Lipid Metabolism and Improvement in Oilseed Crops: Recent Advances in Multi-Omics Studies
Abstract
1. Introduction
2. Current Knowledge on Plant Lipids
2.1. Function of Lipids
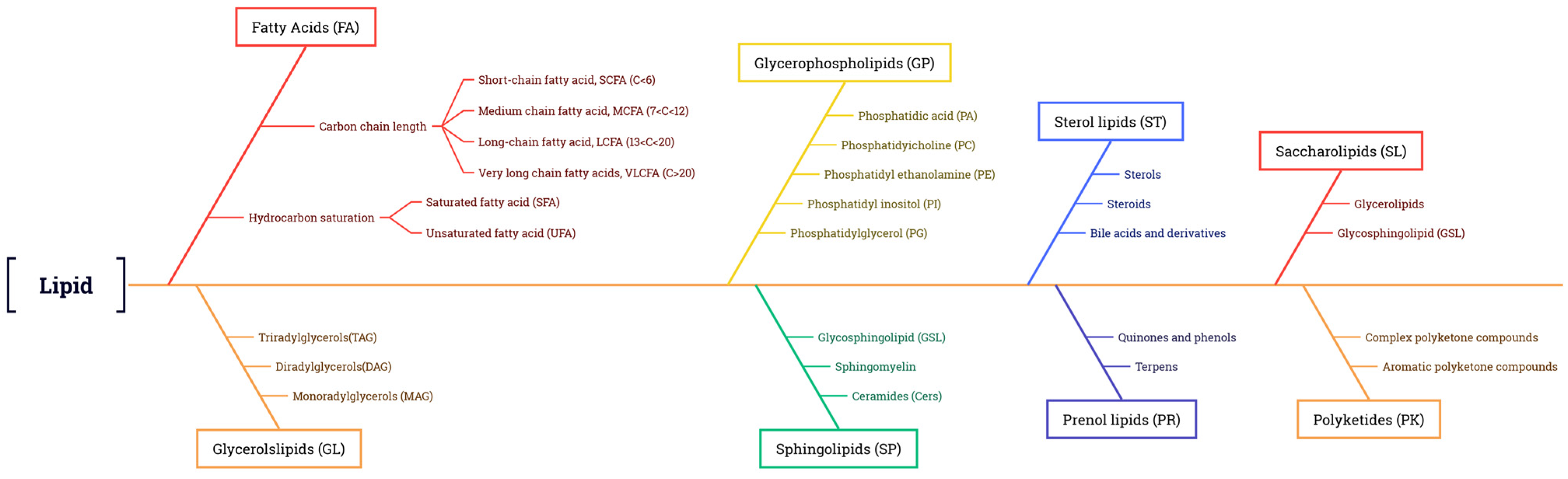
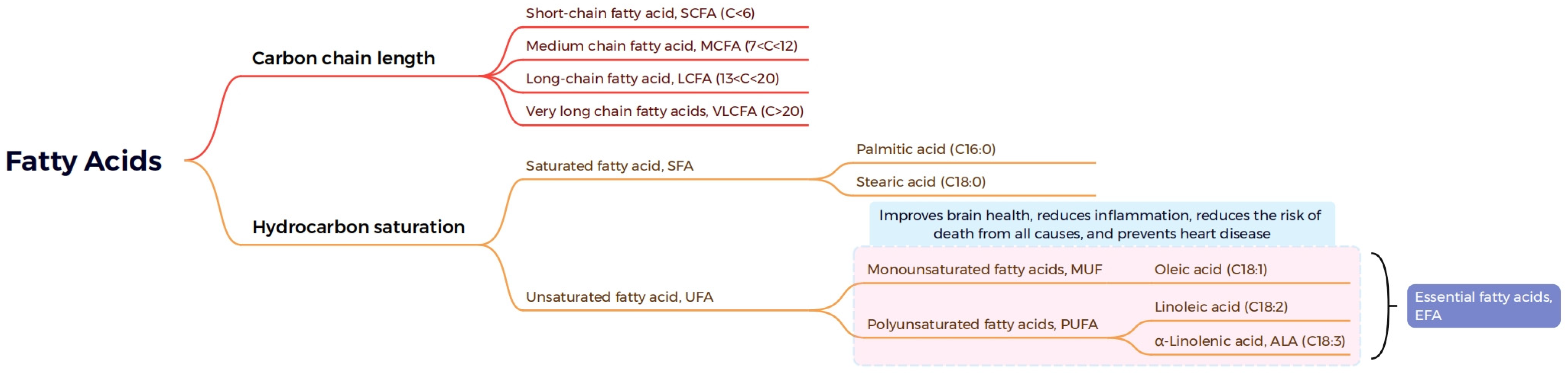
2.2. Classification of Plant Lipids
2.3. Lipid Accumulation and Storage in Plants
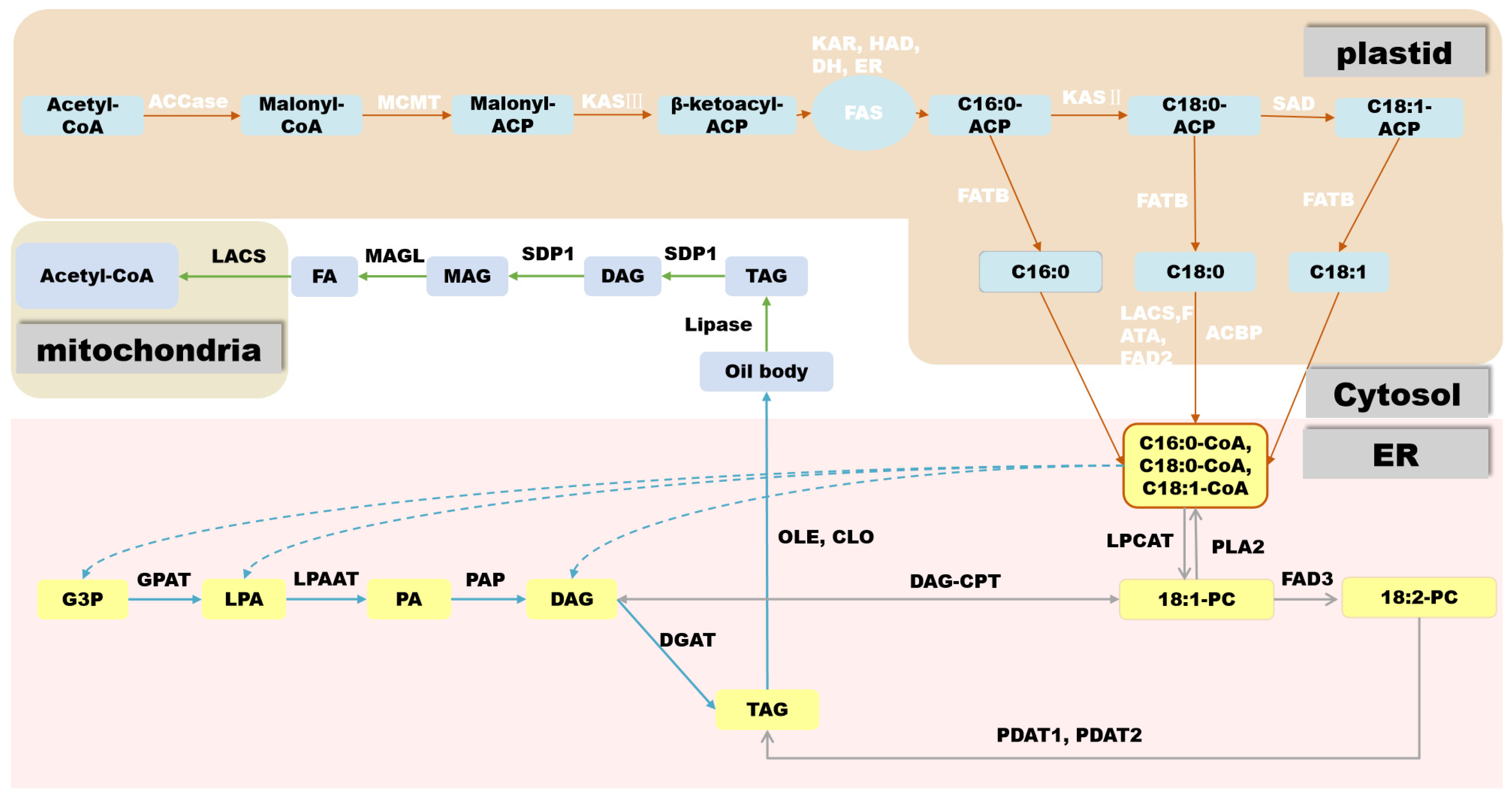
2.3.1. De Novo Biosynthesis of FAs
2.3.2. TAG Biosynthesis
2.3.3. Formation of Lipid Droplets
2.3.4. Inhibition of Lipid Degradation
3. Progress in the Identification of Key Genes behind Lipid Metabolism in Plants
3.1. Identification and Functional Characterization of Key Genes
3.2. Transcription Factors Involved in Regulation
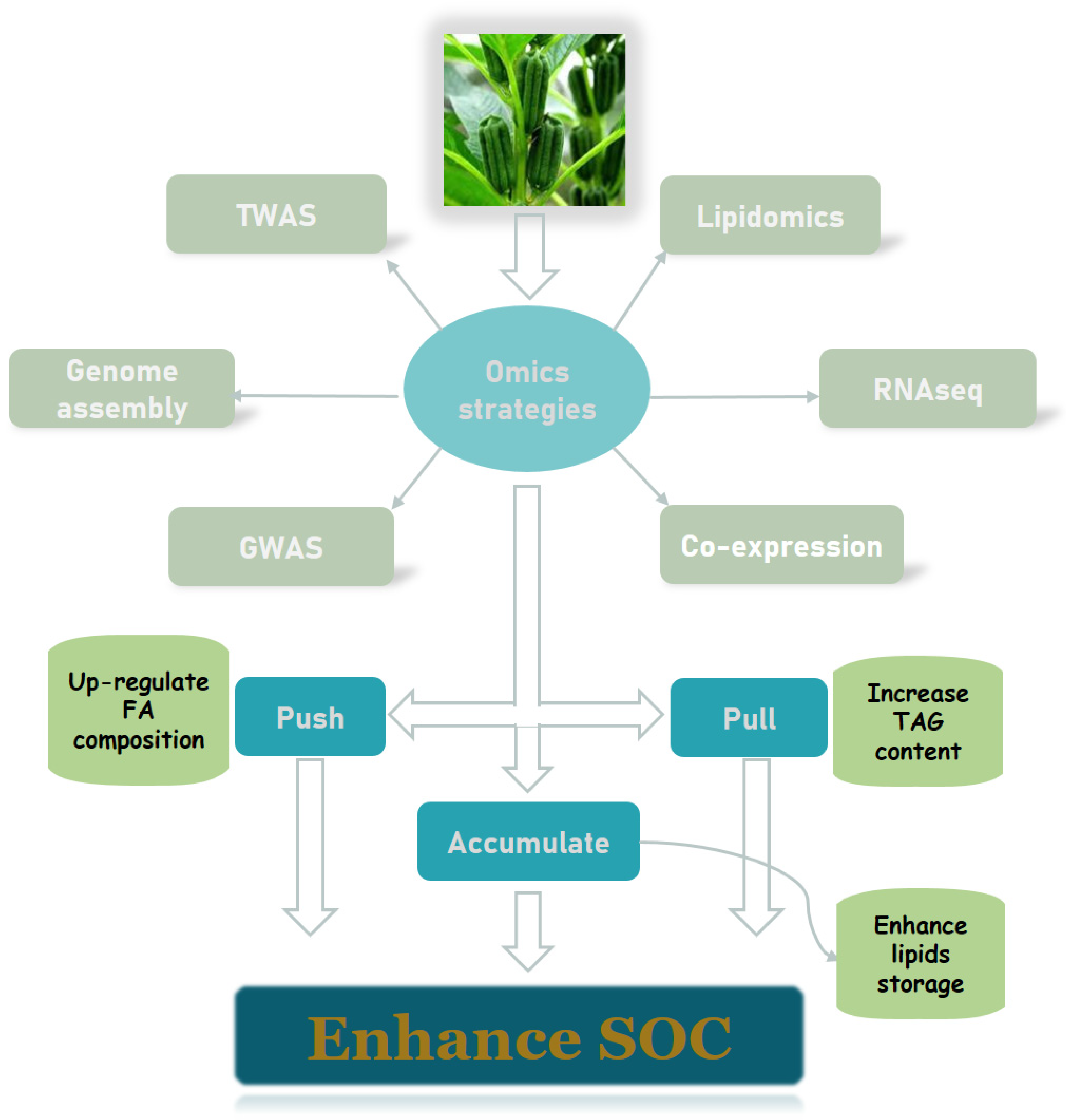
3.3. Advances in Multi-Omics Studies of Oilseed Crops
3.3.1. De Novo Genome Sequencing and Annotation
3.3.2. Identification of Differentially Expressed Genes
3.3.3. Construction of Oil Co-Expression Networks
3.3.4. Genome-Wide Association Studies to Map Oil Content–Related Loci
3.3.5. Lipidomics for Oil Structure and Quality Identification
4. Conclusions and Future Prospects
Author Contributions
Funding
Conflicts of Interest
References
- Mielke, T. World Markets for Vegetable Oils: Status and Prospects. In Encyclopedia of Sustainability Science and Technology; Springer: Berlin/Heidelberg, Germany, 2017; pp. 1–38. [Google Scholar]
- Caine, W.R.; Verstegen, M.W.A.; Sauer, W.C.; Tamminga, S.; Schulze, H. Effect of protease treatment of soybean meal on content of total soluble matter and crude protein and level of soybean trypsin inhibitors. Anim. Feed. Sci. Technol. 1998, 71, 177–183. [Google Scholar] [CrossRef]
- Correa, S.M.; Fernie, A.R.; Nikoloski, Z.; Brotman, Y. Towards model-driven characterization and manipulation of plant lipid metabolism. Prog. Lipid Res. 2020, 80, 101051. [Google Scholar] [CrossRef] [PubMed]
- Kepczynski M, Rog T: Functionalized lipids and surfactants for specific applications. Biochim. Biophys. Acta 2016, 1858, 2362–2379. [CrossRef]
- Mailer, R.J. Oilseeds, Overview. In Reference Module in Food Science; Elsevier: Amsterdam, The Netherlands, 2016. [Google Scholar]
- Jorge, N.; Goncalves, L.A.G.; Dobarganes, M.C. Influence of fatty acid composition on the formation of polar glycerides and polar fatty acids in sunflower oils heated at frying temperatures. Grasas Aceites 1997, 48, 17–24. [Google Scholar] [CrossRef]
- De Carvalho, C.; Caramujo, M. The Various Roles of Fatty Acids. Molecules 2018, 23, 2583. [Google Scholar] [CrossRef] [PubMed]
- Slocombe, S.P.; Cornah, J.; Pinfield-Wells, H.; Soady, K.; Zhang, Q.Y.; Gilday, A.; Dyer, J.M.; Graham, I.A. Oil accumulation in leaves directed by modification of fatty acid breakdown and lipid synthesis pathways. Plant Biotechnol. J. 2009, 7, 694–703. [Google Scholar] [CrossRef] [PubMed]
- Lewandowska, M.; Keyl, A.; Feussner, I. Wax biosynthesis in response to danger: Its regulation upon abiotic and biotic stress. New Phytol. 2020, 227, 698–713. [Google Scholar] [CrossRef]
- Voelker, T.; Kinney, A.T. Variations in the biosynthesis of seed-storage lipids. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001, 52, 335–361. [Google Scholar] [CrossRef]
- Sud, M.; Fahy, E.; Cotter, D.; Brown, A.; Dennis, E.; Glass, C.; Merrill, A.H.; Murphy, R.; Raetz, C.; Russell, D.; et al. LMSD: LIPID MAPS structure database. Nucleic Acids Res. 2007, 35, D527–D532. [Google Scholar] [CrossRef]
- Fahy, E.; Subramaniam, S.; Brown, H.A.; Glass, C.K.; Merrill, A.H., Jr.; Murphy, R.C.; Raetz, C.R.; Russell, D.W.; Seyama, Y.; Shaw, W.; et al. A comprehensive classification system for lipids. J. Lipid Res. 2005, 46, 839–861. [Google Scholar] [CrossRef]
- Ohlrogge, J.; Thrower, N.; Mhaske, V.; Stymne, S.; Baxter, M.; Yang, W.; Liu, J.; Shaw, K.; Shorrosh, B.; Zhang, M.; et al. PlantFAdb: A resource for exploring hundreds of plant fatty acid structures synthesized by thousands of plants and their phylogenetic relationships. Plant J. 2018, 96, 1299–1308. [Google Scholar] [CrossRef]
- Harwood, J.L. Fatty Acid Metabolism. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1988, 39, 101–138. [Google Scholar] [CrossRef]
- Lunn, D.; Wallis, J.G.; Browse, J. A multigene approach secures hydroxy fatty acid production in Arabidopsis. J. Exp. Bot. 2021, 73, 2875–2888. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Huang, Y.; Gao, J.; Pu, Y.; Wang, N.; Shen, W.; Wen, J.; Yi, B.; Ma, C.; Tu, J.; et al. CIPK9 is involved in seed oil regulation in Brassica napus L. and Arabidopsis thaliana (L.) Heynh. Biotechnol. Biofuels 2018, 11, 124. [Google Scholar] [CrossRef] [PubMed]
