Abstract
Early and accurate detection of plant diseases is critical for ensuring global food security and agricultural resilience. Ratoon stunting disease (RSD), caused by the bacterium Leifsonia xyli subsp. xyli (Lxx), is among the most economically significant diseases of sugarcane worldwide. Its cryptic nature—characterized by an absence of visible symptoms—renders timely diagnosis particularly difficult, contributing to substantial undetected yield losses across major sugar-producing regions. Here, we report the development of a potential-induced electrochemical (EC) nanobiosensor platform for the rapid, low-cost, and field-deployable detection of Lxx DNA directly from crude sugarcane sap. This method eliminates the need for conventional nucleic acid extraction and thermal cycling by integrating the following: (i) a boiling lysis-based DNA release from xylem sap; (ii) sequence-specific magnetic bead-based purification of Lxx DNA using immobilized capture probes; and (iii) label-free electrochemical detection using a potential-driven DNA adsorption sensing platform. The biosensor shows exceptional analytical performance, achieving a detection limit of 10 cells/µL with a broad dynamic range spanning from 105 to 1 copy/µL (r = 0.99) and high reproducibility (SD < 5%, n = 3). Field validation using genetically diverse sugarcane cultivars from an inoculated trial demonstrated a strong correlation between biosensor signals and known disease resistance ratings. Quantitative results from the EC biosensor also showed a robust correlation with qPCR data (r = 0.84, n = 10, p < 0.001), confirming diagnostic accuracy. This first-in-class EC nanobiosensor for RSD represents a major technological advance over existing methods by offering a cost-effective, equipment-free, and scalable solution suitable for on-site deployment by non-specialist users. Beyond sugarcane, the modular nature of this detection platform opens up opportunities for multiplexed detection of plant pathogens, making it a transformative tool for early disease surveillance, precision agriculture, and biosecurity monitoring. This work lays the foundation for the development of a universal point-of-care platform for managing plant and crop diseases, supporting sustainable agriculture and global food resilience in the face of climate and pathogen threats.
1. Introduction
Ratoon stunting disease (RSD) remains one of the most insidious and economically damaging diseases affecting sugarcane production globally. First identified in Queensland, Australia, in the mid-1940s in the Q28 sugarcane cultivar [1], RSD can cause significant yield reductions of 12–37% under normal conditions and up to 60% in drought-stressed crops, while also compromising varietal quality [2,3,4]. The causative agent, Leifsonia xyli subsp. xyli (Lxx) is a xylem-limited, slow-growing bacterium [5,6] that causes non-specific physiological symptoms—such as reduced tillering, stalk diameter, and plant height—making RSD visually indistinguishable from drought or poor agronomic practices. This diagnostic ambiguity leads to unchecked propagation of Lxx through infected planting materials and farm machinery, contributing to widespread and persistent disease transmission [7].
The minute size (195–220 nm diameter) and fastidious nature of Lxx present considerable challenges in terms of isolation and culture [8,9]. As a result, various serological [10,11] and molecular detection platforms—including PCR [12,13], multiplex PCR [14], nested PCR [15], real-time PCR [16,17], and loop-mediated isothermal amplification (LAMP) [18]—have been developed. However, these tools remain restricted to centralized laboratories due to their dependence on sophisticated equipment, labor-intensive DNA extraction steps, and reliance on commercial kits or complex chemical reagents [18,19,20]. Diagnostic delays, logistical barriers, and high operational costs continue to limit widespread adoption, especially in remote or resource-limited farming contexts. To address this gap, we introduce a novel amplification-free electrochemical (EC) nanobiosensor platform that leverages potential-induced affinity (adsorption) between the gold surface and nucleic acid for direct detection of Lxx DNA from sugarcane xylem sap. This platform integrates a one-step, boiling-based DNA lysis with sequence-specific magnetic capture and label-free EC signal readout. Unlike existing molecular assays, our method eliminates the need for column- or solvent-based nucleic acid extraction; it employs a sequence-specific magnetic bead capture step, which constitutes a non-conventional yet effective form of nucleic acid purification. It also avoids enzymatic amplification and specialized laboratory infrastructure, offering a portable, low-cost, and rapid diagnostic solution suitable for on-farm deployment by non-specialist users. While affinity interactions-based EC DNA sensors have shown promise for pathogen detection in medical diagnostics [21,22,23,24,25,26], their application in plant pathogen diagnostics remains in its infancy [26,27,28,29,30]. Recent advances in electrochemical biosensing have established DNA-based detection platforms as powerful tools for the rapid and sensitive identification of plant pathogens. These methods typically involve probe-functionalized electrodes coupled with redox-active indicators to convert nucleic acid recognition events into quantifiable electrochemical signals. For instance, methylene blue-labeled probes have enabled sensitive detection of bacterial and fungal pathogens through signal amplification upon hybridization [26,27,28]. In parallel, label-free strategies have gained traction, leveraging changes in charge transfer resistance or suppression of redox current resulting from DNA adsorption at the electrode interface [29,30]. Collectively, these studies underscore the expanding potential of electrochemical DNA sensors for field-deployable, amplification-free diagnostics and provide a strong conceptual framework for the approach presented in this work. To our knowledge, this is the first report of an EC nanobiosensor designed for the direct, amplification-free detection of Lxx DNA from plant samples, achieving high sensitivity, quantitative resolution, and excellent correlation with qPCR. Importantly, this method represents more than a disease-specific diagnostic—it establishes a generalizable biosensing framework for early detection of diverse plant pathogens. While it was developed to detect Leifsonia xyli subsp. xyli in sugarcane, the same underlying approach could be adapted to detect other plant pathogens (bacterial, viral, or fungal) in other crops, just by changing the DNA probe sequence. As such, it offers immense potential as a platform technology for crop health monitoring, biosecurity surveillance, and climate-resilient agriculture. By enabling rapid identification and intervention, our biosensor contributes directly to enhancing food security, reducing yield losses, and supporting sustainable agricultural practices in the face of increasing pathogen pressure and environmental stress.
2. Materials and Methods
2.1. Reagents and Materials
Screen-printed gold electrodes (SPGEs, DRP-220AT) with integrated three-electrode systems were obtained from Metrohm Dropsens (Llanera, Spain). Gold (III) chloride trihydrate, synthetic DNA sequences, and primers (Supplementary Table S1) were purchased from Sigma Aldrich (Castle Hill, Australia) and Integrated DNA Technologies (Coralville, IA, USA), respectively. UltraPureTM DNase/RNase-free distilled water was from Invitrogen (Melbourne, Australia). Electrochemical measurements were carried out using a CH1040C potentiostat (CH Instruments, Inc., Austin, TX, USA).
2.2. Source of Inoculum and Culture Conditions
Lxx was isolated from infected sugarcane stalks at Sugar Research Australia (SRA), Woodford, following sap extraction using positive pressure methods as described in Ngo et al. [11]. Sap was filtered (0.2 µm), inoculated into modified S8 medium, and cultured at 28 °C in the dark for four weeks [5,19]. Presence of Lxx was confirmed by qPCR. Negative controls included Xanthomonas albilineans (Xalb) and Ceratocystis paradoxa (Cpar), cultured using standard methods [31,32].
