Abstract
Dendrobium huoshanense, an endangered orchid species, is renowned for its polysaccharides with vast pharmacological significance in stems. Phosphomannomutase (PMM) critically regulates polysaccharide accumulation. Transcriptional regulation of DhPMM remains poorly characterized. This study employed a yeast one-hybrid (Y1H) system to identify upstream regulators of DhPMM. The 2.15 kb DhPMM promoter was cloned, revealing multiple stress- and hormone-responsive cis-elements (e.g., ABRE, MYC, ERF). A high-complexity Y1H library (3.60 × 109 CFU) was constructed with insert sizes averaging 1–2 kb. Screening using aureobasidin A (AbA)-resistant Y1HGold [pAbAi-DhPMM] identified 11 candidate clones, including four transcription factor families (DOF, NAC, ERF, BES1). Interactions were rigorously confirmed by pairwise Y1H showing AbA-resistant growth and dual-luciferase assays demonstrating DhPMM activation. This represents the first functional cDNA library resource for D. huoshanense and identification of TFs interacting with DhPMM. The discovery of TFs belonging to DOF, NAC, ERF, and BES1 families as DhPMM regulators elucidated the transcriptional network underlying polysaccharide biosynthesis. This establishes a transcriptional framework for engineering polysaccharide biosynthesis in D. huoshanense.
1. Introduction
Dendrobium huoshanense C. Z. Tang et S. J. Cheng, classified within the second largest genus in the family Orchidaceae, is a precious traditional Chinese materia medica [1]. Due to its outstanding medicinal values, it was officially compiled in the Chinese Pharmacopeia as expected in 2020 [2]. The stems of D. huoshanense traditionally used as the medicinal part have shown significant benefits in disease prevention and treatment [3]. Studies have revealed that there are numerous active components in the stem, among which polysaccharide has been the most extensively studied [4,5]. However, wild D. huoshanense populations face critical endangerment due to habitat loss and excessive harvesting despite high market demand accompanied by its listing as a national first-class protected species [6]. Understanding the molecular mechanisms governing polysaccharide biosynthesis is therefore essential for both enhancing metabolite production through metabolic engineering and developing sustainable cultivated germplasm.
Enzymes and their corresponding encoding genes have irreplaceable roles in polysaccharide biosynthesis and metabolism pathways [7]. Phosphomannomutase (PMM), which catalyzes the isomerization of mannose-6-phosphate to mannose-1-phosphate, represents a rate-limiting enzyme in GDP-mannose synthesis—the universal precursor for plant polysaccharides [8]. He et al. [9] conducted transcriptional analyses of DoPMM gene from Dendrobium officinale. DoPMM showed peak expression levels in stems. Similarly, CpPMM expression in Codonopsis pilosula showed spatiotemporal differentiation positively associated with polysaccharide content [10]. D. huoshanense exhibited analogous tissue-specific DhPMM expression patterns, with the highest transcript levels occurring in stems [11]. Collectively, these studies confirm PMM as a phylogenetically conserved regulator of polysaccharide biosynthesis in medicinal plants [8].
Transcriptional regulation of metabolite biosynthesis occurs through cis-elements within enzyme gene promoters. These regulatory sequences serve as binding sites for transcription factors (TFs) that modulate gene expression in response to developmental and environmental stimuli [12]. TFs play pivotal roles in regulating plant metabolite biosynthesis through enzyme-mediated pathways in response to external signals [13]. Accumulating evidence demonstrates that overexpression of TFs is an effective strategy to increase sugar and starch contents [14,15]. The yeast one-hybrid (Y1H) system has been developed as a tool to screen prey proteins interacted with bait DNA promoter sequences, enabling TF identification [16]. Successful applications of Y1H screening have identified candidate TFs regulating secondary metabolites biosynthesis in medicinal species including Madagascar periwinkle [17], Taxus chinensis [18], Ganoderma lucidum [19], and Artemisia annua [20]. However, systematic investigations on transcriptional networks governing pharmacologically valuable polysaccharide biosynthesis in D. huoshanense remain unreported.
Here, we isolated the DhPMM promoter, constructed a Y1H bait vector, and identified TFs binding this regulatory sequence. Our findings may not only contribute to further understanding of the role of PMM in polysaccharide biosynthesis, but also provide validated molecular targets for metabolic engineering to enhance polysaccharide production in D. huoshanense.
2. Materials and Methods
2.1. Plant Materials
Dendrobium huoshanense C. Z. Tang et S. J. Cheng, cultivated under forest from Huoshan County, Anhui Province, China was identified by Professor Nianjun Yu from Anhui University of Chinese Medicine [21]. The other type of greenhouse-cultivated D. huoshanense was also collected and mixed up with that under forest for the samples. Voucher specimens of plants were deposited at the Herbarium Center, Anhui University of Chinese Medicine, Hefei, China (Daiyin Peng, Pengdy@ahtcm.edu.cn, Voucher Nos. 20221104 and 20221105). Nicotiana benthamiana was grown in chambers (22 °C, 16 h light/8 h dark cycle) with three-week-old plants used for transient expression assays. No permission or license was required in this study. The sample was legally collected in accordance with guidelines provided by the national or international regulations. Field studies complied with local legislation.
2.2. Cloning of the Promoter of Gene DhPMM and Cis-Element Analysis
Genomic DNA was isolated from leaves of samples with Ezup Column Plant Genomic DNA Kit (B518262–0050, Sangon Biotech, Shanghai, China). The D. huoshanense genome was searched in the NCBI database (https://www.ncbi.nlm.nih.gov/, accessed on 15 July 2025) [22] for the ATG upstream sequence of PMM gene promoter [23]. Primers were designed to amplify this sequence by polymerase chain reaction (PCR) amplification, and products were verified by 1% agarose gel electrophoresis and sequencing. Predicted cis-acting elements within the promoter were analyzed using PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/, accessed on 15 May 2024) [24].
2.3. Construction of a Recombinant pAbAi-DhPMM Bait Plasmid
The vector pAbAi provided by OEbiotech (Shanghai, China) [25] and purified DhPMM promoter fragment were digested concurrently with SacI and XhoI. Digested fragments were gel-purified using PureLink Quick Gel Extraction Kit (K220001, Invitrogen, Carlsbad, CA, USA) and ligated using T4 DNA Ligase (EL0014, Thermo Fisher Scientific, Waltham, MA, USA). The ligation product was transformed into Escherichia coli (E. coli) DH10β competent cells and plated on Luria–Bertani (LB) agar (ampicillin resistant) to screen positive recombinant clones.
