Abstract
Four colistin susceptibility testing methods were compared with the standard broth microdilution (BMD) in a collection of 75 colistin-susceptible and 75 mcr-positive E. coli, including ST131 isolates. Taking BMD as reference, all methods showed similar categorical agreement rates (CA) of circa 90%, and a low number of very major errors (VME) (0% for the MicroScan system and Etest®, 0.7% for UMIC®), except for the disc diffusion assay (breakpoint ≤ 11 mm), which yielded false-susceptible results for 8% of isolates. Of note is the number of mcr-positive isolates (17.3%) categorized as susceptible (≤2 mg/L) by the BMD method, but as resistant by the MicroScan system. ST131 mcr-positive E. coli were identified as colistin-resistant by all MIC-based methods. Our results show that applying the current clinical cut-off (>2 mg/L), many mcr-positive E. coli remain undetected, while applying a threshold of >1 mg/L the sensitivity of detection increases significantly without loss of specificity. We propose two possible workflows, both starting with the MicroScan system, since it is automated and, importantly, it categorized all mcr-positive isolates as colistin-resistant. MicroScan should be followed by either BMD or MIC-based commercial methods for colistin resistance detection; or, alternatively, MicroScan, followed by PCR for the mcr screening.
1. Introduction
Colistin is one of the so-called last line antimicrobials. Due to the continuous emergence of multidrug resistance among Gram-negative bacteria, the scientific community has renewed interest in polymyxins as a therapeutic option, despite its toxicity, exemplified by the significant increase of it being prescribed in human medicine in the recent years [1,2]. Resistance to polymyxins has been reported in bacterial isolates recovered from humans, animals, food, and the environment [2,3]. Farm animals, poultry, and swine in particular, have been identified as the main reservoir of resistance to these drugs worldwide [2,4,5,6]. Colistin resistance primarily occurs through chromosomal mutations (in the genes encoding two-component systems PmrAB and PhoPQ, or in the MgrB regulator), and/or by plasmid-borne mobile colistin resistance genes (mcr), which have shown a high capacity for dissemination [2]. To date, ten mcr genes (mcr-1 to mcr-10) have been described, mainly in Enterobacteriaceae species [7]. Furthermore, colistin resistance has been strongly linked to the globally mobilized gene mcr-1, presumably due to the over-use of polymyxin in human and veterinary medicine [8]. By contrast, mcr-4 is reported to be widespread in swine and poultry populations, and, in several cases, mcr-4 is found along with mcr-1, mcr-3, or mcr-5 [9]. Similar to the situation in other countries, a high prevalence of mcr-1 and mcr-4 within E. coli isolated from food-producing animals, especially in poultry and pig farming, with mcr-4.5 and mcr-4.2 as the most frequent mcr-4 variants, has been described in Spain [5,6]. García-Meniño et al., (2018) [6] also reported, for the first time, mcr-1 E. coli isolates recovered in pigs belonging to the pandemic lineage ST131.
Due to the difficulties with the performance, reproducibility, and precision of the currently available methods for colistin susceptibility testing [9], evaluation and comparison of these methods on wide collections can help to define more practical algorithms. It is also critical to avoid inaccuracies, especially very major errors (VME), which could lead to therapeutic failures. Currently, both the Clinical and Laboratory Standards Institute (CLSI) and European Committee on Antimicrobial Susceptibility Testing (EUCAST) advise against the use of the disc diffusion method for polymyxin susceptibility testing, and identify the broth microdilution method (BMD) as the only recommended method for this purpose (https://eucast.org/). Other studies have evaluated the efficiency of different susceptibility-testing methodologies for the detection of colistin-resistant isolates, however, most of them were performed on a limited number of isolates and lacked variety of mcr types [10,11,12,13]. In the present study, we analyzed different colistin susceptibility testing methods on a large collection of colistin-susceptible and mcr-positive E. coli of types 1, 2, 4, and 5, including ST131 mcr-positive and negative isolates. The aim was to perform a comprehensive statistical evaluation of four colistin susceptibility testing methods versus the standard BMD, in order to provide a suitable workflow for the accurate detection of colistin-resistant and mcr-positive E. coli.