2.3. Field Trial and Sample Collection
A randomized complete block field trial was established at SRA Woodford (S 26.93°, E 152.78°) in September 2020, using 10 sugarcane cultivars with known RSD resistance ratings (CP72-2086, Ho06-537, Q232, Q253, SRA20, Q208, WSRA24, Q242, SRA26, and SRA22) as shown in Table 1 [11,33,34,35]. One-budded setts were hot-water treated (52 °C, 30 min), inoculated with Lxx [35], germinated in moistened vermiculite, and transplanted to the field. At 53 weeks post-inoculation, xylem sap was extracted from three stalks per cultivar and stored at –20 °C until use.

Table 1.
List of sugarcane varieties and their ratoon stunting disease resistance rating and rating categories (Ngo et al. [35]).
2.4. DNA Isolation
For electrochemical analysis, rapid and simple DNA isolation was achieved by boiling sap spiked with serially diluted Lxx cultures or field samples (95 °C, 2 min), as previously described [21,22]. Supernatants were directly used as templates (10 µL per assay). For qPCR, genomic DNA was extracted using the PureLinkTM Microbiome DNA kit (Thermo Fisher Scientific, Scoresby, VIC, Australia) following the manufacturer’s instructions. All assays were performed in triplicate and repeated independently three times.
2.5. Target Selection and Primer Design
A 34 bp region within the 16S–23S intergenic spacer (IGS) of Lxx (GenBank: AE016822.1) was targeted using a capture probe (LxxCP1) [36,37,38]. Primers for qPCR (Lxx_EC_FP/RP) targeting a 133 bp segment within the same region were designed using NCBI Primer-BLAST [39] and validated for specificity using BLASTn [40]. Structural integrity of oligonucleotides was assessed using OligoAnalyzerTM (Integrated DNA Technologies, Inc., Coralville, IA, USA).
2.6. Probe Hybridization and Magnetic Isolation
Target DNA was captured using sequence-specific magnetic beads following previously established protocols [41,42]. Purified products were stored at −20 °C for subsequent EC quantification. All experiments included triplicate biological and technical replicates.
2.7. Sensor Fabrication and Assay Optimization
To enhance performance, the screen-printed gold electrode (SPGE) surfaces were pretreated and modified with gold nanoparticles (AuNPs) following the protocol described by Zhang et al. [43]. Briefly, a droplet of gold nanoparticle solution was applied to the working electrode surface, and electrochemical deposition was carried out by applying a potential of 0.2 V (vs. Ag/AgCl) to ensure uniform immobilization of the nanoparticles onto the gold surface. The electroactive surface area was calculated using the Randles–Sevcik equation [44]. Target DNA was deposited onto the SPGEs by applying a constant potential of 0.6 V (vs. Ag/AgCl) for 60 s to adsorb the DNA onto the electrode surface. The deposition conditions (potential and time) were optimized to achieve maximum adsorption while minimizing nonspecific binding, based on previous studies [45,46]. After deposition, the electrodes were washed three times with 10 mM PBS.
2.8. Electrochemical Detection
Differential pulse voltammetry (DPV) was performed in 2 mM [Fe(CN)6]3− solution (–0.2 V to 0.45 V; amplitude 0.05 V) to quantify DNA adsorption. The current response was normalized using the following equation:
where iBare and iAdsorbed represent current densities before and after DNA deposition, respectively.
2.9. qPCR Validation and Gel Electrophoresis
Quantitative PCR was conducted using a CFX96 Touch system (Bio-Rad Laboratories, Inc., Gladesville, NSW, Australia) with reaction conditions per manufacturer protocol (New England Biolabs, Inc., Ipswich, MA, USA): 98 °C (15 s), 52 °C (30 s), and 72 °C (30 s) for 40 cycles. Positive results were defined as Cq < 40 or undetermined. qPCR standard curves were generated from serially diluted Lxx DNA (105–100 cells/µL) in spiked sap. Gel electrophoresis was performed on 1% agarose gels and visualized using SYBR Safe DNA stain under UV illumination (90 V, 40 min), with 133 bp amplicons visualized using GeneRuler ladder (Thermo Fisher Scientific, Scoresby, VIC, Australia).
2.10. Statistical Analysis
All data are presented as mean ± standard error (n = 3). No statistical LoD (e.g., 3σ/s, where σ is the standard deviation of the blank and s is the slope of the calibration curve) was calculated; instead, the lowest detectable concentration was used based on the experimentally observed signal distinguishability from the blank. Statistical analyses and visualizations were performed using OriginPro v9.9.0.225 (OriginLab, Northampton, MA, USA), Microsoft Excel 365, and RStudio v4.2.3 [47]. Correlations between EC and qPCR results were assessed using Spearman’s rank correlation. Figures were created using BioRender [48], SnapGene (www.snapgene.com), and Microsoft PowerPoint 365.
3. Results and Discussion
3.1. Assay Design
Figure 1 presents the outline of the newly developed Lxx detection assay. The assay begins with the release of bacterial DNA from sugarcane xylem sap using a simple boiling lysis technique, followed by direct capture of Lxx target DNA through a complementary biotinylated probe (LxxCP1) immobilized on streptavidin-coated magnetic beads (Figure 1A). The captured Lxx targets were magnetically purified through multiple washing steps to remove non-target molecules while retaining the nanoparticle/probe/target complexes. Subsequently, heat treatment was applied to dissociate the Lxx targets from the capture probe/nanoparticle complexes, allowing the probe-bound target sequences to be released into solution. The released Lxx targets were then adsorbed onto the AuNPs-modified SPGE surface via a potential-induced gold–DNA affinity interaction (Figure 1B). Finally, electrochemical detection was performed by voltammetric interrogation using [Fe(CN) 6]3−, generating a differential pulse voltammetry (DPV) readout to quantify the captured target (Figure 1C). Adsorption of Lxx DNA on the electrode surface reduces the electrochemical signal due to increased coulombic repulsion between the negatively charged ferricyanide ions and the electrode surface.
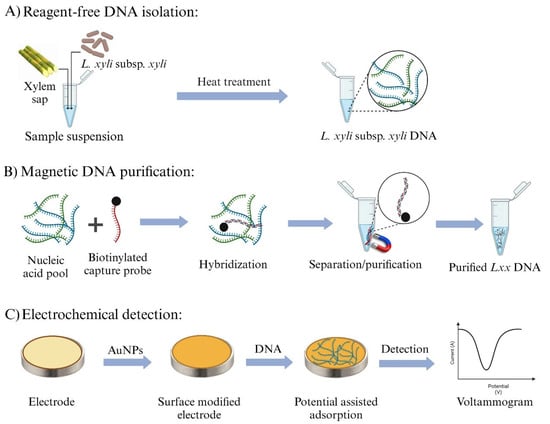
Figure 1.