2.4. Generation of the Yeast Bait Strain
The recombinant pAbAi-DhPMM plasmid was linearized with BstBI. Y1HGold yeast competent cells were transformed with 1 μg linearized DNA via the lithium acetate/single-stranded carrier DNA/polyethylene glycol (LiAc/SS-DNA/PEG) method [26]. Transformants were selected on solid agar synthetic defined (SD)/-Ura plates at 30 °C for 72 h. Individual colonies were screened by PCR to confirm genomic integration of the bait construct. Aureobasidin A (AbA) sensitivity was assayed by spotting serial dilutions of a saturated culture of a single PCR-confirmed bait strain onto SD/-Ura plates containing AbA at concentrations of 0, 100, 150, 200, 300, 500, 700, and 900 ng/mL. The minimal AbA concentration that completely inhibited growth after 3 days at 30 °C was selected for library screening.
2.5. Construction of the Prey cDNA Library
Total RNA was isolated from pooled greenhouse- and forest-cultivated D. huoshanense tissues using RNAiso Plus (9108Q, Takara, Kyoto, Japan) combined with Fruitmate for RNA purification kits (9192, Takara). mRNA was enriched according to FastTrack MAG mRNA Isolation Kit (K1580-02; Invitrogen). After that, first-strand cDNA synthesis was performed using the SMART™ technology (CloneMiner™ cDNA Library Construction Kit, A11180, Invitrogen), incorporating attB adapters via gene-specific primers during PCR amplification of ds cDNA. Purified ds cDNA was size fractionated using a CHROMA SPIN-1000-TE (Clontech, Mountain View, CA, USA). The attB-flanked ds cDNA library was recombined into the pDONR222 vector via BP Clonase II Enzyme Mix (Thermo Fisher Scientific) to generate the primary library plasmid. Then the secondary cDNA library was constructed by LR recombination reaction. The primary library plasmids were cloned into pGADT7-DEST and incubated with LR Clonase II enzyme mix (Invitrogen) [27] to create the AD fusion prey expression library (pGADT7-Rec plasmid). The final normalized prey plasmid library was amplified in E. coli competent cells DH10β and plasmid DNA was extracted using the PureLink HQ Kit (K210001, Invitrogen) for subsequent experiments.
2.6. Screening of a Y1H Library
The 5 μg normalized pGADT7 cDNA prey library was transformed into the AbA-sensitive bait strain (Y1HGold [pAbAi-DhPMM]) via the LiAc/SS-DNA/PEG method. Transformants were resuspended in 0.9% NaCl and subjected to serial 10-fold dilutions (100–10−3) and 100 μL diluents were coated onto SD-Leu plates to calculate the titer and the number of colonies: total number of colonies = colony-forming units (CFU)/(volume plated × dilution factor) × total suspension volume (mL) [28]. Then, yeast cell suspensions were coated on corresponding SD-Leu/AbA (containing the predetermined minimal inhibitory concentration of AbA).
2.7. Identification and Sequencing of Positive Clones
Putative positive clones from the primary screening were restreaked onto SD/-Leu/AbA plates (300 ng/mL AbA) for 72 h as the secondary screening to eliminate false positives. Surviving clones were cultured in SD/-Leu liquid medium at OD600 = 0.6–0.8 and validated by the Matchmaker Insert Check PCR Mix 2 (630497, Takara). Purified plasmids were extracted and Sanger-sequenced with vector-specific primers.
2.8. Pairwise Validation of the Promoter of DhPMM and Candidate TFs
After all the positive clones were analyzed by BLASTn (v2.17.0) against NCBI, several typical TFs were screened out. To validate interactions with the DhPMM promoter, pairwise yeast one-hybrid retransformation assays were performed. Competent Y1HGold [pAbAi-DhPMM] bait cells were generated using the LiAc/SS Carrier DNA/PEG method. Prey plasmids encoding candidate TFs were extracted from yeast clones. Purified prey plasmids (200 ng) were individually transformed into Y1HGold [pAbAi-DhPMM] strains. Transformants were plated on SD/-Leu and SD/-Leu/AbA (300 ng/mL AbA). TF-DhPMM promoter interactions were confirmed by growth on the SD/-Leu/AbA plates.
2.9. Dual-Luciferase (Dual-LUC) Assay
Putative TF-promoter interactions identified by Y1H screening were independently verified through dual-LUC assays to exclude false positives [29]. Target promoter sequences of DhPMM were cloned into the pGreenII 0800-LUC reporter vector, while candidate TFs were inserted into the pGreenII 62-SK effector vector, which were separately transformed into Agrobacterium tumefaciens GV3101. Vectors were part of our laboratory’s established vector collection. Bacterial cultures were centrifuged, resuspended in MES buffer to an OD600 = 0.6, and dark-adapted for 3 h at 22 °C. Effector strains (pGreenII 62-SK-TFs) and reporter strains (pGreenII 0800-DhPMM-pro-LUC) were combined at a 1:1 ratio, with pGreenII 62-SK empty vector controls. Suspensions were infiltrated into N. benthamiana leaves using 1 mL needless syringes. After 48 h of incubation, LUC imaging was detected using a chemiluminescence imaging system (IVIS Lumina Series, PerkinElmer, Waltham, MA, USA). LUC/Renilla (Ren) activity ratios were quantified using the Dual Luciferase Reporter Gene Assay Kit (KTA8010, Abbkine, Mountain View, CA, USA) with three biological replicates performed. Statistical analyses were performed with Bonferroni correction for multiple testing correction [30]. For dual-luciferase assays testing 4 transcription factors, the significance threshold was adjusted to 0.0125.
3. Results
3.1. Cloning of DhPMM Gene Promoter
Using D. huoshanense genomic DNA as template, PCR amplification and agarose gel electrophoresis were performed to isolate the DhPMM promoter sequence (Figure 1a). The PCR product was cloned into pMD19-T vector and transformed into E. coli DH5α, among which positive clones were verified and sequenced with a length of 2145 bp (Figure 1b). The promoter sequences were analyzed on the online website PlantCARE, which showed that varieties of cis-elements were involved such as ABRE, CAAT-box, MYC, and other uncharacterized regions (Table 1, Figure 1c).
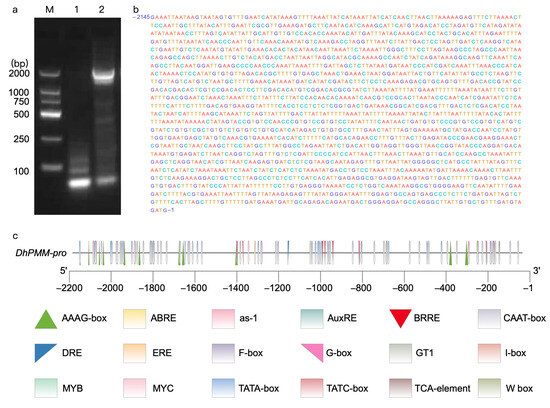
Figure 1.