2. Results
Tables S1 and S2 show the colistin susceptibility results for each isolate and method. The performance of each testing method was represented by receiver operating characteristic (ROC) curves, as shown in Figure 1, except for the MicroScan system, for which the number of dilutions assayed is not enough to build these curves. As a result, the UMIC®, Etest® and disc diffusion displayed good area under curve (AUC) values (0.96 for UMIC® and Etest®, 0.97 for disc diffussion), without significant differences between them.
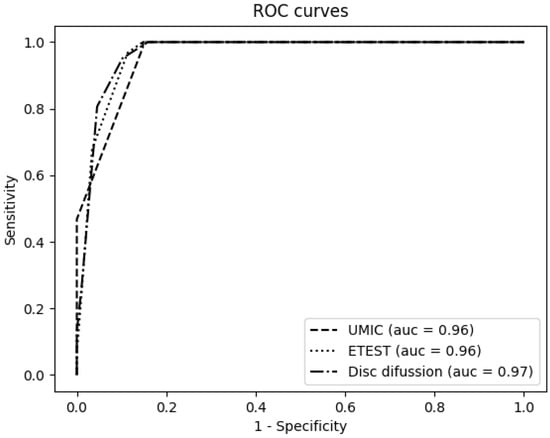
Figure 1.
Receiver operating characteristic (ROC) curves for the Etest®, UMIC® and disc diffusion using the broth microdilution method (BMD) as reference.
However, the results obtained in the lineal regression analysis were varied and did not show good correlation with the reference method (BMD) (Figure 2), being the R-squared values low, especially for the disc diffusion method (0.74).
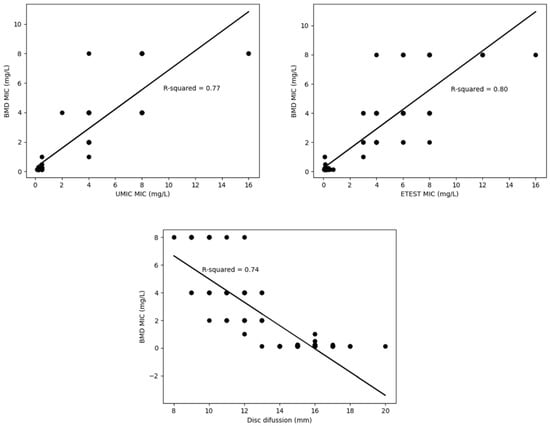
Figure 2.
Correlation of each testing method versus the reference (BMD) represented by regression curves and R-squared.
The essential agreement (EA), categorical agreement (CA), major error (ME), and very major error (VME) for all tests, in comparison with the standard BMD, are summarized in Table 1. Taking BMD as reference, all methods showed similar CA of circa 90%, and a low number of VME (0% for the MicroScan system and Etest®, and 0.7% for UMIC®), except for the disc diffusion assay with the previously recommended breakpoint of ≤11 mm [14], which yielded false-susceptible results for 12 (8%) isolates (Table S1). All methods exceeded the ME rates recommended by CLSI (≤3%) [15], with the exception of the disc diffusion (using the breakpoint of ≤11 mm). However, when applying a breakpoint of ≤13 mm for the disc diffusion method, the CA and the VME values were the same as for the MicroScan system (90.7% and 0%, respectively), although the ME was higher than recommended. The MIC values for mcr-positive isolates obtained by the BMD, UMIC® and Etest® methods are represented in Figure 3.

Table 1.
Results of essential agreement (EA), categorical agreement (CA), major error (ME), very major errors (VMEs), sensitivity and specificity for all tests in comparison with the standard BMD.
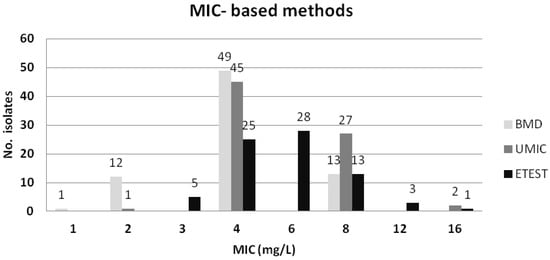
Figure 3.
Colistin MICs for the 75 mcr-positive E. coli evaluated with BMD, UMIC® and Etest®.
