Metagenomics Reveal Dynamic Coastal Ocean Reservoir of Antibiotic Resistance Genes
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and DNA Extraction
2.2. Library Preparation, Sequencing, and Analysis
2.3. Antibiotic Resistance Analysis
2.4. Environmental Analysis
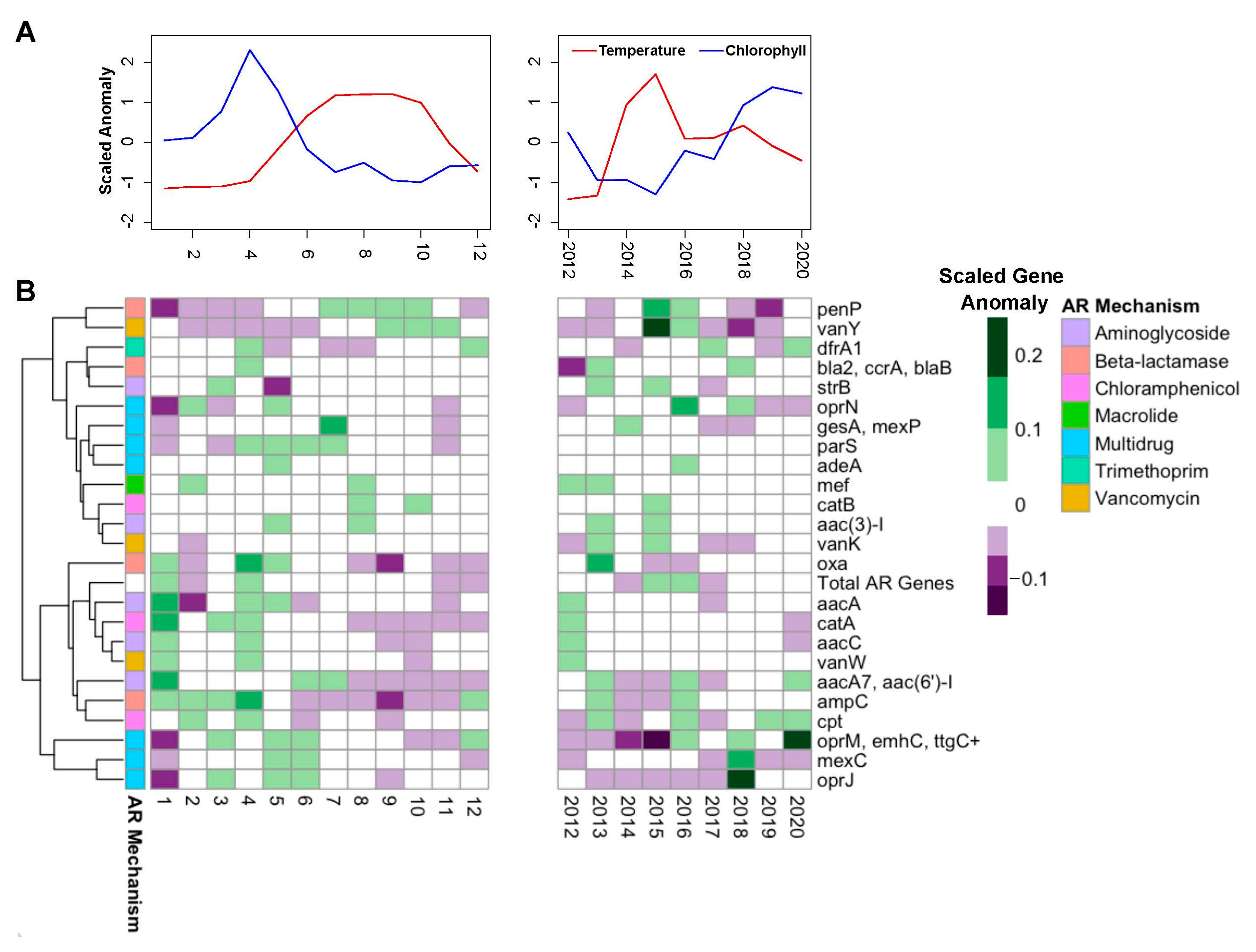
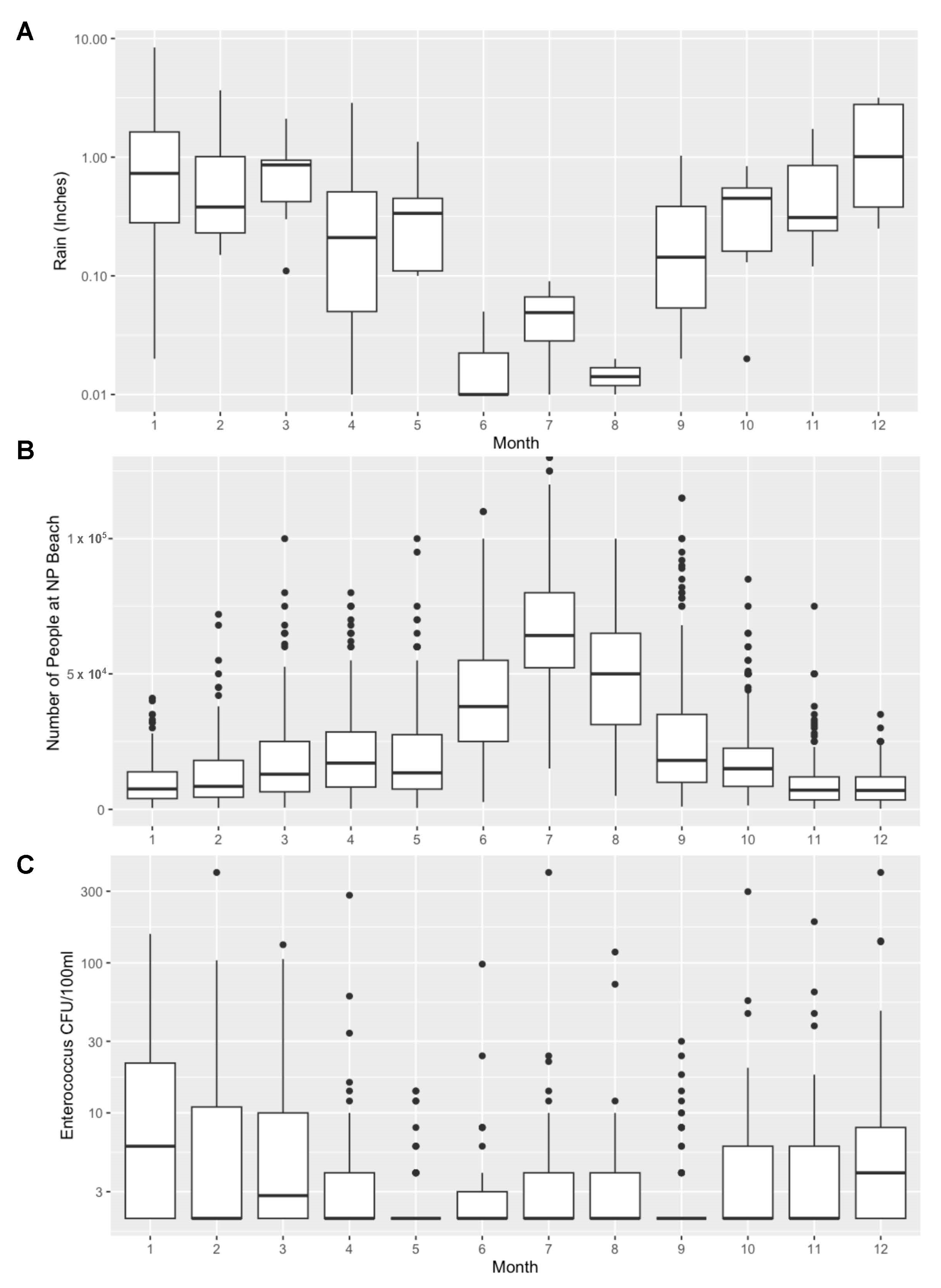
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Leonard, A.F.; Morris, D.; Schmitt, H.; Gaze, W.H. Natural recreational waters and the risk that exposure to antibiotic resistant bacteria poses to human health. Curr. Opin. Microbiol. 2022, 65, 40–46. [Google Scholar] [CrossRef] [PubMed]
- Leonard, A.F.; Zhang, L.; Balfour, A.J.; Garside, R.; Gaze, W.H. Human recreational exposure to antibiotic resistant bacteria in coastal bathing waters. Environ. Int. 2015, 82, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Burnham, J.P. Climate change and antibiotic resistance: A deadly combination. Ther. Adv. Infect. Dis. 2021, 8, 1–7. [Google Scholar] [CrossRef] [PubMed]
- MacFadden, D.R.; McGough, S.F.; Fisman, D.; Santillana, M.; Brownstein, J.S. Antibiotic resistance increases with local temperature. Nat. Clim. Chang. 2018, 8, 510–514. [Google Scholar] [CrossRef]
- Philipsborn, R.; Ahmed, S.M.; Brosi, B.J.; Levy, K. Climatic Drivers of Diarrheagenic Escherichia coli Incidence: A Systematic Review and Meta-analysis. J. Infect. Dis. 2016, 214, 6–15. [Google Scholar] [CrossRef]
- Rodríguez-Verdugo, A.; Gaut, B.S.; Tenaillon, O. Evolution of Escherichia coli rifampicin resistance in an antibiotic-free environment during thermal stress. BMC Evol. Biol. 2013, 13, 50. [Google Scholar] [CrossRef]
- Yang, Y.; Li, Z.; Song, W.; Du, L.; Ye, C.; Zhao, B.; Liu, W.; Deng, D.; Pan, Y.; Lin, H.; et al. Metagenomic insights into the abundance and composition of resistance genes in aquatic environments: Influence of stratification and geography. Environ. Int. 2019, 127, 371–380. [Google Scholar] [CrossRef]
- Nesme, J.; Cécillon, S.; Delmont, T.O.; Monier, J.-M.; Vogel, T.M.; Simonet, P. Large-Scale Metagenomic-Based Study of Antibiotic Resistance in the Environment. Curr. Biol. 2014, 24, 1096–1100. [Google Scholar] [CrossRef]
- Williams, N.L.; Siboni, N.; McLellan, S.L.; Potts, J.; Scanes, P.; Johnson, C.; James, M.; McCann, V.; Seymour, J.R. Rainfall leads to elevated levels of antibiotic resistance genes within seawater at an Australian beach. Environ. Pollut. 2022, 307, 119456. [Google Scholar] [CrossRef]
- Hernandez, R.; Acedo, I.; Dillon, J.G. Impact of wave action and rainfall on incidence and antibiotic resistance of total coliforms in Southern California beaches. J. Water Health 2020, 18, 766–775. [Google Scholar] [CrossRef]