Cloning of the promoter of DhPMM. (a) M: DNA marker, lane 1: the blank control, lane 2: the amplified fragment of the promoter of DhPMM. (b) Sequencing results of cloned promoter of DhPMM. (c) Cis-elements in the sequence of the promoters.

Table 1.
Cis-elements in sequence of DhPMM promoter.
3.2. Identification of Bait Yeast Strain and Determination of AbA Basal Expression
The DhPMM promoter was cloned into pAbAi via restriction digestion, generating recombinant pAbAi-DhPMM. BstBI digestion yielded fragments of 7000 bp, consistent with the expected size (2145 bp insert + 4870 bp vector, Figure 2a). Finally, PCR identification confirmed the bait plasmid pAbAi-DhPMM was successfully transformed into Y1HGold competent cells (Figure 2b).
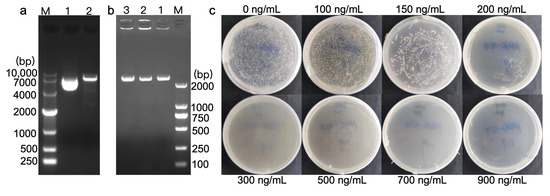
Figure 2.
Bait strain validation and AbA autoactivation determination. (a) BstBI digestion of pAbAi-DhPMM. Lane 1: undigested plasmid, lane 2: digestion products (4.87 kb vector + 2.15 kb insert). (b) PCR verification of genomic integration of linearized pAbAi-DhPMM in Y1HGold (397 bp + 2145 bp). Lanes 1–3: three positive clones. (c) Determination of the minimum inhibitory AbA concentration for Y1HGold [pAbAi-DhPMM] on the SD/-Ura plates.
Prior to cDNA library screening on SD/-Leu/AbA plates, the bait strain’s autoactivation potential was assessed to ascertain that interacting proteins were derived from the library rather than endogenous yeast proteins [31]. Figure 2c illustrated that strains grew vigorously on plates with AbA concentrations of 0, 100, 150, and 200 ng/mL, but showed complete growth inhibition at 300 ng/mL. Thus, the minimum AbA concentration 300 ng/mL was established for subsequent cDNA library screening.
3.3. cDNA Library Construction and Validation
The integrity of mRNA and ds cDNA was confirmed by agarose gel electrophoresis, on which bands showed continuous smears within 0.5–5 kb, (Supplementary Figure S1a,b). The primary library (pDONR222-cDNA) exhibited a volume of 1.44 × 107 CFU with 100% recombination efficiency and ≥1 kb inserts, as determined by colony PCR analysis of 24 randomly selected clones (Supplementary Figure S2a,b). The second library (pGADT7-cDNA) showed a volume of 1.20 × 107 CFU and consistent insert lengths (Supplementary Figure S2c,d). The results of volumes and recombination rates of the cDNA library stood for the favorable capacity and quality of the constructed library.
3.4. Screening of the Yeast One-Hybrid Library and Plasmids Extraction
The constructed Y1H expression library (pGADT7-cDNA) plasmid of D. huoshanense was introduced into the Y1HGold [pAbAi-DhPMM]. There were 1200 clones growing on the SD/-Leu plate with a titer of 1.20 × 107 CFU/mL (Figure 3a). The total number of screened clones was calculated as 3.60 × 109. Colony PCR and electrophoretic identification declared insert fragments ranging from 1 kb to 2 kb in length (Figure 3b).
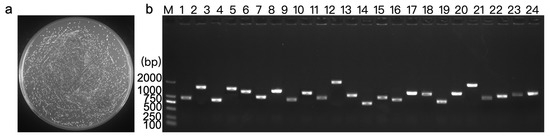
Figure 3.
Quality assessment of the construction of the pAbAi-DhPMM Y1H library. (a) pAbAi-DhPMM Y1H library met stringent quality criteria (CFU = 3.60 × 109). (b) Twenty-four clones were randomly chosen to determine the average size of the inserted fragments using colony PCR. Lanes 1–24: 24 randomly selected clones.
Y1H system represents a well-established strategy for identifying transcriptional regulatory roles. However, it is associated with a high frequency of false positives and sequence replication issues, which can be mitigated by employing two rounds of screening [32]. Following the first screening, 14 clones tested positive were found on the SD/-Leu/AbA (300 ng/mL) plate (Figure 4a). Eleven of the fourteen positive clones underwent further growth after being transferred on the SD/-Leu/AbA (300 ng/mL) medium for a second screening (Table 2, Figure 4b). Ultimately, following sequencing and identification, the roles and functions of the 11 identified genes were clarified. Among these sequences, four were annotated as transcription factors based on domains identification: no apical meristem, Arabidopsis thaliana transcription activation factor, cup-shaped cotyledon (NAC), DNA binding with one C2-C2 zinc finger domain (DOF), ethylene-responsive factors (ERF), and Bri1-ems-suppressors (BES1) families (Table 2).
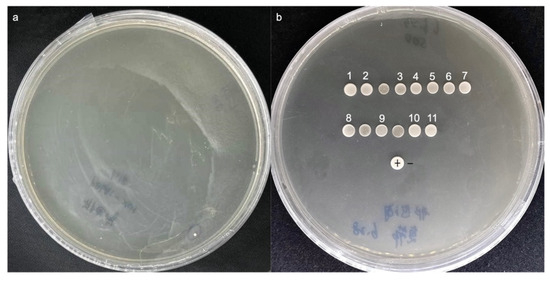
Figure 4.
Screening of the pAbAi-DhPMM Y1H library. Growth of positive mono-clone in SD/-Leu/AbA (300 ng/mL) solid medium (a) and their secondary culture (b).

Table 2.
Annotation of positive sequences from the Y1HGold [pAbAi-DhPMM].
3.5. Validation of the Interaction of the Promoter of DhPMM and Candidate TFs
DhPMM serves as a key catalytic enzyme in the polysaccharide biosynthesis pathway of D. huoshanense [33], while no transcription factor has been reported to regulate the expression of the DhPMM gene. Therefore, we focused on four TFs among those 11 candidate sequences to further confirm whether there were interactions between the candidate TFs with the 5′ transcriptionally active region (−2145 to −1 bp) of DhPMM. Each TF was individually transformed into Y1HGold [pAbAi-DhPMM] for one-to-one verification. All transformants were able to grow and spread on SD/-Leu plates, as demonstrated by the findings (Figure 5), suggesting that the Y1H transformation system was working well. Crucially, strains of D1, D2, D4, and D6 belonging to TFs families demonstrated robust growth on SD/-Leu/AbA (300 ng/mL) plates. The AbA-resistant growth of these four TFs on the plates indicated specific protein–DNA interactions.
