TNFα Effects on Adipocytes Are Influenced by the Presence of Lysine Methyltransferases, G9a (EHMT2) and GLP (EHMT1)
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Small Interfering RNA (siRNA)-Mediated Knockdown
2.3. RNA Analysis
2.4. Whole-Cell Extract Preparation and Subcellular Fractionation
2.5. Gel Electrophoresis and Immunoblotting
2.6. Immunoprecipitation (IP)
2.7. Measurement of Glycerol and Free Fatty Acid Release
2.8. Antibodies
2.9. Statistical Analysis
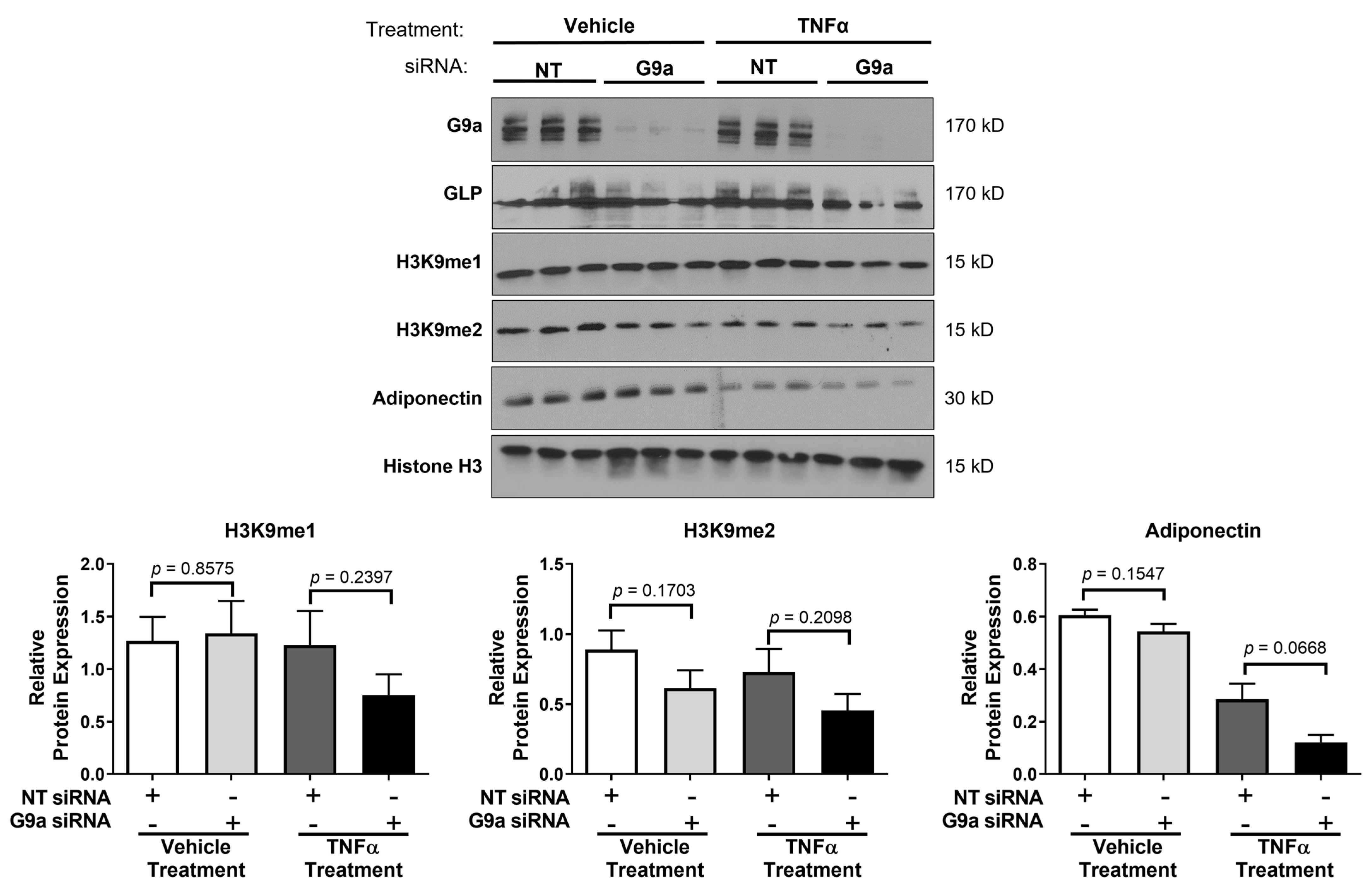
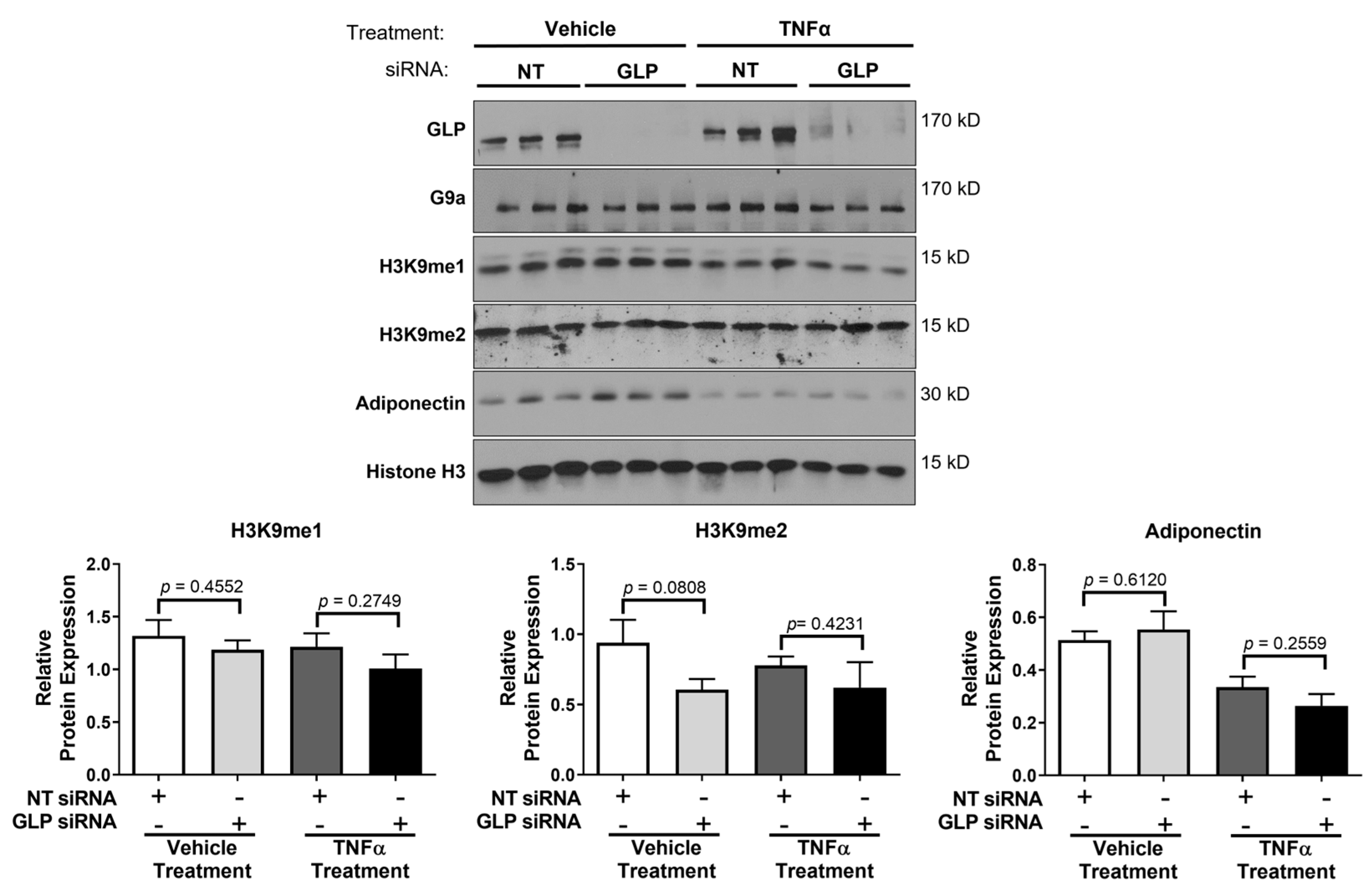
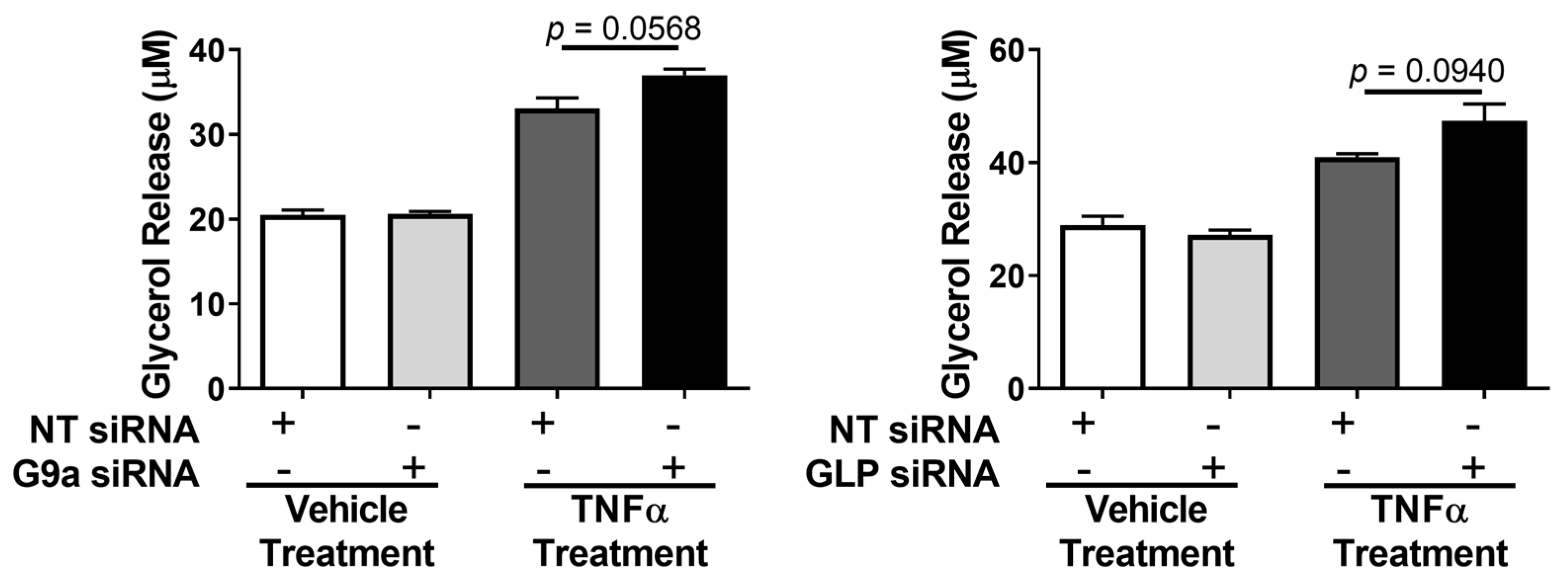
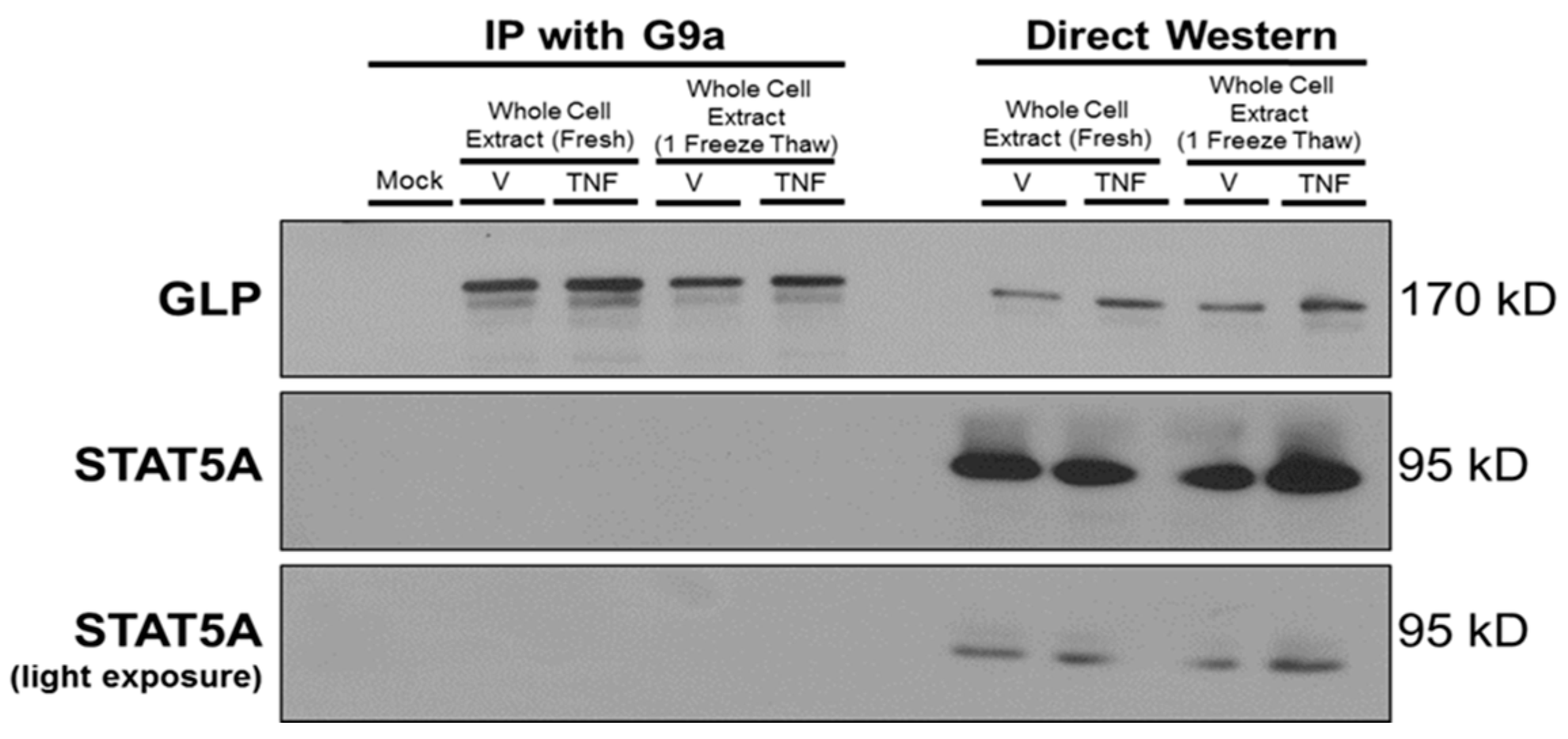
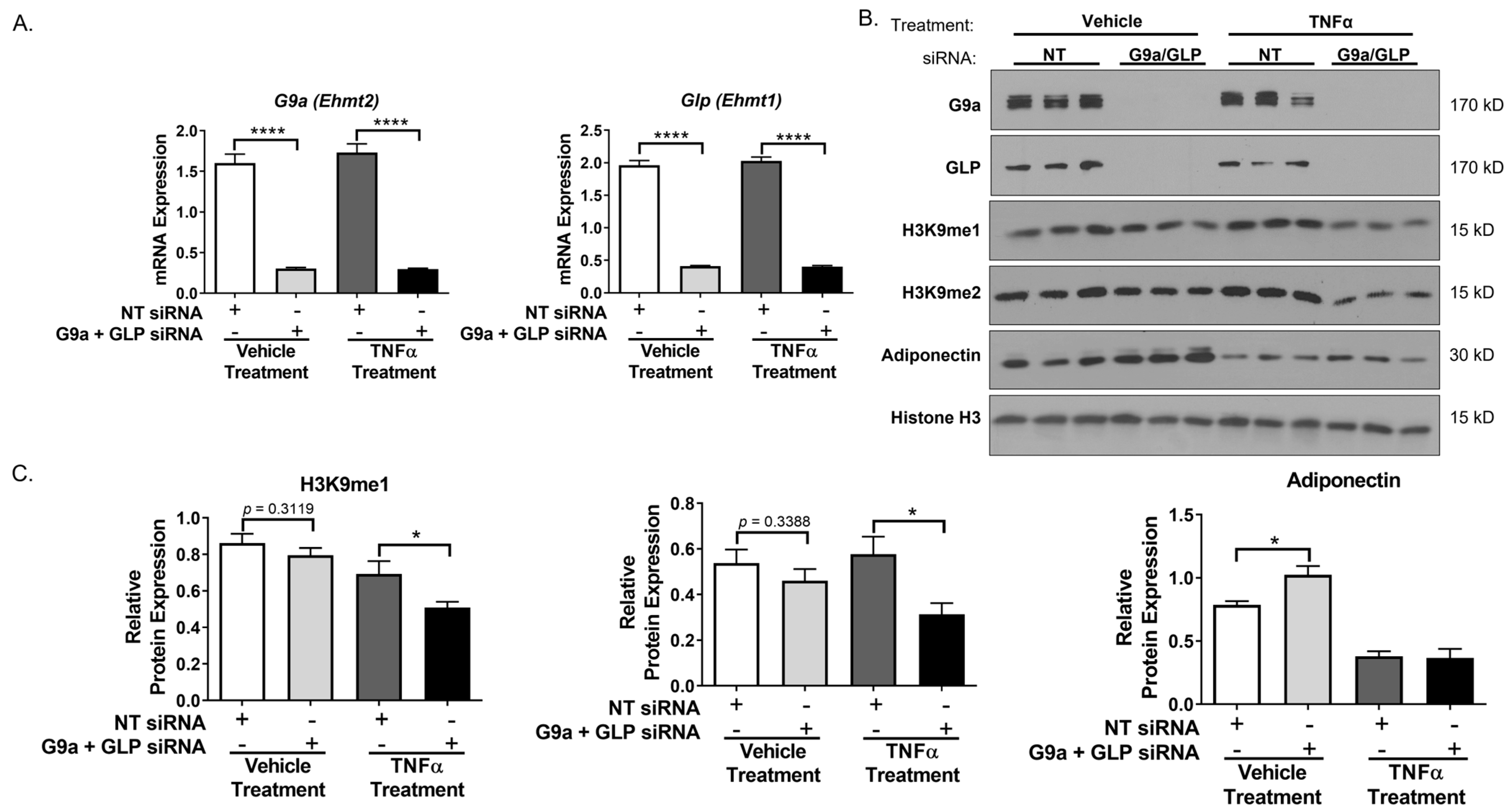
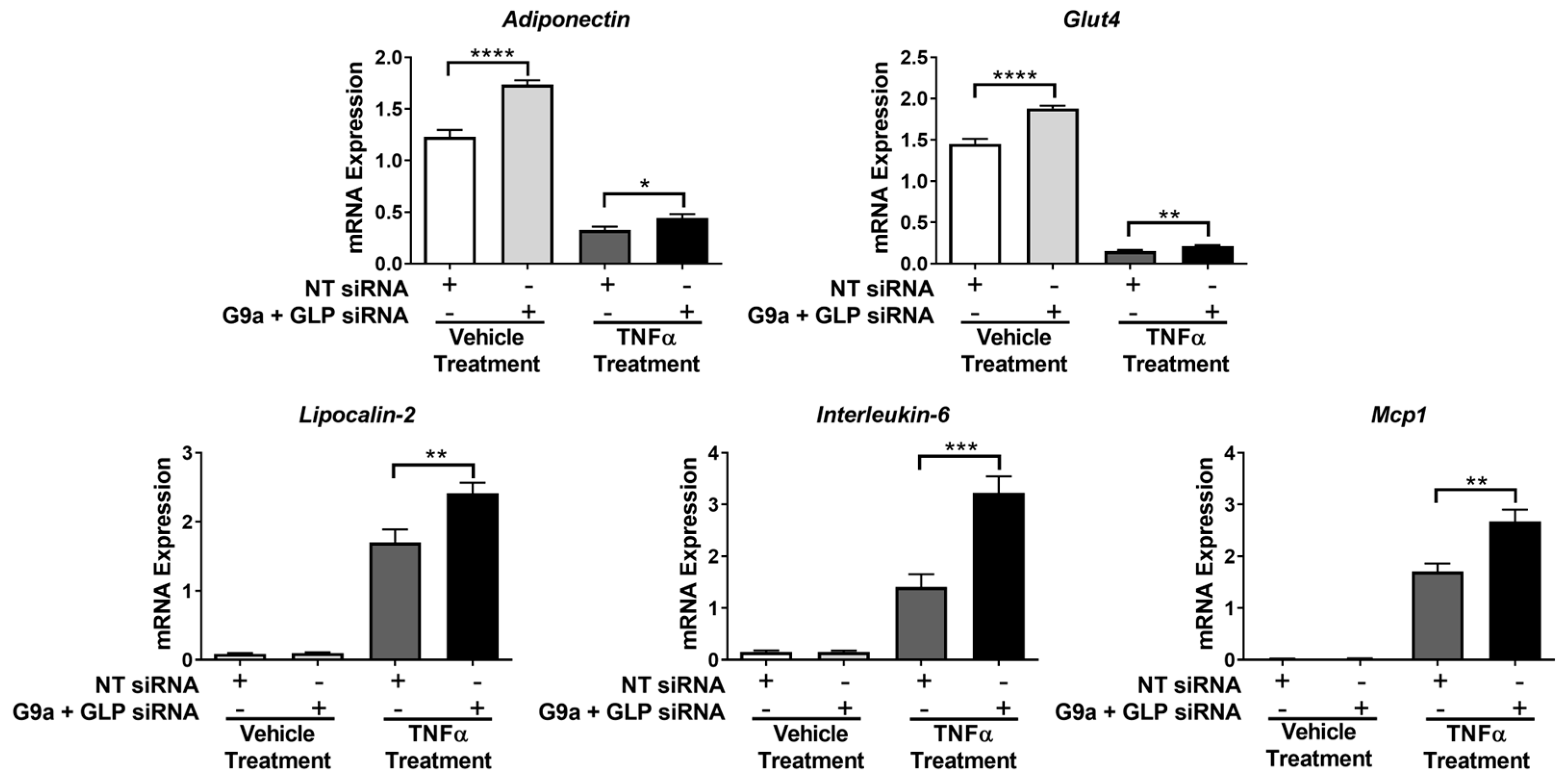
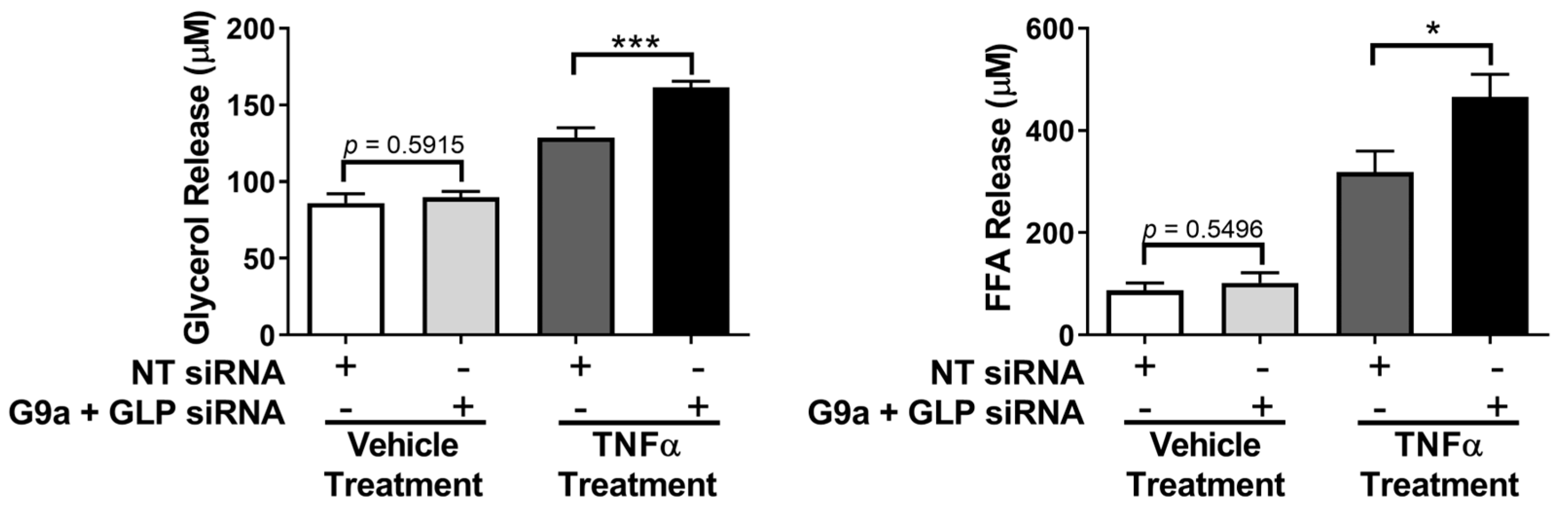
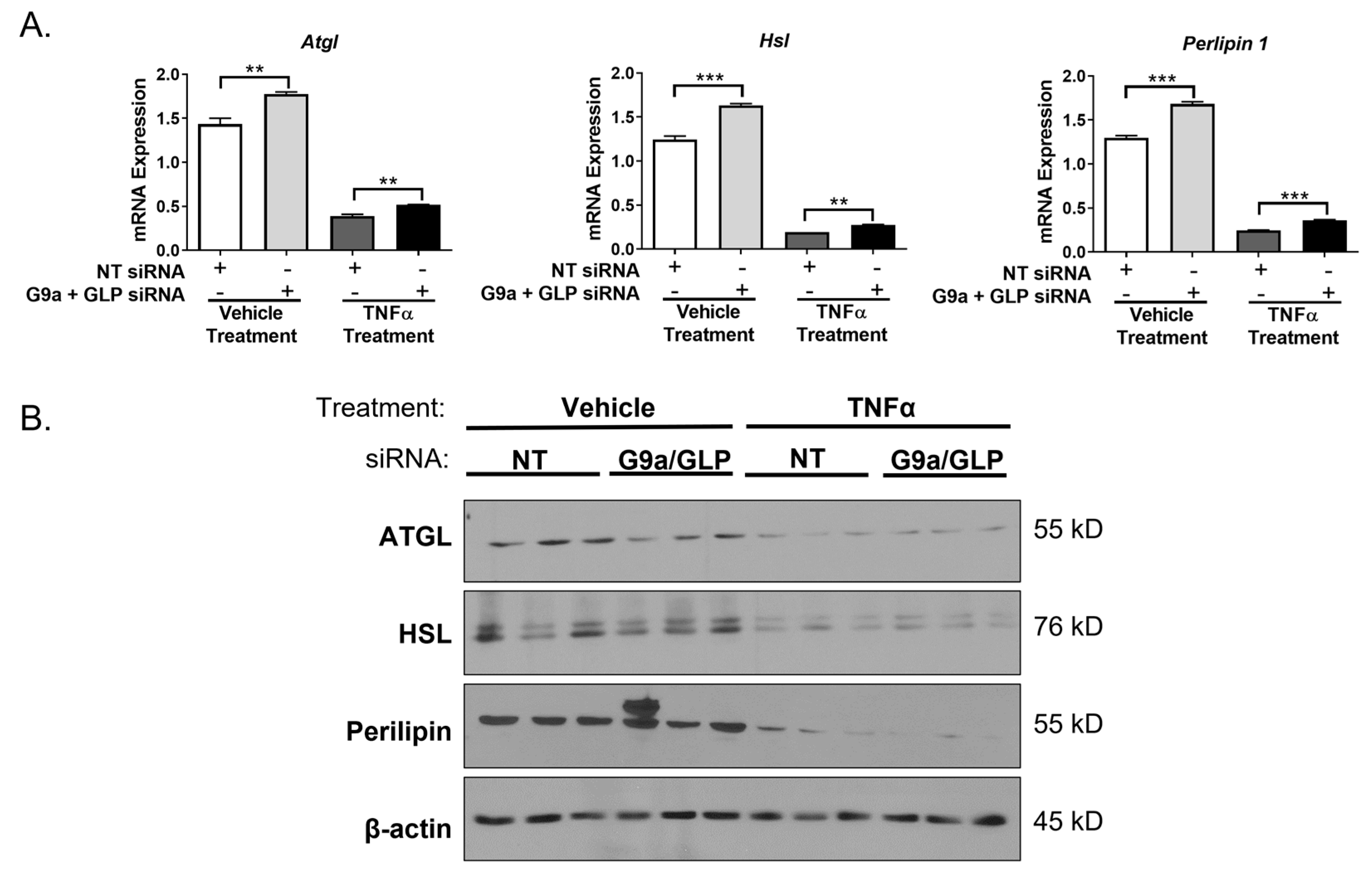
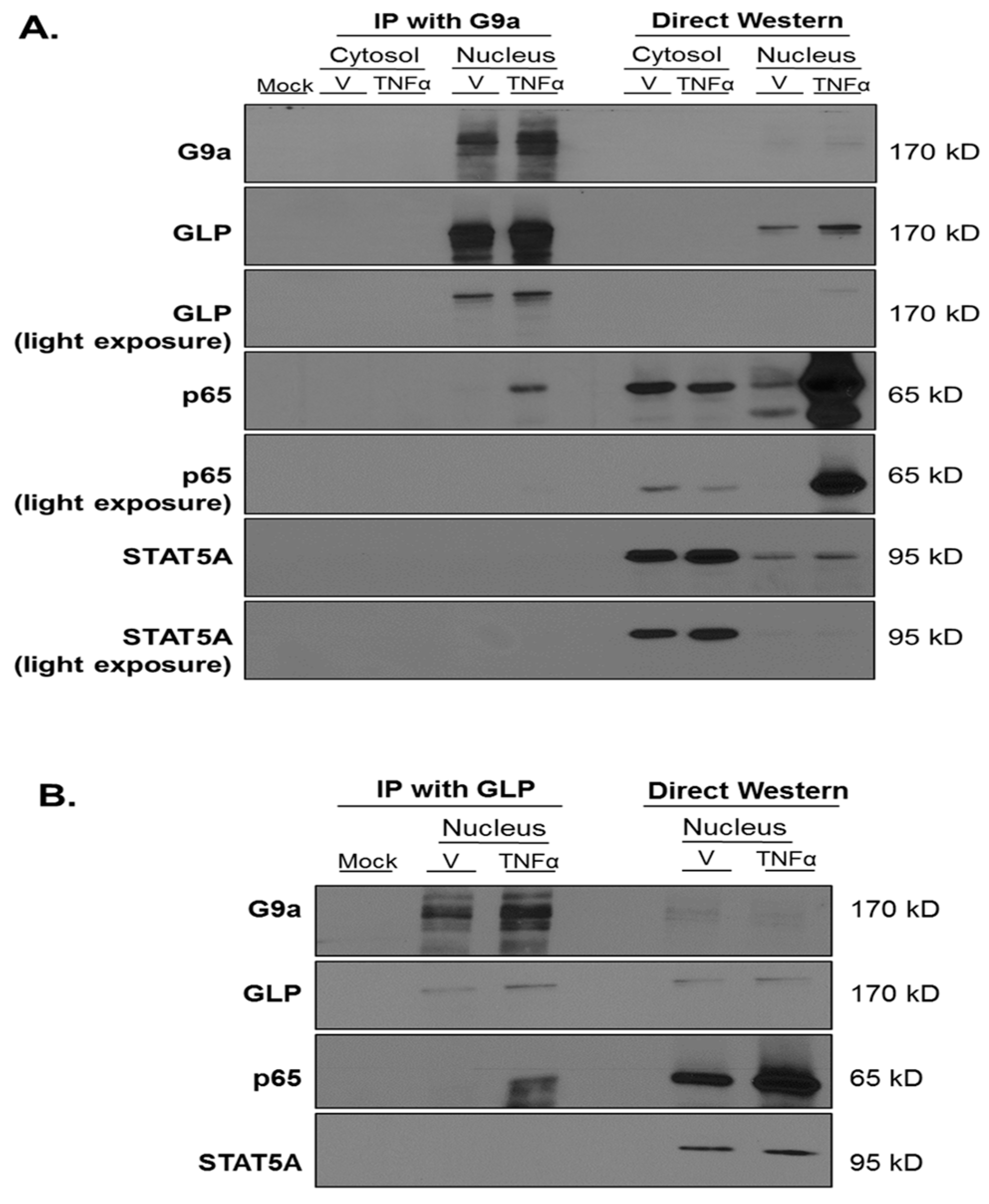
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tachibana, M.; Sugimoto, K.; Nozaki, M.; Ueda, J.; Ohta, T.; Ohki, M.; Fukuda, M.; Takeda, N.; Niida, H.; Kato, H.; et al. G9a Histone Methyltransferase Plays a Dominant Role in Euchromatic Histone H3 Lysine 9 Methylation and Is Essential for Early Embryogenesis. Genes Dev. 2002, 16, 1779–1791. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Zhang, Z.; Wu, H.; Jiang, Y.; Meng, L.; Xiong, J.; Zhao, Z.; Zhou, X.; Li, J.; Li, H.; et al. Recognition of H3K9 Methylation by GLP Is Required for Efficient Establishment of H3K9 Methylation, Rapid Target Gene Repression, and Mouse Viability. Genes Dev. 2015, 29, 379–393. [Google Scholar] [CrossRef] [PubMed]
- Xue, W.; Huang, J.; Chen, H.; Zhang, Y.; Zhu, X.; Li, J.; Zhang, W.; Yuan, Y.; Wang, Y.; Zheng, L.; et al. Histone Methyltransferase G9a Modulates Hepatic Insulin Signaling via Regulating HMGA1. Biochim. Biophys. Acta-Mol. Basis Dis. 2018, 1864, 338–346. [Google Scholar] [CrossRef]
- Huang, J.; Dorsey, J.; Chuikov, S.; Pérez-Burgos, L.; Zhang, X.; Jenuwein, T.; Reinberg, D.; Berger, S.L. G9a and Glp Methylate Lysine 373 in the Tumor Suppressor P53. J. Biol. Chem. 2010, 285, 9636–9641. [Google Scholar] [CrossRef] [PubMed]
- Rathert, P.; Dhayalan, A.; Murakami, M.; Zhang, X.; Tamas, R.; Jurkowska, R.; Komatsu, Y.; Shinkai, Y.; Cheng, X.; Jeltsch, A. Protein Lysine Methyltransferase G9a Acts on Non-Histone Targets. Nat. Chem. Biol. 2008, 4, 344–346. [Google Scholar] [CrossRef]