- Choi, S.; Chu, W.; Brown, J.; Becker, S.J.; Harwood, V.J.; Jiang, S.C. Application of enterococci antibiotic resistance patterns for contamination source identification at Huntington Beach, California. Mar. Pollut. Bull. 2003, 46, 748–755. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Su, H.; Pan, Y.-F.; Xu, X.-R. Spatial and seasonal variations of antibiotics and antibiotic resistance genes and ecological risks in the coral reef regions adjacent to two typical islands in South China Sea. Mar. Pollut. Bull. 2020, 158, 111424. [Google Scholar] [CrossRef]
- Alves, M.S.; Pereira, A.; Araãºjo, S.M.; Castro, B.B.; Correia, A.C.M.; Henriques, I. Seawater is a reservoir of multi-resistant Escherichia coli, including strains hosting plasmid-mediated quinolones resistance and extended-spectrum beta-lactamases genes. Front. Microbiol. 2014, 5, 426. [Google Scholar] [CrossRef]
- Cohen, R.; Paikin, S.; Rokney, A.; Rubin-Blum, M.; Astrahan, P. Multidrug-resistant enterobacteriaceae in coastal water: An emerging threat. Antimicrob. Resist. Infect. Control. 2020, 9, 169. [Google Scholar] [CrossRef] [PubMed]
- Hatosy, S.M.; Martiny, A.C. The Ocean as a Global Reservoir of Antibiotic Resistance Genes. Appl. Environ. Microbiol. 2015, 81, 7593–7599. [Google Scholar] [CrossRef]
- Port, J.A.; Cullen, A.C.; Wallace, J.C.; Smith, M.N.; Faustman, E.M. Metagenomic Frameworks for Monitoring Antibiotic Resistance in Aquatic Environments. Environ. Heal. Perspect. 2014, 122, 222–228. [Google Scholar] [CrossRef]
- Belding, C.; Boopathy, R. Presence of antibiotic-resistant bacteria and antibiotic resistance genes in coastal recreational waters of southeast Louisiana, USA. J. Water Supply Res. Technol. 2018, 67, 800–809. [Google Scholar] [CrossRef]
- Carney, R.L.; Labbate, M.; Siboni, N.; Tagg, K.A.; Mitrovic, S.M.; Seymour, J.R. Urban beaches are environmental hotspots for antibiotic resistance following rainfall. Water Res. 2019, 167, 115081. [Google Scholar] [CrossRef]
- Lu, J.; Zhang, Y.; Wu, J. Continental-scale spatio-temporal distribution of antibiotic resistance genes in coastal waters along coastline of China. Chemosphere 2020, 247, 125908. [Google Scholar] [CrossRef]
- Makkaew, P.; Kongprajug, A.; Chyerochana, N.; Sresung, M.; Precha, N.; Mongkolsuk, S.; Sirikanchana, K. Persisting antibiotic resistance gene pollution and its association with human sewage sources in tropical marine beach waters. Int. J. Hyg. Environ. Health 2021, 238, 113859. [Google Scholar] [CrossRef]
- Šamanić, I.; Kalinić, H.; Fredotović, Ž.; Dželalija, M.; Bungur, A.-M.; Maravić, A. Bacteria tolerant to colistin in coastal marine environment: Detection, microbiome diversity and antibiotic resistance genes’ repertoire. Chemosphere 2021, 281, 130945. [Google Scholar] [CrossRef] [PubMed]
- Gerken, T.J.; Roberts, M.C.; Dykema, P.; Melly, G.; Lucas, D.; Santos, V.D.L.; Gonzalez, J.; Butaye, P.; Wiegner, T.N. Environmental Surveillance and Characterization of Antibiotic Resistant Staphylococcus aureus at Coastal Beaches and Rivers on the Island of Hawai‘i. Antibiotics 2021, 10, 980. [Google Scholar] [CrossRef] [PubMed]
