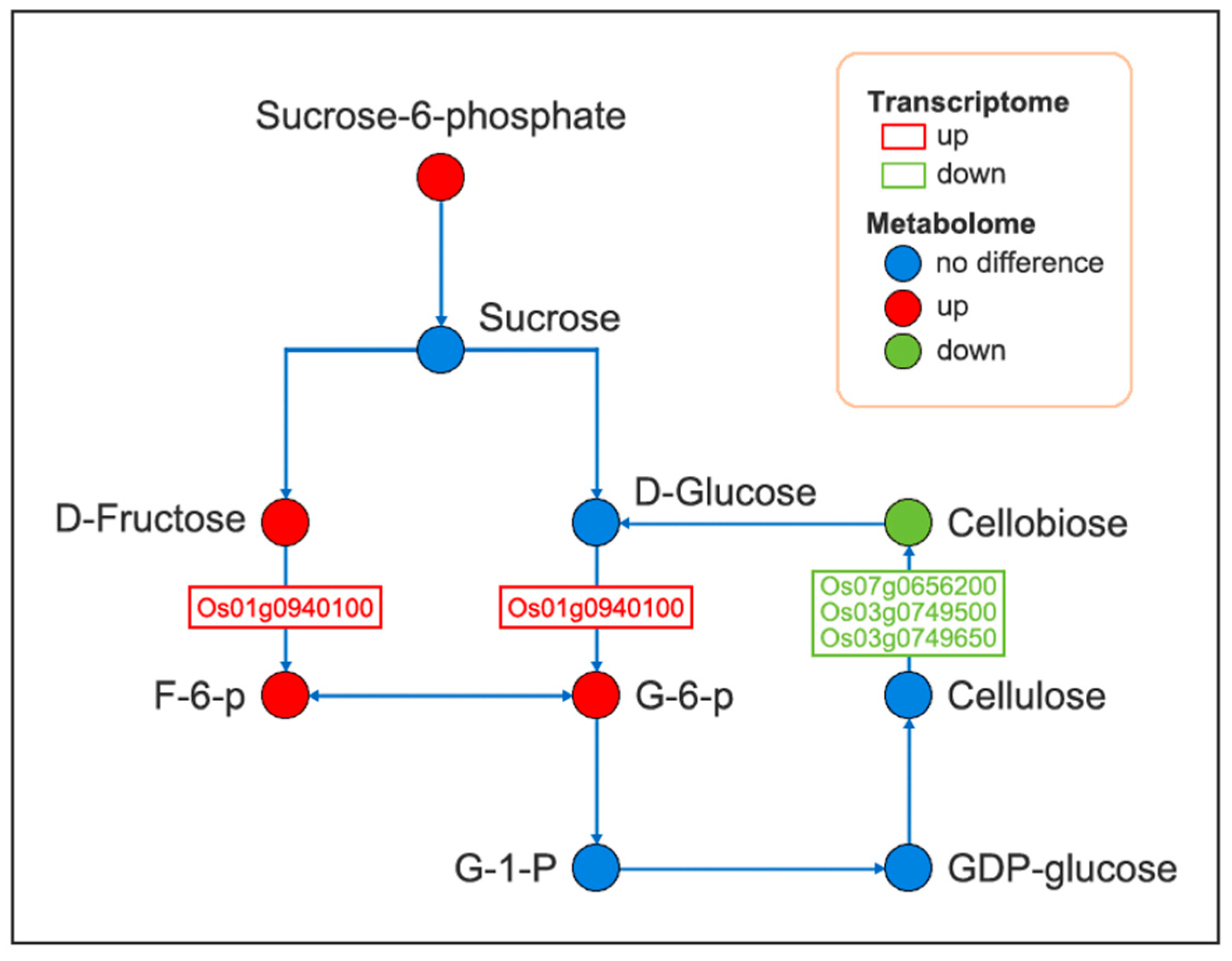
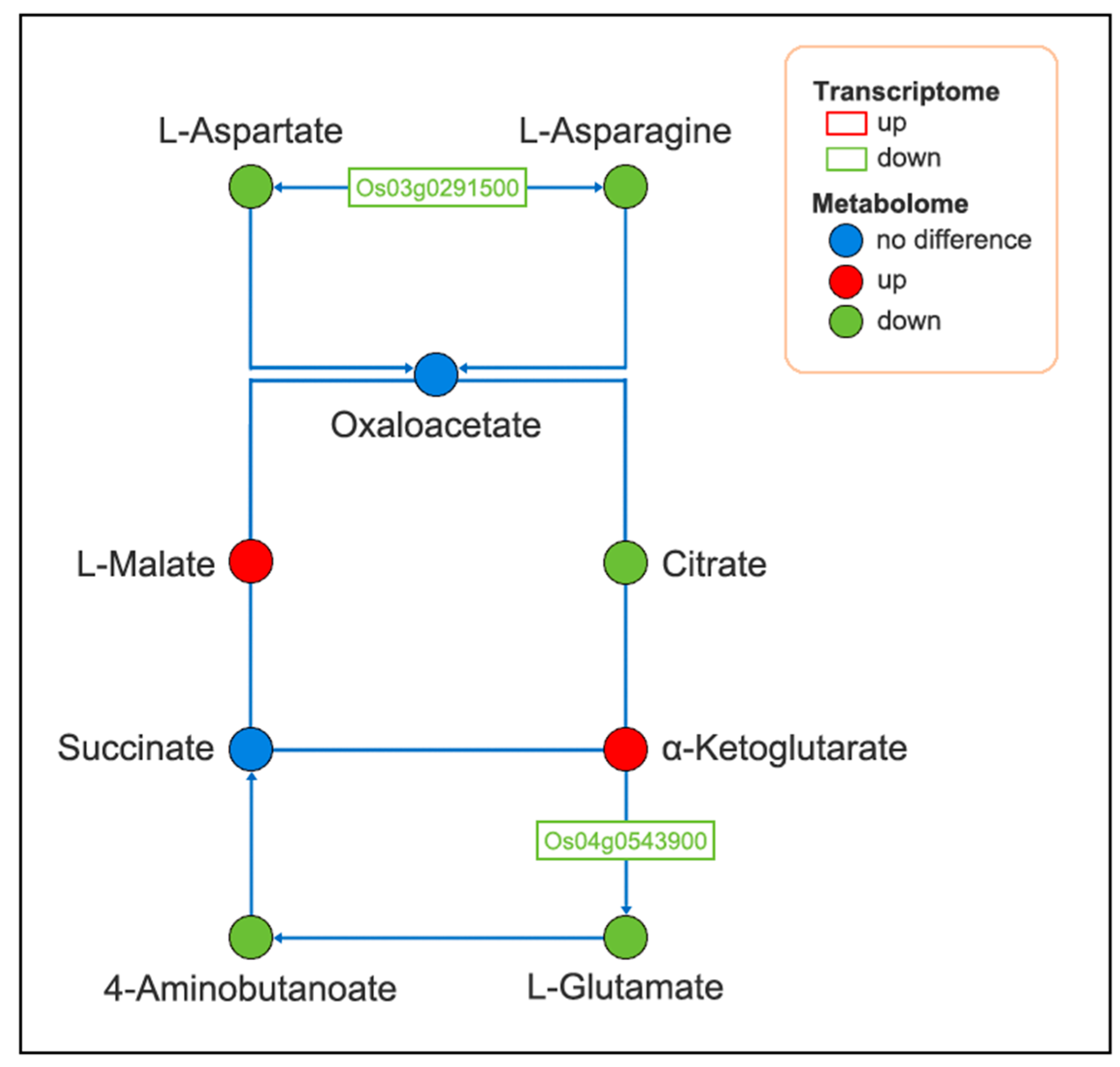
2.6. Integrated Transcriptome and Metabolome Analysis
3. Discussion
4. Materials and Methods
4.1. Endophyte Culture and Rice Treatment
4.2. Determination of Growth Parameters
4.3. Determination of Sucrose Metabolism-Related Enzyme Activities
4.4. Metabolome Analysis
4.5. Transcriptome Analysis
4.6. Quantitative Real-Time PCR Analysis
4.7. Data Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Shahid, M.A.; Balal, R.M.; Khan, N.; Zotarelli, L.; Liu, G.D.D.; Sarkhosh, A.; Fernández-Zapata, J.C.; Martínez, N.J.J.; Garcia-Sanchez, F. Selenium impedes cadmium and arsenic toxicity in potato by modulating carbohydrate and nitrogen metabolism. Ecotoxicol. Environ. Saf. 2019, 180, 588–599. [Google Scholar] [CrossRef] [PubMed]
- Morton, M.J.L.; Awlia, M.; Al-Tamimi, N.; Saade, S.; Pailles, Y.; Negrão, S.; Tester, M. Salt stress under the scalpel-dissecting the genetics of salt tolerance. Plant J. 2019, 97, 148–163. [Google Scholar] [CrossRef] [PubMed]
- Zahra, N.; Uzair, M.; Zaid, I.U.; Attia, K.A.; Inam, S.; Fiaz, S.; Abdallah, R.M.; Naeem, M.K.; Farooq, U.; Rehman, N.; et al. The comparative transcriptome analysis of two green super rice genotypes with varying tolerance to salt stress. Mol. Biol. Rep. 2024, 51, 22. [Google Scholar] [CrossRef]
- Liu, X.L.; Xie, X.Z.; Zheng, C.K.; Wei, L.X.; Li, X.W.; Jin, Y.Y.; Zhang, G.H.; Jiang, C.J.; Liang, Z.W. RNAi-mediated suppression of the abscisic acid catabolism gene OsABA8ox1 increases abscisic acid content and tolerance to saline-alkaline stress in rice (Oryza sativa L.). Crop. J. 2022, 10, 354–367. [Google Scholar] [CrossRef]
- Shaheen, S.; Baber, M.; Aslam, S.; Aslam, S.; Shaheen, M.; Waheed, R.; Seo, H.; Azhar, M.T. Effect of NaCl on morphophysiological and biochemical responses in Gossypium hirsutum L. Agronomy 2023, 13, 1012. [Google Scholar] [CrossRef]
- Wang, H.; Wu, Z.; Chen, Y.; Yang, C.; Shi, D. Effects of salt and alkali stresses on growth and ion balance in rice (Oryza sativa L.). Plant Soil Environ. 2011, 57, 286–294. [Google Scholar] [CrossRef]
- Amirbakhtiar, N.; Ismaili, A.; Ghaffari, M.-R.; Mirdar, M.R.; Sanjari, S.; Shobbar, Z.-S. Transcriptome analysis of bread wheat leaves in response to salt stress. PLoS ONE 2021, 16, e0254189. [Google Scholar] [CrossRef] [PubMed]
