Abstract
Background: Patient blood management (PBM) is a patient-centered, evidence-based approach for optimizing anemia management, minimizing blood loss, and ensuring appropriate transfusion. Artificial intelligence (AI) provides powerful tools for prediction, diagnosis, and decision support across PBM, but current evidence remains emerging and not yet consolidated. Objectives: This review synthesizes AI applications in PBM, summarizing predictive, diagnostic, and decision support models; highlighting methodological trends; and discussing challenges for clinical translation. Methods: PubMed, Scopus, and Web of Science were searched from inception to 31 March 2025. Eligible studies reported AI models addressing the three established PBM pillars. Studies on transfusion safety and blood bank operations relevant to PBM were also included. Extracted data covered study characteristics, predictors, models, validation strategies, and performance. The findings were narratively synthesized given study heterogeneity. Results: A total of 338 studies were included, spanning anemia detection, bleeding risk stratification, transfusion prediction, transfusion safety, and inventory management. Deep learning (DL) predominated in image-based anemia detection, while ensemble and gradient boosting methods frequently outperformed baselines in bleeding and transfusion risk prediction. Recurrent and hybrid architectures proved effective for blood supply forecasting. Across domains, machine learning and DL models generally surpassed logistic regression, clinical scores, and expert judgment. Despite strong internal performance, external validation and clinical deployment remain limited. Conclusions: AI is advancing PBM by enabling earlier anemia detection, more accurate bleeding and transfusion prediction, and smarter resource allocation. Translation into practice requires standardized reporting, robust external validation, explainability, and workflow integration. Future work should emphasize multimodal learning, prospective evaluation, and cost-effectiveness.
1. Introduction
Patient blood management (PBM) has emerged as a global standard of care recognized by the World Health Organization (WHO) for optimizing transfusion practice [1]. By reducing unnecessary transfusions and correcting anemia or bleeding disorders before interventions, PBM lowers complication risks, accelerates recovery, and decreases healthcare costs [1]. At the system level, PBM also safeguards blood supply adequacy by promoting efficient use and minimizing wastage [1].
In parallel, artificial intelligence (AI) has rapidly advanced across healthcare, providing data-driven solutions for prediction, decision support, and resource management [2]. Within PBM, AI holds promise. Algorithms can integrate diverse patient data—including demographics, laboratory results, and imaging—to predict anemia and transfusion requirements, stratify bleeding risk, and personalize transfusion thresholds [2]. At the operational level, AI-based tools can forecast blood product demand, optimize inventory allocation, and streamline logistics, thereby reducing costs and enhancing supply resilience [2].
Integrating AI into PBM offers dual benefits: improved clinical outcomes and more efficient use of healthcare resources [2]. Nevertheless, despite a growing body of research, the field remains heterogeneous, and the translation of AI tools into routine practice is still limited [2]. A scoping review by Meier and Tschoellitsch synthetized available studies on AI in PBM up to October 2021, providing an important first overview of this emerging field [2]. Since then, the literature has expanded substantially, with new studies employing advanced machine learning (ML)/deep learning (DL) architectures and multimodal datasets [3].
The objective of this review is to map and critically appraise the current landscape of AI applications in PBM, building on previous scoping evidence and incorporating recent advances. We aim to highlight methodological trends, identify persisting gaps, and discuss the translational potential of AI-driven PBM strategies for clinical practice.
1.1. Current Landscape in PBM
PBM represents a comprehensive, evidence-based strategy tailored to optimize patient outcomes while minimizing unnecessary blood transfusions and their associated risks [4]. Key guidelines from professional organizations—including the American Association of Blood Banks (AABB) and the Society for the Advancement of Blood Management (SABM)—offer recommendations on transfusion thresholds, clinical indications, and alternatives to transfusion, thereby promoting consistency in clinical practice [5,6]. At a global policy level, the WHO published its Guidance on Implementing Patient Blood Management to Improve Global Blood Health Status in 2025 [7]. This landmark document urges healthcare systems to adopt PBM as an essential element of safe and sustainable care, particularly in resource-limited settings. It outlines a phase-wise implementation strategy, including system-level preparation, pilot program rollout, and national scale-up, and provides contextualized toolkits adapted to varying resource levels. The WHO emphasizes that PBM has the potential to enhance blood health, reduce anemia-related morbidity, and narrow global health inequities [7].
Despite these advances, several challenges persist. Variability in transfusion practice across institutions leads to inconsistent decision-making and, at times, suboptimal outcomes [8]. Determining appropriate transfusion thresholds can be complex and requires careful consideration of individual patient factors, underlying comorbidities, and contextual clinical circumstances [9]. Achieving the balance between avoiding unnecessary transfusions and ensuring adequate oxygen delivery remains a key clinical dilemma. Moreover, blood products are finite and costly resources. Securing a reliable supply, particularly under emergency or resource-constrained scenarios, is both essential and challenging. The COVID-19 pandemic, for instance, starkly illuminated the potential fragility of blood systems amid supply disruptions [10]. These multifaceted challenges underscore the need for ongoing research, structured implementation, and coordinated collaboration among healthcare stakeholders. PBM’s multidisciplinary and patient-centered approach could enhance the quality, safety, and efficiency of blood management practices, ultimately improving patient outcomes, reducing costs, and conserving essential resources. In this context, AI offers new opportunities to address persistent challenges in PBM by reducing practice variability, refining transfusion thresholds, enhancing decision support, and optimizing blood resource utilization, thereby complementing and strengthening evidence-based transfusion practices [11,12].
1.2. AI Models Relevant to PBM
AI applications in PBM have evolved rapidly, supporting anemia detection, bleeding risk stratification, transfusion prediction, transfusion safety, and blood supply optimization [12]. Across the literature, models can be grouped into five broad categories:
- (i)
- Traditional machine learning models: Logistic regression (LR), decision trees (DT), support vector machines (SVM), naïve Bayes (NB), and k-nearest neighbors (KNN) remain widely applied to structured data. These are often benchmarked against clinical scores and are valued for their interpretability in practice [13].