- Martiny, H.-M.; Munk, P.; Brinch, C.; Szarvas, J.; Aarestrup, F.M.; Petersen, T.N. Global Distribution of mcr Gene Variants in 214K Metagenomic Samples. mSystems 2022, 7, e0010522. [Google Scholar] [CrossRef]
- Rippy, M.A.; Stein, R.; Sanders, B.F.; Davis, K.; McLaughlin, K.; Skinner, J.F.; Kappeler, J.; Grant, S.B. Small Drains, Big Problems: The Impact of Dry Weather Runoff on Shoreline Water Quality at Enclosed Beaches. Environ. Sci. Technol. 2014, 48, 14168–14177. [Google Scholar] [CrossRef]
- Larkin, A.A.; Moreno, A.R.; Fagan, A.J.; Fowlds, A.; Ruiz, A.; Martiny, A.C. Persistent El Niño driven shifts in marine cyanobacteria populations. PLoS ONE 2020, 15, e0238405. [Google Scholar] [CrossRef]
- Martiny, A.C.; Talarmin, A.; Mouginot, C.; Lee, J.A.; Huang, J.S.; Gellene, A.G.; Caron, D.A. Biogeochemical interactions control a temporal succession in the elemental composition of marine communities. Limnol. Oceanogr. 2016, 61, 531–542. [Google Scholar] [CrossRef]
- Fagan, A.J.; Moreno, A.R.; Martiny, A.C. Role of ENSO Conditions on Particulate Organic Matter Concentrations and Elemental Ratios in the Southern California Bight. Front. Mar. Sci. 2019, 6, 368. [Google Scholar] [CrossRef]
- Larkin, A.A.; Brock, M.L.; Fagan, A.J.; Moreno, A.R.; Gerace, S.D.; Lees, L.E.; Suarez, S.A.; Eloe-Fadrosh, E.A.; Martiny, A.C. Climate-driven succession in marine microbiome biodiversity and biogeochemical function. Nat. Commun. 2025, 16, 3926. [Google Scholar] [CrossRef]
- Clum, A.; Huntemann, M.; Bushnell, B.; Foster, B.; Foster, B.; Roux, S.; Hajek, P.P.; Varghese, N.; Mukherjee, S.; Reddy, T.B.K.; et al. DOE JGI Metagenome Workflow. mSystems 2021, 6, e00804-20. [Google Scholar] [CrossRef]
- Hyatt, D.; Chen, G.-L.; Locascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef]
- Lomsadze, A.; Gemayel, K.; Tang, S.; Borodovsky, M. Modeling leaderless transcription and atypical genes results in more accurate gene prediction in prokaryotes. Genome Res. 2018, 28, 1079–1089. [Google Scholar] [CrossRef] [PubMed]
- Chan, P.P.; Lin, B.Y.; Mak, A.J.; Lowe, T.M. tRNAscan-SE 2.0: Improved detection and functional classification of transfer RNA genes. Nucleic Acids Res. 2021, 49, 9077–9096. [Google Scholar] [CrossRef]
- Kalvari, I.; Nawrocki, E.P.; Ontiveros-Palacios, N.; Argasinska, J.; Lamkiewicz, K.; Marz, M.; Griffiths-Jones, S.; Toffano-Nioche, C.; Gautheret, D.; Weinberg, Z.; et al. Rfam 14: Expanded coverage of metagenomic, viral and microRNA families. Nucleic Acids Res. 2021, 49, D192–D200. [Google Scholar] [CrossRef]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef]
- Ryu, J.Y.; Kim, H.U.; Lee, S.Y. Deep learning enables high-quality and high-throughput prediction of enzyme commission numbers. Proc. Natl. Acad. Sci. USA 2019, 116, 13996–14001. [Google Scholar] [CrossRef] [PubMed]