- Lee, D.Y.; Northrop, J.P.; Kuo, M.-H.; Stallcup, M.R. Histone H3 Lysine 9 Methyltransferase G9a Is a Transcriptional Coactivator for Nuclear Receptors. J. Biol. Chem. 2006, 281, 8476–8485. [Google Scholar] [CrossRef] [PubMed]
- Rada, M.; Vasileva, E.; Lezina, L.; Marouco, D.; Antonov, A.V.; Macip, S.; Melino, G.; Barlev, N.A. Human EHMT2/G9a Activates P53 through Methylation-Independent Mechanism. Oncogene 2017, 36, 922–932. [Google Scholar] [CrossRef] [PubMed]
- Purcell, D.J.; Jeong, K.W.; Bittencourt, D.; Gerke, D.S.; Stallcup, M.R. A Distinct Mechanism for Coactivator versus Corepressor Function by Histone Methyltransferase G9a in Transcriptional Regulation. J. Biol. Chem. 2011, 286, 41963–41971. [Google Scholar] [CrossRef]
- Chaturvedi, C.-P.; Hosey, A.M.; Palii, C.; Perez-Iratxeta, C.; Nakatani, Y.; Ranish, J.A.; Dilworth, F.J.; Brand, M. Dual Role for the Methyltransferase G9a in the Maintenance of -Globin Gene Transcription in Adult Erythroid Cells. Proc. Natl. Acad. Sci. USA 2009, 106, 18303–18308. [Google Scholar] [CrossRef]
- Demond, H.; Hanna, C.W.; Castillo-Fernandez, J.; Santos, F.; Papachristou, E.K.; Segonds-Pichon, A.; Kishore, K.; Andrews, S.; D’Santos, C.S.; Kelsey, G. Multi-Omics Analyses Demonstrate a Critical Role for EHMT1 Methyltransferase in Transcriptional Repression during Oogenesis. Genome Res. 2023, 33, 18–31. [Google Scholar] [CrossRef]
- Nguekeu-Zebaze, L.; Hanini, N.; Noll, A.; Wadier, N.; Amé, J.C.; Roegel, L.; Dantzer, F. PARP3 Supervises G9a-Mediated Repression of Adhesion and Hypoxia-Responsive Genes in Glioblastoma Cells. Sci. Rep. 2022, 12, 15534. [Google Scholar] [CrossRef] [PubMed]
- Vinson, D.A.; Stephens, K.E.; O’Meally, R.N.; Bhat, S.; Dancy, B.C.R.; Cole, R.N.; Yegnasubramanian, S.; Taverna, S.D. De Novo Methylation of Histone H3K23 by the Methyltransferases EHMT1/GLP and EHMT2/G9a. Epigenetics Chromatin 2022, 15, 36. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Xu, S.; Lee, J.-E.; Baldridge, A.; Grullon, S.; Peng, W.; Ge, K. Histone H3K9 Methyltransferase G9a Represses PPARγ Expression and Adipogenesis. EMBO J. 2012, 32, 45–59. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.-C.; Liu, Y.; Li, S.-F.; Guo, L.; Zhao, Y.; Qian, S.-W.; Wen, B.; Tang, Q.-Q.; Li, X. Suv39h1 Mediates AP-2 -Dependent Inhibition of C/EBP Expression during Adipogenesis. Mol. Cell. Biol. 2014, 34, 2330–2338. [Google Scholar] [CrossRef]