- Beike, A.K.; Jaeger, C.; Zink, F.; Decker, E.L.; Reski, R. High contents of very long-chain polyunsaturated fatty acids in different moss species. Plant Cell Rep. 2014, 33, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Xue, Z.; Sharpe, P.L.; Hong, S.-P.; Yadav, N.S.; Xie, D.; Short, D.R.; Damude, H.G.; Rupert, R.A.; Seip, J.E.; Wang, J.; et al. Production of omega-3 eicosapentaenoic acid by metabolic engineering of Yarrowia lipolytica. Nat. Biotechnol. 2013, 31, 734–740. [Google Scholar] [CrossRef]
- Gill, I.; Valivety, R. Polyunsaturated fatty acids, part 1: Occurrence, biological activities and applications. Trends Biotechnol. 1997, 15, 401–409. [Google Scholar] [CrossRef]
- Severino, L.S.; Auld, D.L.; Baldanzi, M.; Cândido, M.J.D.; Chen, G.; Crosby, W.; Tan, D.; He, X.; Lakshmamma, P.; Lavanya, C.; et al. A Review on the Challenges for Increased Production of Castor. Agron. J. 2012, 104, 853–880. [Google Scholar] [CrossRef]
- Ma, S.; Du, C.; Taylor, D.C.; Zhang, M. Concerted increases of FAE1 expression level and substrate availability improve and singularize the production of very-long-chain fatty acids in Arabidopsis seeds. Plant Direct 2021, 5, e00331. [Google Scholar] [CrossRef]
- Pfeuffer, M.; Jaudszus, A. Pentadecanoic and Heptadecanoic Acids: Multifaceted Odd-Chain Fatty Acids. Adv. Nutr. 2016, 7, 730–734. [Google Scholar] [CrossRef] [PubMed]
- Batsale, M.; Bahammou, D.; Fouillen, L.; Mongrand, S.; Joubes, J.; Domergue, F. Biosynthesis and Functions of Very-Long-Chain Fatty Acids in the Responses of Plants to Abiotic and Biotic Stresses. Cells 2021, 10, 1284. [Google Scholar] [CrossRef] [PubMed]
- Delude, C.; Fouillen, L.; Bhar, P.; Cardinal, M.J.; Pascal, S.; Santos, P.; Kosma, D.K.; Joubes, J.; Rowland, O.; Domergue, F. Primary Fatty Alcohols Are Major Components of Suberized Root Tissues of Arabidopsis in the Form of Alkyl Hydroxycinnamates. Plant Physiol. 2016, 171, 1934–1950. [Google Scholar] [CrossRef] [PubMed]
- De Bigault Du Granrut, A.; Cacas, J.L. How Very-Long-Chain Fatty Acids Could Signal Stressful Conditions in Plants? Front. Plant Sci. 2016, 7, 1490. [Google Scholar] [CrossRef]
- Alban, C.; Baldet, P.; Douce, R. Localization and characterization of 2 structurally different forms of acetyl-coa carboxylase in young pea leaves, of which one is sensitive to aryloxyphenoxypropionate herbicides. Biochem. J. 1994, 300, 557–565. [Google Scholar] [CrossRef]
- Kozaki, A.K.; Mayumi, K.; Sasaki, Y. Thiol-disulfide exchange between nuclear-encoded and chloroplast-encoded subunits of pea acetyl-CoA carboxylase. J. Biol. Chem. 2001, 276, 39919–39925. [Google Scholar] [CrossRef] [PubMed]
- Bates, P.D. Understanding the control of acyl flux through the lipid metabolic network of plant oil biosynthesis. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2016, 1861, 1214–1225. [Google Scholar] [CrossRef] [PubMed]
- Fahy, E.; Subramaniam, S.; Murphy, R.C.; Nishijima, M.; Raetz, C.R.; Shimizu, T.; Spener, F.; van Meer, G.; Wakelam, M.J.; Dennis, E.A. Update of the LIPID MAPS comprehensive classification system for lipids. J. Lipid Res. 2009, 50, S9–S14. [Google Scholar] [CrossRef]
- Lu, S.; Sturtevant, D.; Aziz, M.; Jin, C.; Li, Q.; Chapman, K.D.; Guo, L. Spatial analysis of lipid metabolites and expressed genes reveals tissue-specific heterogeneity of lipid metabolism in high- and low-oil Brassica napus L. seeds. Plant J. 2018, 94, 915–932. [Google Scholar] [CrossRef]
- Kennedy, E.P. Biosynthesis of complex lipids. Fed. Proc. 1961, 20, 934–940. [Google Scholar]
- Miklaszewska, M.; Zienkiewicz, K.; Inchana, P.; Zienkiewicz, A. Lipid metabolism and accumulation in oilseed crops. OCL Oilseeds Fats Crops Lipids 2021, 28, 50. [Google Scholar] [CrossRef]
- Ischebeck, T.; Krawczyk, H.E.; Mullen, R.T.; Dyer, J.M.; Chapman, K.D. Lipid droplets in plants and algae: Distribution, formation, turnover and function. Semin. Cell Dev. Biol. 2020, 108, 82–93. [Google Scholar] [CrossRef]
- Yin, D.; Wang, Y.; Zhang, X.; Li, H.; Lu, X.; Zhang, J.; Zhang, W.; Chen, S. De Novo Assembly of the Peanut (Arachis hypogaea L.) Seed Transcriptome Revealed Candidate Unigenes for Oil Accumulation Pathways. PLoS ONE 2013, 8, e73767. [Google Scholar] [CrossRef] [PubMed]
- Thiam, A.R.; Ikonen, E. Lipid Droplet Nucleation. Trends Cell Biol. 2021, 31, 108–118. [Google Scholar] [CrossRef] [PubMed]
- Taurino, M.; Costantini, S.; De Domenico, S.; Stefanelli, F.; Ruano, G.; Delgadillo, M.O.; Sanchez-Serrano, J.J.; Sanmartin, M.; Santino, A.; Rojo, E. SEIPIN Proteins Mediate Lipid Droplet Biogenesis to Promote Pollen Transmission and Reduce Seed Dormancy. Plant Physiol. 2018, 176, 1531–1546. [Google Scholar] [CrossRef]