Overview of the electrochemical assay workflow for detecting Lxx DNA. (A) Bacterial DNA is isolated from sugarcane xylem sap using a simple boiling lysis method. (B) The isolated Lxx DNA is selectively captured using biotinylated complementary DNA probes and isolated via streptavidin-coated magnetic beads. (C) Captured Lxx DNA is thermally released and field-induced to adsorb onto AuNPs-modified SPGE, followed by electrochemical detection using DPV.
3.2. Assay Optimization
To enhance assay sensitivity, the SPGE surface was modified with AuNPs. This modification resulted in a twofold increase in current density response (mean current density 8.54 vs. 4.12 mA cm−2; n = 3) (Figure S1). This enhancement aligns with prior reports where AuNPs, owing to their high conductivity, facilitate rapid electron transfer between the electrolyte solution and the transducer [49,50,51,52,53,54,55,56]. The modification reduces the surface impedance of the working electrode, enabling detection of minute changes in electron transfer at the electrode–solution interface [54]. Furthermore, this approach leverages the high biocompatibility of AuNPs, which helps preserve the biological activity of immobilized molecules over extended periods [57,58,59]. The AuNP-modified electrodes in this study were fabricated using a well-established electrochemical deposition method, as described by Zhang et al. [43]. Although direct morphological characterization via SEM or TEM was not conducted, this protocol has been extensively validated in the literature for producing uniform, nanoscale gold coatings with high electrochemical activity. The significant enhancement in current response and sensitivity observed in our system (Figure S1) is consistent with effective AuNP deposition and corroborates previous reports employing similar fabrication strategies. This method was selected for its simplicity, scalability, and reproducibility. Nonetheless, we acknowledge the value of direct structural validation, and future studies will incorporate SEM/TEM imaging to further confirm nanoparticle morphology and surface distribution.
For improved target binding specificity and quantification accuracy, the optimal potential and deposition time for Lxx DNA adsorption were determined to be +600 mV and 60 s, respectively (Figures S2 and S3). The electrochemical signal intensity is directly dependent on the amount of DNA adsorbed onto the electrode surface. Electrodes with low DNA surface coverage exhibited electrochemical signals comparable to those of unmodified electrodes, as ferricyanide ions could readily access the surface, resulting in reduced coulombic repulsion and an increased faradaic current. Therefore, maximizing DNA adsorption under optimized conditions is critical for achieving high assay sensitivity and rapid response [45,46,60].
Assay functionality was evaluated by comparing electrochemical responses in the presence and absence of synthetic Lxx targets (Figure 2A,B). Figure 2B illustrates the calculated percentage change in current density between the blank (no DNA) and test conditions. This normalization emphasizes the relative suppression in electrochemical signal due to target DNA adsorption. The presence of targets caused a significant decrease in current response compared to the no-target control, demonstrating the assay’s effectiveness. This decrease arises from the combined effects of (i) coulombic repulsion between the negatively charged adsorbed DNA and the ferricyanide ions at the AuNP-modified SPGE gold sensor surface via the DNA–gold affinity interaction and (ii) steric hindrance of the active sites—some of which are already occupied by adsorbed DNA—preventing redox species from contacting the electrode surface. These repulsion and steric hindrance effects impede electron transfer during the one-electron redox reaction, leading to a diminished faradaic current relative to the control [61]. In addition to steric hindrance and electrostatic repulsion, it is plausible that DNA adsorption induces interfacial changes, such as conformational rearrangements or partial hybridization-like interactions that modulate the charge transfer resistance at the electrode surface. Prior studies have shown that DNA hybridization can significantly alter electrode impedance, thereby affecting redox probe access and overall signal output [43]. Although this study focused on potential-induced adsorption of single-stranded DNA and did not involve classical hybridization events, such impedance-related effects cannot be entirely ruled out. While electrochemical impedance spectroscopy (EIS) was not performed here, the consistent trend in current suppression with increasing DNA concentration, combined with high reproducibility and strong linearity (R2 = 0.98), supports our hypothesis that the signal decreases are due to DNA blocking the electrode surface and repelling charged molecules. Future studies incorporating EIS measurements will be valuable for dissecting the contributions of impedance modulation alongside steric and electrostatic effects, thereby offering deeper mechanistic insight into DNA-induced signal variation.
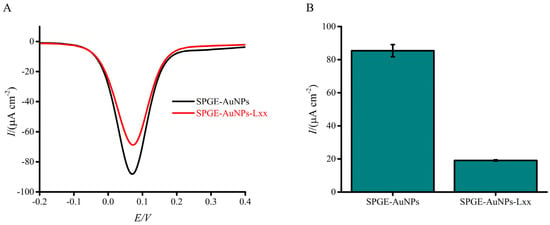
Figure 2.
Electrochemical response of the assay to the presence or absence of synthetic Lxx target DNA (10 pM). (A) DPV signals show a clear reduction in current in the presence of the target due to DNA adsorption at the electrode surface. (B) Bar graph showing the mean percentage change in current density relative to the blank (no target) condition. Error bars represent standard deviations from independent runs with separately prepared electrodes (n = 3).
3.3. Assay Sensitivity and Specificity
The sensitivity of the developed Lxx assay was initially evaluated using a titration series of synthetic Lxx targets spiked into fresh, uncontaminated sugarcane sap (Figure 3A,B). As the concentration of Lxx increased, a corresponding decrease in the DPV current signal was observed, attributed to a greater number of target DNA species adsorbing on the sensor surface [62]. This increased adsorption enhanced the repulsion and steric hindrance effects of approaching [Fe(CN)6]3− ions, hindering their diffusion to the electrode surface and thereby reducing the faradaic current. The detection limit was assessed over a concentration range spanning from 10 nM to 1 fM, and the resulting calibration plot demonstrated excellent linearity. The linear regression equation was y = 9.17 logC + 0.02 (where y represents the change in current and C the concentration of Lxx), with a correlation coefficient (r) of 0.9826 (Figure 3B inset). Compared to the no-template control (NTC), which exhibited a high current response due to an unblocked sensor surface, the assay achieved a detection limit down to 1 fM with a reproducibility of %SD ≤ 5% (n = 3). It is acknowledged that minor variations in current density values, particularly for the blank and 10 pM DNA samples, are observed between Figure 2 and Figure 3. These figures represent results from independent experimental runs, each conducted using separately fabricated screen-printed electrodes and freshly prepared reagents. Such variations are expected in electrochemical biosensing systems and are primarily attributed to inherent differences in electrode surface morphology, ink composition, and handling of biological samples. Despite these run-to-run variations, the data exhibits high internal consistency within each experiment. Specifically, the standard deviation across replicates remained consistently low (SD < 5%, n = 3), reflecting excellent repeatability under identical conditions. Furthermore, the platform demonstrated a strong and reproducible linear relationship between target DNA concentration and electrochemical response (R2 = 0.98), confirming its analytical reliability.