It is of note that the standard BMD categorized as susceptible 13 (17.3%) of the 75 mcr-positive isolates according to the EUCAST 2020 [16] and CLSI [17] breakpoints (resistant >2 mg/L), although 12 of those 13 showed a MIC value = 2 mg/L. In contrast, the MicroScan system and Etest® identified all 75 isolates as colistin-resistant, as displayed in Table 1 and Table S1. On the other hand, the standard BMD, and the compared methods, identified as susceptible the 75 mcr-negative control isolates, with the exception of the MicroScan system which displayed a value of 4 mg/L for a single isolate (Table S2). Importantly, all 11 ST131 mcr-1-positive and the 24 ST131 mcr-negative E. coli were identified correctly as colistin-resistant and susceptible, respectively, by the reference BMD method and the other broth dilution methods. A single ST131 mcr-1 isolate was identified as susceptible by the disc diffusion method.
The sensitivity and specificity of the evaluated methods for detection of mcr-carrying E. coli at different thresholds are shown in Figure 4. Applying a cut-off value of 2 mg/L (which is equivalent to point ≥4 in the graphic representation), the sensitivity and specificity of BMD are 82.67% and 100%, respectively; while, using a threshold of 1 mg/L (point ≥2 in the graphic representation), the sensitivity increases until 98.7% and maintains 100% of specificity. For the disc diffusion method, when applying the Gales et al. [14] cut-off value of resistant ≤11 mm, the sensitivity and specificity achieved are of 72% and 100%, respectively; while, using the cut-off value proposed here of resistant ≤13 mm, the sensitivity increases until 100% with a specificity of 98.7%.
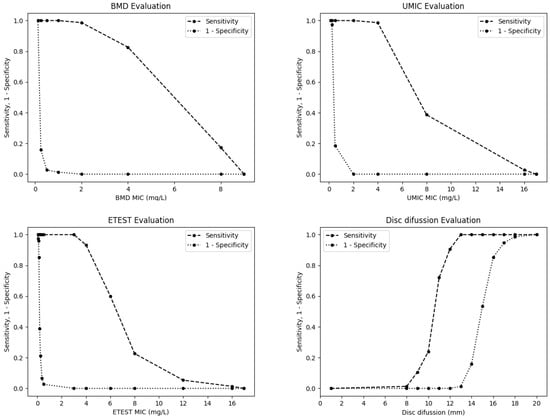
Figure 4.
Sensitivity and specificity of the BMD, UMIC®, Etest® and disc diffusion methods for the detection of mcr-carrying E. coli isolates at different thresholds.
3. Discussion
Despite the increasing use of colistin in human medicine due to the emergence of MDR Gram negative bacteria, susceptibility testing for this antimicrobial is still challenging. Suitable protocols are still needed for the accurate detection of resistant isolates in clinical laboratories. CLSI and EUCAST recommend the BMD as the gold standard for colistin susceptibility testing. Nevertheless, this method is not automated and is very time-consuming, not being available in most hospitals.
As expected, the colistin disc diffusion testing applying the Gales et al. [14] cut-off value of ≤11 mm, showed a poor correlation with BMD (CA of 89.3%) and an unacceptable number of false susceptible isolates, resulting in a significantly high rate of VME (8%). This fact could lead to therapeutic failures, due to the inaccurate administration of this drug. It has been previously reported that agar-based methods perform poorly, being unreliable to measure colistin susceptibility because of its cationic nature and unpredictable diffusion through agar [11]. However, applying the here proposed cut-off value of 13 mm, the VME and ME rates would be 0% and 9.3%, respectively. The results achieved with the broth dilution methods, MicroScan and UMIC®, were significantly more accurate than those obtained with the disc diffusion assay, showing a good correlation with BMD, and with fewer VME (0% for MicroScan and 0.7% for UMIC®). Interestingly, very similar CA and VME rates were observed with the Etest® (91.3% and 0%, respectively).
