Abstract
Mycobacterium bovis is the etiologic agent of bovine tuberculosis (BTB), a serious infectious disease in both humans and animals. BTB is a zoonotic disease primarily affecting cattle and occasionally humans infected through close contact with infected hosts or the consumption of unpasteurized dairy products. Zoonotic tuberculosis is strongly associated with poverty and poor hygiene, and low- and middle-income countries bear the brunt of the disease. BTB has been increasingly recognized as a growing public health threat in developing countries. However, the lack of effective surveillance programs in many of these countries poses a barrier to accurately determining the true burden of this disease. Additionally, the control of BTB is threatened by the emergence of drug-resistant strains that affect the effectiveness of current treatment regimens. Here, we analyzed current trends in the epidemiology of the disease as well as the antimicrobial susceptibility patterns of M. bovis in the Middle East and North Africa (MENA) region, a region that includes several developing countries. Following PRISMA guidelines, a total of 90 studies conducted in the MENA region were selected. Our findings revealed that the prevalence of BTB among humans and cattle varied significantly according to the population size and country in the MENA region. Most of the available studies were based on culture and/or PCR strategies and were published without including data on antimicrobial resistance and molecular typing. Our findings highlighted the paramount need for the use of appropriate diagnostic tools and the implementation of sustainable control measures, especially at the human/animal interface, in the MENA region.
1. Introduction
Bovine tuberculosis (BTB), caused by Mycobacterium bovis, is one of the world’s most neglected zoonotic diseases [1,2]. The prevalence of BTB follows a socioeconomic gradient by being concentrated in low- and middle-income countries (LMICs), mostly affecting poor, marginalized, and rural communities where people live in close contact with animals, and have limited access to sanitation, safe food and health care services [2]. Phylogenetically, M. bovis belongs to the Mycobacterium tuberculosis complex (MTBC), a cluster of genetically related Mycobacterium species that are associated with tuberculosis infections in a wide range of mammals [1]. Of all MTBCs, M. bovis is the most common cause of wildlife tuberculosis, with a morbidity risk reaching up to 15% of tuberculosis cases in humans [1,3]. This species has the widest host range, including domestic animals, livestock, wildlife, and humans [4]. The movement of animals is considered one of the main reasons for the spread of M. bovis, both within a country and across borders [2]. Indeed, the frequent movement (via trade) of cattle within and between countries and continents has facilitated the global spread of BTB [5,6]. Notably, the disease’s zoonotic property and its dynamic distribution has caused severe economic losses for dairy industries worldwide [7].
Although the dissemination of M. bovis has a heterogeneous profile, developed countries reported a significantly lower incidence of BTB infections compared to data from developing countries. The lack of effective policies to control BTB in many LMICs negatively affects the health of livestock, humans, and ecosystems and potentially increases the burden of this disease [6]. The transmission of BTB in humans is bipartite, (i) direct, through inhalation of the etiologic agent when in close contact with infected cattle or their carcasses, and (ii) indirect, associated with the consumption of unpasteurized dairy products or raw meat products from infected cattle [8,9,10,11]. The World Health Organization (WHO) developed the END-TB strategy to substantially reduce the annual number of tuberculosis deaths between 2016 and 2035 [12]. However, the COVID-19 pandemic has potentially adversely impacted progress in reducing the tuberculosis mortality rate, because the pandemic has disrupted access to essential resources for tuberculosis diagnosis and treatment [12]. To reinvigorate and facilitate control efforts, robust surveillance programs are needed, perhaps more than ever [13]. These programs are essential to better understand BTB transmission dynamics, bolster One Health policies, and predict future disease trends. Indeed, closing epidemiologic knowledge gaps is essential for a better understanding of the risk factors for transmission of BTB among vulnerable people, to support infection prevention and control and food safety policies, and to predict future disease trends in the Middle East and North Africa (MENA) region. Therefore, the main objective of this narrative review was to compile and discuss existing epidemiologic data on latent and active BTB in the MENA region, which includes many conflict-affected and economically challenged countries, with notable deficiencies in public health and national surveillance programs for the management and control of infectious diseases, including zoonotic diseases like BTB.
2. Methods
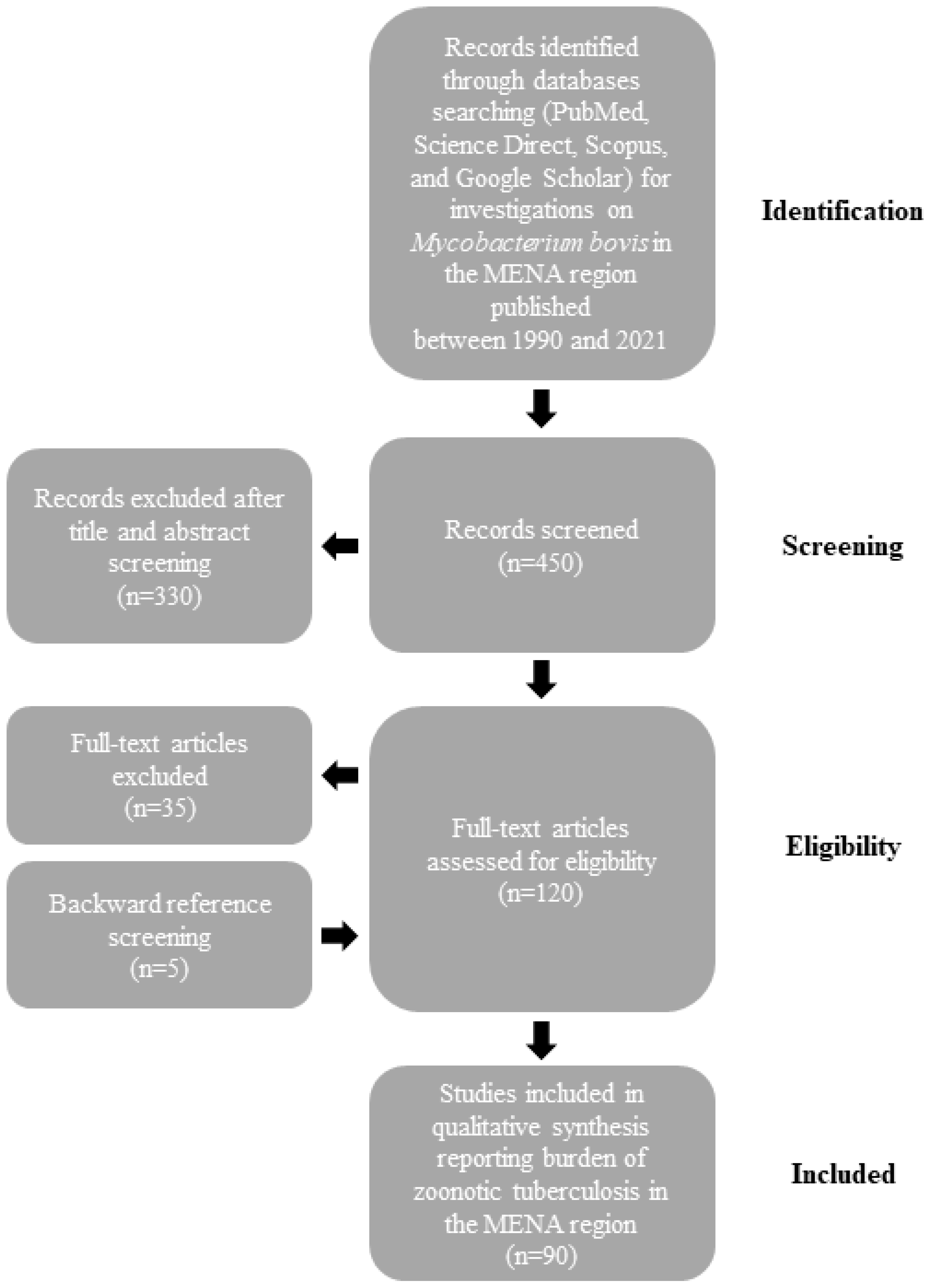
The burden of M. bovis infections in humans and animals is not well defined in the MENA region. Therefore, we searched PubMed, Science Direct, Scopus, and Google Scholar databases for epidemiologic studies on BTB published between 1990 and 2021. We used a combination of words that included “Mycobacterium bovis”, “Bovine”, “MENA countries (as described previously [14])”, “Epidemiology”, “Prevalence”, and “One Health”. After importation of the search results, two authors (D. Kasir and N. Osman) independently screened the citations for their relevance using the title and abstract and all qualified citations were retained for full-text assessment to confirm eligibility. Backward reference screening was performed for all articles. Data extraction was performed by the same authors through a format prepared on a Microsoft Excel workbook (Figure 1). Indexed original articles in English or French, of any epidemiologic design, sampling strategy, and type (case report, longitudinal, case-control, or cross-sectional) were included. All the studies that reported original information on the prevalence of BTB in MENA countries were eligible for inclusion in the review. We excluded narrative and systematic reviews. Given that this manuscript is a narrative review, no quality evaluation of the reviewed studies was performed.
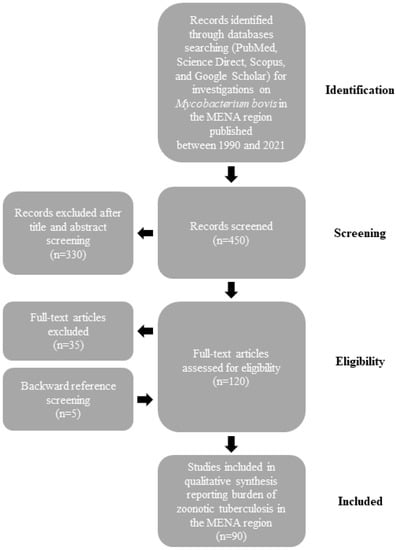
Figure 1.
Flow diagram describing paper selection and inclusion/exclusion process for the review according to PRISMA guidelines.
3. Epidemiology of Mycobacterium bovis in the MENA Region
A total of 90 studies conducted in the MENA region were reviewed. Most studies have a cross-sectional design (87%; 78/90), followed by case reports (10%; 9/90) and longitudinal studies (3%; 3/90). We found that M. bovis has only been reported in humans and animals in eight MENA countries, including Algeria, Egypt, Iraq, Iran, Morocco, Sudan, Turkey, and Tunisia. In other MENA countries, Lebanon, Djibouti, Palestine, and Saudi Arabia, zoonotic tuberculosis has only been reported in humans. The epidemiologic trends of M. bovis infection varied across the MENA countries, likely influenced by the population size, characteristics of the targeted population, the geographical region, and the rigor of the adopted diagnostic tools and investigation methods. Additionally, the heterogeneity of BTB prevalence has been also associated with other factors such as Bacille Calmette-Guérin (BCG) vaccination status, the consumption of unpasteurized dairy products, and the efficiency of national surveillance programs and BTB control measures [15].