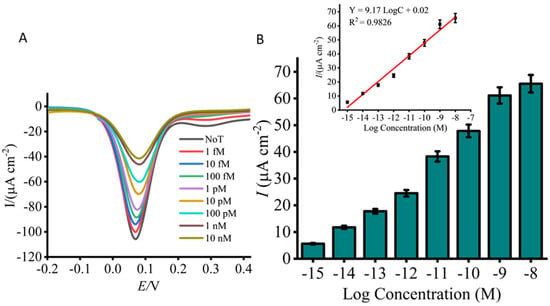
Figure 3.
Analytical sensitivity of the assay using titrated concentrations of synthetic Lxx target DNA (1 nM to 1 fM). (A) DPV signals reveal concentration-dependent decreases in current density relative to the no-target control (NTC). (B) Bar graph summarizes the signal changes across tested concentrations. The inset shows the calibration curve (log concentration vs. current density), indicating strong linearity across the tested range. Error bars represent standard deviations (SD < 5%) from independent runs with separately prepared electrodes (n = 3).
Our assay exhibited sensitivity surpassing previous sugarcane pathogen diagnostic platforms by factors of 100 and 1000, outperforming detection limits reported at 100 fM and 10 pM by Umer et al. [63] and Wongkaew and Poosittisak [28], respectively. In contrast, a recent AuNP-based plant virus diagnostic platform reported by Khater et al. [57] achieved a detection limit of only 100 nM, with a logarithmic response limited to a narrow concentration range (0.1–10 mM). Notably, the platform developed by Siddiquee et al. [64] demonstrated an attomolar-level limit of detection across an extensive dynamic range (1.0 × 10−18 to 1.82 × 10−4 mol L−1). While this approach surpassed the detection limit of our assay, it involved complex sensor fabrication and a labor-intensive phenol-chloroform DNA extraction process. In contrast, our assay offers a straightforward, cost-effective, and quantitative method, employing a simple, field-applicable DNA isolation technique and eliminating the need for complex sensor fabrication. Furthermore, our assay does not require target amplification, labeling, or antibodies. To contextualize the performance of our label-free electrochemical biosensor, a comparison was made with selected electrochemical DNA sensing platforms employing redox reporters such as methylene blue or ferrocene. As shown in Table 2, our method shows comparable or superior sensitivity, a broader dynamic range, and significantly reduced assay time—achieved without the need for probe labeling or amplification steps. This underscores the potential advantages of our approach for rapid and field-deployable diagnostics.

Table 2.
Comparative analysis of electrochemical DNA biosensors using redox reporters (e.g., methylene blue, and ferrocene) vs. the proposed label-free platform.
Achieving a broad detection range and establishing a quantitative relationship between the electrochemical signal and Lxx concentration are crucial for accurate quantification. To assess this, bacterial samples were spiked with known Lxx cell concentrations (105 to 100 cells/μL) in fresh, uncontaminated sugarcane sap, and electrochemical detection was performed. The DPV responses exhibited a robust linear correlation across the tested concentration range (Figure 4A,B). Compared to the NTC, a significant decrease in current was observed with increasing target concentrations, confirming the assay’s capability to detect Lxx sequences. Moreover, the assay showed a dynamic range spanning five orders of magnitude, enabling quantification of Lxx in sap samples with widely varying pathogen loads. The assay detected Lxx at concentrations as low as 10 cells (Figure 4A,B), establishing a simple and robust diagnostic method that compares favorably with other reported electrochemical methods [29,30]. While minor variations were observed between replicate experiments using different electrode batches, all measurements showed high consistency within replicates (SD < 5%, n = 3), indicating good intra-assay reproducibility. Such variability is typical in biosensor systems involving biological samples and surface-adsorbed layers.
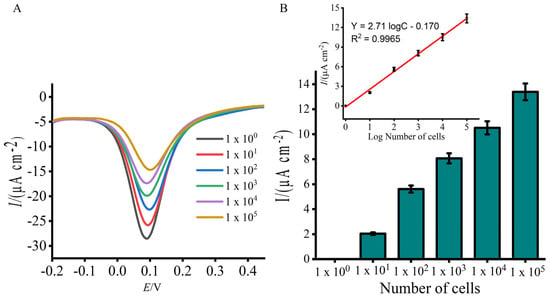
Figure 4.
Sensitivity evaluation using total DNA extracted from known numbers of Lxx cells (105 to 10 cells/μL). (A) DPV signals show decreasing current with increasing input cell numbers, corresponding to higher DNA loads. (B) Mean percentage current response change for each input cell concentration. Inset displays a linear calibration curve for DNA quantity vs. EC signal. Error bars show SD from three replicates.
Specificity was evaluated using 103 cells/µL of other sugarcane pathogen contaminants, Xalb and Cpar. Only marginal decreases in current response (1.39 and 4.18 µA, respectively) were observed relative to the NTC (Figure 5A,B), indicating strong specificity for Lxx. By comparison, the presence of Lxx resulted in a twofold lower current response than the no-target control (7.42 µA) (Figure 5B). These results indicate that the proposed biosensor possesses promising specificity for Lxx detection and holds considerable potential for biological applications. While a full titration of non-target DNA was not conducted, the minimal signal suppression observed in the specificity test (Figure 5B) suggests that increasing concentrations of non-target DNA would not yield significant changes in current response due to the lack of probe–target complementarity.
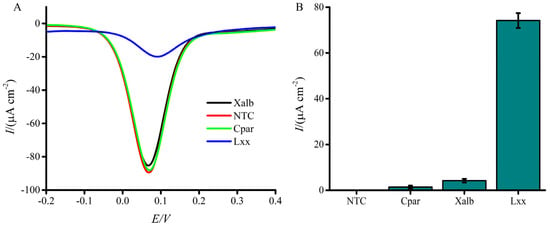
Figure 5.
Specificity of the electrochemical assay for Lxx detection. (A) DPV signals compare current responses to DNA from Lxx, Xanthomonas albilineans (Xalb), and Ceratocystis paradoxa (Cpar) cells. (B) Bar graph illustrates the relative percentage change in current response for each pathogen. A significantly higher signal suppression is observed with Lxx compared to other organisms. NTC = no target control; NG1 = Xalb (negative control 1); NG2 = Cpar (negative control 2). Error bars indicate SD (n = 3).
Reproducibility was confirmed by a relative standard deviation of electrode measurements below 5% (n = 3), demonstrating acceptable assay consistency. Conventional nucleic acid biosensors rely on the specific binding of target sequences to complementary probes immobilized on a two-dimensional transducer surface, which can suffer from interference by non-specific molecules. In contrast, our approach employs selective capture of Lxx DNA via complementary probes followed by magnetic bead-based isolation of the target DNA. The magnetic washing and purification steps effectively remove matrix effects and substantially reduce non-specific interferences, enhancing assay robustness. Reproducibility was evaluated by performing triplicate measurements (n = 3) at each DNA concentration using independently prepared AuNP-modified electrodes. The standard deviation (SD) was calculated for each concentration point and found to be <5%, indicating high reproducibility under the described operating conditions. While this study focused on demonstrating the sensing mechanism and analytical performance, we acknowledge that sensor stability, storage conditions, and long-term usability were not evaluated and remain important areas for future investigation to support real-world deployment.