- Gupta, P.; De, B. Metabolomics analysis of rice responses to salinity stress revealed elevation of serotonin and gentisic acid levels in leaves of tolerant varieties. Plant Signal. Behav. 2017, 12, e1335845. [Google Scholar] [CrossRef]
- Sun, J.; Xie, D.W.; Zhang, E.Y.; Zheng, H.L.; Wang, J.G.; Liu, H.L.; Yang, L.M.; Zhang, S.L.; Wang, L.; Zou, D.T. QTL mapping of photosynthetic-related traits in rice under salt and alkali stresses. Euphytica 2019, 215, 147. [Google Scholar] [CrossRef]
- Gond, S.K.; Bergen, M.S.; Torres, M.S.; White, J.J. Endophytic Bacillus spp. produce antifungal lipopeptides and induce host defence gene expression in maize. Microbiol. Res. 2015, 172, 79–87. [Google Scholar] [CrossRef]
- Usuki, F.; Narisawa, K. A mutualistic symbiosis between a dark septate endophytic fungus, Heteroconium chaetospira, and a nonmycorrhizal plant, Chinese cabbage. Mycologia 2007, 99, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Yadav, V.; Kumar, M.; Deep, D.K.; Kumar, H.; Sharma, R.; Tripathi, T.; Tuteja, N.; Saxena, A.K.; Johri, A.K. A phosphate transporter from the root endophytic fungus Piriformospora indica plays a role in phosphate transport to the host plant. J. Biol. Chem. 2010, 285, 26532–26544. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.H.; Yao, X.F.; Mi, D.D.; Li, Z.J.; Yang, B.Y.; Zheng, Y.; Qi, Y.J.; Guo, J.H. Comparative transcriptome analysis reveals the biocontrol mechanism of Bacillus velezensis F21 against Fusarium wilt on watermelon. Front. Microbiol. 2019, 10, 652. [Google Scholar] [CrossRef] [PubMed]
- Waqas, M.; Khan, A.L.; Shahzad, R.; Ullah, I.; Khan, A.R.; Lee, I.J. Mutualistic fungal endophytes produce phytohormones and organic acids that promote japonica rice plant growth under prolonged heat stress. J. Zhejiang Univ. Sci. B 2015, 16, 1011–1018. [Google Scholar] [CrossRef]
- Khan, A.L.; Lee, I.J. Endophytic Penicillium funiculosum LHL06 secretes gibberellin that reprograms Glycine max L. growth during copper stress. BMC Plant Biol. 2013, 13, 86. [Google Scholar] [CrossRef]
- Li, X.M.; Chen, M.J.; Li, J.; Ma, L.J.; Li, Y.Y.; Bu, N.; Li, Y.Y.; Zhang, L.H. Effect of endophyte infection on chlorophyll a fluorescence in salinity stressed rice. Biol. Plant. 2014, 58, 589–594. [Google Scholar] [CrossRef]
- Li, P.; Liang, H.; Lin, W.T.; Feng, F.; Luo, L.X. Microbiota dynamics associated with environmental conditions and potential roles of cellulolytic communities in traditional Chinese cereal starter solid-state fermentation. Appl. Environ. Microbiol. 2015, 81, 5144–5156. [Google Scholar] [CrossRef]