3.1. Mycobacterium bovis in Animals
Cases of BTB were noted from both pulmonary and extrapulmonary sites in animals (Table 1) and humans (Table 2). In animals, active BTB was usually reported in cattle and buffalo; however, uncommon cases were described among other types of animals. Specifically, M. bovis was reported in a cat and a mongoose in Turkey [16] and Egypt [17], respectively. A deer infected with BTB was also observed in Iran [18], while M. bovis was detected in camels and pigs in Egypt [19,20].
Several risk factors for BTB appear to play an essential role in the spread of M. bovis among animals in the MENA region. Age, gender, animal body condition, immune suppression, crowding, cross-species transmission, grazing practices, feeding system, environment or weather, and physiological and pathological variations are potential factors contributing to the dissemination of zoonotic M. bovis. Female animals are at a greater risk of BTB than males due to lactation, gestation, and parturition [21,22]. Cross-species transmission between goats and cattle and between buffalo and cattle was associated with sharing of drinking and grazing locations in Algeria [23] and Iran, respectively [24,25]. Furthermore, uncontrolled animal migrations and trade within and across countries were noted as key drivers for BTB transmission [26]. People working closely with livestock, particularly dairy cattle (e.g., farmers, veterinarians, slaughterhouse workers) or with wildlife were more susceptible to M. bovis infections [27].
The World Organization for Animal Health (WOAH) has categorized the tuberculin skin test (TST) as a primary screening test for tuberculosis in cattle [28]. TST is the most frequently used test for the diagnosis of BTB in cattle. Typically, TST’s discriminatory power could be improved by combining it with the interferon-gamma release assay (IGRA) which improves both sensitivity and specificity [29]. M. bovis ELISA tests are also available, allowing the detection of antibodies against zoonotic tuberculosis in cattle serum and plasma samples [30]. Although the ELISA assay is not yet recognized as a standard test for tuberculosis in cattle, it has been approved by the WOAH as being complementary to the TST in cattle. It should be noted that when using these diagnostic approaches, it is difficult to distinguish between vaccinated and infected animals and latent and active infections [31]. However, based on these assays, the prevalence of BTB among cattle varied significantly according to the population size and country in the MENA region. In large studies, the prevalence was relatively low, ranging between 0.1 [32] and 16.4% [33] in Egypt, 4.4 [34] and 24.2% [35] in Iraq, 3.5% [36] in Algeria, and 1.4% [37] in Turkey. In contrast, a higher prevalence was reported in studies with small population sizes, ranging from 22.2% [38] to 82.6% [39] in Egypt, 75% [40] in Iraq, and 48% [41] in Tunisia (Table 1). The trends in the prevalence of BTB also changed over time. In Egypt, Iran, Iraq, Morocco and Sudan, the prevalence of infection among cattle varied between 0.2% [42] and 4.3% [43], 8.5% [44] and 26.3% [45], 1.3% [46] and 10.2% [47], 1.7% [48] and 51.3% [49] and 0.2% [50] and 20.8% [51] over the last two decades, respectively.
Regarding M. bovis in milk samples collected in the MENA region, most studies reported a relatively low prevalence, ranging from 0.004% [33] to 10.2% [52]. Notably, Iraq and Tunisia led the list of M. bovis prevalence in milk samples (Table 1). Using the ELISA assay, a higher infection risk (20.2%) among lactating cows was found in rural areas of Waist and Dhi-Qar provinces, Iraq [53]. Despite the challenges in detection of M. bovis in milk samples, available data from the MENA region confirmed that this matrix represents an important source of zoonotic tuberculosis, because milk (1) is still commonly consumed raw, without pasteurization, in many rural regions and (2) is widely used in the manufacturing of popular dairy products such as cheese and yogurt [54]. Taken together, available data underlined the existence of animal and food sources as well as zoonotic risks that escaped common tuberculosis control measures in many MENA countries. Therefore, there is a strong need to increase awareness on food safety and hygiene and strengthen active surveillance programs in food animals and their products [55]. To prevent the further dissemination of BTB infection, effective approaches must be adopted, including early identification, adequate therapy, and contact tracing [56]. Currently, BTB control mostly relies on slaughter policy, postmortem inspection, and slaughterhouse surveillance [57], which do not even address preharvest risks.

Table 1.
Burden of Mycobacterium bovis in animals in the MENA region.
Table 1.
Burden of Mycobacterium bovis in animals in the MENA region.
| Country | Study Period | Study Design | Population (N) | Tuberculosis (TST, IGRA, ELISA) † | Samples (N) for Active Tuberculosis Testing | Health Status | BTB Identification (Culture, PCR) | Prevalence ¶ of BTB (%) | Typing Method | References |
|---|---|---|---|---|---|---|---|---|---|---|
| Algeria | 2007 | Cross-sectional | Cattle (7250) | Tissue (260) | Slaughtered | Culture | 88 (1.2%) | Spoligotyping; MIRU-VNTR | [31] | |
| 2017 | Cross-sectional | Cattle (3848) | Tissue (3848) | Slaughtered | Culture; PCR | 59 (1.5%) | Spoligotyping; MIRU-VNTR | [58] | ||
| 2017–2018 | Cross-sectional | Cattle (928) | Tissue (928) | Slaughtered | Culture; PCR | 13 (1.4%) | WGS | [21] | ||
| 2017–2019 | Cross-sectional | Cattle (3546) | Tissue (3546) | Slaughtered | Culture; PCR | 174 (4.9%) | Spoligotyping | [23] | ||
| 2018–2019 | Cross-sectional | Cattle (516) | 18 (3.5%) | Live | ND | [36] | ||||
| Egypt | 2008–2010 | Cross-sectional | Cattle (3255) Buffalo (2950) | Cattle: 105 (3.2%) Buffalo: 85 (2.9%) | Tissue (190) Milk (520) Blood (190) | Slaughtered | Culture; PCR | 16 (0.2%) | [59] | |
| 2008–2010 | Cross-sectional | Cattle (1180) | 29 (2.5%) | Tissue (29) | Slaughtered | Culture; PCR | 20 (1.7%) | [60] | ||
| 2010–2011 | Cross-sectional | Cattle (3347) | 32 (1%) | Tissue (32) | Live and slaughtered | Culture; PCR | 21 (0.6%) | [61] | ||
| 2013 | Cross-sectional | Cattle | Milk (100) | Healthy | Culture; PCR | 1 (1%) | [62] | |||
| 2014–2015 | Cross-sectional | Cattle (2935) | 63 (2.2%) | Tissue (56) | Slaughtered | Culture | 39 (1.3%) | [63] | ||
| 2008 * | Cross-sectional | Camels (704) | 9 (1.27%) | Tissue (29) | Slaughtered | Culture | 5 (0.7%) | [64] | ||
| 2009 * | Cross-sectional | Cattle (46) | 38 (82.6%) | Milk (23) | Sick | Culture | 1 (2.1%) | [39] | ||
| 2014 * | Cross-sectional | Cattle (422) Buffalo (480) | Cattle: 9 (2.1%) Buffalo: 27 (5.62%) | Tissue (36) | Slaughtered | Culture | 25 (2.8%) | IS6110 RFLP | [65] | |
| 2015 * | Cross-sectional | Cows (420) | 8 (1.9%) | Milk (8) | Healthy | Culture; PCR | 1 (0.2%) | [42] | ||
| 2018 * | Cross-sectional | Sheep (18) | 4 (22.2%) | Tissue (18) | Slaughtered | Culture; PCR | 15 (83.3%) | [38] | ||
| 2009–2013 | Longitudinal | Cows and buffalos (1,186,772) | 1225 (0.1%) | Blood (14) Tissue (34) | Live and slaughtered | Culture; PCR | 29 (0.002%) | [32] | ||
| 2016–2019 | Cross-sectional | Cattle (2200) Buffalo (1500) | Tissue | Culture; PCR | Cattle 36 (1.6%) Buffalo 18 (1.2%) | [66] | ||||
| 2018 | Cross-sectional | Cattle (2650) | 63 (2.4%) | Tissue (63) | Healthy | Culture | 47 (1.8%) | [67] | ||
| 2018–2019 | Cross-sectional | Cattle (569) Buffalo (181) | Tissue (30) | Slaughtered | Culture; PCR | 9 (1.2%) | [68] | |||
| 2011–2016 | Cross-sectional | Cattle (1570) Buffalo (530) | 74 (3.5%) | Tissue (74) | Slaughtered | PCR | 61 (2.9%) | [69] | ||
| 2017 | Cross-sectional | Cattle (2710) | 215 (7.9%) | Milk (245) | Live | Culture; PCR | 68 (2.5%) | [70] | ||
| 2014 * | Cross-sectional | Cattle (300) | 53 (17.6%) | Blood (65) | Live and slaughtered | Culture; PCR | 13 (4.3%) | [43] | ||
| 2011 | Case report | Mongoose (1) | Tissue (1) | Slaughtered | Culture; PCR | 1 | [17] | |||
| 2015–2017 | Longitudinal | Camels (10,903) | 184 (1.7%) | Tissue (184) | Live and slaughtered | Culture; PCR | 112 (1.0%) | [19] | ||
| 2018–2019 | Cross-sectional | Cattle (1464) | Milk (1285); Lymph nodes (179) | Live and slaughtered | Culture; PCR | 127 (8.6%) | [71] | |||
| 2011–2016 | Cross-sectional | Cattle and Buffalo (2100) | 81 (3.8%) | Tissue | Live | Culture; PCR | 61 (2.9%) | MIRU-VNTR | [26] | |
| 2016 * | Cross-sectional | Cattle and Buffalo (6000) | 79 (1.3%) | Tissue | Live and slaughtered | Culture; PCR | 23 (0.4%) | [72] | ||
| 2004–2005 | Cross-sectional | Pigs (745) | Tissue | Slaughtered | Culture; PCR | 12 (1.6%) | [20] | |||
| 2019 * | Cross-sectional | Cattle (2600) | 47 (1.8%) | Tissue | Healthy | Culture; PCR | 40 (1.5%) | [73] | ||
| 2006–2008 | Cross-sectional | Cattle (3000) | 108 (3.6%) | Tissue | Slaughtered | PCR | 90 (3%) | [74] | ||
| 2013 * | Cross-sectional | Cattle (3474) | 78 (2.2%) | Slaughtered | ND | [75] | ||||
| 2013–2015 | Cross-sectional | Cattle (7064) | 242 (3.4%) | Tissue | Slaughtered | Culture; PCR | 31 (0.4%) | MIRU-VNTR; WGS | [76] | |
| 2020 * | Cross-sectional | Cattle (50) | 50 (100%) | Tissue | TST-positive | Culture; PCR | 45 (90%) | [77] | ||