Although this study did not include direct surface characterization techniques such as XPS or FTIR to confirm DNA adsorption onto the AuNP-modified SPGE surface, the electrochemical response patterns observed in this work strongly support adsorption-based interactions. The consistent and concentration-dependent decrease in current density following DNA deposition (Figure 2 and Figure 3), coupled with the linear correlation between target DNA concentration and signal suppression (R2 = 0.98), is well-aligned with prior studies utilizing potential-induced DNA adsorption on gold surfaces [21,22,23,24,25,26,45,46]. These findings, along with the use of a well-established adsorption potential (+600 mV), support the proposed sensing mechanism. Nevertheless, we acknowledge this as a limitation and propose that future work includes surface analytical techniques (e.g., XPS or AFM) to provide direct validation of the adsorption process.
3.4. Detection of Lxx in Sugarcane Xylem Sap Samples
The developed Lxx electrochemical biosensor was applied to detect varying amounts of Lxx in xylem sap samples from 10 sugarcane cultivars collected from a field trial. Detected Lxx DNA levels in most samples fell within the assay’s detection limit of 1 fM, highlighting its suitability for real-world analysis. Detected Lxx levels corresponded well with cultivar disease ratings. For example, samples from cultivars Ho06-537 and CP72-2086 exhibited relatively low current responses, indicative of lower bacterial loads and consistent with their moderate resistance to RSD (Figure 6). Cultivars Q208 and SRA22 were classified as intermediately resistant, while WSRA24, SRA20, and Q232 were considered susceptible. Intermediate susceptible cultivars Q242, Q253, and SRA26 were identified as highly susceptible (Figure 6). These results aligned closely with established industry disease ratings [35].
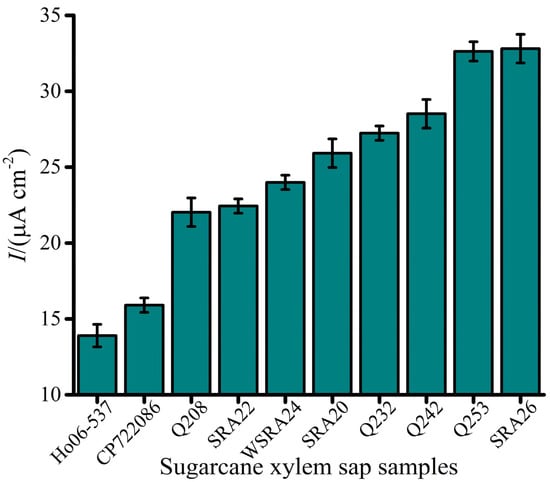
Figure 6.
Electrochemical detection of Lxx in sugarcane xylem sap samples collected from field trials. Bar graph displays current density values for multiple sugarcane cultivars tested during SRA Woodford RSD screening trials. Variations in EC signal correlate with bacterial load and cultivar susceptibility. Error bars represent SD of triplicate measurements.
3.5. Validation with qPCR
A strong correlation was observed between electrochemical and qPCR quantification, confirming the biosensor’s suitability for Lxx detection (Figure 7). qPCR effectively amplified the target intergenic spacer (IGS) region from DNA extracted from 100 cells, with clear and distinct amplicon bands observed via gel electrophoresis (Figure S4). No amplification or bands were detected in the NTC samples. The strong correlation (r = 0.84, p < 0.001; Table 3) between the EC signal and qPCR quantification underscores the assay’s reliability. Notably, the electrochemical assay demonstrated a 10-fold greater sensitivity compared to qPCR (Figure 4 vs. Figure 7A).
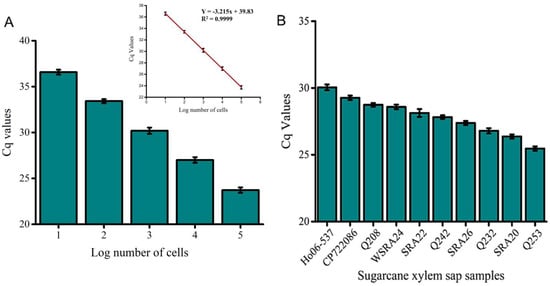
Figure 7.
qPCR validation of the electrochemical assay. (A) Absolute quantification of Lxx DNA in known cell concentrations (105 to 10 cells/µL) confirms the sensitivity of the assay. (B) Quantitative detection of Lxx in selected field samples infected with RSD. qPCR data show good agreement with EC assay trends. Error bars indicate SD across three independent experiments.

Table 3.
Spearman correlation coefficients using Cq Values of EC and qPCR methods for comparing RSD resistance ratings among ten sugarcane varieties.
For field sample validation, qPCR successfully amplified the target region across all analyzed samples with varying cycle thresholds, reflecting differences in pathogen load, susceptibility, and detection limit (Figure 7B). This trend was consistent with the electrochemical assay results (Figure 6). All analyzed samples exhibited clear qPCR amplicon bands (Figure S5), while NTCs showed no amplification. The qPCR cycle threshold (Cq) values and electrochemical signals exhibited negative correlations with RSD resistance ratings (r = −0.78 and −0.87, respectively). Furthermore, qPCR and electrochemical values showed a strong positive correlation (r = 0.87, p < 0.001) (Table 3). The strong agreement between electrochemical current responses and qPCR Cq values (r = 0.84, p < 0.001) confirms the assay’s robustness for detecting and quantifying Lxx in xylem sap samples. Negative control samples exhibited EC signals comparable to background levels, with current changes below 10%, confirming assay specificity. Successful application of the biosensor to real biological samples (Figure 6), coupled with validation through qPCR and gel electrophoresis (Figure 7 and Figure S5), underscores the technique’s potential for practical biological diagnostics. Although slight variability in Cq values was observed at the lowest target concentration (10 cells/µL), the replicates remained consistent with acceptable technical variation. The negative slope confirms the expected inverse relationship between Cq and template quantity. A summary comparison of our biosensor with conventional nucleic acid amplification and hybridization-based techniques is provided in Table 4. This highlights the potential field-deployable advantages of our approach in terms of speed and simplicity.

Table 4.
Comparison of the proposed electrochemical biosensor with conventional nucleic acid extraction and detection methods.