- Ren, X.N.; Shan, Y.; Li, X.; Wang, L.L.; Li, Y.Y.; Ma, L.J.; Li, X.M. Endophytic infection programs the ascorbate-glutathione cycle in rice (Oryza sativa L.) under Na2CO3 stress. Appl. Ecol. Environ. Res. 2021, 19, 1895–1907. [Google Scholar] [CrossRef]
- Ott, K.H.; Aranibar, N.; Singh, B.; Stockton, G.W. Metabonomics classifies pathways affected by bioactive compounds. Artificial neural network classification of NMR spectra of plant extracts. Phytochemistry 2003, 62, 971. [Google Scholar] [CrossRef]
- Sekiyama, Y.; Okazaki, K.; Kikuchi, J.; Ikeda, S. NMR-Based metabolic profiling of field-grown leaves from sugar beet plants harbouring different levels of resistance to cercospora leaf spot disease. Metabolites 2017, 7, 4. [Google Scholar] [CrossRef]
- Ghaffari, M.R.; Ghabooli, M.; Khatabi, B.; Hajirezaei, M.R.; Schweizer, P.; Salekdeh, G.H. Metabolic and transcriptional response of central metabolism affected by root endophytic fungus Piriformospora indica under salinity in barley. Plant Mol. Biol. 2016, 90, 699–717. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.L.; Sheng, Y.Y.; Cai, Z.Y.; Yang, R.; Li, Q.S.; Li, X.M.; Li, D.; Guo, X.Y.; Lu, J.L.; Ye, J.H.; et al. Involvement of salicylic acid in anthracnose infection in tea plants revealed by transcriptome profiling. Int. J. Mol. Sci. 2019, 20, 2439. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.N.; Shan, Y.; Li, X.; Fan, J.H.; Li, Y.Y.; Ma, L.J.; Wang, L.L.; Li, X.M. Application of RNA sequencing to understand the benefits of endophytes in the salt-alkaline resistance of rice seedlings. Environ. Exp. Bot. 2022, 196, 104820. [Google Scholar] [CrossRef]
- Li, M.; Zhou, J.; Lang, X.; Han, D.; Hu, Y.; Ding, Y.; Wang, G.; Guo, J.; Shi, L. Integrating transcriptomic and metabolomic analysis in roots of wild soybean seedlings in response to low-phosphorus stress. Front. Plant Sci. 2022, 13, 1006806. [Google Scholar] [CrossRef]
- Shao, M.; Chen, Y.; Gong, Q.; Miao, S.; Li, C.; Sun, Y.; Qin, D.; Guo, X.; Yan, X.; Cheng, P.; et al. Biocontrol endophytes Bacillus subtilis R31 influence the quality; transcriptome and metabolome of sweet corn. PeerJ 2023, 11, e14967. [Google Scholar] [CrossRef] [PubMed]