| 2017 | Cross-sectional | Cattle (2710) | 444 (16.4%) | Blood and milk (444) | TST-positive | Culture; PCR | Blood: 44 (1.6%); Milk: 12 (0.004%) | [33] | ||
| Iran | 2003–2005 | Cross-sectional | Buffalo (140) | Tissue (140) | Slaughtered | Culture | 0 | RFLP | [45] | |
| 2003–2006 | Cross-sectional | Cattle (213) | Tissue (213) | Slaughtered | Culture; PCR | 56 (26.3%) | RFLP; MIRU-VNTR; Spoligotyping | |||
| 1996–2006 | Cross-sectional | Cattle (488); Buffalo (140) | Tissue | Slaughtered | Culture; PCR | Cattle: 67 (13.7%); Buffalo: 132 (28.1%) | RFLP; RD-PCR; MIRU-VNTR | [24,25] | ||
| 2016 * | Case report | Deer (1) | Tissue | Dead | PCR | 1 | IS6110 RFLP | [18] | ||
| 2016 | Cross-sectional | Cattle (1700) | Tissue | Healthy | PCR | 44 (8.5%) | [44] | |||
| Iraq | 2009 * | Cross-sectional | Cattle | Milk (68) | Culture; PCR | 7 (10.2%) | [47] | |||
| 2016 * | Cross-sectional | Cattle (300) | Tissue | Slaughtered | Culture | 4 (1.3%) | [46] | |||
| 2015–2016 | Cross-sectional | Cows (119) | 24 (20.2%) | Blood and milk | Live | 42 (35.2%) | [53] | |||
| 2019 | Cross-sectional | Cattle (106); Buffalo (90) | Cattle (12.2%); Buffalo (4.4%) | Live | ND | [34] | ||||
| 2010 | Cross-sectional | Cattle | Milk (102) | Healthy | Culture; PCR | 10 (9.8%) | [52] | |||
| 2016 | Cross-sectional | Cattle (186) | 32 (17.2%) | Live | ND | [78] | ||||
| 2014 * | Cross-sectional | Cattle (28) | 21 (75%) | Slaughtered | ND | [40] | ||||
| 2012 * | Cross-sectional | Cows (850) | 206 (24.2%) | Serum (260), Milk (45), swab nasal (45), tissue samples (98), from cattle | Live and slaughtered | Culture | 100 (11.8%) | [35] | ||
| 2016 * | Cross-sectional | Cattle (21) | 4 (19%) | Live | ND | [79] | ||||
| Morocco | 2014–2015 | Cross-sectional | Cattle (8658) | Tissue | Slaughtered | Culture; PCR | 144 (1.7%) | Spoligotyping | [48] | |
| 2018 * | Cross-sectional | Cattle (1087) | 222 (20.4%) | Live | ND | [80] | ||||
| 2000–2001 | Cross-sectional | Cattle (78) | Tissue | Slaughtered | Culture | 40 (51.3%) | [49] | |||
| 1990 | Cross-sectional | Cattle (246) | 114 (46.3%) | Blood and Tissue | Live and slaughtered | Culture | 73 (29.7%) | [81] | ||
| Sudan | 2007–2009 | Cross-sectional | Cattle (6680) | Tissue | Slaughtered | Culture; PCR | 12 (0.2%) | [50] | ||
| 2002 * | Cross-sectional | Cattle (120) | Lymph nodes and tissue | Slaughtered | Culture; PCR | 25 (20.8%) | IS6110 RFLP | [51] | ||
| Turkey | 2019 * | Case report | Cat (1) | Tissue | Slaughtered | Culture; PCR | 1 | [16] | ||
| 2008 | Cross-sectional | Cattle (145) | Milk (145) | Live | Culture; PCR | 1 (0.7%) | Spoligotyping | [82] | ||
| 2011–2012 | Cross-sectional | Cattle (5018) | Tissue (95) | Slaughtered | Culture | 32 (0.6%) | Spoligotyping; MIRU-VNTR | [83] | ||
| 2005 | Cross-sectional | Cattle (210) | 3 (1.4%) | Nasal (198); Milk (146) | Live | PCR | 3(1.42%) | [37] | ||
| 2017–2018 | Cross-sectional | Cattle (ND) | Lymph nodes and tissue | Slaughtered | Culture | 38 (ND) | EIRC-PCR; RAPD-PCR; OUT-PCR; Spoligotyping | [84] | ||
| Tunisia | 2005–2006 | Cross-sectional | Cattle (102) | Milk (306) | TST-positive | Culture; PCR | 5 (4.9%) | IS6110 RFLP; Spoligotyping; MIRU-VNTR | [85] | |
| 2014–2015 | Cross-sectional | Cattle (149) | Tissue (149) | Slaughtered | Culture | 96 (64.4%) | IS6110 RFLP; Spoligotyping; MIRU-VNTR | [86] | ||
| 2010–2011 | Cross-sectional | Cattle (100) | 48 (48%) | Tissue (100) | Slaughtered | Culture; PCR | 27 (27%) | Spoligotyping; MIRU-VNTR | [41] |
* Date of publication; † Based on interferon gamma release assay, ELISA, or tuberculin skin test (TST). If different methods were used, we adopted the results of the TST; ND, Not Determined; MIRU-VNTR, Mycobacterial Interspersed Repetitive Units—Variable Number of Tandem Repeats; MLVA, Multiple Locus Variable Number of Tandem Repeats Analysis; ETR, Exact Tandem Repeats; RFLP-PCR, Restriction Fragment Length Polymorphism-PCR; WGS: Whole Genome Sequencing. ¶ Prevalence = n of M. bovis infected cases/n of total population.
3.2. Mycobacterium bovis in Humans
In humans, Mycobacterium tuberculosis is the primary causative agent of tuberculosis, followed by other MTBC species, including M. bovis. Nationwide estimations in the MENA countries, when available, revealed a relatively low prevalence of M. bovis among tuberculosis patients in some countries (Table 2). The prevalence of M. tuberculosis and M. bovis in Turkey was 94.1% and 4.3%, respectively [87]. Similarly, a study showed that only one tuberculosis case was due to M. bovis out of 67 extrapulmonary [88] and 45 pulmonary [89] tuberculosis cases in Egypt. In Lebanon, a nationwide surveillance study on tuberculosis showed that 3.4% (12/348) of patients were infected with M. bovis, while the remaining cases had human-associated tuberculosis strains (i.e., M. tuberculosis or Mycobacterium africanum) [90]. In contrast, BTB appears to have rapidly increased in comparison to other forms in Tunisia in recent years. Specifically, the estimated prevalence increased from 2.2% in 2009 [91] to 92.4% in 2013 [92]. Additionally, when focusing on at-risk groups such as farmers or slaughterhouse employees, available data showed high proportions of zoonotic tuberculosis, ranging from 8% in Iraq [35] and 5.36% in Egypt [71], to 3.3% in Lebanon [93].
The paucity of data and deficiencies in rigorous monitoring along with inappropriate control measures might cause the disease to spread more within the MENA region and beyond. Several factors might promote the spread of BTB among humans in the MENA region. For example, inappropriate hand washing or disinfection following cow handling appears to be a major risk factor for M. bovis infections among dairy farm workers [42]. The consumption of contaminated raw or unpasteurized milk also plays a crucial role in the transmission of BTB and has been significantly associated with the elevated risk of M. bovis infections in dairy workers [20,42,94]. The latter might also affect other human and animal populations. For instance, in Turkey and Lebanon, raw milk is widely available, which increases the risk of becoming infected from contaminated milk [84]. Furthermore, close quarters and proximity to animals, inadequate ventilation, and cow crowding were significant contributors to an increase in the risk of BTB [42,68]. These conditions are relevant in rural regions and in refugee camps in several Middle Eastern countries (e.g., Lebanon, Jordan, Turkey) and some geographical locations (e.g., the Nile Delta and Valley in Egypt) [63,95,96,97].

Table 2.
Burden of Mycobacterium bovis among humans in the MENA region.
Table 2.
Burden of Mycobacterium bovis among humans in the MENA region.
| Country | Study Period | Study Design | Population (N) | Samples (N) for Active Tuberculosis Testing | Health Status | Identification Method | Prevalence ¶ of BTB (%) | Typing Method | References |
|---|---|---|---|---|---|---|---|---|---|
| Algeria | 2015–2018 | Cross-sectional | ND (98) | Sputum (98) | Pulmonary TB | 4 (4.3%) | WGS | [98] | |
| 2017–2019 | Cross-sectional | ND (1952) | Sputum (51), Bronchial aspiration fluids (7); Gastric aspirations (25); Extra-pulmonary specimens (32) | TB patients | Culture; PCR | 7 (0.3%) | Spoligotyping; PhyloSNP | [23] | |
| Egypt | 1998–2000 | Cross-sectional | ND (67) | Cerebrospinal fluid (67) | Meningitis patients | 1 (1.5%) | IS6110 RFLP; Spoligotyping | [88] | |
| 2010–2011 | Cross-sectional | ND (42) | Sputum (42) | TB patients | Culture; PCR | 0 | [61] | ||
| 2013 | Cross-sectional | Dairy workers | Hand swab (50) | Healthy | Culture; PCR | 0 | [62] | ||
| 2007 * | Cross-sectional | ND (45) | Sputum (45) | Pulmonary TB | PCR | 1 (2.2%) | IS6110 RFLP; Spoligotyping | [89] | |
| 2009 * | Cross-sectional | Farm workers (15) | Sputum (15) | Healthy | Culture | [39] | |||
| 2015 * | Cross-sectional | Farm workers (25) | Sputum (25) | Healthy | Culture; PCR | 1 (4%) | [42] | ||
| 2018 * | Cross-sectional | Farm workers (10) | Blood (10) | Healthy | Culture; PCR | [38] | |||
| 2015–2017 | Longitudinal | Humans in contact with camels (48) | Sputum (48); Serum (48) | Healthy | Culture; PCR | 0 | [19] | ||
| 2018–2019 | Cross-sectional | Farm workers (149) | Sputum (149) | Healthy | Culture; PCR | 8 (5.3%) | [71] | ||
| 2016 * | Cross-sectional | ND (10) | Sputum (3) | Diagnosed human | Culture; PCR | 0 | [72] | ||
| 2020 * | Cross-sectional | ND | Sputum (10) | Tuberculin test positive | Culture; PCR | 90% | Sequencing (Mpb70 genes) | [77] | |
| Iran | 2009 | Cross-sectional | ND | Mycobacteriology bank in MRC (60) | Culture | 1 (1.7%) | MIRU-VNTR Spoligotyping | [99] | |
| 2004–2005 | Cross-sectional | ND (165) | Positives isolates (156) | TB patients | 15 (9.7%) | IS6110-RFLP MIRU-VNTR ETR-VNTR | [100] | ||
| 1995–2004 | Cross-sectional | ND (30) | Serum (30) | Patients with disseminated BCG disease | 17 (56.6%) | [101] | |||
| 2016 | Case report | ND (1) | Tissue | Brain tuberculoma | PCR | 1 | [102] | ||
| Iraq | 2016 * | Cross-sectional | ND (186) | Sputum (186) | Healthy | Culture | 2 (1.1%) | [46] | |
| 2012 * | Cross-sectional | Farm workers and veterinary doctors (25) | Sputum (25); Serum (25) | Healthy | Culture | 2 (8%) | [35] | ||
| Lebanon | 2004–2005 | Cross-sectional | Workers and veterinary doctors (60) | Sputum (60) | Pulmonary TB | Culture; PCR | 2 (3.3%) | Spoligotyping | [93] |