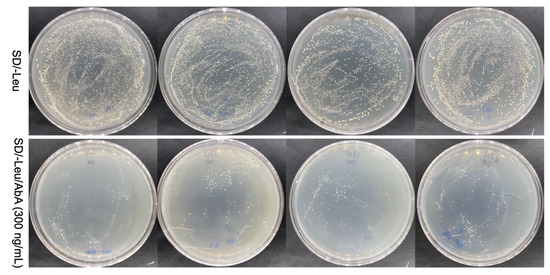
Figure 5.
Validation of candidate transcription factors interacting with the promoter of DhPMM in the SD/-Leu/AbA (300 ng/mL) solid medium. From left to right are positive colonies identified as NAC, DOF, ERF, and BES1 families.
3.6. NAC, DOF, ERF, and BES1 Interact with the Promoter of the DhPMM and Activate Its Expression
In addition, dual-luciferase assays were performed to verify the interactions between the DhPMM promoter and four candidate TFs from NAC, DOF, ERF, and BES1 families identified via Y1H screening. Co-expression of TF effectors (pGreenII 62-SK-TFs) with the promoter-LUC reporter (pGreenII 0800-DhPMM-pro) significantly increased luminescence activity compared to empty vector controls (pGreenII 62-SK). Normalized LUC/REN ratio of activity measurements suggested that NAC, DOF, ERF, and BES1 function as transcriptional activators of DhPMM (Figure 6a–d).
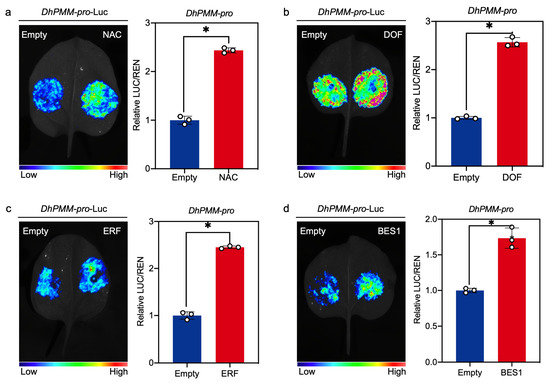
Figure 6.
Interaction of TFs belonging to NAC, DOF, ERF, and BES1 families with DhPMM promoter detected by dual-LUC assay. Luminescence imaging analysis of tobacco leaves and the determination of LUC/REN relative activity ratio of the pGreen-DhPMM-pro-LUC and pGreenII 62-SK-TFs ((a) NAC, (b) DOF, (c) ERF, and (d) BES1). Empty vector pGreenII 62-SK and reporter pGreen-DhPMM-pro-LUC functioned as negative control. Data represent mean ± SD (n = 3). Bonferroni correction: * p < 0.0125.
4. Discussion
Polysaccharides constitute essential bioactive compounds in D. huoshanense, with their biosynthesis involving coordinated enzymatic cascades. PMM, an enzyme of great importance in the mannose metabolic pathway, catalyzes the reversible isomerization of mannose-1-phosphate (M1P) to mannose-6-phosphate (M6P), positioning GDP-mannose as the central precursor for polysaccharide formation [34]. Our previous work verified PMM protein participated in polysaccharide biosynthesis pathway after crotonylation, which may enhance PMM’s catalytic activity [33]. Beyond post-translational modifications, transcription factors represent critical regulators of enzymes, thereby critically governing polysaccharide accumulation. These DNA-binding proteins recognize cis-elements in target gene promoters to orchestrate spatiotemporal gene expression [35]. As trans-acting regulators, TFs orchestrate multifaceted physiological processes, including growth and development [36], secondary metabolite biosynthesis [37], and responses to biotic/abiotic stresses [38] through precise control of gene expression dynamics. Notably, no transcriptional regulators of DhPMM had been characterized prior to this investigation—a significant knowledge gap given PMM’s pivotal metabolic role in D. huoshanense.
The yeast one-hybrid screening system is a gold-standard method of interrogating interactions between DNA sequences and DNA-binding proteins, particularly TFs [39]. The efficacy of cDNA library screening critically depends on two pivotal quality metrics: library complexity and average insert size, which collectively determine the coverage of low-abundance transcripts and functional gene representation [40]. Critical cDNA library parameters include a minimum library complexity of ≥1 × 106 CFU and insert size of ≥ 1 kb to ensure comprehensive detection of rare transcripts. Our cDNA library screening achieved a complexity of 3.60 × 109 CFU and an average insert size of 1.5 kb (range of 1–2 kb determined by colony PCR). These optimized parameters collectively satisfied the technical prerequisites for conducting systematic Y1H screening [41] and enabled successful identification of DhPMM transcriptional regulators.
High-complexity cDNA library screening enables rapid identification of transcription factors in medicinal plants, as established in prior studies [42,43,44]. In this work, we generated a cDNA library of D. huoshanense to capture full-length transcripts of transcription factors. Following BLAST analysis of filtered sequences against the NCBI database, we systematically characterized the domain of candidate genes to identify TFs. It finally revealed four TFs spanning the AAAG-box-combined DOF, G-box-combined NAC, DRE-combined ERF, and BRRE-combined BES1 families, which are key regulatory components warranting prioritization in subsequent studies. Consistent activation of the DhPMM promoter by these four TFs identified through Y1H screening combined with dual-LUC assay enlightened a new perspective of cooperative regulation of polysaccharide biosynthesis in D. huoshanense, potentially through binding to conserved cis-elements.
Based on previous studies in polysaccharide biosynthesis in plants [45,46,47], we proposed polysaccharide biosynthesis in D. huoshanense initiates from sucrose conversion via sucrose phosphate synthase (SPS) and then bifurcates into two metabolic branches (Figure 7): (1) the mannose flux pathway: sucrose synthase (SUS) -mediated fructose, PMM-catalyzed M1P; and (2) uridine diphosphate glucose (UDP-Glc) synthesis via SPS or invertase (INV). Glycosyltransferases integrate these monosaccharide units into polymers, with TF-mediated transcriptional regulation serving as a key control point. Our screening identified four TFs regulating DhPMM in this cascade. In addition, NAC TF identified in Y1H screening shows homology to MdNAC5 [48] and FaNAC035 [49], which bind to promoters of INV, SPS, and SUS to modulate transcriptional activation. Studies also unveiled ClNAC68 knockout reduced sucrose accumulation by suppressing ClINV expression [50], while FvNAC073 overexpression upregulated FvSPS1 and downregulated FvSUS2 to enhance soluble sugar content [51]. Intriguingly, no prior evidence links NAC to PMM transcriptional regulation. Our discovery of a D. huoshanense-specific NAC TF represents the first reported regulatory node in mannose metabolism. Plant-specific DOF TFs contain a highly conserved region of 50 amino acid residues with a C2–C2 zinc finger motif [52]. This motif interacts with the cis-elements containing a common core (A/T) AAAG or the P-box (TTATGG) in the promoter [53,54]. We found there were abundant (A/T) AAAG motifs in the promoters of DhPMM (Table 1). DOFs worked significantly in regulating the expression of genes engaging in saccharides’ metabolism [55], biosynthesis [56], and transport [57]. It has been reported that ZmDof3 upregulated the expression of SUS to administrate the content of sugar in maize [58]. In tomato, SlDof22 can play a role by regulating genes in the carbohydrate metabolism pathway, including PMM gene [59]. As described above, this DOF TF we screened likely modulates DhPMM expression, highlighting its potential as a metabolic engineering target. Additionally, another two TFs (ERF, BES1) showed limited direct polysaccharide links but exhibited metabolic crosstalk. ERF TFs primarily mediated stress responses by binding dehydration-responsive elements (DRE) in target promoters [60]. PtrERF110 activated PtrSPS4 and further regulated sugar and sterol biosynthesis when exposed to cold stress [61]. BES1, governed development via brassinosteroid signaling [62] and stress-induced developmental plasticity [63]. Collectively, the DhPMM promoter contains multiple binding motifs for all four TFs (Figure 1b). This strongly suggests these four TFs combinatorial regulatory potential in modulating DhPMM expression—a prerequisite for enhancing polysaccharide accumulation in D. huoshanense.