- (ii)
- Ensemble methods: Random forests (RF), gradient boosting (GB), AdaBoost, and voting classifiers (VC) frequently outperform single algorithms, particularly when applied to heterogeneous or imbalanced datasets [13].
- (iii)
- DL architectures: Deep neural networks (DNN), convolutional neural networks (CNN), and recurrent models such as long short-term memory (LSTM) and gated recurrent units (GRUs) are well suited to high-dimensional and temporal data, including laboratory time series, imaging, and electronic health records (EHRs). Transformer-based models (e.g., BERT, ViT) and attention mechanisms are emerging for tasks such as radiomics, sequential pattern recognition, and analysis of clinical narratives [14].
- (iv)
- Probabilistic and interpretable models: Bayesian networks, Gaussian processes, and physics-informed neural networks enable uncertainty estimation and offer greater transparency, critical in clinical decision-making contexts such as transfusion thresholds or rare blood group management [13].
- (v)
- Hybrid and meta-learning approaches: Variational autoencoders, stacked generalization, and neuro-fuzzy systems facilitate multimodal integration of laboratory, imaging, and clinical data, aligning well with the multidimensional demands of PBM. In this review, hybrid models are defined as approaches that combine two or more distinct algorithms (e.g., ML with DL, or ensembles of heterogeneous methods) to leverage complementary strengths and improve overall performance [14].
Across categories, AI models consistently outperform traditional regression and established clinical scoring systems. Their ability to integrate diverse data sources and adapt across clinical domains highlights their translational potential for real-world PBM strategies [12].
1.3. Aims and Objectives
This review provides a comprehensive synthesis of AI applications in PBM, focusing on models that perform the following functions:
- (i)
- Predict anemia, transfusion requirements, or bleeding risk;
- (ii)
- Support transfusion decision-making;
- (iii)
- Enhance transfusion safety;
- (iv)
- Optimize blood bank operations.
For each included study, we summarized the PBM domain, sample size and case group characteristics, model inputs (variables and predictors), model types and implementation details, validation approach, and reported performance. This structured synthesis highlights methodological trends, clinical applications, and the translational potential of AI-driven strategies in PBM.
2. Materials and Methods
2.1. Eligibility Criteria
We included original studies that reported the development, validation, or application of AI models within PBM. Eligible domains encompassed the following: (i) prediction of anemia, transfusion requirements, or bleeding risk; (ii) transfusion decision support; (iii) transfusion safety; (iv) blood bank operations and management. Studies were excluded if they did not involve AI methods, were unrelated to PBM, focused solely on laboratory techniques without clinical application, were conducted on animals or in vitro, or were reviews, editorials, commentaries, or conference abstracts. Publications were restricted to English, Portuguese, and Spanish.
This review was not registered in PROSPERO because the protocol was finalized and data extraction completed prior to submission. However, it was conducted in accordance with PRISMA 2020 guidelines [15]. The full PRISMA checklist is provided in the Supplementary Materials.
2.2. Search Strategy
PubMed, Scopus, and Web of Science were searched from inception to 31 March 2025. The PubMed query was (“machine learning” [All Fields] OR “artificial intelligence” [All Fields]) AND (“anemia” [All Fields] OR “bleeding” [All Fields] OR “transfusion” [All Fields] OR “patient blood management” [All Fields]), retrieving 1882 records. Equivalent searches yielded 1534 in Scopus and 265 in Web of Science. In Scopus, the search was limited to articles. After removing duplicates (n = 1691), 1990 records remained. Title/abstract screening excluded 1514, leaving 472 for full-text assessment. Of these, 134 were excluded (beyond PBM scope: 88; no ML/DL model used or developed: 24; outcomes not separated for transfusion/bleeding/anemia: 9; methodological/reporting limitations: 6; no real patient data: 7). The study selection process is summarized in Figure 1. In total, 338 studies were included. No additional records were identified through other sources, and all full texts were available. The complete list of included studies, with full references, is provided in the Supplementary Materials, while Tables S1–S4 summarize the distribution of studies across anemia, bleeding, transfusion, and other PBM domains.
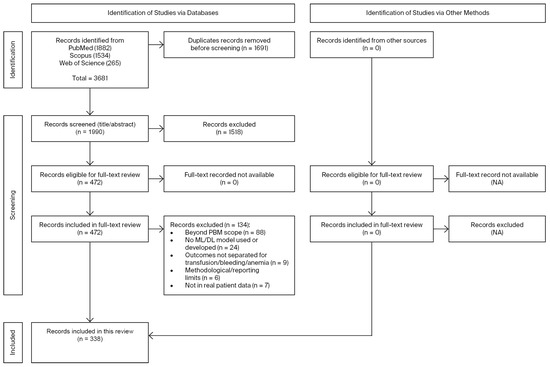
Figure 1.
PRISMA flow diagram of study selection. Of 3681 records identified, 1990 were screened after duplicates were removed, and 338 studies met the inclusion criteria.
2.3. Study Selection
Records retrieved from the databases were imported into Zotero (v7.0.24, Corporation for Digital Scholarship, Fairfax, VA, USA), and duplicates were removed. Two reviewers independently screened titles and abstracts; full texts of potentially eligible articles were assessed against the predefined criteria. Disagreements were resolved through consensus or, when needed, by a third reviewer. The study selection process is illustrated in a PRISMA flow diagram (Figure 1).
2.4. Data Collection Process
Two reviewers independently extracted data using a piloted form. Extracted items included reference (first author, year), PBM domain, sample size and case group characteristics, model inputs and top predictors, AI model(s) used, validation strategy, and reported performance metrics. Discrepancies were resolved through consensus or by a third reviewer.
2.5. Risk of Bias
No single risk-of-bias tool was applied due to the heterogeneity of the models, outcomes, and validation strategies. Instead, two reviewers qualitatively assessed common risks in prediction modeling: small sample size, class imbalance, data leakage, handling of missing data, validation rigor, and adequacy of performance evaluation. Disagreements were resolved through consensus or by a third reviewer.