| 2015–2017 | Cross-sectional | ND (13) | Clinical samples (13) | Suspected TB patients | Culture | 2 (15.4%) | IS6110 insertion; Spoligotyping; MIRU-VNTR; WGS | [103] | |
| 2016–2017 | Cross-sectional | ND (1104) | Clinical samples (1104) | TB patients | Culture; PCR | 12 (1.1%) | Spoligotyping; MIRU-VNTR; Deeplex-TB | [90] | |
| Morocco | 2000–2001 | Cross-sectional | ND (200) | Sputum (200) | Suspected TB patients | Culture | 18 (17.8%) | [49] | |
| 2011 * | Case report | ND (1) | Gastric specimen | Patient with erythema nodosum | Culture | 1 | [104] | ||
| Palestine | 2005–2010 | Cross-sectional | ND (53) | Sputum (53); Smears (31) | TB patients | Culture | 2 (3.7%) | Spoligotyping MIRU-VNTR | [105] |
| Djibouti | 1999 | Cross-sectional | ND (153) | Lymph nodes (196) | Patients with adenopathy | Culture | 1 (0.7%) | [106] | |
| 1997–2011 | Cross-sectional | ND (411) | Sputum (411) | Suspected TB patients | Culture | 1 (0.2%) | Spoligotyping; VNTR-MLVA; WGS | [107] | |
| Saudi-Arabia | 2002–2005 | Cross-sectional | ND (1505) | Clinical isolates (1505) | Healthy | Culture | 13 (0.9%) | Spoligotyping; MIRU-VNTR | [108] |
| 2014–2016 | Cross-sectional | ND (2092) | Extrapulmonary clinical isolates (1003); Pulmonary clinical isolates (1089) | TB patients | Culture | Extrapulmonary: 119 (11.8%); Pulmonary: 32 (2.9%) | MIRU-VNTR | [109] | |
| Sudan | 1998–1999 | Cross-sectional | ND (105) | Sputum (105) | TB patients | PCR | 1 (0.9%) | Spoligotyping | [110] |
| Turkey | 2007–2010 | Cross-sectional | ND (188) | Clinical samples (188) | TB patients | Culture; PCR | 8 (4.3%) | [87] | |
| 2011–2012 | Cross-sectional | ND (10) | Sputum (10) | TB patients | Culture | 5 (50%) | Spoligotyping; MIRU-VNTR | [83] | |
| 2015 * | Case report | ND (1) | Tissue sample | NEMO-deficient patient | Culture; PCR | 1 | GenoType MTBC; Spoligotyping | [111] | |
| 2007–2010 | Cross-sectional | ND (2436) | Clinical samples (188) | TB patients | PCR | 8 (0.3%) | GenoType MTBC | [87] | |
| 2016 | Case report | Slaughterhouse worker (1) | Clinical sample | Skin lesion | Culture | 1 | GenoType MTBC | [112] | |
| 1996 * | Case report | Slaughterhouse worker (1) | Clinical sample | Flexor Tenosynovitis | Culture | 1 | [113] | ||
| 2007 * | Cross-sectional | ND (60) | Sputum (60) | TB patients | PCR | 8 (13.3%) | [114] | ||
| 2004–2014 | Cross-sectional | ND (220) | Clinical samples (220) | TB patients | Culture | 3 (1.4%) | Genotyping MTBC | [115] | |
| 2009–2014 | Cross-sectional | ND (482) | Clinical samples (482) | Pulmonary and extrapulmonary TB patients | Culture | 13 (2.7%) | Spoligotyping | [94] | |
| Tunisia | 2014–2018 | Case report | ND (4) | Tissue (4) | Spondylodiscitis patients | Culture; PCR | 4 | [116] | |
| 2009–2013 | Cross-sectional | ND (181) | Tissues (181) | Patients with adenopathy | Culture | 4 (2.2%) | [91] | ||
| 2013 | Cross-sectional | ND (174) | Lymph node (174) | Patients with adenopathy | Culture; PCR | 60 (34.4%) | [117] | ||
| 2013–2015 | Cross-sectional | ND (170) | Lymph nodes biopsy (144); Pus and abscess (10); Cerebrospinal fluid (8); Pleural fluid (1); Tissue (5); Bone scarping (2) | TB patients | Culture; PCR | 157 (92.4%) | [92] |
* Date of publication; If both were used, we adopted the results of the interferon gamma release assay; ND, Not Determined; MIRU-VNTR, Mycobacterial Interspersed Repetitive Units—Variable Number of Tandem Repeats; MLVA, Multiple Locus Variable Number of Tandem Repeats Analysis; ETR, Exact Tandem Repeats; RFLP-PCR, Restriction Fragment Length Polymorphism-PCR; WGS: Whole Genome Sequencing. ¶ Prevalence = n of M. bovis infected cases/n of total population.
3.3. Laboratory Methods for the Diagnosis and Typing of Mycobacterium bovis Adopted in the MENA Region
Although the reported detection methods in the studies from the MENA countries varied, active tuberculosis infections are still confirmed by mycobacterial culture which is considered the main approach, even for BTB. However, the adoption of molecular assays might be advantageous. For example, comparing molecular assays with microbiological culture revealed that the detection level of PCR-based assays was slightly greater than the conventional culture approach [118]. Moreover, molecular methods provide faster detection and identify the isolates at the species level. To confirm the identification of M. bovis, the detection of polymorphism in pncA or oxyR genes represents a valuable approach [119,120]. Recently, two PCR-based methods, VetMAXTM and GeneXpert®, were developed for M. bovis identification [1,121].
Traditionally, the molecular epidemiology of M. bovis is studied by DNA fingerprinting methods such as IS6110 RFLP (Restriction Fragment Length Polymorphism) [122]. Despite the method’s potential to identify outbreaks in hospitals and communities, its low discriminatory power for strains with low number of IS6110 copies imposes the need for other complementary tools, such as spoligotyping and Mycobacterial Interspersed Repetitive Units/Variable Number Tandem Repeat (MIRU/VNTR) [123,124]. Furthermore, next-generation genome sequencing is receiving significant attention for M. bovis diagnosis because it provides a higher discriminatory power, facilitating the investigation of MTBC molecular epidemiology and genetic diversity with greater resolution [125]. However, when sequencing is unavailable (due to limited resources in LMICs), the combination of MIRU-VNTR with spoligotyping is more suitable for tracking infections and detecting risk factors than either technique alone [99]. Regardless, the application of molecular techniques in the genotyping of M. bovis facilitates infection control and tracking processes. An obvious example of the latter is revealing the effect of the animal movement on the appearance of M. bovis in African countries, which was mainly due to cattle delivered from Europe. This cross-border link was detected by using the spoligotyping approach, with SB0120 and SB0121 spoligotypes being the most abundant of M. bovis [109]. Spoligotype SB0120 is the most common circulating type worldwide while SB0121 mainly exists in Europe [126]. This geographical spillover was also observed in the MENA countries, especially Tunisia, Algeria, Morocco [34,98,116] and Iran [24].
3.4. Antimicrobial Resistance among M. bovis Isolates in the MENA Region
A major factor that might complicate the control of M. bovis in the MENA region is the drug resistant properties of this zoonotic agent. M. bovis has a natural resistance to pyrazinamide, an essential drug for standard short-course anti-tuberculosis therapy in humans. Unfortunately, phenotypic susceptibility to pyrazinamide is often not tested in the MENA region. Since the currently adopted diagnostic tools do not usually differentiate M. bovis from other MTBC species in MENA countries, BTB patients receive inadequate treatment, risking poorer outcomes and enhancing the selection of drug-resistant strains. Additionally, alarming data on drug resistance have been reported recently in the MENA region. Antimicrobial resistance genes were found in isolates retrieved from both infected humans and animals [95,127,128]. M. bovis strains were most commonly resistant to rifampicin and isoniazid in several reports from the MENA region [71,110,111]. A rifampicin (RIF)-resistant M. bovis strain was first reported in a Turkish patient in 2015, an 8-month-old male infant with nuclear factor-kB essential modulator (NEMO) deficiency [111]. Moreover, an Egyptian study reported the spread of multidrug-resistant M. bovis strains among buffaloes [68]. In Sudan, 4% of M. bovis isolates possessed resistance to both rifampicin and isoniazid due to genetic mutations [110], while, in Palestine, mutated rpoB and katG genes were identified in clinical samples from three unrelated individuals who did not respond to the first line of antituberculosis drug therapy [105].
4. Concluding Remarks
To our knowledge, this is the first review regarding the epidemiology of M. bovis in the MENA region. Despite the limited number of studies dealing with the epidemiology of M. bovis in most MENA countries, the currently available data shows that BTB is not negligible. The circulation of M. bovis in the community, even at a comparatively low prevalence, emphasizes the global and regional calls for appropriate diagnosis and control measures at the animal/human interface. In some MENA countries, the literature suggests that the prevailing conditions might be conducive for the spread of M. bovis in humans and other animals. This is facilitated by several factors, including the lack of information and deficient diagnosis and monitoring systems [42,49]. Therefore, it is obvious that the underdiagnosis of M. bovis in the MENA region emphasizes the need to review the current policies and guidelines adopted by public health stakeholders. Particularly, a better detection of zoonotic tuberculosis cases is required via enhancing laboratory capacity, ensuring access to fast and reliable diagnostic tools, raising awareness and expertise of stakeholders, improving food safety, strengthening surveillance (especially in animals), and addressing research gaps. Finally, the implementation of a One Health approach is crucial to control the spread of this disease in the MENA countries and beyond.