- Li, S.-F.; Guo, L.; Qian, S.-W.; Liu, Y.; Zhang, Y.-Y.; Zhang, Z.-C.; Zhao, Y.; Shou, J.-Y.; Tang, Q.-Q.; Li, X. G9a Is Transactivated by C/EBPβ to Facilitate Mitotic Clonal Expansion during 3T3-L1 Preadipocyte Differentiation. Am. J. Physiol. Metab. 2013, 304, E990–E998. [Google Scholar] [CrossRef] [PubMed]
- Ohno, H.; Shinoda, K.; Ohyama, K.; Sharp, L.Z.; Kajimura, S. EHMT1 Controls Brown Adipose Cell Fate and Thermogenesis through the PRDM16 Complex. Nature 2013, 504, 163–167. [Google Scholar] [CrossRef] [PubMed]
- Ling, C.; Rönn, T. Epigenetics in Human Obesity and Type 2 Diabetes. Cell Metab. 2019, 29, 1028–1044. [Google Scholar] [CrossRef]
- Hotamisligil, G.S.; Arner, P.; Caro, J.F.; Atkinson, R.L.; Spiegelman, B.M. Increased Adipose Tissue Expression of Tumor Necrosis Factor-Alpha in Human Obesity and Insulin Resistance. J. Clin. Invest. 1995, 95, 2409–2415. [Google Scholar] [CrossRef]
- Cawthorn, W.P.; Sethi, J.K. TNF-Alpha and Adipocyte Biology. FEBS Lett. 2008, 582, 117–131. [Google Scholar] [CrossRef]
- Libermann, T.A.; Baltimore, D. Activation of Interleukin-6 Gene Expression through the NF-Kappa B Transcription Factor. Mol. Cell. Biol. 1990, 10, 2327–2334. [Google Scholar] [CrossRef]
- Ping, D.; Boekhoudt, G.H.; Rogers, E.M.; Boss, J.M. Nuclear Factor-Kappa B P65 Mediates the Assembly and Activation of the TNF-Responsive Element of the Murine Monocyte Chemoattractant-1 Gene. J. Immunol. 1999, 162, 727–734. [Google Scholar] [CrossRef] [PubMed]
- Ueda, A.; Ishigatsubo, Y.; Okubo, T.; Yoshimura, T. Transcriptional Regulation of the Human Monocyte Chemoattractant Protein-1 Gene. Cooperation of Two NF-KappaB Sites and NF-KappaB/Rel Subunit Specificity. J. Biol. Chem. 1997, 272, 31092–31099. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, B.J.; Griesel, B.A.; King, C.D.; Josey, M.A.; Olson, A.L. Moderate GLUT4 Overexpression Improves Insulin Sensitivity and Fasting Triglyceridemia in High-Fat Diet-Fed Transgenic Mice. Diabetes 2013, 62, 2249–2258. [Google Scholar] [CrossRef]