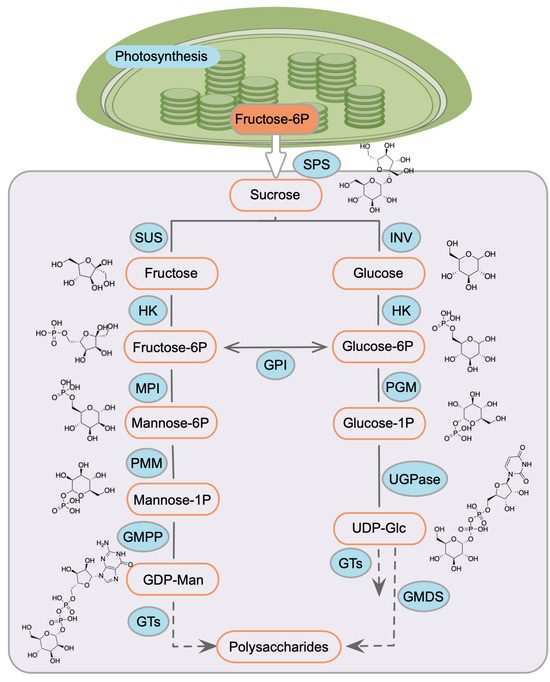
Figure 7.
Putative pathway of polysaccharide biosynthesis in D. huoshanense. SPS: sucrose phosphate synthase, SUS: sucrose synthase, INV: invertase, HK: hexokinase, GPI: glucose-6-phosphate isomerase, MPI: mannose-6-phosphate isomerase, PGM: phosphoglucomutase, PMM: phosphomannomutase, GMPP: GDP-mannose pyrophosphorylase, GMDS: GDP-Mannose 4,6-Dehydratase, UGPase: UDP-Glc pyrophosphorylase, and GTs: glycosyl transferases.
5. Conclusions
This study systematically identified upstream genes including TFs with potential to regulate the expression of DhPMM. The 2.1 kb DhPMM promoter, enriched with various cis-elements, was cloned to construct high-complexity cDNA library (3.60 × 109 CFU). Y1H and dual-LUC assays validated interactions between four TFs (DOF, NAC, ERF, BES1 families) and DhPMM, which may be linked to regulating polysaccharide biosynthesis. This first-reported D. huoshanense cDNA library provides a foundational resource for bridging transcriptional regulation with active constituent’s content, offering insights for the sustainable utilization of non-model medicinal plant.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biom15091251/s1. Figure S1: Electrophoretic diagram of mRNA (a) and cDNA library (b); Figure S2: Quality identification of the constructed primary and secondary cDNA libraries title.
Author Contributions
Conceptualization, J.W. and S.X.; methodology, S.W.; validation, S.W.; data curation, D.P.; writing—original draft preparation, J.W.; writing—review and editing, J.W. and S.X.; supervision, D.P.; project administration, D.P.; funding acquisition, S.X. and D.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Major Special Science and Technology Project of Anhui Province, China (Grant No. 202303a0702005), the National Natural Science Foundation of China (Grant No. 82474037), Traditional Chinese Medicine high-level key discipline construction project of National Administration of Traditional Chinese Medicine—Science of Chinese medicinal material resources (pharmaceutical botany) (Grant No. zyyzdxk-2023095).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The datasets generated during the current study are openly available in Sequence Read Archive (SRA) of NCBI (https://www.ncbi.nlm.nih.gov, accessed on 20 August 2025) under the access numbers SRR32943785, SRR32945542, SRR32946154, SRR32944933, SRR32946815, SRR32946151, SRR32946884, SRR32946904, SRR32946921, SRR32946924, and SRR32947996. The associated BioProject and BioSample numbers are PRJNA1245079 and SAMN47738843 (https://www.ncbi.nlm.nih.gov/sra/PRJNA1245079, accessed on 20 August 2025).
Acknowledgments
We sincerely thank Yumeng Ji for her assistance in validation, Zhaojian Wang for his support in formal analysis, and Nianjun Yu for his help in resources for this study.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| AbA | Aureobasidin |
| ABRE | ABA-responsive element |
| AMP | Ampicillin |
| AP2/ERF | APETALA2/Ethylene Response Factor |
| BES1 | Bri1-ems-suppressor 1 |
| BRRE | Brassinosteroid-responsive element |
| CFU | Colony-forming units |
| cDNA | Complementary DNA |
| DRE | Dehydration-responsive element, |
| DOF | DNA binding with one zinc finger |
| ERF | Ethylene response factor |
| GDP-Man | Guanosine diphosphate mannose |
| GTs | Glycosyltransferases |
| HK | Hexokinase |
| INV | Invertase |
| LB | Luria–Bertani medium |
| M1P | Mannose-1-phosphate |
| M6P | Mannose-6-phosphate |
| MPI | Mannose-6-phosphate isomerase |
| MYC | Myc transcription factor family |
| NAC | Nam, ataf, and cuc |
| PCR | Polymerase chain reaction |
| PEG | Polyethylene glycol |
| PGM | Phosphoglucomutase |
| PMM | Phosphomannomutase |
| SD | Synthetic-defined medium |
| SMART | Switching mechanism at 5′ end of RNA template |
| SPS | Sucrose phosphate synthase |
| SUS | Sucrose synthase |
| TF | Transcription factor |
| UDP-Glc | Uridine diphosphate glucose |
| Y1H | Yeast one-hybrid |
| YPDA | Yeast extract peptone dextrose adenine medium |
References
- Wu, P. Shen Nong Ben Cao Jing; Scientific and Technological Literature Publishing House: Beijing, China, 1996. [Google Scholar]
- Chinese Pharmacopoeia Commission. Pharmacopoeia of the People’s Republic of China; China Medical Science Press: Beijing, China, 2020; pp. 70–71. Available online: https://ydz.chp.org.cn/#/main (accessed on 27 August 2025).