Currently, neither CLSI nor EUCAST recommend the disc-diffusion method and therefore, there is no reference of zone diameter breakpoints. However, this method has been used as a screening test for years, and it is still widely performed in many laboratories. Regarding gradient diffusion methods, their efficacy is controversial for colistin susceptibility testing, although some studies have validated and recommended them as accurate alternative tests, demonstrating good concordances with the reference method [18,19]. In contrast, other authors have highlighted inaccuracies in the gradient diffusion methods, including high rates of VME: 53.6% [10] and 12.0% [20]. Furthermore, depending on the study, it has been claimed that the Etest® overestimates or underestimates MICs [11,19]. In the present study, we did not detect any VME for the Etest®, and the most outstanding fact for both Etest® and MicroScan is that they were able to detect all mcr-positive isolates. In contrast, the BMD method did not identify resistance in 17.3% of the isolates. In agreement with our observation, other authors also reported favorably on the use of MicroScan or other automated systems, such as the BD Phoenix for the screening of mcr-positive E. coli isolates [10,13,19,21], which are currently of high concern. Furthermore, different commercial and user-friendly broth dilution based tests have been assessed (comASp®, colistin MAc test®, Sensititre, Sensitest, MICRONAUT, UMIC®) with promising and reproducible results that correlated well with BMD [11,13].
Regardless of the parameters discussed above, it is important to bear in mind that the regression curves built for the Etest®, UMIC® and the disc diffusion showed low R-squared values, which means that the exact MIC obtained by the reference method cannot be extrapolated from the results obtained by any of these methods, since data are varied with no linear relationship between them.
It is outstanding, as mentioned above, that 13 mcr-carrying isolates were assigned as susceptible by the BMD reference method, although 12 of them showed MIC values close to the breakpoint (2 mg/L). Most studies have reported MIC values to polymyxins of ≥2 mg/L for isolates carrying mcr genes [10,11,12,13,21]. However, the presence of mcr genes does not mean phenotypical expression of colistin resistance or high MIC levels. In fact, mcr-1-positive E. coli isolates usually display a low-level of colistin resistance (2–8 mg/L) [2,3,22]. As an example, 53 (66.3%) of the 80 mcr-1 isolates which were collected from fecal samples of turkeys, chickens, pigs, and cattle in Poland exhibited a colistin MIC of 2 mg/L [23]. Similarly, (17/26; 65.4%) mcr-1-positive E. coli isolates of human clinical origin in Brazil showed a polymyxin B MIC of 2 mg/L [24]. It is worth highlighting that the 11 mcr-1-positive E. coli of the pandemic clone ST131 of our study, were identified as colistin-resistant by the reference method BMD, and all but one also tested as resistant by the disc diffusion method. The acquisition of mcr-1 by this specific clone means a summative risk with respect to treatment failure due to its special feature of congregating virulence, resistance traits, and its high dissemination capability. Since ST131 is implicated in many community- and hospital-acquired urinary tract infections (UTIs) [25], the accurate identification of colistin-resistance in this high-risk lineage is therefore critical.
Low levels of colistin resistance, or even no phenotypic expression, have been observed for other mcr genes, such as mcr-3 and mcr-4 [26]. Although the clinical implications for the polymyxin therapy against the mcr-positive isolates with low MICs remain unclear, it is important to take into account that the mcr-1 gene can facilitate the selection of chromosomal mutations, which eventually leads to high-level of colistin resistance [22]. We underline here that, using the current cut-off values (>2 mg/L), many of the mcr positive isolates that might contribute to the silent dissemination of these genes would remain undetected. On the contrary, if a cut-off value of >1 mg/L is applied, 98.7% of mcr-positive isolates are detected without loss of specificity.
We did not observe differences in MIC values between the 25 mcr-positive genomes with no known chromosomal mutations, and the 16 that had a summative presence of mcr genes and chromosomal mutations in the pmrA (pmrA S39I, 2 isolates) or pmrB (pmrB V161G, 14 isolates) (Table S1). Similarly, a previous study did not found any summative effect in one mcr-1 positive E. coli isolate and carrier of a chromosomal mutation pmrB V161G (MIC = 2 mg/L) [23].
We are conscious that this study has the limitation of being mostly descriptive. Further investigation is needed on the internal molecular mechanisms of mcr-positive isolates to fully understand their phenotype expression of colistin resistance. However, we have to point out in comparison to previous studies, the high number of mcr-positive and negative isolates analyzed here (75 each), including a wide variety of mcr genes (mcr-1, mcr-2, mcr-4, and mcr-5), as well as the comprehensive statistical analysis performed.