- Tatusov, R.L.; Galperin, M.Y.; Natale, D.A.; Koonin, E.V. The COG database: A tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000, 28, 33–36. [Google Scholar] [CrossRef]
- Haft, D.H. The TIGRFAMs database of protein families. Nucleic Acids Res. 2003, 31, 371–373. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef]
- Kiełbasa, S.M.; Wan, R.; Sato, K.; Horton, P.; Frith, M.C. Adaptive seeds tame genomic sequence comparison. Genome Res. 2011, 21, 487–493. [Google Scholar] [CrossRef]
- A Eloe-Fadrosh, E.; Ahmed, F.; Anubhav; Babinski, M.; Baumes, J.; Borkum, M.; Bramer, L.; Canon, S.; Christianson, D.S.; Corilo, Y.E.; et al. The National Microbiome Data Collaborative Data Portal: An integrated multi-omics microbiome data resource. Nucleic Acids Res. 2022, 50, D828–D836. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019, 28, 1947–1951. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Kawashima, M.; Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 2023, 51, D587–D592. [Google Scholar] [CrossRef]
- Cao, Q.; Sun, X.; Rajesh, K.; Chalasani, N.; Gelow, K.; Katz, B.; Shah, V.H.; Sanyal, A.J.; Smirnova, E. Effects of Rare Microbiome Taxa Filtering on Statistical Analysis. Front. Microbiol. 2021, 11, 607325. [Google Scholar] [CrossRef]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.-L.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef]
- Bucher, T.; Keren-Paz, A.; Hausser, J.; Olender, T.; Cytryn, E.; Kolodkin-Gal, I. An active β-lactamase is a part of an orchestrated cell wall stress resistance network of Bacillus subtilis and related rhizosphere species. Environ. Microbiol. 2019, 21, 1068–1085. [Google Scholar] [CrossRef] [PubMed]
- Chow, C.-E.T.; Sachdeva, R.; A Cram, J.; A Steele, J.; Needham, D.M.; Patel, A.; E Parada, A.; A Fuhrman, J. Temporal variability and coherence of euphotic zone bacterial communities over a decade in the Southern California Bight. ISME J. 2013, 7, 2259–2273. [Google Scholar] [CrossRef]
- Jang, J.; Park, J.; Hwang, C.Y.; Choi, J.; Shin, J.; Kim, Y.M.; Cho, K.H.; Kim, J.-H.; Lee, Y.M.; Lee, B.Y. Abundance and diversity of antibiotic resistance genes and bacterial communities in the western Pacific and Southern Oceans. Sci. Total. Environ. 2022, 822, 153360. [Google Scholar] [CrossRef]
- Xu, L.; Wu, Y.-H.; Jian, S.-L.; Wang, C.-S.; Wu, M.; Cheng, L.; Xu, X.-W. Pseudohongiella nitratireducens sp. nov., isolated from seawater, and emended description of the genus Pseudohongiella. Int. J. Syst. Evol. Microbiol. 2016, 66, 5155–5160. [Google Scholar] [CrossRef]
- Du, Z.-J.; Wang, Z.-J.; Zhao, J.-X.; Chen, G.-J. Woeseia oceani gen. nov., sp. nov., a chemoheterotrophic member of the order Chromatiales, and proposal of Woeseiaceae fam. nov. Int. J. Syst. Evol. Microbiol. 2016, 66, 107–112. [Google Scholar] [CrossRef]