- Kelly, A.; Shaw, E.; Powers, S.; Kurup, S.; Eastmond, P. Suppression of the SUGAR-DEPENDENT1 triacylglycerol lipase family during seed development enhances oil yield in oilseed rape (Brassica napus L.). Plant Biotechnol J. 2013, 11, 355–361. [Google Scholar] [CrossRef]
- Pyc, M.; Gidda, S.K.; Seay, D.; Esnay, N.; Kretzschmar, F.K.; Cai, Y.Q.; Doner, N.M.; Greer, M.S.; Hull, J.J.; Coulon, D.; et al. LDIP cooperates with SEIPIN and LDAP to facilitate lipid droplet biogenesis in Arabidopsis. Plant Cell 2021, 33, 3076–3103. [Google Scholar] [CrossRef] [PubMed]
- Chia, T.Y.P.; Pike, M.J.; Rawsthorne, S. Storage oil breakdown during embryo development of Brassica napus (L.). J. Exp. Bot. 2005, 56, 1285–1296. [Google Scholar] [CrossRef]
- Chiofalo, B.; Lo Presti, V. 404-Sampling Techniques for the Determination of Volatile Components in Food of Animal Origin. In Comprehensive Sampling Sample Preparation; Pawliszyn, J., Ed.; Academic Press: Oxford, UK, 2012; pp. 61–85. [Google Scholar]
- Huang, A.H.C. Oleosins and oil bodies in seeds and other organs. Plant Physiol. 1996, 110, 1055–1061. [Google Scholar] [CrossRef] [PubMed]
- Goepfert, S.; Poirier, Y. Beta-oxidation in fatty acid degradation and beyond. Curr. Opin. Plant Biol. 2007, 10, 245–251. [Google Scholar] [CrossRef]
- Gupta, R.; Gupta, N. Lipid Biosynthesis and Degradation; Springer: Singapore, 2021; pp. 491–523. [Google Scholar]
- Li-Beisson, Y.; Shorrosh, B.; Beisson, F.; Andersson, M.X.; Arondel, V.; Bates, P.D.; Baud, S.; Bird, D.; Debono, A.; Durrett, T.P.; et al. Acyl-lipid metabolism. Arab. Book 2013, 11, e0161. [Google Scholar] [CrossRef]
- Ma, J.; Sun, S.; Whelan, J.; Shou, H. CRISPR/Cas9-Mediated Knockout of GmFATB1 Significantly Reduced the Amount of Saturated Fatty Acids in Soybean Seeds. Int. J. Mol. Sci. 2021, 22, 3877. [Google Scholar] [CrossRef] [PubMed]
- Raboanatahiry, N.; Chao, H.; Guo, L.; Gan, J.; Xiang, J.; Yan, M.; Zhang, L.; Yu, L.; Li, M. Synteny analysis of genes and distribution of loci controlling oil content and fatty acid profile based on QTL alignment map in Brassica napus. BMC Genom. 2017, 18, 776. [Google Scholar] [CrossRef]
- Zou, J.; Katavic, V.; Giblin, E.M.; Barton, D.L.; Mackenzie, S.L.; Keller, W.A.; Hu, X.; Taylor, D.C. Modification of seed oil content and acyl composition in the brassicaceae by expression of a yeast sn-2 acyltransferase gene. Plant Cell 1997, 9, 909–923. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.L.; Lei, Y.; Xu, X.; Huang, J.Q.; Jiang, H.F.; Wang, J.; Cheng, Z.S.; Zhang, J.A.; Song, Y.H.; Liao, B.S.; et al. The Peanut (Arachis hypogaea L.) Gene AhLPAT2 Increases the Lipid Content of Transgenic Arabidopsis Seeds. PLoS ONE 2015, 10, e0136170. [Google Scholar] [CrossRef] [PubMed]
- Jako, C.; Kumar, A.; Wei, Y.; Zou, J.; Barton, D.L.; Giblin, E.M.; Covello, P.S.; Taylor, D.C. Seed-Specific Over-Expression of an Arabidopsis cDNA Encoding a Diacylglycerol Acyltransferase Enhances Seed Oil Content and Seed Weight. Plant Physiol. 2001, 126, 861–874. [Google Scholar] [CrossRef]
- Wang, Z.K.; Huang, W.J.; Chang, J.M.; Sebastian, A.; Li, Y.G.; Li, H.Y.; Wu, X.X.; Zhang, B.B.; Meng, F.L.; Li, W.B. Overexpression of SiDGAT1, a gene encoding acyl-CoA:diacylglycerol acyltransferase from Sesamum indicum L. increases oil content in transgenic Arabidopsis and soybean. Plant Cell Tissue Organ Cult. 2014, 119, 399–410. [Google Scholar] [CrossRef]
- Zhao, C.; Li, H.; Zhang, W.; Wang, H.; Xu, A.; Tian, J.; Zou, J.; Taylor, D.C.; Zhang, M. BnDGAT1s Function Similarly in Oil Deposition and Are Expressed with Uniform Patterns in Tissues of Brassica napus. Front. Plant Sci. 2017, 8, 2205. [Google Scholar] [CrossRef]
- Akoh, C.C.; Lee, G.C.; Liaw, Y.C.; Huang, T.H.; Shaw, J.F. GDSL family of serine esterases/lipases. Prog. Lipid Res. 2004, 43, 534–552. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.X.; Du, X.; Zhu, Y.; Wang, Z.; Hua, S.J.; Li, Z.L.; Guo, W.L.; Zhang, G.P.; Peng, J.R.; Jiang, L.X. Seed Fatty Acid Reducer acts downstream of gibberellin signalling pathway to lower seed fatty acid storage in Arabidopsis. Plant Cell Environ. 2012, 35, 2155–2169. [Google Scholar] [CrossRef]
- Zhang, Y.; Bai, B.; Lee, M.; Alfiko, Y.; Suwanto, A.; Yue, G.H. Cloning and characterization of EgGDSL, a gene associated with oil content in oil palm. Sci. Rep. 2018, 8, 11406. [Google Scholar] [CrossRef]
- Brocard-Gifford, I.M.; Lynch, T.J.; Finkelstein, R.R. Regulatory networks in seeds integrating developmental, abscisic acid, sugar, and light signaling. Plant Physiol. 2003, 131, 78–92. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Z.Y.; Keereetaweep, J.; Liu, H.; Feil, R.; Lunn, J.E.; Shanklin, J. Trehalose 6-Phosphate Positively Regulates Fatty Acid Synthesis by Stabilizing WRINKLED1. Plant Cell 2018, 30, 2616–2627. [Google Scholar] [CrossRef]