- Malik, A.; Butt, T.A.; Naqvi, S.; Yousaf, S.; Iqbal, M. Lead tolerant endophyte Trametes hirsuta improved the growth and lead accumulation in the vegetative parts of Triticum aestivum L. Heliyon 2020, 6, e04188. [Google Scholar] [CrossRef]
- Wu, F.; Qu, D.; Tian, W.; Wang, M.; Chen, F.; Li, K.; Sun, Y.; Su, Y.; Yang, L.; Su, H.; et al. Transcriptome analysis for understanding the mechanism of dark septate endophyte S16 in promoting the growth and nitrate uptake of sweet cherry. J. Integr. Agric. 2021, 20, 1819–1831. [Google Scholar] [CrossRef]
- Li, X.M.; Ma, L.J.; Bu, N.; Li, Y.Y.; Zhang, L.H. Endophytic infection modifies organic acid and mineral element accumulation by rice under Na2CO3 stress. Plant Soil 2017, 420, 93–103. [Google Scholar] [CrossRef]
- Rodriguez-Garcia, A.; Sola-Landa, A.; Barreiro, C. RNA-Seq-Based Comparative Transcriptomics, RNA Preparation and Bioinformatics. In Microbial Steroids, Methods and Protocol; Humana Press: New York, NY, USA, 2017; pp. 59–72. [Google Scholar]
- Coueé, I.; Sulmon, C.; Gouesbet, G.; Amrani, A.E. Involvement of soluble sugars in reactive oxygen species balance and responses to oxidative stress in plants. J. Exp. Bot. 2006, 3, 449–459. [Google Scholar] [CrossRef]
- Jammer, A.; Gasperl, A.; Luschin-Ebengreuth, N.; Heyneke, E.; Chu, H.; Cantero-Navarro, E.; Großkinsky, K.D.; Albacete, A.; Stabentheiner, E.; Franzaring, J.; et al. Simple and robust determination of the activity signature of key carbohydrate metabolism enzymes for physiological phenotyping in model and crop plants. J. Exp. Bot. 2015, 66, 5531–5542. [Google Scholar] [CrossRef]
- Krasensky, J.; Jonak, C. Drought, salt and temperature stress-induced metabolic rearrangements and regulatory networks. J. Exp. Bot. 2012, 63, 1593–1608. [Google Scholar] [CrossRef] [PubMed]
- Nemati, F.; Ghanati, F.; Gavlighi, H.A.; Sharifi, M. Comparison of sucrose metabolism in wheat seedlings during drought stress and subsequent recovery. Biol. Plant. 2018, 62, 595–599. [Google Scholar] [CrossRef]
- Xu, Z.; Chen, X.; Lu, X.; Zhao, B.; Yang, Y.; Liu, J. Integrative analysis of transcriptome and metabolome reveal mechanism of tolerance to salt stress in oat (Avena sativa L.). Plant Physiol. Biochem. 2021, 160, 315–328. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Huang, B.; Fry, J. Osmotic potential, sucrose level, and activity of sucrose metabolic enzymes in tall fescue in response to deficit irrigation. J. Am. Soc. Hortic. Sci. 2010, 135, 506–510. [Google Scholar] [CrossRef]