- Shang, Z.Z.; Qin, D.Y.; Li, Q.M.; Zha, X.Q.; Pan, L.H.; Peng, D.Y.; Luo, J.P. Dendrobium huoshanense stem polysaccharide ameliorates rheumatoid arthritis in mice via inhibition of inflammatory signaling pathways. Carbohydr. Polym. 2021, 258, 117657. [Google Scholar] [CrossRef] [PubMed]
- Zha, X.Q.; Zhao, H.W.; Bansal, V.; Pan, L.H.; Wang, Z.M.; Luo, J.P. Immunoregulatory activities of Dendrobium huoshanense polysaccharides in mouse intestine, spleen and liver. Int. J. Biol. Macromol. 2014, 64, 377–382. [Google Scholar] [CrossRef]
- Ng, T.B.; Liu, J.; Wong, J.H.; Ye, X.; Wing Sze, S.C.; Tong, Y.; Zhang, K.Y. Review of research on Dendrobium, a prized folk medicine. Appl. Microbiol. Biotechnol. 2012, 93, 1795–1803. [Google Scholar] [CrossRef]
- Yi, S.; Kang, C.; Wang, W.; Song, X.; Xu, T.; Lu, H.; Luo, S.; Liu, D.; Guo, L.; Han, B. Comparison of planting modes of Dendrobium huoshanense and analysis of advantages of simulated cultivation. Chin. J. Chin. Mater. Medica 2021, 46, 1864–1868. [Google Scholar] [CrossRef]
- Wang, Z.; Chen, J.H.; Wang, L.S.; Ding, J.; Zhao, M.W.; Liu, R. GlPP2C1 silencing increases the content of Ganodermalingzhi polysaccharide (GL-PS) and enhances Slt2 phosphorylation. J. Fungi 2022, 8, 949. [Google Scholar] [CrossRef]
- Qian, W.; Yu, C.; Qin, H.; Liu, X.; Zhang, A.; Johansen, I.E.; Wang, D. Molecular and functional analysis of phosphomannomutase (PMM) from higher plants and genetic evidence for the involvement of PMM in ascorbic acid biosynthesis in Arabidopsis and Nicotiana benthamiana. Plant J. 2007, 49, 399–413. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Zeng, S.; Teixeira da Silva, J.A.; Yu, Z.; Tan, J.; Duan, J. Molecular cloning and functional analysis of the phosphomannomutase (PMM) gene from Dendrobium officinale and evidence for the involvement of an abiotic stress response during germination. Protoplasma 2017, 254, 1693–1704. [Google Scholar] [CrossRef]
- Cao, S.; Wang, X.; Ji, J.; Wang, Y.; Gao, J. Cloning and expression analysis of CpPMM gene in Codonopsis pilosula. Chin. J. Chin. Mater. Medica 2020, 45, 4382–4391. [Google Scholar] [CrossRef]
- Lin, R.; Zhong, H.; Ye, X.; Huang, M. Cloning and quantitative expression analysis of PMM gene from Dendrobium huoshanense. Chin. J. Trop. Crops 2017, 38, 2326–2333. Available online: https://kns.cnki.net/kcms2/article/abstract?v=Y2E-z2Sa5COTnQe2NmizorGkz2DIPEmVtmY1MPPd9PUPctdvDouYHeBAHf0tk3zwVEmtRlwzRw7WZDOjqLz7AxbYpi1g-PFQzp4FC0gnp6fIWQBUFKUnLg4J9Ev3ely3Eu-JIK7D-J4HYZ84ELYFI1_8mGbm8mQUuy9u86yqnci4FaXduoxC7djskNK3OCUa&uniplatform=NZKPT&language=CHS (accessed on 27 August 2025).
- Kang, J.Y.; Choi, H.I.; Im, M.Y.; Kim, S.Y. Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell 2002, 14, 343–357. [Google Scholar] [CrossRef] [PubMed]
- Shu, G.; Tang, Y.; Yuan, M.; Wei, N.; Zhang, F.; Yang, C.; Lan, X.; Chen, M.; Tang, K.; Xiang, L.; et al. Molecular insights into AabZIP1-mediated regulation on artemisinin biosynthesis and drought tolerance in Artemisia annua. Acta Pharm. Sin. B 2022, 12, 1500–1513. [Google Scholar] [CrossRef]
- Dong, Q.; Xu, Q.; Kong, J.; Peng, X.; Zhou, W.; Chen, L.; Wu, J.; Xiang, Y.; Jiang, H.; Cheng, B. Overexpression of ZmbZIP22 gene alters endosperm starch content and composition in maize and rice. Plant Sci. 2019, 283, 407–415. [Google Scholar] [CrossRef]
- Sagor, G.H.; Berberich, T.; Tanaka, S.; Nishiyama, M.; Kanayama, Y.; Kojima, S.; Muramoto, K.; Kusano, T. A novel strategy to produce sweeter tomato fruits with high sugar contents by fruit-specific expression of a single bZIP transcription factor gene. Plant Biotechnol. J. 2016, 14, 1116–1126. [Google Scholar] [CrossRef]
- Bush, S.M.; Folta, S.; Lannigan, D.A. Use of the yeast one-hybrid system to screen for mutations in the ligand-binding domain of the estrogen receptor. Steroids 1996, 61, 102–109. [Google Scholar] [CrossRef]
- Chebbi, M.; Ginis, O.; Courdavault, V.; Glevarec, G.; Lanoue, A.; Clastre, M.; Papon, N.; Gaillard, C.; Atanassova, R.; St-Pierre, B.; et al. ZCT1 and ZCT2 transcription factors repress the activity of a gene promoter from the methyl erythritol phosphate pathway in Madagascar periwinkle cells. J. Plant Physiol. 2014, 171, 1510–1513. [Google Scholar] [CrossRef]
- Li, S.; Zhang, P.; Zhang, M.; Fu, C.; Yu, L. Functional analysis of a WRKY transcription factor involved in transcriptional activation of the DBAT gene in Taxus chinensis. Plant Biol. 2013, 15, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Zhu, F.; Lai, R.; Shi, L.; Chen, S. Construction of yeast one-hybrid library and screening of transcription factors regulating LS expression in Ganoderma lucidum. Chin. J. Chin. Mater. Medica 2019, 44, 3967–3973. [Google Scholar] [CrossRef]
- Lu, X.; Jiang, W.; Zhang, L.; Zhang, F.; Zhang, F.; Shen, Q.; Wang, G.; Tang, K. AaERF1 positively regulates the resistance to Botrytis cinerea in Artemisia annua. PLoS ONE 2013, 8, e57657. [Google Scholar] [CrossRef]
- Tang, Z.; Cheng, S. A study on the raw plants for the Chinese traditional medicine “Huoshan Shi-Hu”. Bull. Bot. Res. 1984, 4, 141–146. Available online: https://kns.cnki.net/kcms2/article/abstract?v=Y2E-z2Sa5CNKbj19Dg7s8pGHnoto_OqpkOJ1l2uyk2066hx5HDcZee326toYHkZId2ITYMNdaYiN9_sNIaDBzzxEq6jppel6vXviFJv4ao8i_CYOQWO1YS8sThU5ml-TMpZwDsMkwuJwMzsEBERO_GoLQwXPoeTTN5PzYhmCLiWhVx1cwq_pKw==&uniplatform=NZKPT&language=CHS (accessed on 27 August 2025).