- Zafar, S.; Li, Y.-L.; Li, N.-N.; Zhu, K.-M.; Tan, X.-L. Recent advances in enhancement of oil content in oilseed crops. J. Biotechnol. 2019, 301, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Fei, W.J.; Yang, S.Q.; Hu, J.; Yang, F.; Qu, G.Y.; Peng, D.; Zhou, B. Research advances of WRINKLED1 (WRI1) in plants. Funct. Plant Biol. 2020, 47, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Benning, N.F.a.C. wrinkled1: A Novel, Low-Seed-Oil Mutant of Arabidopsis with a Deficiency in the Seed-Specific Regulation of Carbohydrate Metabolism. Plant Physiol. 1998, 118, 91–101. [Google Scholar]
- Ji, X.-J.; Mao, X.; Hao, Q.-T.; Liu, B.-L.; Xue, J.-A.; Li, R.-Z. Splice Variants of the Castor WRI1 Gene Upregulate Fatty Acid and Oil Biosynthesis When Expressed in Tobacco Leaves. Int. J. Mol. Sci. 2018, 19, 146. [Google Scholar] [CrossRef]
- Mu, J.Y.; Tan, H.L.; Zheng, Q.; Fu, F.Y.; Liang, Y.; Zhang, J.; Yang, X.H.; Wang, T.; Chong, K.; Wang, X.J.; et al. LEAFY COTYLEDON1 is a key regulator of fatty acid biosynthesis in Arabidopsis. Plant Physiol. 2008, 148, 1042–1054. [Google Scholar] [CrossRef]
- Tan, H.; Yang, X.; Zhang, F.; Zheng, X.; Qu, C.; Mu, J.; Fu, F.; Li, J.; Guan, R.; Zhang, H.; et al. Enhanced seed oil production in canola by conditional expression of Brassica napus LEAFY COTYLEDON1 and LEC1-LIKE in developing seeds. Plant Physiol. 2011, 156, 1577–1588. [Google Scholar] [CrossRef]
- Manan, S.; Ahmad, M.Z.; Zhang, G.Y.; Chen, B.B.; Haq, B.U.; Yang, J.H.; Zhao, J. Soybean LEC2 Regulates Subsets of Genes Involved in Controlling the Biosynthesis and Catabolism of Seed Storage Substances and Seed Development. Front. Plant Sci. 2017, 8, 1604. [Google Scholar] [CrossRef]
- Elahi, N.; Duncan, R.W.; Stasolla, C. Decreased seed oil production in FUSCA3 Brassica napus mutant plants. Plant Physiol. Biochem. 2015, 96, 222–230. [Google Scholar] [CrossRef]
- Zhang, M.; Cao, X.; Jia, Q.; Ohlrogge, J. FUSCA3 activates triacylglycerol accumulation in Arabidopsis seedlings and tobacco BY2 cells. Plant J. 2016, 88, 95–107. [Google Scholar] [CrossRef]
- Manan, S.; Zhao, J. Role of Glycine max ABSCISIC ACID INSENSITIVE 3 (GmABI3) in lipid biosynthesis and stress tolerance in soybean. Funct. Plant Biol. 2021, 48, 171. [Google Scholar] [CrossRef] [PubMed]
- Pouvreau, B.; Blundell, C.; Vohra, H.; Zwart, A.B.; Arndell, T.; Singh, S.; Vanhercke, T. A Versatile High Throughput Screening Platform for Plant Metabolic Engineering Highlights the Major Role of ABI3 in Lipid Metabolism Regulation. Front. Plant Sci. 2020, 11, 288. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Liu, X.; Wang, K.; Li, Z.; Jia, Q.; Zhao, C.; Zhang, M. ABA-INSENSITIVE 3 with or without FUSCA3 highly up-regulates lipid droplet proteins and activates oil accumulation. J. Exp. Bot. 2022, 73, 2077–2092. [Google Scholar] [CrossRef] [PubMed]
- Santos-Mendoza, M.; Dubreucq, B.; Baud, S.; Parcy, F.; Caboche, M.; Lepiniec, L. Deciphering gene regulatory networks that control seed development and maturation in Arabidopsis. Plant J. 2008, 54, 608–620. [Google Scholar] [CrossRef]
- Chen, Y.H.; Yang, X.Y.; He, K.; Liu, M.H.; Li, J.G.; Gao, Z.F.; Lin, Z.Q.; Zhang, Y.F.; Wang, X.X.; Qiu, X.M.; et al. The MYB transcription factor superfamily of arabidopsis: Expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol. Biol. 2006, 60, 107–124. [Google Scholar]
- To, A.; Joubes, J.; Thueux, J.; Kazaz, S.; Lepiniec, L.; Baud, S. AtMYB92 enhances fatty acid synthesis and suberin deposition in leaves of Nicotiana benthamiana. Plant J. 2020, 103, 660–676. [Google Scholar] [CrossRef] [PubMed]
- Song, G.; Li, X.; Munir, R.; Khan, A.R.; Azhar, W.; Yasin, M.U.; Jiang, Q.; Bancroft, I.; Gan, Y. The WRKY6 transcription factor affects seed oil accumulation and alters fatty acid compositions in Arabidopsis thaliana. Physiol. Plant. 2020, 169, 612–624. [Google Scholar] [CrossRef]
- Liu, J.; Hua, W.; Yang, H.L.; Zhan, G.M.; Li, R.J.; Deng, L.B.; Wang, X.F.; Liu, G.H.; Wang, H.Z. The BnGRF2 gene (GRF2-like gene from Brassica napus) enhances seed oil production through regulating cell number and plant photosynthesis. J. Exp. Bot. 2012, 63, 3727–3740. [Google Scholar] [CrossRef]
- Chen, N.; Veerappan, V.; Abdelmageed, H.; Kang, M.; Allen, R.D. HSI2/VAL1 Silences AGL15 to Regulate the Developmental Transition from Seed Maturation to Vegetative Growth in Arabidopsis. Plant Cell 2018, 30, 600–619. [Google Scholar] [CrossRef]