- Yan, H.; Nie, Y.; Cui, K.; Sun, J. Integrative transcriptome and metabolome profiles reveal common and unique pathways involved in seed initial imbibition under artificial and natural salt stresses during germination of halophyte quinoa. Front. Plant Sci. 2022, 13, 853326. [Google Scholar] [CrossRef]
- Granot, D.; Kelly, G.; Stein, O.; David-Schwartz, R. Substantial roles of hexokinase and fructokinase in the effects of sugars on plant physiology and development. J. Exp. Bot. 2014, 65, 809–819. [Google Scholar] [CrossRef]
- Richardson, M.D.; Chapman, G.W.; Hoveland, C.S.; Bacon, C.W. Sugar alcohols in endophyte-infected tall fescue under drought. Crop Sci. 1992, 32, 1060–1061. [Google Scholar] [CrossRef]
- SkZ, A.; Vardharajula, S.; Vurukonda, S.S.K.P. Transcriptomic profiling of maize (Zea mays L.) seedlings in response to Pseudomonas putida stain FBKV2 inoculation under drought stress. Ann. Microbiol. 2018, 68, 331–349. [Google Scholar] [CrossRef]
- Heinemann, B.; Hildebrandt, T.M. The role of amino acid metabolism in signaling and metabolic adaptation to stress-induced energy deficiency in plants. J. Exp. Bot. 2021, 72, 4634–4645. [Google Scholar] [CrossRef]
- Huang, T.; Jander, G. Abscisic acid-regulated protein degradation causes osmotic stress-induced accumulation of branched-chain amino acids in Arabidopsis thaliana. Planta 2017, 246, 737. [Google Scholar] [CrossRef]
- Zhao, X.; Huang, L.J.; Sun, X.F.; Zhao, L.L.; Wang, P.C. Transcriptomic and metabolomic analyses reveal key metabolites; pathways and candidate genes in Sophora davidii (franch.) skeels seedlings under drought stress. Front. Plant Sci. 2022, 13, 785702. [Google Scholar] [CrossRef] [PubMed]
- Dey, S.; Kundu, R.; Gopal, G.; Mukherjee, A.; Nag, A.; Paul, S. Enhancement of nitrogen assimilation and photosynthetic efficiency by novel iron pulsing technique in Oryza sativa L. var Pankaj. Plant Physiol. Biochem. 2019, 144, 207–221. [Google Scholar] [CrossRef] [PubMed]
- Less, H.; Galili, G. Principal transcriptional programs regulating plant amino acid metabolism in response to abiotic stresses. Plant Physiol. 2008, 147, 316. [Google Scholar] [CrossRef] [PubMed]
- Rashmi, D.; Barvkar, V.T.; Nadaf, A.; Mundhe, S.; Kadoo, N.Y. Integrative omics analysis in Pandanus odorifer (Forssk.) Kuntze reveals the role of asparagine synthetase in salinity tolerance. Sci. Rep. 2019, 9, 932. [Google Scholar] [CrossRef]