2.6. Synthesis
As part of this systematic review, we conducted a structured narrative synthesis due to substantial clinical and methodological heterogeneity across studies. Included studies were grouped by PBM domain: anemia detection or prediction, bleeding risk, transfusion prediction and decision support, transfusion safety, and blood bank operations. Within each domain, we summarized the study setting, data modality, model type, validation approach (internal, external, or temporal), and performance. When multiple models were tested on the same dataset, the most rigorously validated or best-performing model was prioritized to avoid redundancy. Calibration findings and explainability techniques were reported when available. Statistical summaries were performed using IBM SPSS Statistics, version 30.0 (IBM Corp., Armonk, NY, USA).
All 338 included studies are listed with full citation details in Supplementary Tables S1–S4. To maintain clarity and conciseness in the main text, only representative or flagship studies discussed in Section 3 are included in the reference list. A complete list of studies excluded at the full-text screening stage, along with reasons for exclusion, is provided in Supplementary Table S5.
3. Results: AI Applications in PBM
AI has been applied across the full spectrum of PBM, targeting both clinical decision-making and operational optimization. The studies reviewed here illustrate applications aligned with the three classical PBM pillars—optimizing red cell mass, minimizing blood loss and bleeding, and harnessing tolerance of anemia—as well as transfusion safety and blood bank operations. Evidence is organized into five domains: (i) anemia detection and prediction, where AI supports early diagnosis and proactive management; (ii) bleeding prediction, encompassing high-risk settings such as cardiology, gastroenterology, and critical care; (iii) prediction of transfusion requirements, including perioperative and ICU contexts; (iv) decision support and transfusion safety, where AI augments or surpasses clinical scores and enhances laboratory compatibility testing; and (v) blood bank operations and resource optimization, where AI improves demand forecasting, inventory management, and waste reduction. This structure reflects both the clinical and operational aspects of PBM, underscoring AI’s potential to improve patient outcomes while promoting sustainability in blood product use.
3.1. Anemia Detection and Prediction
AI has been increasingly applied to anemia detection and prediction using imaging, physiological signals, laboratory data, and multimodal approaches. DL applied to ocular and facial images achieved strong non-invasive performance. In a dataset of >5800 retinal images, InceptionV3 achieved 98% accuracy and an AUC (area under the curve) of 0.98 for anemia detection, with a mean absolute error (MAE) of 0.58 g/dL for hemoglobin (Hb) estimation, validated in an external cohort [16]. Stacked ensemble models trained on palmar or conjunctival images reported near-perfect discrimination, with AUCs ranging from 0.97 to 1.0 and accuracies close to 100% [17,18]. Similarly, CNNs applied to palmar, fingernail, and conjunctival images consistently achieved excellent results, with reported accuracies above 97% and AUCs up to 0.999 [19].
Signal- and physiology-based approaches have expanded the scope of non-invasive anemia assessment. CNNs trained on electrocardiogram features from nearly 40,000 patients achieved AUCs of 0.92 internally and 0.90 externally, outperforming LR [20]. Nailbed optical spectra analyzed with ANN correlated strongly with laboratory Hb (r = 0.987) [21]. Fingertip video-based CNNs and fusion models also showed promise: the best Fusion-1 model achieved an RMSE (root mean square error) of 0.63 and an MSE (mean square error) of 0.61, with sensitivity of 0.97 and specificity of 0.68 [22]. A multimodal CNN fusion model integrating fingertip video signals achieved accuracy of 98.4% and an AUC of 0.988, demonstrating excellent discriminatory ability in anemia detection [23].
Multimodal models have demonstrated added value for early prediction; in the UK Biobank, combining fundus images with demographic metadata improved discrimination compared with metadata alone (AUC 0.88 vs. 0.74) [24]. Recurrent neural networks (RNN) trained on laboratory time-series predicted iron deficiency anemia (IDA) up to six months before conventional diagnosis, with GRUs achieving an AUC of 0.89 [25]. In dialysis populations, GRU and LSTM models forecasted Hb trajectories with MAEs of 0.5–0.7 g/dL, values comparable to routine laboratory variability, supporting their potential role in proactive erythropoiesis-stimulating agent titration [26,27].
AI has also been applied to hematologic conditions with direct implications for PBM. Differentiating IDA from thalassemia is clinically critical, as the two require distinct management strategies. SVM and XGB (XGBoost, Extreme Gradient Boosting) models achieved AUCs of 0.88–0.96 and consistently outperformed classical discriminant formulas [28,29,30]. RF and CNN models achieved AUCs around 0.95, with one study reporting a 60% reduction in unnecessary genetic testing [31,32]. For sickle cell disease, CNN and SVM classifiers achieved accuracies ranging from ~90% to >95% using blood smears, digital scans, and microfluidic imaging [33,34,35].
3.2. Bleeding Prediction
In cardiology, AI models trained on national PCI (percutaneous coronary intervention) registries have consistently outperformed established clinical scores. In a dataset of more than 3.3 million PCI procedures, XGB improved bleeding prediction compared with the NCDR risk model (C-statistic 0.82 vs. 0.78) [36]. Validation in over 100,000 PCI cases confirmed superior performance, with AUCs of 0.887 for bleeding and 0.917 for transfusion, both exceeding LR [37]. Another large-scale study demonstrated that an AI-based bleeding risk model achieved an AUC of 0.873, outperforming the ACC-BR score [38]. Collectively, these findings highlight AI’s ability to enhance peri-procedural bleeding risk stratification in interventional cardiology.
In gastroenterology, AI methods have consistently outperformed guideline-based scores and expert assessment. Gradient boosting models predicted variceal bleeding in cirrhosis with AUCs of 0.94 in internal and 0.86 in external validation, surpassing endoscopic classification [39]. A multimodal stacking ensemble achieved an external AUC of 0.975, outperforming MELD and Child–Pugh scores [40]. DL applied to endoscopic images alone reached an external accuracy of 0.893, exceeding the performance of experienced endoscopists [41].