Author Contributions
Conceptualization, M.O.; methodology, D.K., A.A. and I.E.R.; validation, I.I.K. and M.O.; formal analysis, D.K. and N.O.; investigation, D.K., N.O., A.A., I.E.R., R.R., I.A.K., D.E.S., R.S., K.E.O., K.J.C., I.I.K. and M.O.; resources, M.O.; data curation, D.K. and N.O.; writing—original draft preparation, D.K., N.O., I.I.K. and M.O.; writing—review and editing, A.A., I.E.R., R.R., I.A.K., D.E.S., R.S., K.E.O. and K.J.C.; supervision, M.O.; project administration, M.O.; All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
M.O. is supported by the Atkinson Postdoctoral Fellowship (Cornell University).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Bernitz, N.; Kerr, T.J.; Goosen, W.J.; Chileshe, J.; Higgitt, R.L.; Roos, E.O.; Meiring, C.; Gumbo, R.; de Waal, C.; Clarke, C.; et al. Review of Diagnostic Tests for Detection of Mycobacterium bovis Infection in South African Wildlife. Front. Vet. Sci. 2021, 8, 588697. [Google Scholar] [CrossRef] [PubMed]
- Inlamea, O.F.; Soares, P.; Ikuta, C.Y.; Heinemann, M.B.; Achá, S.J.; Machado, A.; Ferreira Neto, J.S.; Correia-Neves, M.; Rito, T. Evolutionary analysis of Mycobacterium bovis genotypes across Africa suggests co-evolution with livestock and humans. PLoS Negl. Trop. Dis. 2020, 14, e0008081. [Google Scholar] [CrossRef] [PubMed]
- de la Rua-Domenech, R. Human Mycobacterium bovis infection in the United Kingdom: Incidence, risks, control measures and review of the zoonotic aspects of bovine tuberculosis. Tuberculosis 2006, 86, 77–109. [Google Scholar] [CrossRef] [PubMed]
- Michel, A.L.; Bengis, R.G.; Keet, D.F.; Hofmeyr, M.; Klerk, L.M.; Cross, P.C.; Jolles, A.E.; Cooper, D.; Whyte, I.J.; Buss, P.; et al. Wildlife tuberculosis in South African conservation areas: Implications and challenges. Vet. Microbiol. 2006, 112, 91–100. [Google Scholar] [CrossRef]
- El-Sayed, A.; El-Shannat, S.; Kamel, M.; Castañeda Vazquez, M.; Vázquez, H. Molecular Epidemiology of Mycobacterium bovis in Humans and Cattle. Zoonoses Public Health 2016, 63, 251–264. [Google Scholar] [CrossRef]
- Michel, A.L.; Müller, B.; van Helden, P.D. Mycobacterium bovis at the animal-human interface: A problem, or not? Vet. Microbiol. 2010, 140, 371–381. [Google Scholar] [CrossRef]
- Ejeh, E.F.; Raji, M.A.; Bello, M.; Lawan, F.A.; Francis, M.I.; Kudi, A.C.; Cadmus, S.I.B. Prevalence and direct economic losses from bovine tuberculosis in makurdi, Nigeria. Vet. Med. Int. 2014, 2014, 904861. [Google Scholar] [CrossRef]
- Silva, M.R.; Rocha, A.D.S.; Araujo, F.R.; Fonseca-Junior, A.A.; Alencar, A.P.; Suffys, P.N.; Costa, R.R.D.; Moreira, M.A.S.; Guimaraes, M.D.C. Risk factors for human Mycobacterium bovis infections in an urban area of Brazil. Mem. Inst. Oswaldo Cruz 2018, 113, e170445. [Google Scholar] [CrossRef]
- Cosivi, O.; Grange, J.M.; Daborn, C.J.; Raviglione, M.C.; Fujikura, T.; Cousins, D.; Robinson, R.A.; Huchzermeyer, H.F.; de Kantor, I.; Meslin, F.X. Zoonotic tuberculosis due to Mycobacterium bovis in developing countries. Emerg. Infect. Dis. 1998, 4, 59–70. [Google Scholar] [CrossRef]
- Moiane, I.; Machado, A.; Santos, N.; Nhambir, A.; Inlamea, O.; Hattendorf, J.; Källenius, G.; Zinsstag, J.; Correia-Neves, M. Prevalence of bovine tuberculosis and risk factor assessment in cattle in rural livestock areas of Govuro District in the Southeast of Mozambique. PLoS ONE 2014, 9, e91527. [Google Scholar] [CrossRef]
- Thoen, C.; LoBue, P.; de Kantor, I. The importance of Mycobacterium bovis as a zoonosis. Vet. Microbiol. 2006, 112, 339–345. [Google Scholar] [CrossRef]
- WHO. Global Tuberculosis Report 2021. 2021. Available online: https://www.who.int/publications/i/item/9789240037021 (accessed on 22 February 2023).
- Chapman, H.J.; Veras-Estévez, B.A. Lessons Learned during the COVID-19 Pandemic to Strengthen TB Infection Control: A Rapid Review. Glob. Health Sci. Pract. 2021, 9, 964–977. [Google Scholar] [CrossRef]
- Osman, M.; Kasir, D.; Rafei, R.; Kassem, I.I.; Ismail, M.B.; El Omari, K.; Dabboussi, F.; Cazer, C.; Papon, N.; Bouchara, J.-P.; et al. Trends in the epidemiology of dermatophytosis in the Middle East and North Africa region. Int. J. Dermatol. 2022, 61, 935–968. [Google Scholar] [CrossRef]
- Bapat, P.R.; Dodkey, R.S.; Shekhawat, S.D.; Husain, A.A.; Nayak, A.R.; Kawle, A.P.; Daginawala, H.F.; Singh, L.K.; Kashyap, R.S. Prevalence of zoonotic tuberculosis and associated risk factors in Central Indian populations. J. Epidemiol. Glob. Health 2017, 7, 277–283. [Google Scholar] [CrossRef]
- Eroksuz, Y.; Baydar, E.; Otlu, B.; Dabak, M.; Eroksuz, H.; Karabulut, B.; Incili, C.A.; Timurkan, M.O. Case report: Systemic tuberculosis caused by Mycobacterium bovis in a cat. BMC Vet. Res. 2019, 15, 9. [Google Scholar] [CrossRef]
- Matos, A.C.; Figueira, L.; Martins, M.H.; Matos, M.; Morais, M.; Dias, A.P.; Coelho, A.C.; Pinto, M.L. Mycobacterium bovis in an Egyptian mongoose. Vet. Rec. 2013, 173, 376–377. [Google Scholar] [CrossRef]
- Mombeni, E.G.; Mosavari, N.; Gravand, M.M.; Amir Rezai, A.; Keshavarz, R.; Tadayon, K.; Bakhshi, R.; Behmanesh, R. First Report of Mycobacterium bovis Isolation from a European Fallow Deer (Dama Dama Dama) in Iran. Iran. J. Public Health 2016, 45, 814–816. [Google Scholar]
- Elnaker, Y.F.; Diab, M.S.; Ibrahim, N.A.; El-Gedawy, A.; Zaki, R.S.; Radwan, A. Seroprevalence and molecular characterization of Mycobacterium bovis infection in camels (Camelus dromedarius) in the Delta region, Egypt. Vet. World 2019, 12, 1180. [Google Scholar] [CrossRef]
- Mohamed, A.M.; El-Ella, G.A.A.; Nasr, E.A. Phenotypic and molecular typing of tuberculous and nontuberculous Mycobacterium species from slaughtered pigs in Egypt. J. Vet. Diagn. Investig. 2009, 21, 48–52. [Google Scholar] [CrossRef]
- Tazerart, F.; Saad, J.; Sahraoui, N.; Yala, D.; Niar, A.; Drancourt, M. Whole Genome Sequence Analysis of Mycobacterium bovis Cattle Isolates, Algeria. Pathogens 2021, 10, 802. [Google Scholar] [CrossRef]
- Ghebremariam, M.K.; Rutten, V.P.; Vernooij, J.C.; Uqbazghi, K.; Tesfaalem, T.; Butsuamlak, T.; Idris, A.M.; Nielen, M.; Michel, A.L. Prevalence and risk factors of bovine tuberculosis in dairy cattle in Eritrea. BMC Vet. Res. 2016, 12, 80. [Google Scholar] [CrossRef] [PubMed]
- Damene, H.; Tahir, D.; Diels, M.; Berber, A.; Sahraoui, N.; Rigouts, L. Broad diversity of Mycobacterium tuberculosis complex strains isolated from humans and cattle in Northern Algeria suggests a zoonotic transmission cycle. PLoS Negl. Trop. Dis. 2020, 14, e0008894. [Google Scholar] [CrossRef] [PubMed]
- Tadayon, K.; Mosavari, N.; Shahmoradi, A.H.; Sadeghi, F.; Azarvandi, A.; Forbes, K.J. The Epidemiology of Mycobacterium bovis in Buffalo in Iran. J. Vet. Med. 2006, 53, 41–42. [Google Scholar] [CrossRef]
- Tadayon, K.; Mosavari, N.; Sadeghi, F.; Forbes, K.J. Mycobacterium bovis infection in Holstein Friesian cattle, Iran. Emerg. Infect. Dis. 2008, 14, 1919–1921. [Google Scholar] [CrossRef] [PubMed]
- Elsayed, M. A first insight into the application of high discriminatory MIRU-VNTR typing using QIAxcel technology for genotyping Mycobacterium bovis isolated from the Delta area in Egypt. Infect. Genet. Evol. 2019, 71, 211–214. [Google Scholar] [CrossRef]
- Lombard, J.E.; Patton, E.A.; Gibbons-Burgener, S.N.; Klos, R.F.; Tans-Kersten, J.L.; Carlson, B.W.; Keller, S.J.; Pritschet, D.J.; Rollo, S.; Dutcher, T.V.; et al. Human-to-cattle Mycobacterium tuberculosis complex transmission in the United States. Front. Vet. Sci. 2021, 8, 691192. [Google Scholar] [CrossRef]
- WOAH. Bovine Tuberculosis. Available online: https://www.woah.org/en/disease/bovine-tuberculosis/ (accessed on 22 February 2023).