- He, Y.; Lu, L.; Wei, X.; Jin, D.; Qian, T.; Yu, A.; Sun, J.; Cui, J.; Yang, Z. The Multimerization and Secretion of Adiponectin Are Regulated by TNF-Alpha. Endocrine 2016, 51, 456–468. [Google Scholar] [CrossRef] [PubMed]
- Stephens, J.M.; Lee, J.; Pilch, P.F. Tumor Necrosis Factor-Alpha-Induced Insulin Resistance in 3T3-L1 Adipocytes Is Accompanied by a Loss of Insulin Receptor Substrate-1 and GLUT4 Expression without a Loss of Insulin Receptor-Mediated Signal Transduction. J. Biol. Chem. 1997, 272, 971–976. [Google Scholar] [CrossRef]
- Able, A.A. Characterizing the Roles of Nuclear Proteins DBC1, G9A, and GLP in Adipocyte Function and Metabolic Health. Ph.D. Dissertation, Louisiana State University, Baton Rouge, LA, USA, 2019. [Google Scholar] [CrossRef]
- Richard, A.J.; Hang, H.; Stephens, J.M. Pyruvate Dehydrogenase Complex (PDC) Subunits Moonlight as Interaction Partners of Phosphorylated STAT5 in Adipocytes and Adipose Tissue. J. Biol. Chem. 2017, 292, 19733–19742. [Google Scholar] [CrossRef] [PubMed]
- Able, A.A.; Richard, A.J.; Stephens, J.M. Loss of DBC1 (CCAR2) Affects TNFα-Induced Lipolysis and Glut4 Gene Expression in Murine Adipocytes. J. Mol. Endocrinol. 2018, 61, 195. [Google Scholar] [CrossRef]
- Tachibana, M.; Ueda, J.; Fukuda, M.; Takeda, N.; Ohta, T.; Iwanari, H.; Sakihama, T.; Kodama, T.; Hamakubo, T.; Shinkai, Y. Histone Methyltransferases G9a and GLP Form Heteromeric Complexes and Are Both Crucial for Methylation of Euchromatin at H3-K9. Genes Dev. 2005, 19, 815–826. [Google Scholar] [CrossRef]
- Barski, A.; Cuddapah, S.; Cui, K.; Roh, T.-Y.; Schones, D.E.; Wang, Z.; Wei, G.; Chepelev, I.; Zhao, K. High-Resolution Profiling of Histone Methylations in the Human Genome. Cell 2007, 129, 823–837. [Google Scholar] [CrossRef]
- Antignano, F.; Burrows, K.; Hughes, M.R.; Han, J.M.; Kron, K.J.; Penrod, N.M.; Oudhoff, M.J.; Wang, S.K.H.; Min, P.H.; Gold, M.J.; et al. Methyltransferase G9A Regulates T Cell Differentiation during Murine Intestinal Inflammation. J. Clin. Investig. 2014, 124, 1945. [Google Scholar] [CrossRef]