4. Materials and Methods
A set of 150 E. coli was chosen from a collection of isolates recovered from food (meat), food-producing animals (pigs, chicken, and cattle), and wildlife in the period 2006–2020 [4,5,6,27,28]. The selection included 75 colistin-susceptible and 75 mcr-positive isolates carrying different mcr types: mcr-1 (43 isolates), mcr-2 (1 isolate), mcr-4 (mcr-4.1, 1 isolate; mcr-4.2, 17 isolates; and mcr-4.5, 6 isolates), mcr-5 (2 isolates), mcr-1/mcr-4 (3 isolates), and mcr-4/mcr-5 (2 isolates) (Table S1).
The accuracy of antimicrobial susceptibility tests (ASTs) was measured using categorical agreement (CA) and essential agreement (EA). CA is the total number of isolates tested using the ASTs that yields a MIC result, and the same categorical interpretation as the BMD MIC result (used as reference). CA discrepancies were subdivided into two types of errors: major error (ME), defined as those isolates found to be resistant using one of the alternative methods but categorized as susceptible by the reference BMD method; and very major error (VME), defined as those isolates found to be susceptible using one of the alternative methods but categorized as resistant by the BMD method. EA is defined as obtaining a MIC value with the evaluated AST that is within 1 log2 dilution of the reference BMD MIC value, and it is applicable only to methods that determine MIC values [29]. In the present study, the CA, EA, ME, and VME of four different methods (disc diffusion, Etest®, bioMérieux, Marcy l’Etoile, France; UMIC®, Biocentric, Bandol, France; and MicroScan WalkAway System, Beckman coulter, Inc., Brea, CA, USA) were calculated for colistin susceptibility testing in comparison with the gold standard BMD method. The diffusion and MIC-based methods (the manual Etest® and UMIC®; and the automated MicroScan system) were performed according to the manufacturer’s recommendations, all on the same day, and from the same bacterial inoculums adjusted to the standard 0.5 McFarland. BMD was performed according to ISO standard 20776-1. In brief, 96-well polystyrene plates were filled with cation-adjusted Mueller-Hinton broth (Sigma-Aldrich®, Merck, Germany) with colistin concentrations adjusted (Sigma-Aldrich®, Merck, Germany) and ranging from 0.125 to 128 mg/L, and then inoculated with a bacterial concentration of ~106 CFU/mL. The results obtained were read and interpreted after 18–24 h of incubation, according to the colistin clinical cut-off values established by EUCAST [16], which considers MICs >2 mg/L as resistant. Following the recommendations of EUCAST, a drug-susceptible E. coli strain (ATCC 25922) was included as a quality-control for all the assays (MIC values for colistin = 0.5–1). The disc diffusion susceptibility testing was performed using colistin discs of 10 µg (BIO-RAD). The zone inhibition diameters were interpreted according to Gales et al. [14] (resistant ≤ 11 mm), and according to the new criteria proposed in this paper (resistant ≤ 13 mm), since there is not a standardized cut-off value for colistin in either the EUCAST or the CLSI guidelines.
In order to perform a comprehensive statistical evaluation, receiver operating characteristic (ROC) curves were built to visualize the performance of each testing method, showing the diagnostic ability to identify colistin resistance and their discrimination thresholds. BMD results were used as the sample’s result reference. True positive rate (sensitivity) versus false positive rate (1-specificity) was plotted in the ROC curves, together with the area under the curve (AUC) for each testing method. AUC represents a measure of separability, which shows how much a testing method is capable of distinguishing between classes (colistin-resistant versus colistin-susceptible), and the best method is the one with the highest AUC. Additionally, in order to identify the optimal cut-off value of each testing method to discriminate mcr-carrying isolates, a figure showing the evolution of sensitivity and 1-specificity in relation to the threshold changes was plot for each testing method. Finally, regression curves were built to measure the correlation of each testing method versus the reference (BMD) using R-squared (coefficient of determination) and showing the variability in the samples. All these statistical tests were performed using Python programming language (Python Software Foundation. Python Language Reference, version 3.7. Available at http://www.python.org), scikit-learn library and visualize using matplotlib [30].