- de la Rua-Domenech, R.; Goodchild, A.T.; Vordermeier, H.M.; Hewinson, R.G.; Christiansen, K.H.; Clifton-Hadley, R.S. Ante mortem diagnosis of tuberculosis in cattle: A review of the tuberculin tests, gamma-interferon assay and other ancillary diagnostic techniques. Res. Vet. Sci. 2006, 81, 190–210. [Google Scholar] [CrossRef]
- Ortega, J.; Infantes-Lorenzo, J.A.; Bezos, J.; Roy, Á.; de Juan, L.; Romero, B.; Moreno, I.; Gómez-Buendía, A.; Agulló-Ros, I.; Domínguez, L.; et al. Evaluation of P22 ELISA for the detection of Mycobacterium bovis-specific antibody in the oral fluid of goats. Front. Vet. Sci. 2021, 8, 674636. [Google Scholar] [CrossRef]
- Sahraoui, N.; Müller, B.; Guetarni, D.; Boulahbal, F.; Yala, D.; Ouzrout, R.; Berg, S.; Smith, N.H.; Zinsstag, J. Molecular characterization of Mycobacterium bovis strains isolated from cattle slaughtered at two abattoirs in Algeria. BMC Vet. Res. 2009, 5, 4. [Google Scholar] [CrossRef]
- Abdellrazeq, G.; Elnaggar, M.; Osman, H.; Davis, W.; Singh, M. Prevalence of Bovine Tuberculosis in Egyptian Cattle and the Standardization of the Interferon-gamma Assay as an Ancillary Test. Transbound. Emerg. Dis. 2016, 63, 497–507. [Google Scholar] [CrossRef]
- Elsohaby, I.; Ahmed, H.A.; El-Diasty, M.M.; Elgedawy, A.A.; Mahrous, E.; El Hofy, F.I. Serological and molecular evidence of Mycobacterium bovis in dairy cattle and dairy farm workers under the intensive dairy production system in Egypt. J. Appl. Microbiol. 2020, 129, 1207–1219. [Google Scholar] [CrossRef]
- Aliraqi, O.M.M.; Al-Jammaly, M.; AL-Hankawi, O.; Al-Farwachi, M.I.; Dahl, M.O. Preliminary Prevalence and Risk Factors of Mycobacterium bovis in Local and Imported Breeds of Cattle and Buffaloes in Mosul city, Iraq. Egypt. J. Vet. Sci. 2020, 51, 83–88. [Google Scholar] [CrossRef]
- Barak, S. The incidence of bovine tuberculosis and its public health hazards in a dairy cattle station in Iraq. Al-Anbar J. Vet. Sci. 2012, 5, 23–29. [Google Scholar]
- Djafar, Z.R.; Benazi, N.; Bounab, S.; Sayhi, M.; Diouani, M.F.; Benia, F. Distribution of seroprevalence and risk factors for bovine tuberculosis in east Algeria. Prev. Vet. Med. 2020, 183, 105127. [Google Scholar] [CrossRef]
- Solmaz, H.; İlhan, Z.; Aksakal, A.; Gülhan, T.; Ekin, I.H. Detection of bovine tuberculosis by tuberculin test and polymerase chain reaction in Van, Turkey. Turk. J. Vet. Anim. Sci. 2009, 33, 229–233. [Google Scholar] [CrossRef]
- Elnaker, Y.F.; ElSharawy, N.T.; Daib, M.S.; ElGedawy, A.A.; Ibrahim, N.A. Conventional and molecular detection of Mycobacterium bovis in Aburden angus cattle and human contact in the new valley governorate, Egypt. Assiut Vet. Med. 2018, 64, 179–186. [Google Scholar]
- Hassanain, N.A.; Hassanain, M.A.; Soliman, Y.A.; Ghazy, A.A.; Ghazyi, Y.A. Bovine tuberculosis in a dairy cattle farm as a threat to public health. Afr. J. Microbiol. Res. 2009, 3, 446–450. [Google Scholar]
- Kalaf, J.M.; Salbouk, A.J.; Salman, S.S. Detection of bovine tuberculosis in Wasit city by the use of comparative intradermal tuberculin test and antigen rapid bovine TB Ab test. AL-Qadisiya J. Vet. Med. Sci. 2014, 13, 58–62. [Google Scholar]
- Lamine-Khemiri, H.; Martínez, R.; García-Jiménez, W.L.; Benítez-Medina, J.M.; Cortés, M.; Hurtado, I.; Abassi, M.S.; Khazri, I.; Benzarti, M.; Hermoso-de-Mendoza, J. Genotypic characterization by spoligotyping and VNTR typing of Mycobacterium bovis and Mycobacterium caprae isolates from cattle of Tunisia. Trop. Anim. Health Prod. 2014, 46, 305–311. [Google Scholar] [CrossRef]
- Lobna, M.A.S.; Nasr, E.A. Conventional and molecular detection of Mycobacterium bovis in milk of cows and its public health hazard. Int. J. Adv. Res. 2015, 3, 693–701. [Google Scholar]
- Sabry, M.; Elkerdasy, A. A polymerase chain reaction and enzyme linked immunosorbent assay based approach for diagnosis and differentiation between vaccinated and infected cattle with Mycobacterium bovis. J. Pharm. Bioallied Sci. 2014, 6, 115–121. [Google Scholar] [CrossRef] [PubMed]
- Karamian, M.; Soleimanzadeh, A.; Tukmechi, A.; Batavani, R. PCR investigation of the vertical transmission of Mycobacterium bovis in montbéliarde cattle in gonbad, northeast of Iran. Bulg. J. Vet. Med. 2022, 25, 586–592. [Google Scholar] [CrossRef]
- Mosavari, N.; Feizabadi, M.M.; Jamshidian, M.; Shahpouri, M.R.S.; Forbes, K.J.; Pajoohi, R.A.; Keshavarz, R.; Taheri, M.M.; Tadayon, K. Molecular genotyping and epidemiology of Mycobacterium bovis strains obtained from cattle in Iran. Vet. Microbiol. 2011, 151, 148–152. [Google Scholar] [CrossRef] [PubMed]
- Al-Thwani, A.N.; Al-Mashhadani, M.S. Tuberculosis in slaughtered cattle and workers in some abattoirs of Baghdad governorate. Int. J. Mycobacteriol. 2016, 5 (Suppl. 1), S250–S251. [Google Scholar] [CrossRef] [PubMed]
- Al-Saqur, I.; Al-Thwani, A.; Al-Attar, I. Detection of Mycobacteria spp in cows milk using conventional methods and PCR. Iraqi J. Vet. Sci. 2009, 23, 259–262. [Google Scholar]
- Yahyaoui-Azami, H.; Aboukhassib, H.; Bouslikhane, M.; Berrada, J.; Rami, S.; Reinhard, M.; Gagneux, S.; Feldmann, J.; Borrell, S.; Zinsstag, J. Molecular characterization of bovine tuberculosis strains in two slaughterhouses in Morocco. BMC Vet. Res. 2017, 13, 272. [Google Scholar] [CrossRef]
- Bendadda, O. Tuberculose Humaine à Mycobacterium bovis: Enquête Bactériologique et Application de la PCR à la Détection et l’identification du Complexe Mycobacterium tuberculosis. Université Sidi Mohamed Ben Abdellah Faculté des Sciences Dhar Mehraz -Fès-. 2003. Available online: https://opac.imist.ma/cgi-bin/koha/opac-detail.pl?biblionumber=24777 (accessed on 22 February 2023).
- Asil, E.T.A.; El Sanousi, S.M.; Gameel, A.; El Beir, H.; Fathelrahman, M.; Terab, N.M.; Muaz, M.A.; Hamid, M.E. Bovine tuberculosis in South Darfur State, Sudan: An abattoir study based on microscopy and molecular detection methods. Trop. Anim. Health Prod. 2013, 45, 469–472. [Google Scholar] [CrossRef]
- Sulieman, M.S.; Hamid, M.E. Identification of acid fast bacteria from caseous lesions in cattle in Sudan. J. Vet. Med. B Infect. Dis. Vet. Public Health 2002, 49, 415–418. [Google Scholar] [CrossRef]
- Al-Rubiaii, E.; Alwan, M.; Al-Thwani, A. Bovine tuberculosis: Diagnosis by PCR technique in bovine milk samples. Iraqi J. Vet. Med. 2013, 37, 156–159. [Google Scholar]
- Al-Fattli, H.H.H. The clinical and serological diagnosis of Mycobacterium bovis in blood and milk serums of lactating cows by IDEXX ELISA test in Wasit and Dhi-Qar provinces/Iraq. J. Contemp. Med. Sci. 2016, 2, 70–73. [Google Scholar] [CrossRef]
- Early, R. 17—Dairy products and milk-based food ingredients. In Natural Food Additives, Ingredients and Flavourings; Baines, D., Seal, R., Eds.; Woodhead Publishing: Sawston, UK, 2012; pp. 417–445. [Google Scholar] [CrossRef]
- Mackenzie, J.S.; McKinnon, M.; Jeggo, M. One Health: From Concept to Practice; Springer Japan: Tokyo, Japan, 2014; pp. 163–189. [Google Scholar] [CrossRef]
- Merker, M.; Kohl, T.A.; Niemann, S.; Supply, P. The evolution of strain typing in the Mycobacterium tuberculosis complex. Adv. Exp. Med. Biol. 2017, 1019, 43–78. [Google Scholar] [CrossRef]
- de Kantor, I.N.; Ritacco, V. An update on bovine tuberculosis programmes in Latin American and Caribbean countries. Vet. Microbiol. 2006, 112, 111–118. [Google Scholar] [CrossRef]
- Belakehal, F.; Barth, S.A.; Menge, C.; Mossadak, H.T.; Malek, N.; Moser, I. Evaluation of the discriminatory power of spoligotyping and 19-locus mycobacterial interspersed repetitive unit-variable number of tandem repeat analysis (MIRU-VNTR) of Mycobacterium bovis strains isolated from cattle in Algeria. PLoS ONE 2022, 17, e0262390. [Google Scholar] [CrossRef]
- Alwathnani, H.; Ashgan, M.; Ihab, M. Nested polymerase chain reaction as a molecular tool for detection of Mycobacterium tuberculosis complex recovered from milk samples. Afr. J. Microbiol. Res. 2012, 6, 1338–1344. [Google Scholar] [CrossRef]
- Moussa, I.M.; Mohamad, K.H.F.; Marwah, M.; Nasr, E.A.; Shibl, A.M.; Salem-Bekhit, M.M.; Hatem, M.E. Comparaison between conventional and modern techniques used for identification of Mycobacterium tuberculosis complex. Afr. J. Microbiol. Res. 2011, 5, 4334–4343. [Google Scholar] [CrossRef]
- Ramadan, H.H.; El-Gohary, A.H.N.; Mohamed, A.A.; Nasr, E.A. Detection of Mycobacterium bovis and Mycobacterium tuberculosis from clinical samples by conventional and molecular techniques in Egypt. Glob. Vet. 2012, 9, 648–654. [Google Scholar]
- El-Gedawy, A.; Ahmed, H.; Awadallah, M. Occurrence and molecular characterization of some zoonotic bacteria in bovine milk, milking equipments and humans in dairy farms, Sharkia, Egypt. Int. Food Res. J. 2014, 21, 1813–1823. [Google Scholar]
- Shereen, A.; Sobhy, G.; El-Maghraby, A.S. Assessment of routine and detailed inspection of tuberculous lesions in tuberculin reactor cattle. Intern. J. Microbiol. Res. 2015, 6, 155–159. [Google Scholar]