- Sayers, E.W.; Beck, J.; Bolton, E.E.; Brister, J.R.; Chan, J.; Connor, R.; Feldgarden, M.; Fine, A.M.; Funk, K.; Hoffman, J.; et al. Database resources of the National Center for Biotechnology Information in 2025. Nucleic Acids Res. 2025, 53, D20–D29. [Google Scholar] [CrossRef] [PubMed]
- Han, B.; Jing, Y.; Dai, J.; Zheng, T.; Gu, F.; Zhao, Q.; Zhu, F.; Song, X.; Deng, H.; Wei, P.; et al. A chromosome-level genome assembly of Dendrobium huoshanense using long reads and Hi-C data. Genome Biol. Evol. 2020, 12, 2486–2490. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhu, J.; Khan, M.; Wang, Y.; Xiao, W.; Fang, T.; Qu, J.; Xiao, P.; Li, C.; Liu, J.H. Transcription factors ABF4 and ABR1 synergistically regulate amylase-mediated starch catabolism in drought tolerance. Plant Physiol. 2023, 191, 591–609. [Google Scholar] [CrossRef] [PubMed]
- Gietz, R.D. Yeast transformation by the LiAc/SS carrier DNA/PEG method. Methods Mol. Biol. 2014, 1205, 1–12. [Google Scholar] [CrossRef]
- Degnin, M.; Tognon, C.E.; Eide, C.A.; Wilmot, B.; McWeeney, S.K.; Bottomly, D.; Smith, R.; Druker, B.J. High-throughput validation of mutations identified in primary leukemia cells. Blood 2016, 128, 4725. [Google Scholar] [CrossRef]
- Fernández-Rodríguez, D.; Cho, J.E.; Parvizi, N.; Khan, A.Z.; Parvizi, J.; Namdari, S. Next-generation sequencing results require higher inoculum for detection than conventional anaerobic culture. Clin. Orthop. Relat. Res. 2023, 481, 2484–2491. [Google Scholar] [CrossRef]
- Pu, X.L.; Zhang, L.Y.; Zhang, J.Y.; Ye, X.; Lin, L.; Wu, D.R.; Kang, J.J.; Hu, H.; Chen, J.; Guo, K.; et al. Identification of a bZIP transcription factor PpTGA3 regulating polyphyllin biosynthesis in Paris polyphylla. Ind. Crops Prod. 2025, 225, 120492. [Google Scholar] [CrossRef]
- Armstrong, R.A. When to use the Bonferroni correction. Ophthalic Physl. Opt. 2014, 34, 502–508. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Li, Y.A.; Zhang, Y. Yeast two-hybrid screening for proteins that interact with the extracellular domain of amyloid precursor protein. Neurosci. Bull. 2016, 32, 171–176. [Google Scholar] [CrossRef] [PubMed]
- Xue, J.; Zhang, H.; Zhao, Q.; Cui, S.; Yu, K.; Sun, R.; Yu, Y. Construction of yeast one-hybrid library of Alternaria oxytropis and screening of transcription factors regulating swnK gene expression. J. Fungi 2023, 9, 822. [Google Scholar] [CrossRef]
- Wu, J.; Meng, X.; Jiang, W.; Wang, Z.; Zhang, J.; Meng, F.; Yao, X.; Ye, M.; Yao, L.; Wang, L.; et al. Qualitative proteome-wide analysis reveals the diverse functions of lysine crotonylation in Dendrobium huoshanense. Front. Plant Sci. 2022, 13, 822374. [Google Scholar] [CrossRef]
- Pablo, O.; Miguel, R.; Sandra, D.; Francisco, J. Stress response requires an efficient connection between glycogen and central carbon metabolism by phosphoglucomutases in cyanobacteria. J. Exp. Bot. 2023, 74, 1532–1550. [Google Scholar] [CrossRef]
- Koyama, T. Regulatory mechanisms of transcription factors in plant morphology and function. Int. J. Mol. Sci. 2023, 24, 7039. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Chen, W.; Xu, Z.; Chen, M.; Yu, D. Functions of WRKYs in plant growth and development. Trends Plant Sci. 2023, 28, 630–645. [Google Scholar] [CrossRef]
- Jia, H.; Xu, Y.; Deng, Y.; Xie, Y.; Gao, Z.; Lang, Z.; Niu, Q. Key transcription factors regulate fruit ripening and metabolite accumulation in tomato. Plant Physiol. 2024, 195, 2256–2273. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhang, Z.; Wang, P.; Qin, C.; He, L.; Kong, L.; Ren, W.; Liu, X.; Ma, W. Genome-wide identification of the NAC transcription factors family and regulation of metabolites under salt stress in Isatis indigotica. Int. J. Biol. Macromol. 2023, 240, 124436. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Vijaychander, S.; Stile, J.; Zhu, L. Cloning and analysis of DNA-binding proteins by yeast one-hybrid and one-two-hybrid systems. Biotechniques 1996, 20, 564–568. [Google Scholar] [CrossRef]
- Xu, Y.; Zhou, J.; Liu, Q.; Li, K.; Zhou, Y. Construction and characterization of a high-quality cDNA library of Cymbidium faberi suitable for yeast one- and two-hybrid assays. BMC Biotechnol. 2020, 20, 4. [Google Scholar] [CrossRef]
- Serra, T.S.; Figueiredo, D.D.; Cordeiro, A.M.; Almeida, D.M.; Lourenço, T.; Abreu, I.A.; Sebastián, A.; Fernandes, L.; Contreras-Moreira, B.; Oliveira, M.M.; et al. OsRMC, a negative regulator of salt stress response in rice, is regulated by two AP2/ERF transcription factors. Plant Mol. Biol. 2013, 82, 439–455. [Google Scholar] [CrossRef]
- Jingwen, W.; Jingxin, W.; Ye, Z.; Yan, Z.; Caozhi, L.; Yanyu, C.; Fanli, Z.; Su, C.; Yucheng, W. Building an improved transcription factor-centered yeast one hybrid system to identify DNA motifs bound by protein comprehensively. BMC Plant Biol. 2023, 23, 236. [Google Scholar] [CrossRef]
