Correction: Chakraborty et al. A DNA Adsorption-Based Biosensor for Rapid Detection of Ratoon Stunting Disease in Sugarcane. Biosensors 2025, 15, 518
Error in Figure
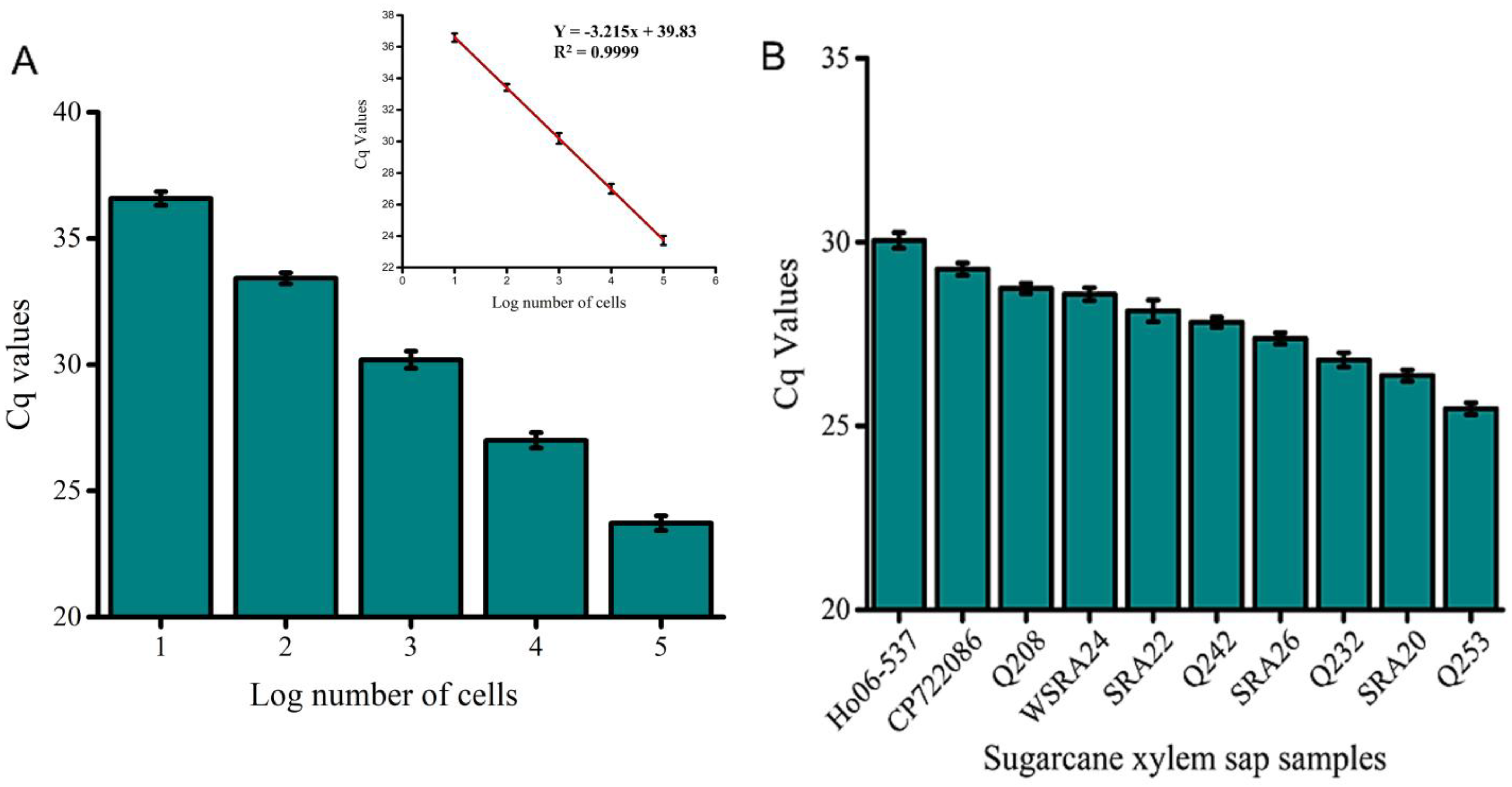
- Slope (m): −3.215 ± 0.011.
- Intercept (b): 39.828 ± 0.037.
- Coefficient of determination (R2): 0.99996.
- Amplification efficiency: 104.7%.
Reference
- Chakraborty, M.; Bhuiyan, S.A.; Strachan, S.; Shiddiky, M.J.A.; Nguyen, N.-T.; Soda, N.; Ford, R. A DNA Adsorption-Based Biosensor for Rapid Detection of Ratoon Stunting Disease in Sugarcane. Biosensors 2025, 15, 518. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chakraborty, M.; Bhuiyan, S.A.; Strachan, S.; Shiddiky, M.J.A.; Nguyen, N.-T.; Soda, N.; Ford, R. Correction: Chakraborty et al. A DNA Adsorption-Based Biosensor for Rapid Detection of Ratoon Stunting Disease in Sugarcane. Biosensors 2025, 15, 518. Biosensors 2025, 15, 646. https://doi.org/10.3390/bios15100646
Chakraborty M, Bhuiyan SA, Strachan S, Shiddiky MJA, Nguyen N-T, Soda N, Ford R. Correction: Chakraborty et al. A DNA Adsorption-Based Biosensor for Rapid Detection of Ratoon Stunting Disease in Sugarcane. Biosensors 2025, 15, 518. Biosensors. 2025; 15(10):646. https://doi.org/10.3390/bios15100646
Chicago/Turabian StyleChakraborty, Moutoshi, Shamsul Arafin Bhuiyan, Simon Strachan, Muhammad J. A. Shiddiky, Nam-Trung Nguyen, Narshone Soda, and Rebecca Ford. 2025. "Correction: Chakraborty et al. A DNA Adsorption-Based Biosensor for Rapid Detection of Ratoon Stunting Disease in Sugarcane. Biosensors 2025, 15, 518" Biosensors 15, no. 10: 646. https://doi.org/10.3390/bios15100646
APA StyleChakraborty, M., Bhuiyan, S. A., Strachan, S., Shiddiky, M. J. A., Nguyen, N.-T., Soda, N., & Ford, R. (2025). Correction: Chakraborty et al. A DNA Adsorption-Based Biosensor for Rapid Detection of Ratoon Stunting Disease in Sugarcane. Biosensors 2025, 15, 518. Biosensors, 15(10), 646. https://doi.org/10.3390/bios15100646