5. Conclusions
Our results denote that applying the current clinical cut-off (>2 mg/L), many mcr-positive E. coli remain undetected, while applying a threshold of >1mg/L, the sensitivity of detection increases significantly, without loss of specificity. For this reason, we propose two possible workflows, both starting with the MicroScan system, since it is automated, available in numerous clinical microbiology laboratories, and, importantly, it categorized all 75 mcr-positive as colistin-resistant. MicroScan should be followed by either BMD or broth dilution commercial methods for colistin resistance detection; or, alternatively, MicroScan, followed by PCR for the mcr screening.
Supplementary Materials
The following are available online at https://www.mdpi.com/2079-6382/9/12/861/s1, Table S1: Results of colistin susceptibility for 75 mcr-positive E. coli isolates using BMD, UMIC, MicroScan, gradient diffusion strip (Etest®) and disc diffusion. Table S2: Results of colistin susceptibility for 75 mcr-negative E. coli isolates using BMD, UMIC, MicroScan, gradient diffusion strip (Etest®) and disc diffusion.
Author Contributions
Conceptualization, J.F. and A.M.; methodology, J.F., A.M. and I.G.-M.; formal analysis, I.G.-M., P.L., P.V., D.D.-J. and L.L.; investigation, J.F., A.M., I.G.-M., P.L. and D.D.-J.; writing—original draft preparation, I.G.-M.; writing—review and editing, J.F., A.M., I.G.-M. and P.V.; visualization, J.F., A.M., I.G.-M., P.V., P.L., D.D.-J. and L.L.; supervision, J.F. and A.M.; funding acquisition, J.F. and A.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded through: Project FIS PI17-00728 (Fondo de Investigación Sanitaria, Instituto de Salud Carlos III, Ministerio de Economía y Competitividad, Spain), cofunded by the European Regional Development Fund of the European Union: a Way to Making Europe (FEDER); Projects AGL2016-79343-R, PID2019-104439RB-C21/AEI/10.13039/501100011033 from the Agencia Estatal de Investigación and FEDER; ED431C 2017/57 from the Consellería de Cultura, Educación e Ordenación Universitaria, Xunta de Galicia and FEDER; and by the Strategic Researcher Cluster BioReDeS funded by the Regional Government Xunta de Galicia under the project no. ED431E 2018/09.
Acknowledgments
D.D.-J. and I.G.-M. acknowledge support of the Consellería de Cultura, Educación e Ordenación Universitaria, Xunta de Galicia for their pre-doctoral grants (ED481A-2019/022 and ED481A-2015/149, respectively). L.L. acknowledges the Ministry of Education of Spain for her pre-doctoral grant FPU19/01127.The Research stay of I.G.-M at the Hospital Universitario Central de Asturias was funded by a grant from the Sociedad Española de Enfermedades Infecciosas y Microbiología Clínica (SEIMC).
Conflicts of Interest
The authors declare no conflict of interest.
References
- European Centre for Disease Prevention and Control. Antimicrobial Consumption in the EU/EEA, Annual Epidemiological Report for 2018; European Centre for Disease Prevention and Control: Solna, Sweden, 2019.
- Poirel, L.; Jayol, A.; Nordmann, P. Polymyxins: Antibacterial Activity, Susceptibility Testing, and Resistance Mechanisms Encoded by Plasmids or Chromosomes. Clin. Microbiol. Rev. 2017, 30, 557–596. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-Y.; Wang, Y.; Walsh, T.R.; Yi, L.-X.; Zhang, R.; Spencer, J.D.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef]