- Wang, L.; Shao, Z. Aerobic Denitrification and Heterotrophic Sulfur Oxidation in the Genus Halomonas Revealed by Six Novel Species Characterizations and Genome-Based Analysis. Front. Microbiol. 2021, 12, 652766. [Google Scholar] [CrossRef] [PubMed]
- Ren, W.-T.; Cheng, H.; Wang, C.-S.; Xu, X.-W.; Zhou, P.; Wu, Y.-H. Physiological and genomic features of Henriciella with the description of Henriciella mobilis sp. nov. Int. J. Syst. Evol. Microbiol. 2021, 71, 004889. [Google Scholar] [CrossRef] [PubMed]
- Nedashkovskaya, O.I.; Kim, S.B.; Vancanneyt, M.; Snauwaert, C.; Lysenko, A.M.; Rohde, M.; Frolova, G.M.; Zhukova, N.V.; Mikhailov, V.V.; Bae, K.S.; et al. Formosa agariphila sp. nov., a budding bacterium of the family Flavobacteriaceae isolated from marine environments, and emended description of the genus Formosa. Int. J. Syst. Evol. Microbiol. 2006, 56, 161–167. [Google Scholar] [CrossRef] [PubMed]
- Baker-Austin, C.; Oliver, J.D.; Alam, M.; Ali, A.; Waldor, M.K.; Qadri, F.; Martinez-Urtaza, J. Vibrio spp. infections. Nat. Rev. Dis. Prim. 2018, 4, 1–19. [Google Scholar] [CrossRef]
- Dvořák, P.; Casamatta, D.A.; Poulíčková, A.; Hašler, P.; Ondřej, V.; Sanges, R. Synechococcus: 3 billion years of global dominance. Mol. Ecol. 2014, 23, 5538–5551. [Google Scholar] [CrossRef]
- Loch, T.P.; Faisal, M. Emerging flavobacterial infections in fish: A review. J. Adv. Res. 2015, 6, 283–300. [Google Scholar] [CrossRef]
- Sharma, M.; Khurana, H.; Singh, D.N.; Negi, R.K. The genus Sphingopyxis: Systematics, ecology, and bioremediation potential—A review. J. Environ. Manag. 2021, 280, 111744. [Google Scholar] [CrossRef]
- Talwar, C.; Nagar, S.; Kumar, R.; Scaria, J.; Lal, R.; Negi, R.K. Defining the Environmental Adaptations of Genus Devosia: Insights into its Expansive Short Peptide Transport System and Positively Selected Genes. Sci. Rep. 2020, 10, 1151. [Google Scholar] [CrossRef]
- González, V.; Santamaría, R.I.; Bustos, P.; Pérez-Carrascal, O.M.; Vinuesa, P.; Juárez, S.; Martínez-Flores, I.; Cevallos, M.; Brom, S.; Martinez-Romero, E.; et al. Phylogenomic Rhizobium Species Are Structured by a Continuum of Diversity and Genomic Clusters. Front. Microbiol. 2019, 10, 910. [Google Scholar] [CrossRef]
- Durand, G.; Lagier, J.-C.; Khelaifia, S.; Armstrong, N.; Robert, C.; Rathored, J.; Fournier, P.-E.; Raoult, D. Drancourtella massiliensis gen. nov., sp. nov. isolated from fresh healthy human faecal sample from South France. New Microbes New Infect. 2016, 11, 34–42. [Google Scholar] [CrossRef]
- Criscuolo, A.; Issenhuth-Jeanjean, S.; Didelot, X.; Thorell, K.; Hale, J.; Parkhill, J.; Thomson, N.R.; Weill, F.-X.; Falush, D.; Brisse, S. The speciation and hybridization history of the genus Salmonella. Microb. Genom. 2019, 5, e000284. [Google Scholar] [CrossRef]