- Wang, C.; Huang, R.; Wang, J.; Jin, J.; Malik, K.; Niu, X.; Tang, R.; Hou, W.; Cheng, C.; Liu, Y.; et al. Comprehensive analysis of transcriptome and metabolome elucidates the molecular regulatory mechanism of salt resistance in roots of Achnatherum inebrians mediated by Epichloë gansuensis. J. Fungi 2022, 8, 1092. [Google Scholar] [CrossRef]
- Ren, X.N.; Fan, J.H.; Li, X.; Shan, Y.; Wang, L.L.; Ma, L.J.; Li, Y.Y.; Li, X.M. Application of RNA sequencing to understand the response of rice seedlings to salt-alkali stress. BMC Genom. 2023, 24, 21. [Google Scholar] [CrossRef]
- Zhou, Z.; Liu, J.; Meng, W.; Sun, Z.; Tan, Y.; Liu, Y.; Tan, M.; Wang, B.; Yang, J. Integrated analysis of transcriptome and metabolome reveals molecular mechanisms of rice with different salinity tolerances. Plants 2023, 12, 3359. [Google Scholar] [CrossRef]
- Li, X.M.; Bu, N.; Li, Y.Y.; Ma, L.J.; Xin, S.G.; Zhang, L.H. Growth, photosynthesis and antioxidant responses of endophyte infected and non-infected rice under lead stress conditions. J. Hazard. Mater. 2012, 213, 55–61. [Google Scholar] [CrossRef]
- Liu, R.J.; Chen, Y.L. Mycorrhizology; Science Press: Beijing, China, 2007. [Google Scholar]
- Zhang, Z.L.; Qu, W.J.; Li, X.F. Plant Physiology Laboratory Guide; Higher Education Press: Beijing, China, 2009. [Google Scholar]

| Metabolite | Name | N | EN | FC |
|---|---|---|---|---|
| Glycolysis and | l-Malic acid | 123.43 ± 8.37 | 216.20 ± 15.91 | 0.81 |
| TCA cycles | Glucose 6-phosphate | 0.49 ± 0.13 | 0.76 ± 0.10 | 6.52 |
| Fructose 6-phosphate | 1.81 ± 0.43 | 2.49 ± 0.21 | 0.46 | |
| Cis-Aconitic acid | 0.15 ± 0.03 | 0.21 ± 0.03 | 0.43 | |
| Alpha-Ketoglutarate | 25.09 ± 5.94 | 40.39 ± 11.26 | 0.68 | |
| Citric acid | 729.92 ± 66.67 | 627.08 ± 59.73 | −0.21 | |
| Amino acid | l-Asparagine | 3.18 ± 0.74 | 0.00 ± 0.00 | −14.8 |
| l-Glutamate | 38.99 ± 6.88 | 11.07 ± 3.21 | −1.82 | |
| l-Aspartic acid | 4.43 ± 0.89 | 3.36 ± 1.96 | −0.83 | |
| 5-Aminopentanoic acid | 0.16 ± 0.03 | 0.71 ± 0.23 | 1.27 | |
| Pyroglutamic acid | 445.79 ± 43.75 | 281.20 ± 22.41 | −0.66 | |
| l-Threonine | 95.25 ± 14.58 | 49.21 ± 11.62 | −0.95 | |
| l-Phenylalanine | 3.47 ± 0.62 | 1.73 ± 0.45 | −1.01 | |
| 3-Aminoisobutanoic acid | 0.42 ± 0.02 | 0.33 ± 0.04 | −0.36 | |
| l-Isoleucine | 7.95 ± 1.35 | 5.72 ± 1.32 | −0.47 | |