- Kralisch, S.; Klein, J.; Lossner, U.; Bluher, M.; Paschke, R.; Stumvoll, M.; Fasshauer, M. Isoproterenol, TNFα, and Insulin Downregulate Adipose Triglyceride Lipase in 3T3-L1 Adipocytes. Mol. Cell. Endocrinol. 2005, 240, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Rydén, M.; Arvidsson, E.; Blomqvist, L.; Perbeck, L.; Dicker, A.; Arner, P. Targets for TNF-α-Induced Lipolysis in Human Adipocytes. Biochem. Biophys. Res. Commun. 2004, 318, 168–175. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Elks, C.M.; Stephens, J.M. The Induction of Lipocalin-2 Protein Expression in Vivo and in Vitro. J. Biol. Chem. 2014, 289, 5960–5969. [Google Scholar] [CrossRef]
- Floyd, Z.E.; Segura, B.M.; He, F.; Stephens, J.M. Degradation of STAT5 Proteins in 3T3-L1 Adipocytes Is Induced by TNF-α and Cycloheximide in a Manner Independent of STAT5A Activation. Am. J. Physiol. Metab. 2007, 292, E461–E468. [Google Scholar] [CrossRef]
- Tachibana, M.; Matsumura, Y.; Fukuda, M.; Kimura, H.; Shinkai, Y. G9a/GLP Complexes Independently Mediate H3K9 and DNA Methylation to Silence Transcription. EMBO J. 2008, 27, 2681–2690. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, A.; Sampath, S.C.; Intrator, A.; Min, A.; Gertler, T.S.; Surmeier, D.J.; Tarakhovsky, A.; Greengard, P. Control of Cognition and Adaptive Behavior by the GLP/G9a Epigenetic Suppressor Complex. Neuron 2009, 64, 678–691. [Google Scholar] [CrossRef]
- Inagawa, M.; Nakajima, K.; Makino, T.; Ogawa, S.; Kojima, M.; Ito, S.; Ikenishi, A.; Hayashi, T.; Schwartz, R.J.; Nakamura, K.; et al. Histone H3 Lysine 9 Methyltransferases, G9a and GLP Are Essential for Cardiac Morphogenesis. Mech. Dev. 2013, 130, 519–531. [Google Scholar] [CrossRef]
- Zhang, Y.; Xue, W.; Zhang, W.; Yuan, Y.; Zhu, X.; Wang, Q.; Wei, Y.; Yang, D.; Yang, C.; Chen, Y.; et al. Histone Methyltransferase G9a Protects against Acute Liver Injury through GSTP1. Cell Death Differ. 2019, 27, 1243–1258. [Google Scholar] [CrossRef]
- Peters, A.H.F.M.; Kubicek, S.; Mechtler, K.; O’Sullivan, R.J.; Derijck, A.A.H.A.; Perez-Burgos, L.; Kohlmaier, A.; Opravil, S.; Tachibana, M.; Shinkai, Y.; et al. Partitioning and Plasticity of Repressive Histone Methylation States in Mammalian Chromatin. Mol. Cell 2003, 12, 1577–1589. [Google Scholar] [CrossRef]