In critical care and surgery, DL models have been evaluated for bleeding surveillance and perioperative risk assessment. GRU-based RNN predicted ICU bleeding risk in patients on antithrombotic therapy more accurately and more rapidly than clinicians (AUC 0.83) [42]. In cardiothoracic surgery, analysis of more than 660,000 cases showed that NN achieved an AUC of 0.97 for postoperative bleeding [43]. Additional ML studies in ICU and surgical populations have further confirmed robust predictive performance in high-risk perioperative contexts [44].
Across multiple surgical specialties, ensemble methods have further demonstrated strong generalizability. In a dataset of 48,543 perioperative cases, Light Gradient Boosting Machine (LightGBM) models achieved an AUC of 0.933 for intraoperative bleeding, validated externally [45]. These findings underscore the ability of AI to generalize across heterogeneous surgical populations, supporting hospital-wide PBM strategies for perioperative bleeding risk prediction.
3.3. Prediction of Transfusion Requirements
In surgical populations, ensemble methods such as GB and LGBM consistently outperformed LR and established transfusion scores, achieving strong discrimination in very large datasets. In a study of over four million surgeries, GB reduced unnecessary type-and-screen orders by one-third, translating into measurable cost savings, while LGBM achieved an AUC of 0.93 in more than 100,000 procedures with external validation [46,47].
In intraoperative settings, DL has enabled real-time prediction of massive transfusion. Recurrent architectures such as GRUs anticipated transfusion needs up to ten minutes in advance, maintaining AUCs above 0.94 across both internal and external validation cohorts. These models demonstrate the feasibility of integrating streaming physiological and laboratory data into perioperative decision support [48,49].
In critical care, ML ensembles have been used to support transfusion decisions for patients with gastrointestinal bleeding. By combining LR with RF, these models achieved high sensitivity and strong performance across external validation cohorts of more than 4000 patients, reinforcing their potential for deployment in acute care settings [50].
3.4. Decision Support and Transfusion Safety
AI-based decision support systems have demonstrated superiority to traditional risk scores in transfusion-related decision-making. In upper gastrointestinal bleeding, XGB models achieved AUCs of around 0.90 and significantly outperformed established clinical tools such as Rockall, AIMS65, and the Glasgow Blatchford Score, with consistent performance after external validation. These findings suggest that AI can provide more accurate triage and intervention support than guideline-based scoring systems [51].
Causal inference methods using ML have also been applied to evaluate transfusion strategies and donor characteristics. Targeted maximum likelihood estimation and SuperLearner ensembles provided insights into differential risks associated with restrictive versus liberal transfusion strategies in acute myocardial infarction, the impact of donor sex on recipient outcomes, and the role of plasma-to-RBC ratios in trauma resuscitation. These applications highlight AI’s potential not only for prediction but also for estimating causal effects, thereby informing transfusion policy and clinical practice [52,53,54].
In transfusion safety, AI has been applied to genomic, phenotypic, and imaging data. RF and XGB models predicted blood group antigens in over 110,000 donors with near-perfect accuracy, substantially improving precision in donor–recipient matching. DL approaches enhanced RH genotyping concordance, while CNN ensembles automated Coombs test interpretation with >99% accuracy, surpassing expert immunologists. These advances demonstrate the feasibility of embedding AI into laboratory workflows to strengthen compatibility testing and reduce adverse events [55,56,57].
3.5. Blood Bank Operations and Resource Optimization
AI models have been increasingly applied to blood bank operations, where efficient demand forecasting and inventory management are central to PBM. GB and ensemble methods trained on large surgical datasets improved transfusion risk prediction and reduced unnecessary testing. In a study of more than four million procedures, gradient boosting reduced type-and-screen orders by one-third, generating substantial cost savings [46]. Similarly, in over 100,000 surgeries, LGBM achieved an AUC of 0.93 with external validation, supporting the generalizability of such models [47].
Time-series approaches have also shown strong performance in blood inventory management. A hybrid model combining seasonal trend decomposition with XGB reduced red cell inventory by 38%, eliminated urgent orders, and lowered costs by 43% [58]. For platelet demand, LSTM models tailored to hematology/oncology patients achieved high accuracy (AUC 0.98), supporting early identification of shortages in vulnerable populations [59]. Ensemble learning has also proven effective for daily RBC demand forecasting, improving alignment with actual usage and reducing supply chain risk [60].
AI has also been applied to identify inefficiencies and minimize wastage in blood product supply chains. Association rule mining and RNN uncovered drivers of product disposal and returns, offering actionable insights for optimizing blood product utilization. These studies illustrate AI’s role not only in predictive modeling but also in uncovering hidden patterns that inform stewardship and sustainability in blood banking [61,62].
3.6. Comparative Distribution of Model Performance
The distribution of reported model performance is summarized in Figure 2, using boxplots of AUC values stratified by model category (ML, DL, and hybrid). To avoid redundancy, only the best-performing model from each study was extracted. Thus, the plots reflect the upper performance bound for each domain rather than the full range of algorithms tested.
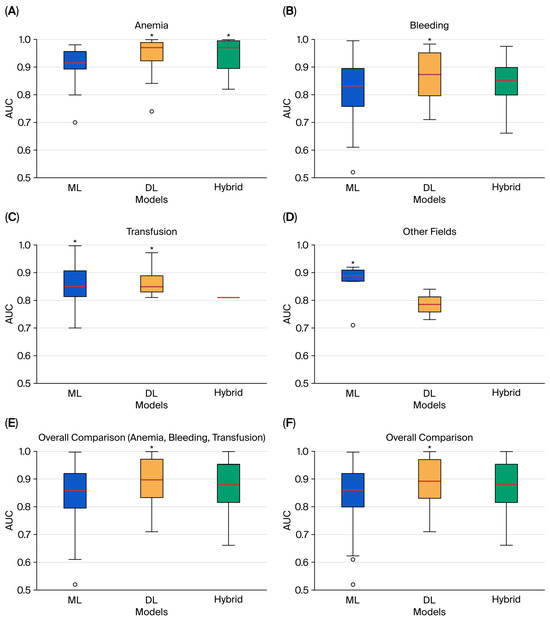
Figure 2.