| l-Lysine | 2.20 ± 0.26 | 4.69 ± 2.39 | 1.09 | |
| Sugars and polyols | d-Glucuronic acid | 0.23 ± 0.03 | 0.31 ± 0.07 | 8.62 |
| Sucrose-6-phosphate | 0.00 ± 0.00 | 0.09 ± 0.06 | 6.52 | |
| Threonic acid | 1.21 ± 0.22 | 2.15 ± 0.23 | 0.83 | |
| Allose | 0.48 ± 0.03 | 0.23 ± 0.08 | −1.03 | |
| d-Mannose | 146.01 ± 25.61 | 187.22 ± 43.21 | 0.34 | |
| d-Fructose | 242.96 ± 42.73 | 357.43 ± 57.83 | 0.53 | |
| 6-Phosphogluconic acid | 0.91 ± 0.03 | 0.78 ± 0.07 | −0.22 | |
| 2-Deoxygalactopyranose | 7.60 ± 2.92 | 2.12 ± 1.84 | −1.84 | |
| d-Threitol | 0.10 ± 0.04 | 0.17 ± 0.03 | 0.76 | |
| Cellobiose | 0.90 ± 0.28 | 0.54 ± 0.11 | −0.74 | |
| d-Xylose | 0.09 ± 0.01 | 0.11 ± 0.02 | 0.34 | |
| Isomaltose | 0.21 ± 0.12 | 0.66 ± 0.17 | 1.66 | |
| Organic acid | 4-Hydroxycinnamic acid | 0.10 ± 0.02 | 0.29 ± 0.09 | 1.58 |
| Oxalic acid | 368.55 ± 54.49 | 456.90 ± 13.54 | 0.31 | |
| Quinic acid | 2.62 ± 0.38 | 3.02 ± 0.30 | 0.2 | |
| Gentisic acid | 0.24 ± 0.02 | 0.47 ± 0.06 | 0.98 | |
| Salicylic acid | 5.98 ± 0.43 | 7.11 ± 0.58 | 0.3 | |
| Protocatechuic acid | 0.15 ± 0.03 | 0.04 ± 0.06 | −0.66 | |
| Ribonolactone | 00.14 ± 0.03 | 0.08 ± 0.00 | −0.74 | |
| Glutaric acid | 0.09 ± 0.03 | 0.13 ± 0.03 | 0.72 | |
| 4-Aminobutyric acid | 95.74 ± 27.83 | 69.45 ± 23.55 | −0.46 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Li, Y.; Sun, H.; Zhang, L.; Li, X. Integrative Transcriptomics and Metabolomics Reveal the Key Metabolic Pathways in Endophyte-Infected Rice Seedlings Resistance to Na2CO3 Stress. Plants 2025, 14, 1524. https://doi.org/10.3390/plants14101524
Wang X, Li Y, Sun H, Zhang L, Li X. Integrative Transcriptomics and Metabolomics Reveal the Key Metabolic Pathways in Endophyte-Infected Rice Seedlings Resistance to Na2CO3 Stress. Plants. 2025; 14(10):1524. https://doi.org/10.3390/plants14101524
Chicago/Turabian StyleWang, Xinnan, Yanan Li, Hefei Sun, Lihong Zhang, and Xuemei Li. 2025. "Integrative Transcriptomics and Metabolomics Reveal the Key Metabolic Pathways in Endophyte-Infected Rice Seedlings Resistance to Na2CO3 Stress" Plants 14, no. 10: 1524. https://doi.org/10.3390/plants14101524
APA StyleWang, X., Li, Y., Sun, H., Zhang, L., & Li, X. (2025). Integrative Transcriptomics and Metabolomics Reveal the Key Metabolic Pathways in Endophyte-Infected Rice Seedlings Resistance to Na2CO3 Stress. Plants, 14(10), 1524. https://doi.org/10.3390/plants14101524