- Manal, M.Y.; Gobran, R.A. Some studies on tuberculosis in camel. Egypt. J. Comp. Path. Clin. Path. 2008, 21, 58–74. [Google Scholar]
- Zahran, R.N.; El Behiry, A.; Marzouk, E.; Askar, T. Comparison of LCD array and IS6110-PCR with conventional techniques for detection of Mycobacterium bovis isolated from Egyptian cattle and Buffaloes. Int. J. Mycobacteriol. 2014, 3, 197–204. [Google Scholar] [CrossRef]
- Elsayed, M.S.A.E.; Salah, A.; Elbadee, A.A.; Roshdy, T. Real-time PCR using atpE, conventional PCR targeting different regions of difference, and flow cytometry for confirmation of Mycobacterium bovis in buffaloes and cattle from the Delta area of Egypt. BMC Microbiol. 2022, 22, 154. [Google Scholar] [CrossRef] [PubMed]
- Hassan, W.H.; Nasr, E.A.; Moussa, H.M. A qualitative immunoassay as complementary test with tuberculin skin test for detection of tuberculosis in dairy cattle. J. Vet. Med. Res. 2018, 25, 182–190. [Google Scholar] [CrossRef]
- Borham, M.; Oreiby, A.; El-Gedawy, A.; Hegazy, Y.; Hemedan, A.; Al-Gaabary, M. Abattoir survey of bovine tuberculosis in tanta, centre of the Nile delta, with in silico analysis of gene mutations and protein-protein interactions of the involved Mycobacteria. Transbound Emerg. Dis. 2022, 69, 434–450. [Google Scholar] [CrossRef] [PubMed]
- Elsayed, M.; Amer, A. The rapid detection and differentiation of Mycobacterium tuberculosis complex members from cattle and water buffaloes in the delta area of Egypt, using a combination of real-time and conventional PCR. Mol. Biol. Rep. 2019, 46, 3909–3919. [Google Scholar] [CrossRef] [PubMed]
- Elsohaby, I.; Mahmmod, Y.S.; Mweu, M.M.; Ahmed, H.A.; El-Diasty, M.M.; Elgedawy, A.A.; Mahrous, E.; El Hofy, F.I. Accuracy of PCR, mycobacterial culture and interferon-γ assays for detection of Mycobacterium bovis in blood and milk samples from Egyptian dairy cows using Bayesian modelling. Prev. Vet. Med. 2020, 181, 105054. [Google Scholar] [CrossRef]
- Abdelsadek, H.A.; Sobhy, H.M.; Mohamed, K.F.; Hekal, S.H.A.; Dapgh, A.N.; Hakim, A.S. Multidrug-resistant strains of Mycobacterium complex species in Egyptian farm animals, veterinarians, and farm and abattoir workers. Vet. World 2020, 13, 2150–2155. [Google Scholar] [CrossRef]
- Elsayed, M.; Elkerdasy, A.; Akeila, M.; Elsayed, A. Comparison between immunological and molecular based methods for diagnosis of Mycobacterium infections in cattle, buffaloes and human in Egypt. Cell Mol. Biol 2016, 62, 125. [Google Scholar]
- Algammal, A.M.; Wahdan, A.; Elhaig, M.M. Potential efficiency of conventional and advanced approaches used to detect Mycobacterium bovis in cattle. Microb. Pathog. 2019, 134, 103574. [Google Scholar] [CrossRef]
- Mossad, A.; Akeila, M.; Radwan, G.; Samaha, H.; Nasr, E.; El-Battawy, E. Prevalence of bovine infection with Mycobacterium bovis in some Egyptian governorates. Vet. Med. J. Giza 2009, 57, 35–52. [Google Scholar]
- Elsify, A.; Nayel, M.; Hazem, S.; Tarabess, R.; Akram, S.; Allaam, M.; Hassan, H.; El Garhy, M. Sero-diagnosis of bovine tuberculosis by ELISA using bovine PPD and ST.CF. Vet. Med. J. 2013, 22, 126–129. [Google Scholar]
- Abdelaal, H.F.M.; Spalink, D.; Amer, A.; Steinberg, H.; Hashish, E.A.; Nasr, E.A.; Talaat, A.M. Genomic Polymorphism Associated with the Emergence of Virulent Isolates of Mycobacterium bovis in the Nile Delta. Sci. Rep. 2019, 9, 11657. [Google Scholar] [CrossRef]
- Wahdan, A.; Riad, E.M.; Enany, S. Genetic differentiation of Mycobacterium bovis and Mycobacterium tuberculosis isolated from cattle and human sources in, Egypt (Suez Canal area). Comp. Immunol. Microbiol. Infect. Dis. 2020, 73, 101553. [Google Scholar] [CrossRef]
- AL-Taee, H.S. Comparative Identification of Mycobacterium bovis by Using A Serological ELISA (IDEXX) and Tuberculin Test in Cattle in Wasit Province/Iraq. J. Educ. Coll. Wasit Univ. 2016, 1, 459–472. [Google Scholar] [CrossRef]
- Ahmed, W.A. Performance of Comparative Cervical Tuberculin Test and Serological Methods with Culturing of Nasal Swab in Diagnosis of Bovine Tuberculosis in Cross Breed Cattle Baghdad Iraq: A Comparative Evaluation. Adv. Microbiol. 2016, 06, 867–878. [Google Scholar] [CrossRef]
- Yahyaoui Azami, H.; Ducrotoy, M.J.; Bouslikhane, M.; Hattendorf, J.; Thrusfield, M.; Conde-Álvarez, R.; Moriyón, I.; Zúñiga-Ripa, A.; Muñoz Álvaro, P.M.; Mick, V.; et al. The prevalence of brucellosis and bovine tuberculosis in ruminants in Sidi Kacem Province, Morocco. PLoS ONE 2018, 13, e0203360. [Google Scholar] [CrossRef]
- Berrada, J. Mycobacterium bovis Infection in Cattle in Morocco: Preparation and Evaluation of Chemical Extracts for Use in Detection of Immune Responses; Iowa State University: Ames, IA, USA, 1993. [Google Scholar]
- Aydın, F.E.; Ulger, M.; Emekdaş, G.; Aslan, G.; Günal, S. Isolation and identification of Mycobacterium bovis and non-tuberculous mycobacteria in raw milk samples in Mersin province. Mikrobiyoloji Bul. 2012, 46, 283–289. [Google Scholar]
- Tuzcu, N.; Köksal, F. Genetic evaluation of Mycobacteriumbovis isolates with MIRU-VNTR and spoligotyping. Turk. J. Med. Sci. 2020, 50, 2017–2023. [Google Scholar] [CrossRef]
- Sahin, F.; Tarhan, G.L.; Cinoglu, H.; Kayar, M.B.M.; Yakici, G.L. Spoligotyping and polymerase chain reaction based Mycobacterium bovis strains typing with methods (enterobacterial repetitive intergenic consensus-polymerase chain reaction, randomly amplified polymorphic dnas-polymerase chain reaction and out polymerase chain reaction). Int. J. Mycobacteriol. 2022, 11, 88–94. [Google Scholar] [CrossRef]
- Ben Kahla, I.; Boschiroli, M.L.; Souissi, F.; Cherif, N.; Benzarti, M.; Boukadida, J.; Hammami, S. Isolation and molecular characterisation of Mycobacterium bovis from raw milk in Tunisia. Afr. Health Sci. 2011, 11 (Suppl. 1), S2–S5. [Google Scholar] [CrossRef]
- Djemal, S.E.; Siala, M.; Smaoui, S.; Kammoun, S.; Marouane, C.; Bezos, J.; Messadi-Akrout, F.; Romero, B.; Gdoura, R. Genetic diversity assessment of Tunisian Mycobacterium bovis population isolated from cattle. BMC Vet. Res. 2017, 13, 393. [Google Scholar] [CrossRef]
- Bayraktar, B.; Bulut, E.; Bariş, A.B.; Toksoy, B.; Dalgic, N.; Celikkan, C.; Sevgi, D. Species distribution of the Mycobacterium tuberculosis complex in clinical isolates from 2007 to 2010 in Turkey: A prospective study. J. Clin. Microbiol. 2011, 49, 3837–3841. [Google Scholar] [CrossRef] [PubMed]
- Cooksey, R.C.; Abbadi, S.H.; Woodley, C.L.; Sikes, D.; Wasfy, M.; Crawford, J.T.; Mahoney, F. Characterization of Mycobacterium tuberculosis complex isolates from the cerebrospinal fluid of meningitis patients at six fever hospitals in Egypt. J. Clin. Microbiol. 2002, 40, 1651–1655. [Google Scholar] [CrossRef] [PubMed]
- Abbadi, S.; El Hadidy, G.; Gomaa, N.; Cooksey, R. Strain differentiation of Mycobacterium tuberculosis complex isolated from sputum of pulmonary tuberculosis patients. Int. J. Infect. Dis. 2009, 13, 236–242. [Google Scholar] [CrossRef] [PubMed]
- El Achkar, S.; Demanche, C.; Osman, M.; Rafei, R.; Ismail, M.B.; Gaudin, C.; Duthoy, S.; Matos, F.D.; Yaacoub, H.; Pinçon, C.; et al. Zoonotic tuberculosis in humans assessed by next-generation sequencing: An 18-month nationwide study in Lebanon. Eur. Respir. J. 2020, 55, 1900513. [Google Scholar] [CrossRef] [PubMed]
- Smaoui, S.; Mezghanni, M.A.; Hammami, B.; Zalila, N.; Marouane, C.; Kammoun, S.; Ghorbel, A.; Ben Jemaa, M.; Messadi-Akrout, F. Tuberculosis lymphadenitis in a southeastern region in Tunisia: Epidemiology, clinical features, diagnosis and treatment. Int. J. Mycobacteriol. 2015, 4, 196–201. [Google Scholar] [CrossRef]
- Siala, M.; Smaoui, S.; Taktak, W.; Hachicha, S.; Ghorbel, A.; Marouane, C.; Kammoun, S.; Gamara, D.; Slim, L.; Gdoura, R.; et al. First-time detection and identification of the Mycobacterium tuberculosis Complex members in extrapulmonary tuberculosis clinical samples in south Tunisia by a single tube tetraplex real-time PCR assay. PLoS Negl. Trop. Dis. 2017, 11, e0005572. [Google Scholar] [CrossRef]
- Bedrossian, N.; Hamze, M.; Rahmo, A.K.; Jurjus, A.; Saliba, J.; Dabboussi, F.; Karam, W. Mycobacterium tuberculosis spoligotypes circulating in the Lebanese population: A retrospective study. East. Mediterr. Health J. 2013, 19, 119–124. [Google Scholar] [CrossRef]
- Çavuşoğlu, C.; Yılmaz, F.F. [Molecular epidemiology of human Mycobacterium bovis infection in Aegean Region, Turkey]. Mikrobiyoloji Bul. 2017, 51, 165–170. [Google Scholar] [CrossRef]
- Osman, M.; Rafei, R.; Ismail, M.B.; Omari, S.A.; Mallat, H.; Dabboussi, F.; Cazer, C.; Karah, N.; Abbara, A.; Hamze, M. Antimicrobial resistance in the protracted Syrian conflict: Halting a war in the war. Future Microbiol. 2021, 16, 825–845. [Google Scholar] [CrossRef]