- Yang, Y.; Kong, Q.; Lim, A.R.Q.; Lu, S.; Zhao, H.; Guo, L.; Yuan, L.; Ma, W. Transcriptional regulation of oil biosynthesis in seed plants: Current understanding, applications and perspectives. Plant Commun. 2022, 3, 100328. [Google Scholar] [CrossRef]
- Cai, G.; Kim, S.-C.; Li, J.; Zhou, Y.; Wang, X. Transcriptional Regulation of Lipid Catabolism during Seedling Establishment. Mol. Plant 2020, 13, 984–1000. [Google Scholar] [CrossRef]
- Libault, M.; Farmer, A.; Joshi, T.; Takahashi, K.; Langley, R.J.; Franklin, L.D.; He, J.; Xu, D.; May, G.; Stacey, G. An integrated transcriptome atlas of the crop model Glycine max, and its use in comparative analyses in plants. Plant J. 2010, 63, 86–99. [Google Scholar] [CrossRef]
- Severin, A.J.; Woody, J.L.; Bolon, Y.-T.; Joseph, B.; Diers, B.W.; Farmer, A.D.; Muehlbauer, G.J.; Nelson, R.T.; Grant, D.; Specht, J.E.; et al. RNA-Seq Atlas of Glycine max: A guide to the soybean transcriptome. BMC Plant Biol. 2010, 10, 160. [Google Scholar] [CrossRef]
- Huang, X.; Han, B. Natural Variations and Genome-Wide Association Studies in Crop Plants. Annu. Rev. Plant Biol. 2014, 65, 531–551. [Google Scholar] [CrossRef] [PubMed]
- Ardui, S.; Ameur, A.; Vermeesch, J.R.; Hestand, M.S. Single molecule real-time (SMRT) sequencing comes of age: Applications and utilities for medical diagnostics. Nucleic Acids Res. 2018, 46, 2159–2168. [Google Scholar] [CrossRef] [PubMed]
- Lieberman-Aiden, E.; Van Berkum, N.L.; Williams, L.; Imakaev, M.; Ragoczy, T.; Telling, A.; Amit, I.; Lajoie, B.R.; Sabo, P.J.; Dorschner, M.O.; et al. Comprehensive Mapping of Long-Range Interactions Reveals Folding Principles of the Human Genome. Science 2009, 326, 289–293. [Google Scholar] [CrossRef] [PubMed]
- Chalhoub, B.; Denoeud, F.; Liu, S.; Parkin, I.A.P.; Tang, H.; Wang, X.; Chiquet, J.; Belcram, H.; Tong, C.; Samans, B.; et al. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 2014, 345, 950–953. [Google Scholar] [CrossRef]
- Bayer, P.E.; Hurgobin, B.; Golicz, A.A.; Chan, C.K.K.; Yuan, Y.; Lee, H.; Renton, M.; Meng, J.; Li, R.; Long, Y.; et al. Assembly and comparison of two closely related Brassica napus genomes. Plant Biotechnol. J. 2017, 15, 1602–1610. [Google Scholar] [CrossRef]
- Sun, F.; Fan, G.; Hu, Q.; Zhou, Y.; Guan, M.; Tong, C.; Li, J.; Du, D.; Qi, C.; Jiang, L.; et al. The high-quality genome of Brassica napuscultivar ‘ZS11’ reveals the introgression history in semi-winter morphotype. Plant J. 2017, 92, 452–468. [Google Scholar] [CrossRef]
- Zou, J.; Mao, L.; Qiu, J.; Wang, M.; Jia, L.; Wu, D.; He, Z.; Chen, M.; Shen, Y.; Shen, E.; et al. Genome-wide selection footprints and deleterious variations in young Asian allotetraploid rapeseed. Plant Biotechnol. J. 2019, 17, 1998–2010. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Tong, C.; Zhang, X.; Song, A.; Hu, M.; Dong, W.; Chen, F.; Wang, Y.; Tu, J.; Liu, S.; et al. A high-quality Brassica napus genome reveals expansion of transposable elements, subgenome evolution and disease resistance. Plant Biotechnol. J. 2021, 19, 615–630. [Google Scholar] [CrossRef]
- Zhuang, W.; Chen, H.; Yang, M.; Wang, J.; Pandey, M.K.; Zhang, C.; Chang, W.-C.; Zhang, L.; Zhang, X.; Tang, R.; et al. The genome of cultivated peanut provides insight into legume karyotypes, polyploid evolution and crop domestication. Nat. Genet. 2019, 51, 865–876. [Google Scholar] [CrossRef]
- Chen, X.; Lu, Q.; Liu, H.; Zhang, J.; Hong, Y.; Lan, H.; Li, H.; Wang, J.; Liu, H.; Li, S.; et al. Sequencing of Cultivated Peanut, Arachis hypogaea, Yields Insights into Genome Evolution and Oil Improvement. Mol. Plant 2019, 12, 920–934. [Google Scholar] [CrossRef]
- Badouin, H.; Gouzy, J.; Grassa, C.J.; Murat, F.; Staton, S.E.; Cottret, L.; Lelandais-Brière, C.; Owens, G.L.; Carrère, S.; Mayjonade, B.; et al. The sunflower genome provides insights into oil metabolism, flowering and Asterid evolution. Nature 2017, 546, 148–152. [Google Scholar] [CrossRef]
- Zhang, H.; Miao, H.; Wang, L.; Qu, L.; Liu, H.; Wang, Q.; Yue, M. Genome sequencing of the important oilseed crop Sesamum indicum L. Genome Biol. 2013, 14, 401. [Google Scholar] [CrossRef]
- Wang, L.; Yu, S.; Tong, C.; Zhao, Y.; Liu, Y.; Song, C.; Zhang, Y.; Zhang, X.; Wang, Y.; Hua, W.; et al. Genome sequencing of the high oil crop sesame provides insight into oil biosynthesis. Genome Biol. 2014, 15, R39. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Liu, K.; Zhang, Y.; Feng, Q.; Wang, L.; Zhao, Y.; Li, D.; Zhao, Q.; Zhu, X.; Zhu, X.; et al. Genetic discovery for oil production and quality in sesame. Nat. Commun. 2015, 6, 8609. [Google Scholar] [CrossRef]