- Rea, S.; Eisenhaber, F.; O’Carroll, D.; Strahl, B.D.; Sun, Z.-W.; Schmid, M.; Opravil, S.; Mechtler, K.; Ponting, C.P.; Allis, C.D.; et al. Regulation of Chromatin Structure by Site-Specific Histone H3 Methyltransferases. Nature 2000, 406, 593–599. [Google Scholar] [CrossRef]
- Wang, H.; An, W.; Cao, R.; Xia, L.; Erdjument-Bromage, H.; Chatton, B.; Tempst, P.; Roeder, R.G.; Zhang, Y. MAM Facilitates Conversion by ESET of Dimethyl to Trimethyl Lysine 9 of Histone H3 to Cause Transcriptional Repression. Mol. Cell 2003, 12, 475–487. [Google Scholar] [CrossRef] [PubMed]
- Fritsch, L.; Robin, P.; Mathieu, J.R.R.; Souidi, M.; Hinaux, H.; Rougeulle, C.; Harel-Bellan, A.; Ameyar-Zazoua, M.; Ait-Si-Ali, S. A Subset of the Histone H3 Lysine 9 Methyltransferases Suv39h1, G9a, GLP, and SETDB1 Participate in a Multimeric Complex. Mol. Cell 2010, 37, 46–56. [Google Scholar] [CrossRef] [PubMed]
- Fasshauer, M.; Klein, J.; Neumann, S.; Eszlinger, M.; Paschke, R. Hormonal Regulation of Adiponectin Gene Expression in 3T3-L1 Adipocytes. Biochem. Biophys. Res. Commun. 2002, 290, 1084–1089. [Google Scholar] [CrossRef]
- Degawa-Yamauchi, M.; Moss, K.A.; Bovenkerk, J.E.; Shankar, S.S.; Morrison, C.L.; Lelliott, C.J.; Vidal-Puig, A.; Jones, R.; Considine, R.V. Regulation of Adiponectin Expression in Human Adipocytes: Effects of Adiposity, Glucocorticoids, and Tumor Necrosis Factor α. Obes. Res. 2005, 13, 662–669. [Google Scholar] [CrossRef] [PubMed]
- Cornett, E.M.; Ferry, L.; Defossez, P.A.; Rothbart, S.B. Lysine Methylation Regulators Moonlighting Outside the Epigenome. Mol. Cell 2019, 75, 1092. [Google Scholar] [CrossRef]
- Carvalho, E.; Kotani, K.; Peroni, O.D.; Kahn, B.B. Adipose-Specific Overexpression of GLUT4 Reverses Insulin Resistance and Diabetes in Mice Lacking GLUT4 Selectively in Muscle. Am. J. Physiol. Metab. 2005, 289, E551–E561. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Luo, N.; Klein, R.L.; Garvey, W.T. Adiponectin Promotes Adipocyte Differentiation, Insulin Sensitivity, and Lipid Accumulation. J. Lipid Res. 2005, 46, 1369–1379. [Google Scholar] [CrossRef]
- Ea, C.-K.; Hao, S.; Yeo, K.S.; Baltimore, D. EHMT1 Protein Binds to Nuclear Factor-ΚB P50 and Represses Gene Expression. J. Biol. Chem. 2012, 287, 31207. [Google Scholar] [CrossRef]