- Díaz-Jiménez, D.; García-Meniño, I.; Fernández, J.; García, V.; Mora, A. Chicken and turkey meat: Consumer exposure to multidrug-resistant Enterobacteriaceae including mcr-carriers, uropathogenic E. coli and high-risk lineages such as ST131. Int. J. Food Microbiol. 2020, 331, 108750. [Google Scholar] [CrossRef] [PubMed]
- García, V.; García-Meniño, I.; Mora, A.; Flament-Simon, S.C.; Díaz-Jiménez, D.; Blanco, J.E.; Alonso, M.P.; Blanco, J. Co-occurrence of mcr-1, mcr-4 and mcr-5 genes in multidrug-resistant ST10 Enterotoxigenic and Shiga toxin-producing Escherichia coli in Spain (2006–2017). Int. J. Antimicrob. Agents 2018, 52, 104–108. [Google Scholar] [CrossRef] [PubMed]
- García-Meniño, I.; García, V.; Mora, A.; Díaz-Jiménez, D.; Flament-Simon, S.C.; Alonso, M.P.; Blanco, J.E.; Blanco, M.; Blanco, J. Swine Enteric Colibacillosis in Spain: Pathogenic Potential of mcr-1 ST10 and ST131 E. coli Isolates. Front. Microbiol. 2018, 9, 2659. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Feng, Y.; Liu, L.; Wei, L.; Kang, M.; Zong, Z. Identification of novel mobile colistin resistance gene mcr-10. Emerg. Microbes Infect. 2020, 9, 508–516. [Google Scholar] [CrossRef]
- Wang, R.; Van Dorp, L.; Shaw, L.; Bradley, P.; Wang, Q.; Wang, X.; Jin, L.; Zhang, Q.; Liu, Y.; Rieux, A.; et al. The global distribution and spread of the mobilized colistin resistance gene mcr-1. Nat. Commun. 2018, 9, 1–9. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, J.; Wang, J.; Butaye, P.; Kelly, P.; Li, M.; Yang, F.; Gong, J.; Yassin, A.K.; Guo, W.; et al. Newly identified colistin resistance genes, mcr-4 and mcr-5, from upper and lower alimentary tract of pigs and poultry in China. PLoS ONE 2018, 13, e0193957. [Google Scholar] [CrossRef]
- Lutgring, J.D.; Kim, A.; Campbell, D.; Karlsson, M.; Brown, A.C.; Burd, E.M. Evaluation of the MicroScan Colistin Well and Gradient Diffusion Strips for Colistin Susceptibility Testing in Enterobacteriaceae. J. Clin. Microbiol. 2019, 57, e01866-18. [Google Scholar] [CrossRef]
- Matuschek, E.; Åhman, J.; Webster, C.; Kahlmeter, G. Antimicrobial susceptibility testing of colistin—evaluation of seven commercial MIC products against standard broth microdilution for Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Acinetobacter spp. Clin. Microbiol. Infect. 2018, 24, 865–870. [Google Scholar] [CrossRef]
- Sekyere, J.O.; Sephofane, A.K.; Mbelle, N.M. Comparative Evaluation of CHROMagar COL-APSE, MicroScan Walkaway, ComASP Colistin, and Colistin MAC Test in Detecting Colistin-resistant Gram-Negative Bacteria. Sci. Rep. 2020, 10, 6221. [Google Scholar] [CrossRef] [PubMed]
- Pfennigwerth, N.; Kaminski, A.; Korte-Berwanger, M.; Pfeifer, Y.; Simon, M.; Werner, G.; Jantsch, J.; Marlinghaus, L.; Gatermann, S.G. Evaluation of six commercial products for colistin susceptibility testing in Enterobacterales. Clin. Microbiol. Infect. 2019, 25, 1385–1389. [Google Scholar] [CrossRef] [PubMed]
- Gales, A.C.; Reis, A.O.; Jones, R.N. Contemporary Assessment of Antimicrobial Susceptibility Testing Methods for Polymyxin B and Colistin: Review of Available Interpretative Criteria and Quality Control Guidelines. J. Clin. Microbiol. 2001, 39, 183–190. [Google Scholar] [CrossRef] [PubMed]
- CLSI. Verification of Commercial Microbial Identification and Antimicrobial Susceptibility Testing Systems, 1st ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2015. [Google Scholar]
- EUCAST. The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters. Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_10.0_Breakpoint_Tables.pdf (accessed on 3 August 2020).