4. Conclusions
In this work, we present a proof-of-concept electrochemical assay capable of detecting Lxx, the causative agent of ratoon stunting disease in sugarcane, with ultrahigh sensitivity and excellent analytical performance. The assay enabled quantitative detection of Lxx DNA at concentrations as low as 1 fM, with a wide dynamic range (1 fM to 10 nM), strong linearity (r = 0.99), and high reproducibility (SD < 5%, n = 3). Key features of this platform include its compatibility with a simplified boiling lysis DNA extraction protocol, eliminating the need for labor-intensive or kit-based methods, and its use of low-cost, disposable SPGE (~US$5 per sensor), which brings the total assay cost to under US$10 per sample. Notably, the sensor requires no complex surface modification or amplification steps, distinguishing it from many existing biosensing platforms. Beyond sensitivity and cost-effectiveness, the modular design of this assay provides a highly adaptable framework for broader applications. By tailoring the capture probes, the platform could be readily extended to detect a range of plant pathogens in diverse sample matrices, including soil, sap, or leaf extracts. The integration of multi-well electrodes further enables potential high-throughput or multiplexed analyses. Taken together, this study lays the foundation for the development of field-deployable diagnostic devices for on-site agricultural pathogen monitoring. With future integration into automated or smartphone-assisted platforms, this assay could support rapid disease surveillance, geospatial risk mapping, and early warning systems for exotic pathogens, especially at biosecurity checkpoints such as ports and border entries. This approach contributes to the growing need for portable, sensitive, and cost-effective diagnostics in sustainable agriculture and plant biosecurity. In summary, this work presents a simple, rapid, and label-free electrochemical biosensor for the detection of Leifsonia xyli subsp. xyli, demonstrating high sensitivity, minimal sample preparation, and suitability for field application. While the electrochemical data strongly support DNA adsorption onto the SPGE surface, future studies could incorporate complementary surface analytical techniques, such as XPS, atomic force microscopy (AFM), or quartz crystal microbalance (QCM), to directly visualize and chemically characterize the DNA layer. Further mechanistic investigations using electrochemical impedance spectroscopy (EIS) can also help distinguish between physical adsorption and DNA hybridization-induced impedance changes at the electrode interface. Most importantly, systematic studies are required to assess the sensor’s stability and shelf life under conditions relevant to real-world field applications.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/bios15080518/s1.
Author Contributions
Conceptualization, M.J.A.S.; Methodology, M.C. and N.S.; Formal analysis, M.C. and N.S.; Investigation, M.C. and N.S.; Resources, M.C. and S.S.; Data curation, M.C. and S.S.; Writing—original draft, M.C.; Writing—review & editing, S.A.B., S.S., M.J.A.S., N.-T.N. and R.F.; Supervision, R.F.; Project administration, N.-T.N.; Funding acquisition, M.C. and N.-T.N. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Australian Research Council through the Linkage Project scheme (LP200100016) and co-funded by Sugar Research Australia (SRA). Additional support was provided by Griffith University, including a Griffith University International Postgraduate Research Scholarship and a PhD Competitive Research Grant from the Centre for Planetary Health and Food Security (CPHFS).
Data Availability Statement
Data will be made available upon request.
Acknowledgments
The authors gratefully acknowledge Sugar Research Australia for providing both infected and disease-free plant materials essential to this study.
Conflicts of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Abbreviations
| RSD | Ratoon Stunting Disease |
| Lxx | Leifsonia xyli subsp. xyli |
| Xalb | Xanthomonas albilineans |
| Cpar | Ceratocystis paradoxa |
| PCR | Polymerase Chain Reaction |
| qPCR | Quantitative Polymerase Chain Reaction |
| LAMP | Loop-Mediated Isothermal Amplification |
| EC | Eectrochemical |
| DNA | Deoxyribonucleic Acid |
| RNA | Ribonucleic Acid |
| SPGE | Screen-Printed Gold Electrode |
| PDA | Potato Dextrose Agar |
| RCB | Randomized Complete Block |
| mL | Milliliter |
| EC | Electrochemical |
| Bp | Base pair |
| IGS | Intergenic Spacer |
| NCBI | National Center for Biotechnology Information |
| BLASTn | Basic Local Alignment Search Tool for nucleotides |
| [Fe(CN)6]3– | Ferricyanide ion |
| cm2 | Square centimeter |
| Fe3+ | Ferric ion |
| Fe2+ | Ferrous ion |
| n | Number |
| s−1 | Per second |
| Vs−1 | Volts per second |
| mol cm−3 | Mole per cubic centimeter |
| Ag/AgCl | Silver/Silver Chloride |
| mV | Millivolts |
| AuNPs | Gold Nanoparticles |
| CV | Cyclic Voltammetry |
| DPV | Differential Pulse Voltammetry |
| mM | Millimolar |
| V | Volt |
| i | Current |
| Cq | Cycle Quantification |
| µL | Microliter |
| SD | Standard Deviation |
| r | Correlation coefficient |
| TAE | Tris-Acetate-EDTA |
| LxxCP1 | Lxx-specific Capture probe-1 |
| mA cm−2 | Milliamps per square centimeter |
| nM | Nanomolar |
| fM | Femtomolar |
| pM | Picomolar |
References
- Gillaspie, A.G., Jr. Ratoon Stunting Disease, 1st ed.; Elsevier: Amsterdam, The Netherlands, 1989; pp. 16–55. [Google Scholar]
- James, G. A review of ratoon stunting disease. Int. Sugar J. 1996, 1174, 532–541. [Google Scholar]
- Bailey, R.A.; Bechet, G.R. Further evidence of the effects of ratoon stunting disease on production under irrigated and rainfed conditions. Proc. South Afr. Sugar Technol. Assoc. 1997, 71, 97–101. [Google Scholar]
- Que, Y.X.; Xu, J.S.; Xu, L.P.; Gao, S.J.; Chen, R.K. PCR detection for Leifsonia xyli subsp. xyli, pathogen of the sugarcane ratoon stunting disease. Fujian J. Agric. Sci. 2008, 23, 364–367. [Google Scholar]