- Kitts, P.A.; Church, D.M.; Thibaud-Nissen, F.; Choi, J.; Hem, V.; Sapojnikov, V.; Smith, R.G.; Tatusova, T.; Xiang, C.; Zherikov, A.; et al. Assembly: A resource for assembled genomes at, N.C.B.I. Nucleic Acids Res. 2016, 44, D73–D80. [Google Scholar] [CrossRef]
- Zhang, H.; Miao, H.; Li, C.; Wei, L.; Duan, Y.; Ma, Q.; Kong, J.; Xu, F.; Chang, S. Ultra-dense SNP genetic map construction identification of SiDt gene controlling the determinate growth habit in Sesamum indicum L. Sci. Rep. 2016, 6, 31556. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Xia, Q.; Zhang, Y.; Zhu, X.; Zhu, X.; Li, D.; Ni, X.; Gao, Y.; Xiang, H.; Wei, X.; et al. Updated sesame genome assembly and fine mapping of plant height and seed coat color QTLs using a new high-density genetic map. BMC Genom. 2016, 17, 31. [Google Scholar] [CrossRef]
- Li, Y.-H.; Zhou, G.; Ma, J.; Jiang, W.; Jin, L.-G.; Zhang, Z.; Guo, Y.; Zhang, J.; Sui, Y.; Zheng, L.; et al. De novo assembly of soybean wild relatives for pan-genome analysis of diversity and agronomic traits. Nat. Biotechnol. 2014, 32, 1045–1052. [Google Scholar] [CrossRef]
- Liu, Y.C.; Du, H.L.; Li, P.C.; Shen, Y.T.; Peng, H.; Liu, S.L.; Zhou, G.A.; Zhang, H.K.; Liu, Z.; Shi, M.; et al. Pan-Genome of Wild and Cultivated Soybeans. Cell 2020, 182, 162–176. [Google Scholar] [CrossRef]
- Song, J.-M.; Guan, Z.; Hu, J.; Guo, C.; Yang, Z.; Wang, S.; Liu, D.; Wang, B.; Lu, S.; Zhou, R.; et al. Eight high-quality genomes reveal pan-genome architecture and ecotype differentiation of Brassica napus. Nat. Plants 2020, 6, 34–45. [Google Scholar] [CrossRef]
- Hübner, S.; Bercovich, N.; Todesco, M.; Mandel, J.R.; Odenheimer, J.; Ziegler, E.; Lee, J.S.; Baute, G.J.; Owens, G.L.; Grassa, C.J.; et al. Sunflower pan-genome analysis shows that hybridization altered gene content and disease resistance. Nat. Plants 2019, 5, 54–62. [Google Scholar] [CrossRef]
- Yu, J.; Golicz, A.A.; Lu, K.; Dossa, K.; Zhang, Y.; Chen, J.; Wang, L.; You, J.; Fan, D.; Edwards, D.; et al. Insight into the evolution and functional characteristics of the pan-genome assembly from sesame landraces and modern cultivars. Plant Biotechnol. J. 2019, 17, 881–892. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, S.-B.; Li, Q.-G.; Song, J.; Hao, Y.-Q.; Zhou, L.; Zheng, H.-Q.; Dunwell, J.M.; Zhang, Y.-M. An Integrated Bioinformatics Analysis Reveals Divergent Evolutionary Pattern of Oil Biosynthesis in High- and Low-Oil Plants. PLoS ONE 2016, 11, e0154882. [Google Scholar] [CrossRef]
- Wang, J.; Singh, S.K.; Du, C.; Li, C.; Fan, J.; Pattanaik, S.; Yuan, L. Comparative Transcriptomic Analysis of Two Brassica napus Near-Isogenic Lines Reveals a Network of Genes That Influences Seed Oil Accumulation. Front. Plant Sci. 2016, 7, 1498. [Google Scholar] [CrossRef]
- O’Rourke, J.A.; Bolon, Y.T.; Bucciarelli, B.; Vance, C.P. Legume genomics: Understanding biology through DNA and RNA sequencing. Ann. Bot. 2014, 113, 1107–1120. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Qi, F.Y.; Zheng, Z.; Sun, Z.Q.; Tian, M.D.; Wang, X.; Huang, B.Y.; Dong, W.Z.; Zhang, X.Y. Global Transcriptome Analyses Provide Into Several Fatty Acid Biosynthesis-related Genes in Peanut (Arachis hypogaea L.). Trop. Plant Biol. 2021, 14, 267–282. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, Y.; Li, D.; Dossa, K.; Wang, M.L.; Zhou, R.; Yu, J.; Zhang, X. Gene expression profiles that shape high and low oil content sesames. BMC Genet. 2019, 20, 45. [Google Scholar] [CrossRef]
- Cegielska-Taras, T.; Nogala-Kalucka, M.; Szala, L.; Siger, A. Study of variation of tocochromanol and phytosterol contents in black and yellow seeds of Brassica napus L. doubled haploid populations. Acta Sci. Pol. Technol. Aliment. 2016, 15, 321–332. [Google Scholar] [CrossRef]
- Zhou, F.; Liu, Y.; Wang, W.; Wu, L.; Yuan, H.; Liu, X.; Ma, J.; Wang, J.; Yao, Y.; Zhang, L.; et al. Comparative transcriptomic analyses of high and low oleic acid content sunflower (Helianthus annuus L.) seed development. Pak. J. Bot. 2022, 54, 2131–2141. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wen, H.; Jin, Q.; Yu, W.; Li, G.; Wu, M.; Bai, H.; Shen, L.; Wu, C. Comparative transcriptome analysis on candidate genes involved in lipid biosynthesis of developing kernels for three walnut cultivars in Xinjiang. Food Sci. Hum. Wellness 2022, 11, 1201–1214. [Google Scholar] [CrossRef]
- Wong, Y.C.; Teh, H.F.; Mebus, K.; Ooi, T.E.K.; Kwong, Q.B.; Koo, K.L.; Ong, C.K.; Mayes, S.; Chew, F.T.; Appleton, D.R.; et al. Differential gene expression at different stages of mesocarp development in high- and low-yielding oil palm. BMC Genom. 2017, 18, 470. [Google Scholar] [CrossRef]