- Rivera-Mendoza, D.; Martínez-Flores, I.; Santamaría, R.I.; Lozano, L.; Bustamante, V.H.; Pérez-Morales, D. Genomic Analysis Reveals the Genetic Determinants Associated with Antibiotic Resistance in the Zoonotic Pathogen Campylobacter spp. Distributed Globally. Front. Microbiol. 2020, 11, 513070. [Google Scholar] [CrossRef] [PubMed]
- Ngom, I.I.; Hasni, I.; Lo, C.I.; Traore, S.I.; Fontanini, A.; Raoult, D.; Fenollar, F. Taxono-genomics and description of Gordonibacter massiliensis sp. nov., a new bacterium isolated from stool of healthy patient. New Microbes New Infect. 2020, 33, 100624. [Google Scholar] [CrossRef]
- Wurdemann, D.; Tindall, B.J.; Pukall, R.; Lunsdorf, H.; Strompl, C.; Namuth, T.; Nahrstedt, H.; Wos-Oxley, M.; Ott, S.; Schreiber, S.; et al. Gordonibacter pamelaeae gen. nov., sp. nov., a new member of the Coriobacteriaceae isolated from a patient with Crohn’s disease, and reclassification of Eggerthella hongkongensis Lau et al. 2006 as Paraeggerthella hongkongensis gen. nov., comb. nov. Int. J. Syst. Evol. Microbiol. 2009, 59, 1405–1415. [Google Scholar] [CrossRef] [PubMed]
- Hage, J.E.; Schoch, P.E.; Cunha, B.A. Pseudomonas Pseudoalcaligenes Peritoneal Dialysis–Associated Peritonitis. Perit. Dial. Int. J. Int. Soc. Perit. Dial. 2013, 33, 223–224. [Google Scholar] [CrossRef] [PubMed]
- Yumoto, I.; Yamazaki, K.; Hishinuma, M.; Nodasaka, Y.; Suemori, A.; Nakajima, K.; Inoue, N.; Kawasaki, K. Pseudomonas alcaliphila sp. nov., a novel facultatively psychrophilic alkaliphile isolated from seawater. Int. J. Syst. Evol. Microbiol. 2001, 51, 349–355. [Google Scholar] [CrossRef]
- Peter, S.; Oberhettinger, P.; Schuele, L.; Dinkelacker, A.; Vogel, W.; Dörfel, D.; Bezdan, D.; Ossowski, S.; Marschal, M.; Liese, J.; et al. Genomic characterisation of clinical and environmental Pseudomonas putida group strains and determination of their role in the transfer of antimicrobial resistance genes to Pseudomonas aeruginosa. BMC Genom. 2017, 18, 859. [Google Scholar] [CrossRef]
- Salvà-Serra, F.; Pérez-Pantoja, D.; Donoso, R.A.; Jaén-Luchoro, D.; Fernández-Juárez, V.; Engström-Jakobsson, H.; Moore, E.R.B.; Lalucat, J.; Bennasar-Figueras, A. Comparative genomics of Stutzerimonas balearica (Pseudomonas balearica): Diversity, habitats, and biodegradation of aromatic compounds. Front. Microbiol. 2023, 14, 1159176. [Google Scholar] [CrossRef]
- Diggle, S.P.; Whiteley, M. Microbe Profile: Pseudomonas aeruginosa: Opportunistic pathogen and lab rat. Microbiology 2020, 166, 30–33. [Google Scholar] [CrossRef]
- Alcalde-Rico, M.; Olivares-Pacheco, J.; Alvarez-Ortega, C.; Cámara, M.; Martínez, J.L. Role of the Multidrug Resistance Efflux Pump MexCD-OprJ in the Pseudomonas aeruginosa Quorum Sensing Response. Front. Microbiol. 2018, 9, 2752. [Google Scholar] [CrossRef]
- Luna, R.A.; Fasciano, L.R.; Jones, S.C.; Boyanton, B.L.; Ton, T.T.; Versalovic, J. DNA Pyrosequencing-Based Bacterial Pathogen Identification in a Pediatric Hospital Setting. J. Clin. Microbiol. 2007, 45, 2985–2992. [Google Scholar] [CrossRef] [PubMed]