- Osman, M.; Cummings, K.J.; El Omari, K.; Kassem, I.I. Catch-22: War, Refugees, COVID-19, and the Scourge of Antimicrobial Resistance. Front. Med. 2022, 9, 921921. [Google Scholar] [CrossRef]
- Kassem, I.I.; Osman, M.; Jaafar, H.; El Omari, K. Refugee settlements, COVID-19, pollution, and the unfolding cholera outbreak in Lebanon. J. Travel Med. 2022, 29, taac142. [Google Scholar] [CrossRef]
- Tazerart, F.; Saad, J.; Niar, A.; Sahraoui, N.; Drancourt, M. Mycobacterium bovis Pulmonary Tuberculosis, Algeria. Emerg. Infect. Dis. 2021, 27, 972–974. [Google Scholar] [CrossRef]
- Jafarian, M.; Aghali-Merza, M.; Farnia, P.; Ahmadi, M.; Masjedi, M.R.; Velayati, A.A. Synchronous Comparison of Mycobacterium tuberculosis Epidemiology Strains by “MIRU-VNTR” and “MIRU-VNTR and Spoligotyping” Technique. Avicenna J. Med. Biotechnol. 2010, 2, 145–152. [Google Scholar]
- Asgharzadeh, M.; Kafil, H.S.; Roudsary, A.A.; Hanifi, G.R. Tuberculosis transmission in Northwest of Iran: Using MIRU-VNTR, ETR-VNTR and IS6110-RFLP methods. Infect. Genet. Evol. 2011, 11, 124–131. [Google Scholar] [CrossRef]
- Paiman, S.A.; Siadati, A.; Mamishi, S.; Tabatabaie, P.; Khotaee, G. Disseminated Mycobacterium bovis infection after BCG vaccination. Iran. J. Allergy Asthma Immunol. 2006, 5, 133–137. [Google Scholar]
- Sharifi, G.; Mousavinejad, S.A.; Moradian, K.; Ebrahimzadeh, K.; Samadian, M.; Zerehpoosh, F.B.; Rezaei, O. Pineal Region Tuberculoma Caused by Mycobacterium bovis as a Complication of Bacille Calmette-Guérin Vaccine: Case Report and Review of the Literature. World Neurosurg. 2020, 133, 416–418. [Google Scholar] [CrossRef]
- Panossian, B.; Salloum, T.; Araj, G.F.; Khazen, G.; Tokajian, S. First insights on the genetic diversity of MDR Mycobacterium tuberculosis in Lebanon. BMC Infect. Dis. 2018, 18, 710. [Google Scholar] [CrossRef]
- Méchaï, F.; Soler, C.; Aoun, O.; Fabre, M.; Mérens, A.; Imbert, P.; Rapp, C. Primary Mycobacterium bovis infection revealed by erythema nodosum. Int. J. Tuberc. Lung Dis. 2011, 15, 1131–1132. [Google Scholar] [CrossRef]
- Ereqat, S.; Nasereddin, A.; Azmi, K.; Abdeen, Z.; Greenblatt, C.L.; Spigelman, M.; Rastogi, N.; Bar-Gal, G.K. Genetic characterization of Mycobacterium tuberculosis in the West Bank, Palestinian Territories. BMC Res. Notes 2012, 5, 270. [Google Scholar] [CrossRef]
- Koeck, J.L.; Bernatas, J.J.; Gerome, P.; Fabre, M.; Houmed, A.; Herve, V.; Teyssou, R. Epidemiology of resistance to antituberculosis drugs in Mycobacterium tuberculosis complex strains isolated from adenopathies in Djibouti. Prospective study carried out in 1999. Med. Trop. Rev. Du Corps Sante Colon. 2002, 62, 70–72. [Google Scholar]
- Blouin, Y.; Hauck, Y.; Soler, C.; Fabre, M.; Vong, R.; Dehan, C.; Cazajous, G.; Massoure, P.L.; Kraemer, P.; Jenkins, A.; et al. Significance of the identification in the Horn of Africa of an exceptionally deep branching Mycobacterium tuberculosis clade. PLoS ONE 2012, 7, e52841. [Google Scholar] [CrossRef] [PubMed]
- Al-Hajoj, S.A.; Zozio, T.; Al-Rabiah, F.; Mohammad, V.; Al-Nasser, M.; Sola, C.; Rastogi, N. First insight into the population structure of Mycobacterium tuberculosis in Saudi Arabia. J. Clin. Microbiol. 2007, 45, 2467–2473. [Google Scholar] [CrossRef] [PubMed]
- Varghese, B.; Enani, M.; Alrajhi, A.; Al Johani, S.; Albarak, A.; Althawadi, S.; Elkhizzi, N.; AlGhafli, H.; Shoukri, M.; Al-Hajoj, S. Impact of Mycobacterium tuberculosis complex lineages as a determinant of disease phenotypes from an immigrant rich moderate tuberculosis burden country. Respir. Res. 2018, 19, 259. [Google Scholar] [CrossRef] [PubMed]
- Sharaf-Eldin, G.S.; Saeed, N.S.; Hamid, M.E.; Jordaan, A.M.; Van der Spuy, G.D.; Warren, R.M.; Van Helden, P.D.; Victor, T.C. Molecular analysis of clinical isolates of Mycobacterium tuberculosis collected from patients with persistent disease in the Khartoum region of Sudan. J. Infect. 2002, 44, 244–251. [Google Scholar] [CrossRef] [PubMed]
- Çavuşoğlu, C.; Edeer Karaca, N.; Azarsız, E.; Ulusoy, E.; Kütükçüler, N. Rifampicin-resistant Mycobacterium bovis BCG strain isolated from an infant with NEMO mutation. Mikrobiyoloji Bul. 2015, 49, 272–277. [Google Scholar] [CrossRef]
- Mertoğlu, A.; Biçmen, C.; Karaarslan, S.; Buğdayci, M.H. Pulmonary tuberculosis due to Mycobacterium bovis revealed by skin lesion in slaughterhouse worker. Clin. Respir. J. 2018, 12, 317–321. [Google Scholar] [CrossRef]
- Bagatur, E.; Bayramiçli, M. Flexor tenosynovitis caused by Mycobacterium bovis: A case report. J. Hand Surg. 1996, 21, 700–702. [Google Scholar] [CrossRef]
- Ağaçayak, A.; Bulut, Y.; Seyrek, A. Detection of Mycobacterium species distribution in the sputum samples of tuberculosis patients by PCR-RFLP method in Elazig province. Mikrobiyoloji Bul. 2007, 41, 203–209. [Google Scholar]
- Öztürk, C.E.; Şahin, İ.; Öksüz, Ş.; Kılıç, N.; Kılınçel, Ö.; Aydın, L.; Atik, D.; Afşin, E. Investigation of Mycobacterium bovis subsp. bovis among the strains of Mycobacterium tuberculosis complex isolated in Düzce Province, Turkey. Mikrobiyoloji Bul. 2016, 50, 392–400. [Google Scholar] [CrossRef]
- Chebbi, Y.; Riahi, H.; Bouaziz, M.C.; Romdhane, E.; Mhiri, E.; Rammeh, S.; Saidi, L.S.; Achour, W.; Ladeb, M.F. Mycobacterium bovis Spondylodiscitis: Report of 4 Cases. J. Clin. Rheumatol. Pract. Rep. Rheum. Musculoskelet. Dis. 2019, 27, S546–S549. [Google Scholar] [CrossRef]
- Ghariani, A.; Jaouadi, T.; Smaoui, S.; Mehiri, E.; Marouane, C.; Kammoun, S.; Essalah, L.; Driss, M.; Messadi, F.; Slim-Saidi, L. Diagnosis of lymph node tuberculosis using the GeneXpert MTB/RIF in Tunisia. Int. J. Mycobacteriol. 2015, 4, 270–275. [Google Scholar] [CrossRef]
- Negi, S.S.; Khan, S.F.; Gupta, S.; Pasha, S.T.; Khare, S.; Lal, S. Comparison of the conventional diagnostic modalities, bactec culture and polymerase chain reaction test for diagnosis of tuberculosis. Indian J. Med. Microbiol. 2005, 23, 29–33. [Google Scholar] [CrossRef] [PubMed]
- Al-Thani, R.; Al Khal, A.L.; AL-Suwaidi, Z.D.; Elshafie, S.S.; El-Sheikh, N. Drug resistant Mycobacterium tuberculosis In Qatar: Comparison between the conventional and DNA-based methods. Mol. Diagn. Vaccines 2003, 1, 111–118. [Google Scholar]
- de los Monteros, L.E.E.; Galán, J.C.; Gutiérrez, M.; Samper, S.; García Marín, J.F.; Martín, C.; Domínguez, L.; de Rafael, L.; Baquero, F.; Gómez-Mampaso, E.; et al. Allele-specific PCR method based on pncA and oxyR sequences for distinguishing Mycobacterium bovis from Mycobacterium tuberculosis: Intraspecific M. bovis pncA sequence polymorphism. J. Clin. Microbiol. 1998, 36, 239–242. [Google Scholar] [CrossRef] [PubMed]
- Hlokwe, T.M.; Mogano, R.M. Utility of xpert® MTB/RIF ultra assay in the rapid diagnosis of bovine tuberculosis in wildlife and livestock animals from South Africa. Prev. Vet. Med. 2020, 177, 104980. [Google Scholar] [CrossRef]
- Moström, P.; Gordon, M.; Sola, C.; Ridell, M.; Rastogi, N. Methods used in the molecular epidemiology of tuberculosis. Clin. Microbiol. Infect. 2002, 8, 694–704. [Google Scholar] [CrossRef]
- Lee, A.S.; Tang, L.L.; Lim, I.H.; Bellamy, R.; Wong, S.Y. Discrimination of single-copy IS6110 DNA fingerprints of Mycobacterium tuberculosis isolates by high-resolution minisatellite-based typing. J. Clin. Microbiol. 2002, 40, 657–659. [Google Scholar] [CrossRef]
- Kam, K.M.; Yip, C.W.; Tse, L.W.; Wong, K.L.; Lam, T.K.; Kremer, K.; Au, B.K.; van Soolingen, D. Utility of mycobacterial interspersed repetitive unit typing for differentiating multidrug-resistant Mycobacterium tuberculosis isolates of the Beijing family. J. Clin. Microbiol. 2005, 43, 306–313. [Google Scholar] [CrossRef]
- Dippenaar, A.; Parsons, S.D.C.; Miller, M.A.; Hlokwe, T.; van Pittius, N.C.G.; Adroub, S.A.; Abdallah, A.M.; Pain, A.; Warren, R.M.; Michel, A.L.; et al. Progenitor strain introduction of Mycobacterium bovis at the wildlife-livestock interface can lead to clonal expansion of the disease in a single ecosystem. Infect. Genet. Evol. 2017, 51, 235–238. [Google Scholar] [CrossRef]
- Ghavidel, M.; Mansury, D.; Nourian, K.; Ghazvini, K. The most common spoligotype of Mycobacterium bovis isolated in the world and the recommended loci for VNTR typing; A systematic review. Microb. Pathog. 2018, 118, 310–315. [Google Scholar] [CrossRef]
- Zakaria, A.; Osman, M.; Dabboussi, F.; Rafei, R.; Mallat, H.; Papon, N.; Bouchara, J.P.; Hamze, M. Recent trends in the epidemiology, diagnosis, treatment, and mechanisms of resistance in clinical Aspergillus species: A general review with a special focus on the Middle Eastern and North African region. J. Infect. Public Health 2020, 13, 1–10. [Google Scholar] [CrossRef]
- Rafei, R.; Hawli, M.; Osman, M.; Dabboussi, F.; Hamze, M. Distribution of emm types and macrolide resistance determinants among group A streptococci in the Middle East and North Africa. J. Glob. Antimicrob. Resist. 2020, 22, 334–348. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).