- Liu, C.; Yu, Y.; Liu, F.; Wei, X.; Wrobel, J.A.; Gunawardena, H.P.; Zhou, L.; Jin, J.; Chen, X. A Chromatin Activity-Based Chemoproteomic Approach Reveals a Transcriptional Repressome for Gene-Specific Silencing. Nat. Commun. 2014, 5, 5733. [Google Scholar] [CrossRef]
- Chen, X.; El Gazzar, M.; Yoza, B.K.; McCall, C.E. The NF-ΚB Factor RelB and Histone H3 Lysine Methyltransferase G9a Directly Interact to Generate Epigenetic Silencing in Endotoxin Tolerance. J. Biol. Chem. 2009, 284, 27857–27865. [Google Scholar] [CrossRef]
- Zhang, J.; Wu, Y.; Zhang, Y.; Leroith, D.; Bernlohr, D.A.; Chen, X. The Role of Lipocalin 2 in the Regulation of Inflammation in Adipocytes and Macrophages. Mol. Endocrinol. 2008, 22, 1416–1426. [Google Scholar] [CrossRef]
- Yan, Q.-W.; Yang, Q.; Mody, N.; Graham, T.E.; Hsu, C.-H.; Xu, Z.; Houstis, N.E.; Kahn, B.B.; Rosen, E.D. The Adipokine Lipocalin 2 Is Regulated by Obesity and Promotes Insulin Resistance. Diabetes 2007, 56, 2533–2540. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.; Stark, G.R. NF-ΚB: Regulation by Methylation. Cancer Res. 2015, 75, 3692–3695. [Google Scholar] [CrossRef] [PubMed]
- Laurencikiene, J.; Van Harmelen, V.; Nordström, E.A.; Dicker, A.; Blomqvist, L.; Näslund, E.; Langin, D.; Arner, P.; Rydén, M. NF-KB Is Important for TNF-a-Induced Lipolysis in Human Adipocytes. J. Lipid Res 2007, 48, 1069–1077. [Google Scholar] [CrossRef] [PubMed]
- Arner, P.; Rydén, M. Fatty Acids, Obesity and Insulin Resistance. Obes. Facts 2015, 8, 147–155. [Google Scholar] [CrossRef]
- Frayn, K.N.; Arner, P.; Yki-Järvinen, H. Fatty Acid Metabolism in Adipose Tissue, Muscle and Liver in Health and Disease. Essays Biochem. 2006, 42, 89–103. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Zhang, X.; Heckmann, B.L.; Lu, X.; Liu, J. Relative Contribution of Adipose Triglyceride Lipase and Hormone-Sensitive Lipase to Tumor Necrosis Factor-α (TNF-α)-Induced Lipolysis in Adipocytes. J. Biol. Chem. 2011, 286, 40477–40485. [Google Scholar] [CrossRef] [PubMed]
| Gene; Abbreviation (GenBank Accession No.) | Primer 1 Sequence | Primer 2 Sequence |
|---|---|---|
| Cyclophilin A; PpiA (NM_008907) | 5′-CCACTGTCGCTTTTCGCCGC-3 | 5′-TGCAAACAGCTCGAAGGAGACGC-3′ |
| Cyclophilin B; PpiB (NM_0011149) | 5′-CCGTAGTGCTTCAGCTTGA-3′ | 5′-AGCAAGTTCCATCGTGTCATC-3′ |