- Davis, M.J.; Gillaspie, A.G.R., Jr.; Harris, W.; Lawson, R.H. Ratoon stunting disease of sugarcane: Isolation of the causal bacterium. Science 1980, 210, 1365–1367. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, M.; Soda, N.; Strachan, S.; Ngo, C.N.; Bhuiyan, S.A.; Shiddiky, M.J.A.; Ford, R. Ratoon stunting disease (RSD) of sugarcane: A review emphasizing detection strategies and challenges. Phytopathology 2024, 114, 7–20. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.S.; Xu, L.P.; Que, Y.X.; Gao, S.J.; Chen, R.K. Advances in the ratoon stunting disease of sugarcane. J. Trop. Subtrop. Bot. 2008, 16, 184–188. [Google Scholar]
- Brumbley, S.M.; Petrasovits, L.A.; Hermann, S.R.; Young, A.J.; Croft, B.J. Recent advances in the molecular biology of Leifsonia xyli subsp. xyli, causal organism of ratoon stunting disease. Australas. Plant Pathol. 2006, 35, 681–689. [Google Scholar] [CrossRef]
- Luo, L.F.; Wei, C.Z.; Tang, J.H. Progress on detection technologies of sugarcane ratoon stunting disease. Agric. Res. Appl. 2011, 6, 25–27. [Google Scholar]
- Harrison, N.; Davis, M. Comparison of serological techniques for diagnosis of ratoon stunting disease. Sugar Cane 1990, 1, 5–9. [Google Scholar]
- Croft, B.J.; Greet, A.D.; Lehmann, T.M.; Teakle, D.S. RSD diagnosis and varietal resistance screening in sugarcane using the EB-EIA technique. Proc. Aust. Soc. Sugar Cane Technol. 1994, 16, 143–151. [Google Scholar]
- Pan, Y.; Grisham, M.; Burner, D.; Wei, Q.; Damann, K., Jr. Detecting Clavibacter xyli subsp. xyli by tissue blot DNA hybridization. Sugar Cane 1998, 3, 3–8. [Google Scholar]
- Taylor, P.; Petrasovits, L.; Vall der Velde, R.; Birch, R.; Croft, B.; Fegan, M.; Smith, G.; Brumbley, S. Development of PCR-based markers for detection of Leifsonia xyli subsp. xyli in fibrovascular fluid of infected sugarcane plants. Australas. Plant Pathol. 2003, 32, 367–375. [Google Scholar] [CrossRef]
- Davis, M.J.; Rott, P.; Astua Monge, G. Nested, multiplex PCR for detection of both Clavibacter xyli subsp. xyli and Xanthomonas albilineans in sugarcane. Int. Cong. Plant Pathol. 1998, 3, 9–16. [Google Scholar]
- Farahani, A.S.; Taghavi, S.; Taher-Khani, K. Comparison of conventional, nested, and real-time PCR for detection of the causal agent of ratoon stunt in Iran. J. Plant Pathol. 2015, 97, 10–23. [Google Scholar]
- Carvalho, G.; da Silva, T.; Munhoz, A.; Monteiro-Vitorello, C.; Azevedo, R.; Melotto, M.; Camargo, L.E.A. Development of a qPCR for Leifsonia xyli subsp. xyli and quantification of the effects of heat treatment of sugarcane cuttings on Lxx. Crop Prot. 2016, 80, 51–55. [Google Scholar] [CrossRef]
- Grisham, M.; Pan, Y.-B.; Richard, E., Jr. Early detection of Leifsonia xyli subsp. xyli in sugarcane leaves by real-time polymerase chain reaction. Plant Dis. 2007, 91, 430–434. [Google Scholar] [CrossRef]
- Ghai, M.; Singh, V.; Martin, L.; McFarlane, S.; van Antwerpen, T.; Rutherford, R. A rapid and visual loop-mediated isothermal amplification assay to detect Leifsonia xyli subsp. xyli targeting a transposase gene. Lett. Appl. Microbiol. 2014, 59, 648–657. [Google Scholar] [CrossRef]
- Brumbley, S.M.; Petrasovits, L.A.; Birch, R.G.; Taylor, P.W. Transformation and transposon mutagenesis of Leifsonia xyli subsp. xyli, causal organism of ratoon stunting disease of sugarcane. Mol. Plant-Microbe Interact. 2002, 15, 262–268. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, Z.; Liu, Y.; Wang, X.; Li, Y.; Ma, P.; Gu, B.; Li, H. Recent advances in rapid pathogen detection method based on biosensors. Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 1021–1037. [Google Scholar] [CrossRef]
- Shiddiky, M.J.A.; Rahman, M.A.; Shim, Y.-B. Hydrazine-catalyzed ultrasensitive detection of DNA and proteins. Anal. Chem. 2007, 79, 6886–6890. [Google Scholar] [CrossRef]
- Boriachek, K.; Umer, M.N.; Islam, M.N.; Gopalan, V.; Lam, A.K.; Nguyen, N.-T.; Shiddiky, M.J.A. An amplification-free electrochemical detection of exosomal miRNA-21 in serum samples. Analyst 2018, 143, 1662–1669. [Google Scholar] [CrossRef]
- Bilkiss, M.; Shiddiky, M.J.A.; Ford, R. Advanced diagnostic approaches for necrotrophic fungal pathogens of temperate legumes with a focus on Botrytis spp. Front. Microbiol. 2019, 10, 1889. [Google Scholar]
- Dyussembayev, K.; Sambasivam, P.; Bar, I.; Brownlie, J.C.; Shiddiky, M.J.A.; Ford, R. Biosensor technologies for early detection and quantification of plant pathogens. Front. Chem. 2021, 9, 636245. [Google Scholar] [CrossRef] [PubMed]
- Drummond, T.G.; Hill, M.G.; Barton, J.K. Electrochemical DNA sensors. Nat. Biotechnol. 2003, 21, 1192–1199. [Google Scholar] [CrossRef] [PubMed]
- Ferapontova, E.E. DNA Electrochemistry and electrochemical sensors for nucleic acids. Annu. Rev. Anal. Chem. 2018, 11, 197–218. [Google Scholar] [CrossRef]
- Nezhad, A.S. Future of portable devices for plant pathogen diagnosis. Lab Chip 2014, 14, 2887–2904. [Google Scholar] [CrossRef]
- Wongkaew, P.; Poosittisak, S. Diagnosis of sugarcane white leaf disease using the highly sensitive DNA-based voltammetric electrochemical determination. Am. J. Plant Sci. 2014, 5, 2256–2268. [Google Scholar] [CrossRef]
- Fang, Y.; Ramasamy, R.P. Current and prospective methods for plant disease detection. Biosensors 2015, 5, 537–561. [Google Scholar] [CrossRef]
- Khater, M.; de la Escosura-Muñiz, A.; Merkoci, A. Biosensors for plant pathogen detection. Biosens. Bioelectron. 2017, 93, 72–86. [Google Scholar] [CrossRef]
- Dawson, W.J. Plant Diseases Due to Bacteria, 2nd ed.; Cambridge University Press: Cambridge, UK, 1957; Volume 1, p. 48. [Google Scholar]
- Rahman, M.A.; Begum, M.F.; Alam, M.F. Screening of Trichoderma isolates as a biological control agent against Ceratocystis paradoxa causing pineapple disease of sugarcane. Mycobiology 2009, 37, 277–285. [Google Scholar] [CrossRef]
- Croft, B.; Johnson, A. Ratoon stunting disease resistance of Australian sugarcane varieties. Proc. Conf. Aust. Soc. Sugar Cane Technol. 2013, 1, 13–22. [Google Scholar]
- Croft, B.J. A method for rating sugarcane cultivars for resistance to ratoon stunting disease based on an enzyme-linked immunoassay. Australas. Plant Pathol. 2002, 31, 63–66. [Google Scholar] [CrossRef]
- Ngo, C.N.; Gibbs, L.; Garlic, L.; Bhuiyan, S.A.; Wei, X. Revisiting variety ratings for ratoon stunting disease. Proc. Conf. Aust. Soc. Sugar Cane Technol. 2023, 44, 273–275. [Google Scholar]
- National Center for Biotechnology Information (NCBI). Leifsonia xyli subsp. xyli, Complete Genome; Accession No. AE016822. 1988. Available online: https://www.ncbi.nlm.nih.gov/nuccore/AE016822 (accessed on 8 December 2022).