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing, 30th ed.; CLSI supplement M100; CLSI: Wayne, PA, USA, 2020; ISBN 978-1-68440-066-9. [Google Scholar]
- Lo-Ten-Foe, J.R.; De Smet, A.M.G.A.; Diederen, B.M.W.; Kluytmans, J.A.J.W.; Van Keulen, P.H.J. Comparative Evaluation of the VITEK 2, Disk Diffusion, Etest, Broth Microdilution, and Agar Dilution Susceptibility Testing Methods for Colistin in Clinical Isolates, Including Heteroresistant Enterobacter cloacae and Acinetobacter baumannii Strains. Antimicrob. Agents Chemother. 2007, 51, 3726–3730. [Google Scholar] [CrossRef] [PubMed]
- Maalej, S.; Meziou, M.; Rhimi, F.; Hammami, A. Comparison of disc diffusion, Etest and agar dilution for susceptibility testing of colistin against Enterobacteriaceae. Lett. Appl. Microbiol. 2011, 53, 546–551. [Google Scholar] [CrossRef]
- Chew, K.L.; La, M.-V.; Lin, R.T.P.; Teo, J.W.P. Colistin and Polymyxin B Susceptibility Testing for Carbapenem-Resistant and mcr -Positive Enterobacteriaceae: Comparison of Sensititre, MicroScan, Vitek 2, and Etest with Broth Microdilution. J. Clin. Microbiol. 2017, 55, 2609–2616. [Google Scholar] [CrossRef]
- Jayol, A.; Nordmann, P.; Lehours, P.; Poirel, L.; Dubois, V. Comparison of methods for detection of plasmid-mediated and chromosomally encoded colistin resistance in Enterobacteriaceae. Clin. Microbiol. Infect. 2018, 24, 175–179. [Google Scholar] [CrossRef]
- Zhang, H.; Zhao, D.-D.; Quan, J.; Hua, X.; Yu, Y. mcr-1 facilitated selection of high-level colistin-resistant mutants in Escherichia coli. Clin. Microbiol. Infect. 2019, 25, 517.e1–517.e4. [Google Scholar] [CrossRef]
- Zając, M.; Sztromwasser, P.; Bortolaia, V.; Leekitcharoenphon, P.; Cavaco, L.M.; Ziȩtek-Barszcz, A.; Hendriksen, R.S.; Wasyl, D. Occurrence and Characterization of mcr-1-Positive Escherichia coli Isolated from Food-Producing Animals in Poland, 2011–2016. Front. Microbiol. 2019, 10, 1753. [Google Scholar] [CrossRef]
- Pillonetto, M.; Mazzetti, A.; Becker, G.N.; Siebra, C.A.; Arend, L.N.; Barth, A.L. Low level of polymyxin resistance among nonclonal mcr-1–positive Escherichia coli from human sources in Brazil. Diagn. Microbiol. Infect. Dis. 2019, 93, 140–142. [Google Scholar] [CrossRef]
- Manges, A.R.; Geum, H.M.; Guo, A.; Edens, T.J.; Fibke, C.D.; Pitout, J.D.D. Global Extraintestinal Pathogenic Escherichia coli (ExPEC) Lineages. Clin. Microbiol. Rev. 2019, 32. [Google Scholar] [CrossRef] [PubMed]
- Teo, J.W.P.; Kalisvar, M.; Venkatachalam, I.; Ng, O.T.; Lin, R.T.P.; Octavia, S. mcr-3 and mcr-4 Variants in Carbapenemase-Producing Clinical Enterobacteriaceae Do Not Confer Phenotypic Polymyxin Resistance. J. Clin. Microbiol. 2017, 56, e01562-17. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Jiménez, D.; García-Meniño, I.; Herrera, A.; Lestón, L.; Mora, A. Microbiological risk assessment of Turkey and chicken meat for consumer: Significant differences regarding multidrug resistance, mcr or presence of hybrid aEPEC/ExPEC pathotypes of E. coli. Food Control. 2020, 2020. [Google Scholar] [CrossRef]
- García-Meniño, I.; Díaz-Jiménez, D.; García, V.; De Toro, M.; Flament-Simon, S.C.; Blanco, J.; Mora, A. Genomic Characterization of Prevalent mcr-1, mcr-4, and mcr-5 Escherichia coli Within Swine Enteric Colibacillosis in Spain. Front. Microbiol. 2019, 10, 2469. [Google Scholar] [CrossRef]
- Humphries, R.M.; Ambler, J.; Mitchell, S.L.; Castanheira, M.; Dingle, T.; Hindler, J.A.; Koeth, L.; Sei, K.; Hardy, D.; Zimmer, B.; et al. CLSI Methods Development and Standardization Working Group Best Practices for Evaluation of Antimicrobial Susceptibility Tests. J. Clin. Microbiol. 2018, 56. [Google Scholar] [CrossRef] [PubMed]
- Hunter, J.D. Matplotlib: A 2D Graphics Environment. Comput. Sci. Eng. 2007, 9, 90–95. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).