- Yang, S.; Miao, L.; He, J.; Zhang, K.; Li, Y.; Gai, J. Dynamic Transcriptome Changes Related to Oil Accumulation in Developing Soybean Seeds. Int. J. Mol. Sci. 2019, 20, 2202. [Google Scholar] [CrossRef] [PubMed]
- Niu, Y.; Wu, L.; Li, Y.; Huang, H.; Qian, M.; Sun, W.; Zhu, H.; Xu, Y.; Fan, Y.; Mahmood, U.; et al. Deciphering the transcriptional regulatory networks that control size, color, and oil content in Brassica rapa seeds. Biotechnol. Biofuels 2020, 13, 90. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Liu, J.; Fan, S.; Liu, H.; Zhou, X.-R.; Hua, W.; Zheng, M. An integrated omics analysis reveals the gene expression profiles of maize, castor bean, and rapeseed for seed oil biosynthesis. BMC Plant Biol. 2022, 22, 153. [Google Scholar] [CrossRef]
- Huang, R.; Zhou, Y.; Zhang, J.; Ji, F.; Jin, F.; Fan, W.; Pei, D. Transcriptome Analysis of Walnut (Juglans regia L.) Embryos Reveals Key Developmental Stages and Genes Involved in Lipid Biosynthesis and Polyunsaturated Fatty Acid Metabolism. J. Agric. Food Chem. 2021, 69, 377–396. [Google Scholar] [CrossRef]
- Liu, J.; Dong, L.; Duan, R.; Hu, L.; Zhao, Y.; Zhang, L.; Wang, X. Transcriptomic Analysis Reveals the Regulatory Networks and Hub Genes Controlling the Unsaturated Fatty Acid Contents of Developing Seed in Soybean. Front. Plant Sci. 2022, 13, 876371. [Google Scholar] [CrossRef]
- Zhang, Z.; Dunwell, J.M.; Zhang, Y.M. An integrated omics analysis reveals molecular mechanisms that are associated with differences in seed oil content between Glycine max and Brassica napus. BMC Plant Biol. 2018, 18, 328. [Google Scholar] [CrossRef] [PubMed]
- Yao, M.; Guan, M.; Zhang, Z.; Zhang, Q.; Cui, Y.; Chen, H.; Liu, W.; Jan, H.U.; Voss-Fels, K.P.; Werner, C.R.; et al. GWAS and co-expression network combination uncovers multigenes with close linkage effects on the oleic acid content accumulation in Brassica napus. BMC Genom. 2020, 21, 320. [Google Scholar] [CrossRef] [PubMed]
- Guerin, C.; Joët, T.; Serret, J.; Lashermes, P.; Vaissayre, V.; Agbessi, M.D.T.; Beulé, T.; Severac, D.; Amblard, P.; Tregear, J.; et al. Gene coexpression network analysis of oil biosynthesis in an interspecific backcross of oil palm. Plant J. 2016, 87, 423–441. [Google Scholar] [CrossRef]
- Han, X.; Zhang, Y.W.; Liu, J.Y.; Zuo, J.F.; Zhang, Z.C.; Guo, L.; Zhang, Y.M. 4D genetic networks reveal the genetic basis of metabolites and seed oil-related traits in 398 soybean RILs. Biotechnol. Biofuels Bioprod. 2022, 15, 92. [Google Scholar] [CrossRef]
- Flint-Garcia, S.A.; Thuillet, A.-C.; Yu, J.; Pressoir, G.; Romero, S.M.; Mitchell, S.E.; Doebley, J.; Kresovich, S.; Goodman, M.M.; Buckler, E.S. Maize association population: A high-resolution platform for quantitative trait locus dissection. Plant J. 2005, 44, 1054–1064. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Chen, B.; Zhao, J.; Zhang, F.; Xie, T.; Xu, K.; Gao, G.; Yan, G.; Li, H.; Li, L.; et al. Genomic selection and genetic architecture of agronomic traits during modern rapeseed breeding. Nat. Genet. 2022, 54, 694–704. [Google Scholar] [CrossRef]
- Zhou, Z.; Jiang, Y.; Wang, Z.; Gou, Z.; Lyu, J.; Li, W.; Yu, Y.; Shu, L.; Zhao, Y.; Ma, Y.; et al. Resequencing 302 wild and cultivated accessions identifies genes related to domestication and improvement in soybean. Nat. Biotechnol. 2015, 33, 408–414. [Google Scholar] [CrossRef]
- Wang, X.H.; Xu, P.; Yin, L.; Ren, Y.; Li, S.L.; Shi, Y.M.; Alcock, T.D.; Xiong, Q.; Qian, W.; Chi, X.Y.; et al. Genomic and Transcriptomic Analysis Identified Gene Clusters and Candidate Genes for Oil Content in Peanut (Arachis hypogaea L.). Plant Mol. Biol. Rep. 2018, 36, 518–529. [Google Scholar] [CrossRef]
- Pandey, M.K.; Wang, M.L.; Qiao, L.; Feng, S.; Khera, P.; Wang, H.; Tonnis, B.; Barkley, N.A.; Wang, J.; Holbrook, C.C.; et al. Identification of QTLs associated with oil content and mapping FAD2 genes and their relative contribution to oil quality in peanut (Arachis hypogaea L.). BMC Genet. 2014, 15, 133. [Google Scholar] [CrossRef]
- Chen, F.; Zhang, W.; Yu, K.; Sun, L.; Gao, J.; Zhou, X.; Peng, Q.; Fu, S.; Hu, M.; Long, W.; et al. Unconditional and conditional QTL analyses of seed fatty acid composition in Brassica napus L. BMC Plant Biol. 2018, 18, 49. [Google Scholar] [CrossRef] [PubMed]
- Hobbs, D.H.; Flintham, J.E.; Hills, M.J. Genetic Control of Storage Oil Synthesis in Seeds of Arabidopsis. Plant Physiol. 2004, 136, 3341–3349. [Google Scholar] [CrossRef]