- Fan, S.; Li, X.; Lin, S.; Li, Y.; Ma, H.; Zhang, Z.; Qin, Z. Screening and identification of transcription factors potentially regulating Foxl2 expression in Chlamys farreri Ovary. Biology 2022, 11, 113. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Bi, M.; Yang, P.; Song, M.; He, G.; Wang, J.; Yang, Y.; Xu, L.; Ming, J. Construction of yeast one-hybrid library and screening of transcription factors regulating LhMYBSPLATTER expression in Asiatic hybrid lilies (Lilium spp.). BMC Plant Biol. 2021, 21, 563. [Google Scholar] [CrossRef]
- Li, M.; Li, P.; Ma, F.; Dandekar, A.M.; Cheng, L. Sugar metabolism and accumulation in the fruit of transgenic apple trees with decreased sorbitol synthesis. Hortic. Res. 2018, 5, 60. [Google Scholar] [CrossRef] [PubMed]
- Sturm, A.; Tang, G. The sucrose-cleaving enzymes of plants are crucial for development, growth and carbon partitioning. Trends Plant Sci. 1999, 4, 401–407. [Google Scholar] [CrossRef]
- Zanor, M.; Osorio, S.; Nunes, N.; Carrari, F.; Lohse, M.; Usadel, B.; Kühn, C.; Bleiss, W.; Giavalisco, P.; Willmitzer, L.; et al. RNA interference of LIN5 in tomato confirms its role in controlling brix content, uncovers the influence of sugars on the levels of fruit hormones, and demonstrates the importance of sucrose cleavage for normal fruit development and fertility. Plant Physiol. 2009, 150, 1204–1218. [Google Scholar] [CrossRef]
- Zhang, B.; Yang, H.J.; Li, Y.N.; Zhu, Z.Z.; Zhao, Z.Y.; Yang, Y.Z. MdNAC5: A key regulator of fructose accumulation in apple fruit. New Phytol. 2024, 244, 2458–2473. [Google Scholar] [CrossRef]
- Martin-Pizarro, C.; Vallarino, J.G.; Osorio, S.; Meco, V.; Urrutia, M.; Pillet, J.; Casanal, A.; Merchante, C.; Amaya, I.; Willmitzer, L.; et al. The NAC transcription factor FaRIF controls fruit ripening in strawberry. Plant Cell 2021, 33, 1574–1593. [Google Scholar] [CrossRef]
- Wang, J.; Wang, Y.; Zhang, J.; Ren, Y.; Li, M.; Tian, S.; Yu, Y.; Zuo, Y.; Gong, G.; Zhang, H.; et al. Correction: The NAC transcription factor ClNAC68 positively regulates sugar content and seed development in watermelon by repressing ClINV and ClGH3.6. Hortic. Res. 2021, 8, 265. [Google Scholar] [CrossRef]
- Xiao, K.; Fan, J.M.; Bi, X.Y.; Tu, X.Y.; Li, X.Y.; Cao, M.H.; Liu, Z.; Lin, A.Q.; Wang, C.; Xu, P.B.; et al. A NAC transcription factor and a MADS-box protein antagonistically regulate sucrose accumulation in strawberry receptacles. Plant Physiol. 2025, 197, kiaf043. [Google Scholar] [CrossRef]
- Yanagisawa, S. The Dof family of plant transcription factors. Trends Plant Sci. 2002, 7, 555–560. [Google Scholar] [CrossRef] [PubMed]
- Yanagisawa, S.; Sheen, J. Involvement of maize Dof zinc finger proteins in tissue-specific and light-regulated gene expression. Plant Cell 1998, 10, 75–89. [Google Scholar] [CrossRef] [PubMed]
- Dong, G.; Ni, Z.; Yao, Y.; Nie, X.; Sun, Q. Wheat Dof transcription factor WPBF interacts with TaQM and activates transcription of an alpha-gliadin gene during wheat seed development. Plant Mol. Biol. 2007, 63, 73–84. [Google Scholar] [CrossRef]
- Wei, Z.; Zhang, H.; Fang, M.; Lin, S.; Zhu, M.; Li, Y.; Jiang, L.; Cui, T.; Cui, Y.; Kui, H.; et al. The Dof transcription factor COG1 acts as a key regulator of plant biomass by promoting photosynthesis and starch accumulation. Mol Plant 2023, 16, 1759–1772. [Google Scholar] [CrossRef]
- Du, C.; Sun, W.; Song, Q.; Zuo, K. GhDOFD45 promotes sucrose accumulation in cotton seeds by transcriptionally activating GhSWEET10 expression. Plant J. 2024, 120, 2468–2484. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, L.; Ansah, E.O.; Peng, W.; Zhang, W.; Li, P.; An, G.; Xiong, F. The sucrose transport regulator OsDOF11 mediates cytokinin degradation during rice development. Plant Physiol. 2022, 189, 1083–1094. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.; Li, S.; Zhu, Y.; Zhao, Q.; Zhu, D.; Yu, J. ZmDof3, a maize endosperm-specific Dof protein gene, regulates starch accumulation and aleurone development in maize endosperm. Plant Mol. Biol. 2017, 93, 7–20. [Google Scholar] [CrossRef]
- Cai, X. Genome-Wide Analysis of DOF Family and Functional Characterization of SlDof22, SlDHAR1 and FAGALUR in Tomato Ascorbate Accumulation. Ph.D. Thesis, Huazhong Agricultural University, Wuhan, China, 2014. [Google Scholar]
- Phukan, U.J.; Jeena, G.S.; Tripathi, V.; Shukla, R.K. MaRAP2-4, a waterlogging-responsive ERF from Mentha, regulates bidirectional sugar transporter AtSWEET10 to modulate stress response in Arabidopsis. Plant Biotechnol. J. 2018, 16, 221–233. [Google Scholar] [CrossRef]
- Khan, M.; Dahro, B.; Wang, Y.; Wang, M.; Xiao, W.; Qu, J.; Zeng, Y.; Fang, T.; Xiao, P.; Xu, X.; et al. The transcription factor ERF110 promotes cold tolerance by directly regulating sugar and sterol biosynthesis in citrus. Plant J. 2024, 119, 2385–2401. [Google Scholar] [CrossRef]
- Shi, H.; Li, X.; Lv, M.; Li, J. BES1/BZR1 family transcription factors regulate plant development via brassinosteroid-dependent and independent pathways. Int. J. Mol. Sci. 2022, 23, 10149. [Google Scholar] [CrossRef] [PubMed]
- Nolan, T.M.; Vukašinović, N.; Liu, D.; Russinova, E.; Yin, Y. Brassinosteroids: Multidimensional regulators of plant growth, development, and stress responses. Plant Cell 2020, 32, 295–318. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).