- Jara, D.; Bello-Toledo, H.; Domínguez, M.; Cigarroa, C.; Fernández, P.; Vergara, L.; Quezada-Aguiluz, M.; Opazo-Capurro, A.; Lima, C.A.; González-Rocha, G. Antibiotic resistance in bacterial isolates from freshwater samples in Fildes Peninsula, King George Island, Antarctica. Sci. Rep. 2020, 10, 3145. [Google Scholar] [CrossRef] [PubMed]
- Overbey, K.N.; Hatcher, S.M.; Stewart, J.R. Water quality and antibiotic resistance at beaches of the Galápagos Islands. Front. Environ. Sci. 2015, 3, 64. [Google Scholar] [CrossRef]
- Mote, B.L.; Turner, J.W.; Lipp, E.K. Persistence and Growth of the Fecal Indicator Bacteria Enterococci in Detritus and Natural Estuarine Plankton Communities. Appl. Environ. Microbiol. 2012, 78, 2569–2577. [Google Scholar] [CrossRef] [PubMed]
- Powers, N.C.; Wallgren, H.R.; Marbach, S.; Turner, J.W. Relationship between Rainfall, Fecal Pollution, Antimicrobial Resistance, and Microbial Diversity in an Urbanized Subtropical Bay. Appl. Environ. Microbiol. 2020, 86, e01229-20. [Google Scholar] [CrossRef]
- Dong, C.; Idica, E.Y.; McWilliams, J.C. Circulation and multiple-scale variability in the Southern California Bight. Prog. Oceanogr. 2009, 82, 168–190. [Google Scholar] [CrossRef]
- Allison, S.D.; Chao, Y.; Farrara, J.D.; Hatosy, S.; Martiny, A.C. Fine-Scale Temporal Variation in Marine Extracellular Enzymes of Coastal Southern California. Front. Microbiol. 2012, 3, 31955. [Google Scholar] [CrossRef]
- Hunter-Cevera, K.R.; Neubert, M.G.; Olson, R.J.; Solow, A.R.; Shalapyonok, A.; Sosik, H.M. Physiological and ecological drivers of early spring blooms of a coastal phytoplankter. Science 2016, 354, 326–329. [Google Scholar] [CrossRef]
- Six, C.; Ratin, M.; Marie, D.; Corre, E. Marine Synechococcus picocyanobacteria: Light utilization across latitudes. Proc. Natl. Acad. Sci. USA 2021, 118, e2111300118. [Google Scholar] [CrossRef]
- Mackey, K.R.M.; Hunter-Cevera, K.; Britten, G.L.; Murphy, L.G.; Sogin, M.L.; Huber, J.A. Seasonal Succession and Spatial Patterns of Synechococcus Microdiversity in a Salt Marsh Estuary Revealed through 16S rRNA Gene Oligotyping. Front. Microbiol. 2017, 8, 1496. [Google Scholar] [CrossRef]
- Deeb, R.; Tufford, D.; Scott, G.I.; Moore, J.G.; Dow, K. Impact of Climate Change on Vibrio vulnificus Abundance and Exposure Risk. Estuaries Coasts 2018, 41, 2289–2303. [Google Scholar] [CrossRef] [PubMed]
- Pachiadaki, M.G.; Brown, J.M.; Brown, J.; Bezuidt, O.; Berube, P.M.; Biller, S.J.; Poulton, N.J.; Burkart, M.D.; La Clair, J.J.; Chisholm, S.W.; et al. Charting the Complexity of the Marine Microbiome through Single-Cell Genomics. Cell 2019, 179, 1623–1635.e11. [Google Scholar] [CrossRef] [PubMed]
- Hong, P.-Y.; Mantilla-Calderon, D.; Wang, C. Metagenomics as a Tool to Monitor Reclaimed-Water Quality. Appl. Environ. Microbiol. 2020, 86, e00724-20. [Google Scholar] [CrossRef] [PubMed]