Comparative boxplots of best-performing AUC values for ML, DL, and hybrid models across domains: (A) anemia; (B) bleeding; (C) transfusion; (D) other fields; (E) overall comparison of anemia, bleeding, and transfusion; (F) overall comparison of all domains. Boxes represent the interquartile range, the line inside the box shows the median, and circles denote outliers. An asterisk (*) marks the model category with the highest median AUC.
In anemia detection and treatment, both ML (n = 50) and DL (n = 48) models were frequently applied, while hybrid approaches were less common (n = 16). DL and hybrid models consistently achieved higher AUCs, clustering near or above 0.95 with narrow interquartile ranges, while ML model showed greater dispersion. These findings align with the strong results of image-based DL architectures and ensemble methods highlighted in Supplementary Table S1.
In bleeding prediction, ML approaches were most common (n = 81), compared with DL (n = 20) and hybrid (n = 16). The distributions were more heterogeneous: ML performance displayed a wider spread and included several lower outliers (AUC < 0.80), while DL and hybrid models demonstrated higher medians and reduced variability, consistent with their role in perioperative and interventional risk prediction (Supplementary Table S2).
In transfusion prediction, ML again predominated (n = 62), compared with DL (n = 18) and hybrid (n = 2). DL models dominated among the top performers, while only a single hybrid model reached best-in-class status. ML models exhibited greater variability, suggesting performance was highly dependent on dataset quality and feature selection (Supplementary Table S3).
In the other fields category, which included transfusion safety and blood bank operations, the total number of models was limited (ML: 13; DL: 9; hybrid: 5). Here, no hybrid model was identified as the best performer. Both ML and DL achieved competitive AUCs, though variability was greater than in the anemia domain (Supplementary Table S4).
When aggregating across domains, the overall comparison plots confirmed these trends (Figure 2E,F). DL and hybrid models generally displayed higher central tendencies, while ML results covered a broader range, reflecting both their greater representation in the literature and their sensitivity to dataset heterogeneity. These findings emphasize that, across PBM domains, DL and hybrid approaches more often reached the highest performance levels, particularly in image-rich and multimodal tasks. At the same time, the imbalance in model counts—with ML overwhelmingly more common in bleeding and transfusion—helps explain the wider variability observed for ML in the boxplots. Trends in the publication of ML, DL, and hybrid PBM studies over time are shown in Figure 3.
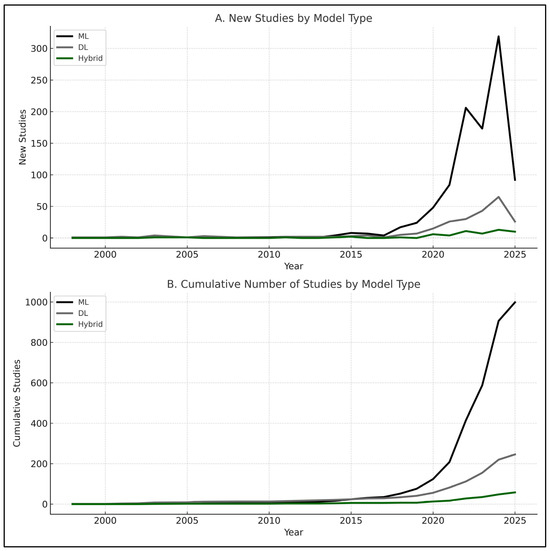
Figure 3.
Trends in studies applying ML, DL, and hybrid models over time: (A) new studies per year by model type (1998–2025); (B) cumulative number of studies by model type (1998–2025). ML dominates in absolute numbers, while DL and hybrid show accelerated growth in recent years. Note: 2025 data include publications up to March only.
Overall, AI applications in PBM have demonstrated robust potential across clinical and operational domains. From early anemia detection and bleeding risk stratification to real-time transfusion forecasting, decision support, and blood bank optimization, AI models frequently outperformed LR, clinical scores, and even expert judgment in selected settings. Large-scale studies with external validation highlight the translational promise of these approaches, while real-time and multimodal models illustrate the feasibility of integration into perioperative and critical care workflows. At the same time, evidence remains heterogeneous, with variable study quality, limited calibration reporting, and a paucity of prospective validations. These findings underscore the need for critical appraisal of current evidence, careful consideration of limitations, and identification of future directions, which are addressed in the following section.
4. Discussion and Future Directions
This review highlights both the progress and the limitations of AI applications in PBM. Most AI models included in this review were trained and validated exclusively on retrospective datasets. While such datasets allow scalable model development, retrospective designs introduce risk of bias, unmeasured confounding, and potential data leakage. As a result, high retrospective performance cannot be assumed to translate into real-world clinical benefit. To determine true utility and safety, future work must incorporate prospective, real-world evaluations—such as silent-mode deployment within EHR or pragmatic clinical trials—to assess how AI models behave in everyday PBM workflows. A further challenge relates to generalizability. Several high-performing AI models were developed using national or multinational datasets, yet such models may not perform equivalently when applied to smaller or demographically distinct institutions. Differences in laboratory methods, transfusion practices, case mix, and disease prevalence can significantly reduce model accuracy when transferred to local contexts. Therefore, external validation alone is insufficient; local calibration, transfer learning, or domain adaptation may be required before these models can be safely implemented in smaller centers. It is also important to emphasize that AI should not be regarded as a universal replacement for traditional statistical modeling. In settings characterized by limited sample size, low event prevalence, or restricted access to robust data infrastructures, classical approaches—such as logistic regression or guideline-based clinical scores—may remain more reliable, interpretable, and clinically appropriate. In such environments, AI may serve as a complementary tool rather than a primary decision support modality. Across domains, AI models frequently outperformed LR and established clinical scores when direct comparisons were performed. In interventional cardiology, XGB improved bleeding risk stratification in more than 3.3 million PCI procedures and outperformed the NCDR model [36], while Hamilton et al. [37] confirmed these findings in over 100,000 PCI cases with external validation. In upper gastrointestinal bleeding, XGB surpassed Rockall, AIMS65, and the Glasgow Blatchford Score [51]. In internal medicine, ML achieved superior discrimination compared with RIETE and VTE-BLEED scores in anticoagulated patients [63,64]. Large-scale studies with external validation further demonstrated generalizability, including transfusion prediction across more than 100,000 surgeries [47], perioperative bleeding in 48,543 procedures [45], and postoperative bleeding in over 660,000 cardiothoracic cases [43].