- Zhao, Q.; Wu, J.; Cai, G.; Yang, Q.; Shahid, M.; Fan, C.; Zhang, C.; Zhou, Y. A novel quantitative trait locus on chromosome A9 controlling oleic acid content in Brassica napus. Plant Biotechnol. J. 2019, 17, 2313–2324. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.Q.; Zhang, Y.Y.; Liu, Y.Y.; Tong, C.B.; Cheng, X.H.; Zhu, W.; Li, Z.Y.; Huang, J.Y.; Liu, S.Y. Mapping loci controlling fatty acid profiles, oil and protein content by genome-wide association study in Brassica napus. Crop J. 2019, 7, 217–226. [Google Scholar] [CrossRef]
- Luo, Z.L.; Wang, M.; Long, Y.; Huang, Y.J.; Shi, L.; Zhang, C.Y.; Liu, X.; Fitt, B.D.L.; Xiang, J.X.; Mason, A.S.; et al. Incorporating pleiotropic quantitative trait loci in dissection of complex traits: Seed yield in rapeseed as an example. Theor. Appl. Genet. 2017, 130, 1569–1585. [Google Scholar] [CrossRef]
- Schaefer, R.J.; Michno, J.-M.; Jeffers, J.; Hoekenga, O.; Dilkes, B.; Baxter, I.; Myers, C.L. Integrating Coexpression Networks with GWAS to Prioritize Causal Genes in Maize. Plant Cell 2018, 30, 2922–2942. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Zeng, X.; Xiong, Q.; Wei, D.; Liao, J.; Xu, Y.; Chen, G.; Zhou, Y.; Dong, H.; Wan, H.; et al. Combining quantitative trait locus and co-expression analysis allowed identification of new candidates for oil accumulation in rapeseed. J. Exp. Bot. 2021, 72, 1649–1660. [Google Scholar] [CrossRef] [PubMed]
- Wenk, M.R. The emerging field of lipidomics. Nat. Rev. Drug Discov. 2005, 4, 725. [Google Scholar] [CrossRef]
- Borisjuk, L.; Neuberger, T.; Schwender, J.; Heinzel, N.; Sunderhaus, S.; Fuchs, J.; Hay, J.O.; Tschiersch, H.; Braun, H.P.; Denolf, P.; et al. Seed architecture shapes embryo metabolism in oilseed rape. Plant Cell 2013, 25, 1625–1640. [Google Scholar] [CrossRef]
- Woodfield, H.K.; Sturtevant, D.; Borisjuk, L.; Munz, E.; Guschina, I.A.; Chapman, K.; Harwood, J.L. Spatial and Temporal Mapping of Key Lipid Species in Brassica napus Seeds. Plant Physiol. 2017, 173, 1998–2009. [Google Scholar] [CrossRef]
- Zhang, C.Y.; Ma, C.L.; Zhu, L.; Yao, M.Z. Simultaneous determination of protoporphyrin IX and magnesium protoporphyrin IX in Arabidopsis thaliana and Camellia sinensis using UPLC-MS/MS. Plant Methods 2023, 19, 34. [Google Scholar] [CrossRef]
- Zhang, G.; Ahmad, M.Z.; Chen, B.; Manan, S.; Zhang, Y.; Jin, H.; Wang, X.; Zhao, J. Lipidomic and transcriptomic profiling of developing nodules reveals the essential roles of active glycolysis and fatty acid and membrane lipid biosynthesis in soybean nodulation. Plant J. 2020, 103, 1351–1371. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Guo, X.; Wang, Q.; Zhao, L.Y.; Sun, Q.C.; Duan, X.L.; Cao, Y.P.; Sun, H. Investigation on lipid profile of peanut oil and changes during roasting by lipidomic approach. Lwt-Food Sci. Technol. 2022, 154, 112594. [Google Scholar] [CrossRef]
- Shi, T.; Wu, G.; Jin, Q.; Wang, X. Detection of camellia oil adulteration using chemometrics based on fatty acids GC fingerprints and phytosterols GC-MS fingerprints. Food Chem. 2021, 352, 129422. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Ma, R.; Xu, Y.; Zhong, K.; Bu, Q.; Gao, H. A Comparison of Lipid Contents in Different Types of Peanut Cultivars Using UPLC-Q-TOF-MS-Based Lipidomic Study. Foods 2021, 11, 4. [Google Scholar] [CrossRef] [PubMed]
- Demiral, I.; Dogan, M.; Bastu, E.; Buyru, F. Genomic, proteomic and lipidomic evaluation of endometrial receptivity. Turk. J. Obstet. Gynecol. 2015, 12, 237–243. [Google Scholar] [CrossRef]
- Przykaza, K.; Nikolaichuk, H.; Kozub, A.; Tomaszewska-Gras, J.; Persuric, Z.; Pavelic, S.K.; Fornal, E. Newly marketed seed oils. What we can learn from the current status of authentication of edible oils. Food Control 2021, 130, 108349. [Google Scholar] [CrossRef]
- Allen, D.K. Assessing compartmentalized flux in lipid metabolism with isotopes. Biochim. Biophys. Acta-Mol. Cell Biol. Lipids 2016, 1861, 1226–1242. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bu, M.; Fan, W.; Li, R.; He, B.; Cui, P. Lipid Metabolism and Improvement in Oilseed Crops: Recent Advances in Multi-Omics Studies. Metabolites 2023, 13, 1170. https://doi.org/10.3390/metabo13121170
Bu M, Fan W, Li R, He B, Cui P. Lipid Metabolism and Improvement in Oilseed Crops: Recent Advances in Multi-Omics Studies. Metabolites. 2023; 13(12):1170. https://doi.org/10.3390/metabo13121170
Chicago/Turabian StyleBu, Mengjia, Wei Fan, Ruonan Li, Bing He, and Peng Cui. 2023. "Lipid Metabolism and Improvement in Oilseed Crops: Recent Advances in Multi-Omics Studies" Metabolites 13, no. 12: 1170. https://doi.org/10.3390/metabo13121170
APA StyleBu, M., Fan, W., Li, R., He, B., & Cui, P. (2023). Lipid Metabolism and Improvement in Oilseed Crops: Recent Advances in Multi-Omics Studies. Metabolites, 13(12), 1170. https://doi.org/10.3390/metabo13121170