| Ubiquitin B; Ubb (NM011664) | 5′-GCTTACCATGCAACAAAACCT-3′ | 5′-CCAGTGGGCAGTGATGG-3′ |
| Non-POU domain containing octamer binding; Nono (NM_001252518) | 5′-TCTTCAGGTCAATAGTCAAGCC-3′ | 5′-CATCATCAGCA TCACCACCA-3′ |
| G9a/Ehmt2 (NM_001286573) | 5′-TCCTCCTCACTCAACTGTTCA-3′ | 5′-CGATGACTTCAGCCTGT ACTATG-3′ |
| Glp/Ehmt1 (NM_001012518) | 5′-TCCATCAACCAGCATGAGAAG-3′ | 5′-GTGCTCTAATCGCTCTAGACTC-3′ |
| suadAdiponectin (NM_009605) | 5′-GCAGGA TTAAGAGGAACAGGAG-3′ | 5′-TGTCTGTACGATTGTCAGTGG-3′ |
| Lipocalin 2; Lcn2 (NM_008491) | 5′-AGTCACATTCGTTGCAGAAGA-3′ | 5′-CAGAGATGTGCCTCCA TACTG-3′ |
| Interleukin 6; Il6 (NM_031168) | 5′-AGTACATCTCCAGTCTCCTCAG-3′ | 5′-ATGCTCTTCAGTTCGTGTGT-3′ |
| Monocyte chemoattractant protein 1; Mcp1/Ccl2 (NM_011333) | 5′-GCAGAGAGCCAGACGGGAGGA-3′ | 5′-TGGGGCGTTAACTGCATCTGG-3′ |
| Glucose transporter 4; Glut4/Slc2a4 (NM_009204) | 5′-TCTT A TTGCAGCGCCTGAG-3′ | 5′-GAGAATACAGCT AGGACCAGTG-3′ |
| Adipose triglyceride lipase; Atgl/Pnpla2 (NM_001163689) | 5′-CTCATAAAGTGGCAAGTTGTCTG-3′ | 5′-GAGCTCATCCAGGCCAA T-3′ |
| Hormone sensitive lipase; Hsl/Lipe (NM_010719) | 5′-CTCGTTGCGTTIGTAGTGC-3′ | 5′-CTGCAAGAGTATGTCACGCTA-3′ |
| Perilipin 1; Plin1 (NM_175640) | 5′-CGTGGAGAGTAAGGATGTCAATG-3′ | 5′-GTGCTGTTGTAGGTCTTCTGG-3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Able, A.A.; Richard, A.J.; Stephens, J.M. TNFα Effects on Adipocytes Are Influenced by the Presence of Lysine Methyltransferases, G9a (EHMT2) and GLP (EHMT1). Biology 2023, 12, 674. https://doi.org/10.3390/biology12050674
Able AA, Richard AJ, Stephens JM. TNFα Effects on Adipocytes Are Influenced by the Presence of Lysine Methyltransferases, G9a (EHMT2) and GLP (EHMT1). Biology. 2023; 12(5):674. https://doi.org/10.3390/biology12050674
Chicago/Turabian StyleAble, Ashley A., Allison J. Richard, and Jacqueline M. Stephens. 2023. "TNFα Effects on Adipocytes Are Influenced by the Presence of Lysine Methyltransferases, G9a (EHMT2) and GLP (EHMT1)" Biology 12, no. 5: 674. https://doi.org/10.3390/biology12050674
APA StyleAble, A. A., Richard, A. J., & Stephens, J. M. (2023). TNFα Effects on Adipocytes Are Influenced by the Presence of Lysine Methyltransferases, G9a (EHMT2) and GLP (EHMT1). Biology, 12(5), 674. https://doi.org/10.3390/biology12050674