Several studies extended beyond prediction to address causal inference and transfusion safety. Portela et al. [54] and Nguyen et al. [53] applied targeted learning to evaluate restrictive versus liberal transfusion strategies and plasma-to-RBC ratios in trauma, while Hyvärinen et al. [56] and Chang et al. [55] used ML and DL for precise antigen prediction and RH genotyping, enhancing transfusion safety. Operational studies showed measurable system-level benefits: Lou et al. [46] demonstrated that GB reduced type-and-screen orders by 33% across more than four million procedures, and Li et al. [58] reported that XGB-based forecasting cut RBC inventory by 38% and costs by 43%. Collectively, these findings underscore AI’s ability to deliver both clinical and operational gains, reinforcing its potential to transform PBM practice.
The comparative boxplots provide an integrated view of model performance across PBM domains. By restricting the analysis to the best-performing model from each study, the figures illustrate the upper bound of achievable AUC. In anemia detection, DL models—tested in nearly equal numbers to ML—consistently clustered above 0.95, confirming their robustness in image-rich tasks. In bleeding prediction, the predominance of ML studies explains the wider variability observed, while DL and hybrid approaches reached higher and more stable medians. In transfusion prediction, DL again outperformed ML, with only a single hybrid model achieving best performer status. For other fields, where study numbers were modest, no hybrid model was the top performer. Taken together, these distributions show that DL and hybrid methods more often achieved the highest and most consistent AUCs, whereas ML displayed broader variability, reflecting both methodological diversity and their numerical predominance across domains. This predominance of ML, despite higher AUCs reported for DL and hybrid in some domains, likely reflects its simplicity, lower computational requirements, and greater interpretability. In addition, ML approaches were introduced earlier in PBM research, leading to a larger accumulated body of studies compared with newer DL and hybrid methods.
Despite these advances, several limitations were consistently observed. Many models were developed on relatively small datasets or highly imbalanced case–control groups, limiting generalizability. Examples include anemia detection in a rare cohort [65,66] and bleeding prediction in orthopedics, where only 293 bleeding cases were reported among >35,000 procedures [67]. Postpartum hemorrhage studies also often involved <2% prevalence, reducing model stability. Larger, multicenter datasets are needed to ensure representativeness, particularly for low-prevalence outcomes.
Validation strategies also limited confidence in many studies. A large proportion relied solely on internal hold-out or k-fold cross-validation [17,18,19]. While these methods mitigate overfitting, they cannot replace independent testing. Stronger designs incorporated external validation, such as Khan et al. [16] for retinal imaging in anemia, Shi et al. [45] for perioperative bleeding, Zapf et al. [47] for transfusion forecasting, and Abbasi et al. [43] for cardiothoracic bleeding. Systematic external and temporal validation should become standard practice.
Benchmarking was also inconsistent. Several models were reported without comparison to LR or established clinical tools, limiting interpretability. Where such comparisons were made, AI consistently outperformed conventional approaches. Notable examples include Mortazavi et al. [36] and Hamilton et al. [37] against NCDR scores, Shung et al. [51] against Rockall and AIMS65, and Mora et al. [63] against RIETE and VTE-BLEED. Ensuring consistent benchmarking against LR and specialty-specific tools will be crucial for clinician confidence.
Performance reporting was heterogeneous. Some studies reported only accuracy, which can be misleading in imbalanced datasets, while others provided AUC, sensitivity, specificity, or F1-score. Calibration, essential for clinical adoption, was rarely assessed; few studies reported Brier scores. Adoption of transparent reporting of a multivariable prediction model for individual prognosis or diagnosis/AI and the prediction model risk-of-bias assessment tool/AI reporting standards will improve transparency and comparability.
Almost all studies were retrospective. Only a few incorporated prospective validations, such as Mora et al. [63] in anticoagulated patients and Almeida et al. [66] in cytomorphology. Furthermore, most papers stopped at model development and did not explore integration into workflows, EHR, or transfusion services. Prospective, pragmatic trials and pilot implementations are urgently needed to demonstrate real-world utility, workflow feasibility, and clinician acceptance.
Explainability was another recurring gap. DL models applied to imaging and time-series data often lacked interpretable outputs. Few applied SHapley Additive exPlanations or Local Interpretable Model-agnostic Explanations to identify drivers of predictions. For PBM, where decisions carry immediate transfusion consequences, explainability is critical for clinician trust and regulatory approval.
Taken together, these limitations highlight both the promise and immaturity of AI in PBM. Addressing them will require larger, more representative datasets, systematic external and prospective validation, consistent benchmarking against established tools, and greater emphasis on explainability and workflow integration. Future research should also explore multimodal and federated learning to improve generalizability while preserving data privacy, and cost-effectiveness analyses to establish system-level value. By advancing along these lines, AI can evolve from proof-of-concept models into clinically embedded tools that enhance safety, efficiency, and sustainability in PBM.
5. Limitations of This Review and Conclusions
This review has several limitations that should be acknowledged. Although we conducted comprehensive searches in PubMed, Scopus, and Web of Science, relevant studies indexed elsewhere or unpublished work may have been missed, introducing potential publication bias. Only studies published in English, Portuguese, and Spanish were included, which may have excluded evidence in other languages. While we systematically extracted study characteristics and performance metrics, heterogeneity in reporting limited direct comparisons across studies. For example, some reported only accuracy, while others provided AUC, sensitivity, specificity, or calibration metrics, precluding formal meta-analysis or pooled synthesis. In addition, our scope was limited to studies that explicitly reported AI applications in PBM; related work in adjacent domains may not have been captured. Finally, although we applied structured search and screening methods, this was not a formal systematic review, and risk-of-bias assessments were conducted qualitatively rather than with standardized scoring tools. Although AI demonstrates a consistently strong retrospective performance across PBM domains, the absence of prospective clinical validation remains a critical limitation. Most models have not yet been assessed in real-time clinical settings, and their performance in smaller or heterogeneous local cohorts is largely unknown. These gaps restrict the immediate translational impact of current AI applications in PBM and highlight the need for multicenter, prospective, and workflow-integrated evaluations before clinical adoption.