- Monteiro-Vitorello, C.B.; Camargo, L.E.; Van Sluys, M.A.; Kitajima, J.P.; Do Amaral, D.; Harakava, A.M.; De Oliveira, R.; Truffi, J.C.; De Oliveira, D.; Wood, M.C. The genome sequence of the gram-positive sugarcane pathogen Leifsonia xyli subsp. xyli. Mol. Plant Microbe Interact. 2004, 17, 827–836. [Google Scholar] [CrossRef]
- Monteiro-Vitorello, C.B.; Zerillo, M.M.; Van Sluys, M.-A.; Camargo, L.E.A.; Kitajima, J.P. Complete genome sequence of Leifsonia xyli subsp. cynodontis strain DSM46306, a gram-positive bacterial pathogen of grasses. Genome Announc. 2013, 1, e00915-13. [Google Scholar] [CrossRef]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef]
- Chen, Y.; Ye, W.; Zhang, Y.; Xu, Y. High-speed BLASTN: An accelerated Mega BLAST search tool. Nucleic Acids Res. 2015, 43, 7762–7768. [Google Scholar] [CrossRef]
- Clack, K.; Soda, N.; Kasetsirikul, S.; Kline, R.; Salomon, C.; Shiddiky, M.J.A. An interfacial affinity interaction-based method for detecting HOTAIR lncRNA in cancer plasma samples. Biosensors 2022, 12, 287. [Google Scholar] [CrossRef]
- Koo, K.M.; Soda, N.; Shiddiky, M.J.A. Magnetic nanomaterial–based electrochemical biosensors for the detection of diverse circulating cancer biomarkers. Curr. Opin. Electrochem. 2021, 25, 100645. [Google Scholar] [CrossRef]
- Zhang, J.; Song, S.; Wang, L.; Pan, D.; Fan, C. A gold nanoparticle-based chronocoulometric DNA sensor for amplified detection of DNA. Nat. Protoc. 2007, 2, 2888–2895. [Google Scholar] [CrossRef]
- Shiddiky, M.J.A.; Torriero, A.A.J.; Zhao, C.; Burgar, I.; Kennedy, G.; Bond, A.M. Nonadditivity of faradaic currents and modification of capacitance currents in the voltammetry of mixtures of ferrocene and the cobaltocenium cation in protic and aprotic ionic liquids. J. Am. Chem. Soc. 2009, 131, 7976–7989. [Google Scholar] [CrossRef]
- Leung, K.K.; Yu, H.Z.; Bizzotto, D. Electrodepositing DNA self-assembled monolayers on Au: Detailing the influence of electrical potential perturbation and surface crystallography. ACS Sens. 2019, 4, 513–520. [Google Scholar] [CrossRef]
- Leung, K.K.; Martens, I.; Yu, H.Z.; Bizzotto, D. Measuring and controlling the local environment of surface-bound DNA in Self-assembled monolayers on gold when prepared using potential-assisted deposition. Langmuir 2020, 36, 6837–6847. [Google Scholar] [CrossRef]
- RStudio Team. RStudio: Integrated Development for R, Version 2022.07.1 “Spotted Wakerobin”; RStudio, PBC: Boston, MA, USA, 2022.
- BioRender, BioRender: Version accessed June 2022; BioRender: Toronto, ON, Canada, 2022.
- Renedo, O.D.; Martínez, M.J.A. Anodic stripping voltammetry of antimony using gold nanoparticle-modified carbon screen-printed electrodes. Anal. Chim. Acta 2007, 589, 255–260. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Wang, E. Synthesis and electrochemical applications of gold nanoparticles. Anal. Chim. Acta 2007, 598, 181–192. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Ye, Y.; Liu, S. Gold nanoparticle-based signal amplification for biosensing. Anal. Biochem. 2011, 417, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Davis, D.; Guo, X.; Musavi, L.; Lin, C.-S.; Chen, S.-H.; Wu, V.C.H. Gold nanoparticle-modified carbon electrode biosensor for the detection of Listeria monocytogenes. Ind. Biotechnol. 2013, 9, 31–36. [Google Scholar] [CrossRef]
- Wang, Y.; Alocilja, E.C. Gold nanoparticle-labeled biosensor for rapid and sensitive detection of bacterial pathogens. J. Biol. Eng. 2015, 9, 16. [Google Scholar] [CrossRef]
- Tian, L.; Liu, L.; Li, Y.; Wei, Q.; Cao, Q.; Al, E. Ultrasensitive sandwich-type electrochemical immunosensor based on trimetallic nanocomposite signal amplification strategy for the ultrasensitive detection of CEA. Sci. Rep. 2016, 6, 30849. [Google Scholar] [CrossRef]
- Khater, M.; de la Escosura-Muñiz, A.; Quesada-González, D.; Merkoçi, A. Electrochemical detection of plant virus using gold nanoparticle-modified electrodes. Anal. Chim. Acta 2019, 1046, 123–131. [Google Scholar] [CrossRef]
- Randviir, E.P. A cross examination of electron transfer rate constants for carbon screen-printed electrodes using electrochemical impedance spectroscopy and cyclic voltammetry. Electrochim. Acta 2018, 286, 179–186. [Google Scholar] [CrossRef]
- Shukla, R.; Bansal, V.; Chaudhary, M.; Basu, A.; Bhonde, R.R.; Sastry, M. Biocompatibility of gold nanoparticles and their endocytotic fate inside the cellular compartment: A microscopic overview. Langmuir 2005, 21, 10644–10654. [Google Scholar] [CrossRef]
- Pengfei, J.; Wang, Y.; Zhao, L.; Ji, C.; Chen, D.; Nie, L. Applications of gold nanoparticles in non-optical biosensors. Nanomaterials 2018, 8, 977. [Google Scholar] [CrossRef]
- Li, Y.; Xu, S.; Schluesener, H.J. Gold nanoparticle-based biosensors. Gold Bull. 2010, 43, 29–41. [Google Scholar] [CrossRef]
- Koo, K.M.; Carrascosa, L.G.; Shiddiky, M.J.A.; Trau, M. Amplification-free detection of gene fusions in prostate cancer urinary samples using mRNA–Gold affinity interactions. Anal. Chem. 2016, 88, 6781–6788. [Google Scholar] [CrossRef]
- Islam, M.N.; Masud, M.K.; Haque, M.S.; Al Hossain, M.S.; Yamauchi, Y.; Nguyen, N.T.; Shiddiky, M.J.A. A PCR-free electrochemical method for messenger RNA detection in cancer tissue samples. Biosens. Bioelectron. 2017, 1, 15–24. [Google Scholar] [CrossRef]
- Murugappan, K.; Sundaramoorthy, U.; Damry, A.M.; Nisbet, D.R.; Jackson, C.J.; Tricoli, A. Electrodetection of small molecules by conformation-mediated signal enhancement. JACS Au 2022, 2, 2481–2490. [Google Scholar] [CrossRef] [PubMed]
- Umer, M.; Aziz, N.B.; Al Jabri, S.; Bhuiyan, S.A.; Shiddiky, M.J.A. Naked eye evaluation and quantitative detection of the sugarcane leaf scald pathogen, Xanthomonas albilineans, in sugarcane xylem sap. Crop Pasture Sci. 2021, 72, 361–371. [Google Scholar] [CrossRef]
- Siddiquee, S.; Rovina, K.; Yusof, N.A.; Rodrigues, K.F.; Suryani, S. Nanoparticle-enhanced electrochemical biosensor with DNA immobilization and hybridization of Trichoderma harzianum gene. Sens. Bio-Sens. Res. 2014, 2, 16–22. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).