| ARG Type | KO a | Gene a | Genus 1 b | Env. b | Genus 2 c | Env. c |
|---|---|---|---|---|---|---|
| Beta-lactamase | K01467 | ampC | Pseudohongiella | A | Thalassomonas | H |
| K17836 | penP | Synechococcus | A | Chryseolinea | E | |
| K17837 | bla2, ccrA, blaB | Woeseia | A | Pseudohongiella | A | |
| K17838 | oxa | Pseudohongiella | A | Unclassified Verrucomicrobiae | A/E/H | |
| Aminoglycoside | K00662 | aacC | Campylobacter | A | Lishizhenia | A |
| K00663 | aacA | Cognatiyoonia | A/E | Candidatus Planktophila | A | |
| K03395 | aac(3)-I | Sphingopyxis | E | Alcanivorax | A | |
| K04343 | strB | Devosia | E | Drancourtella | H | |
| K18816 | aacA7, aac(6′)-I | Rhizobium | E | Salmonella | H | |
| Multidrug | K08721 | oprJ | Pseudomonas | A/E/H | Marinobacter | A |
| K18139 | oprM, emhC, ttgC, cusC, adeK, smeF, mtrE, cmeC, gesC | Pseudomonas | A/E/H | Psychrobacter | A/E/H | |
| K18295 | mexC | Pseudomonas | A/E/H | Halioglobus | A/H | |
| K18072 | parS | Pseudomonas | A/E/H | Bradyrhizobium | E/H | |
| K18300 | oprN | Salinisphaera | A | Pseudomonas | A/E/H | |
| K19595 | gesA, mexP | Unclassified Halieaceae | A | Limibacillus | E/H | |
| K18145 | adeA | SAR116 | A | Halomonas | A | |
| Chloramphenicol | K00638 | catB | Halomonas | A | Acinetobacter | A/E/H |
| K18554 | cpt | Henriciella | A | Streptomyces | A/E/H | |
| K19271 | catA | Formosa | A | Mucilaginibacter | A/E | |
| Macrolide | K08217 | mef | Gordonibacter | H | Rhodoluna | A |
| Trimethoprim | K18589 | dfrA1 | Vibrio | A | ||
| Vancomycin | K07260 | vanY | Synechococcus | A | Prochlorococcus | A |
| K18346 | vanW | Flavobacterium | A | Synechococcus | A | |
| K18354 | vanK | Catenulispora | E | Nocardioides | A/E/H |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Suarez, S.A.; Larkin, A.A.; Brock, M.L.; Moreno, A.R.; Fagan, A.J.; Martiny, A.C. Metagenomics Reveal Dynamic Coastal Ocean Reservoir of Antibiotic Resistance Genes. J. Mar. Sci. Eng. 2025, 13, 1165. https://doi.org/10.3390/jmse13061165
Suarez SA, Larkin AA, Brock ML, Moreno AR, Fagan AJ, Martiny AC. Metagenomics Reveal Dynamic Coastal Ocean Reservoir of Antibiotic Resistance Genes. Journal of Marine Science and Engineering. 2025; 13(6):1165. https://doi.org/10.3390/jmse13061165
Chicago/Turabian StyleSuarez, Stacy A., Alyse A. Larkin, Melissa L. Brock, Allison R. Moreno, Adam J. Fagan, and Adam C. Martiny. 2025. "Metagenomics Reveal Dynamic Coastal Ocean Reservoir of Antibiotic Resistance Genes" Journal of Marine Science and Engineering 13, no. 6: 1165. https://doi.org/10.3390/jmse13061165
APA StyleSuarez, S. A., Larkin, A. A., Brock, M. L., Moreno, A. R., Fagan, A. J., & Martiny, A. C. (2025). Metagenomics Reveal Dynamic Coastal Ocean Reservoir of Antibiotic Resistance Genes. Journal of Marine Science and Engineering, 13(6), 1165. https://doi.org/10.3390/jmse13061165