Despite these limitations, this review provides the most comprehensive and up-to-date synthesis of AI applications in PBM to date. Across anemia detection, bleeding risk stratification, transfusion forecasting, decision support, and blood bank operations, AI models frequently outperformed LR, established clinical scores, and even expert judgment. Large-scale studies with external validation demonstrated strong generalizability, and several reports showed tangible system-level benefits such as reduced type-and-screen orders, lower inventory requirements, and cost savings. At the same time, current evidence remains heterogeneous, limited by small or imbalanced datasets, inconsistent validation, and scarce prospective evaluation. Future research should prioritize multicenter and prospective studies, standardized reporting, explainability, and integration into clinical workflows. By addressing these challenges, AI has the potential to transform PBM into a more precise, efficient, and sustainable practice, improving both patient outcomes and resource stewardship.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/jcm14238479/s1, Table S1: Optimization of red cell mass [16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,65,66,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111,112,113,114,115,116,117,118,119,120,121,122,123,124,125,126,127,128,129,130,131,132,133,134,135,136,137,138,139,140,141,142,143,144,145,146,147,148,149,150,151,152,153]; Table S2: Minimization of blood loss and bleeding [36,37,38,39,40,41,42,43,44,45,63,64,67,154,155,156,157,158,159,160,161,162,163,164,165,166,167,168,169,170,171,172,173,174,175,176,177,178,179,180,181,182,183,184,185,186,187,188,189,190,191,192,193,194,195,196,197,198,199,200,201,202,203,204,205,206,207,208,209,210,211,212,213,214,215,216,217,218,219,220,221,222,223,224,225,226,227,228,229,230,231,232,233,234,235,236,237,238,239,240,241,242,243,244,245,246,247,248,249,250,251,252,253,254,255,256,257,258,259,260,261]; Table S3: Harness and optimize physiological reserve of anemia [37,46,47,48,49,50,211,222,237,262,263,264,265,266,267,268,269,270,271,272,273,274,275,276,277,278,279,280,281,282,283,284,285,286,287,288,289,290,291,292,293,294,295,296,297,298,299,300,301,302,303,304,305,306,307,308,309,310,311,312,313,314,315,316,317,318,319,320,321,322,323,324,325,326,327,328,329]; Table S4: Other fields of indication [51,52,53,54,55,56,57,58,59,60,61,62,330,331,332,333,334,335,336,337,338,339,340,341,342,343,344,345,346,347,348,349]; Table S5: TabList of Abbreviations; PRISMA Checklist [349,350,351,352,353,354,355,356,357,358,359,360,361,362,363,364,365,366,367,368,369,370,371,372,373,374,375,376,377,378,379,380,381,382,383,384,385,386,387,388,389,390,391,392,393,394,395,396,397,398,399,400,401,402,403,404,405,406,407,408,409,410,411,412,413,414,415,416,417,418,419,420,421,422,423,424,425,426,427,428,429,430,431,432,433,434,435,436,437,438,439,440,441,442,443,444,445,446,447,448,449,450,451,452,453,454,455,456,457,458,459,460,461,462,463,464,465,466,467,468,469,470,471,472,473,474,475,476,477,478,479,480,481,482,483].
Author Contributions
Conceptualization, methodology, data curation, writing—original draft preparation, H.C.; writing—review and editing, M.C. and P.M.R.; supervision, F.S., M.C. and P.M.R.; project administration and funding acquisition, P.M.R. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Foundation for Science and Technology (FCT, Portugal) under project number UIDB/50016/2020.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data analyzed in this systematic review were extracted from previously published studies. The full list of included articles and extracted variables is provided in the Supplementary Materials (Tables S1–S5). No new datasets were generated. Additional information is available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Goobie, S.M. Patient blood management is a new standard of care to optimize blood health. Anesth. Analg. 2022, 135, 443–446. [Google Scholar] [CrossRef]
- Meier, J.M.; Tschoellitsch, T. Artificial intelligence and machine learning in patient blood management: A scoping review. Anesth. Analg. 2022, 135, 524–531. [Google Scholar] [CrossRef]
- Alowais, S.A.; Alghamdi, S.S.; Alsuhebany, N.; Alqahtani, T.; Alshaya, A.I.; Almohareb, S.N.; Aldairem, A.; Alrashed, M.; Bin Saleh, K.; Badreldin, H.A.; et al. Revolutionizing healthcare: The role of artificial intelligence in clinical practice. BMC Med. Educ. 2023, 23, 689. [Google Scholar] [CrossRef]
- Carson, J.L.; Stanworth, S.J.; Guyatt, G.; Valentine, S.; Dennis, J.; Bakhtary, S.; Cohn, C.S.; Dubon, A.; Grossman, B.J.; Gupta, G.K.; et al. Red blood cell transfusion: 2023 AABB international guidelines. JAMA 2023, 330, 1892–1902. [Google Scholar] [CrossRef]
- American Association of Blood Banks (AABB). Standards for a Patient Blood Management Program, 5th ed.; AABB: Bethesda, MD, USA, 2023; Available online: https://www.aabb.org/standards-accreditation/standards/patient-blood-management-program (accessed on 1 November 2025).
- Society for the Advancement of Blood Management (SABM). Administrative and Clinical Standards for Patient Blood Management Programs, 5th ed.; SABM: Mt. Royal, NJ, USA, 2021; Available online: https://sabm.memberclicks.net/s20-standard-showcase (accessed on 1 November 2025).
- World Health Organization. Guidance on Implementing Patient Blood Management to Improve Global Health Status; WHO: Geneva, Switzerland, 2025. Available online: https://iris.who.int/bitstream/handle/10665/380784/9789240104662-eng.pdf (accessed on 1 November 2025).
- Irving, A.; Harris, A.; Petrie, D.; Avdic, D.; Smith, J.; Tran, L.; Reid, C.M.; McQuilten, Z.K. Can clinical guidelines reduce variation in transfusion practice? A pre-post study of blood transfusions during cardiac surgery. Vox Sang. 2025, 120, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Delaforce, A.; Duff, J.; Munday, J.; Hardy, J. Overcoming barriers to evidence-based patient blood management: A restricted review. Implement. Sci. 2020, 15, 6. [Google Scholar] [CrossRef] [PubMed]
- Shander, A.; Goobie, S.M.; Warner, M.A.; Aapro, M.; Bisbe, E.; Perez-Calatayud, A.A.; Callum, J.; Cushing, M.M.; Dyer, W.B.; Erhard, J.; et al. Essential role of patient blood management in a pandemic: A call for action. Anesth. Analg. 2020, 131, 74–85. [Google Scholar] [CrossRef] [PubMed]
- Lou, S.S.; Kumar, S.; Goss, C.W.; Avidan, M.S.; Kheterpal, S.; Kannampallil, T. Multicenter validation of a machine learning model for surgical transfusion risk at 45 US hospitals. JAMA Netw. Open 2025, 8, e2517760. [Google Scholar] [CrossRef]
- Li, N.; Goel, R.; Raza, S.; Riazi, K.; Pan, J.; Nguyen, H.Q.; Shih, A.W.; D’Souza, A.; Dubey, R.; Tobian, A.A.R.; et al. Artificial intelligence and machine learning in transfusion practice: An analytical assessment. Transfus. Med. Rev. 2025, 39, 150926. [Google Scholar] [CrossRef]
- Sarker, I.H. Machine learning: Algorithms, real-world applications and research directions. SN Comput. Sci. 2021, 2, 160. [Google Scholar] [CrossRef]
- Sarker, I.H. Deep learning: A comprehensive overview on techniques, taxonomy, applications and research directions. SN Comput. Sci. 2021, 2, 420. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef]
- Khan, R.; Maseedupally, V.; Thakoor, K.A.; Raman, R.; Roy, M. Noninvasive anemia detection and hemoglobin estimation from retinal images using deep learning: A scalable solution for resource-limited settings. Transl. Vis. Sci. Technol. 2025, 14, 20. [Google Scholar] [CrossRef] [PubMed]
- Sehar, N.; Krishnamoorthi, N.; Kumar, C.V. Deep learning model-based detection of anemia from conjunctiva images. Healthc. Inform. Res. 2025, 31, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Appiahene, P.; Dogbe, S.S.D.; Kobina, E.E.Y.; Dartey, P.S.; Afrifa, S.; Donkoh, E.T.; Asare, J.W. Application of ensemble models approach in anemia detection using images of the palpable palm. Med. Nov. Technol. Devices 2023, 20, 100269. [Google Scholar] [CrossRef]
- Asare, J.W.; Appiahene, P.; Donkoh, E.T.; Dimauro, G. Iron deficiency anemia detection using machine learning models: A comparative study of fingernails, palm and conjunctiva of the eye images. Eng. Rep. 2023, 5, e12667. [Google Scholar] [CrossRef]
- Kwon, J.-M.; Cho, Y.; Jeon, K.H.; Cho, S.; Kim, K.H.; Baek, S.D.; Jeung, S.; Park, J.; Oh, B.H. A deep learning algorithm to detect anaemia with ECGs: A retrospective, multicentre study. Lancet Digit. Health 2020, 2, e358–e367. [Google Scholar] [CrossRef]
- Banerjee, A.; Bhattacharyya, N.; Ghosh, R.; Singh, S.; Adhikari, A.; Mondal, S.; Roy, L.; Bajaj, A.; Ghosh, N.; Bhushan, A.; et al. Non-invasive estimation of hemoglobin, bilirubin and oxygen saturation of neonates simultaneously using whole optical spectrum analysis at point of care. Sci. Rep. 2023, 13, 2370. [Google Scholar] [CrossRef]
- Das, S.; Kesarwani, A.; Dalui, M.; Kisku, D.R.; Sen, B.; Roy, S.; Basu, A. Smartphone-based non-invasive haemoglobin level estimation by analyzing nail pallor. Biomed. Signal Process. Control 2023, 85, 104959. [Google Scholar] [CrossRef]
- Valles-Coral, M.A.; Navarro-Cabrera, J.R.; Pinedo, L.; Injante, R.; Quintanilla-Morales, L.K.; Farro-Roque, M.E. Non-invasive detection of iron deficiency anemia in young adults through finger-tip video image analysis. Int. J. Online Biomed. Eng. 2024, 20, 53–70. [Google Scholar] [CrossRef]
- Mitani, A.; Huang, A.; Venugopalan, S.; Corrado, G.S.; Peng, L.; Webster, D.R.; Hammel, N.; Liu, Y.; Varadarajan, A.V. Detection of anaemia from retinal fundus images via deep learning. Nat. Biomed. Eng. 2019, 4, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Garduno-Rapp, N.E.; Ng, Y.S.; Weon, J.L.; Saleh, S.N.; Lehmann, C.U.; Tian, C.; Quinn, A. Early identification of patients at risk for iron-deficiency anemia using deep learning techniques. Am. J. Clin. Pathol. 2024, 162, 243–251. [Google Scholar] [CrossRef] [PubMed]
- Lobo, B.; Abdel-Rahman, E.; Brown, D.; Dunn, L.; Bowman, B. A recurrent neural network approach to predicting hemoglobin trajectories in patients with end-stage renal disease. Artif. Intell. Med. 2020, 104, 101823. [Google Scholar] [CrossRef] [PubMed]
- Pellicer-Valero, O.J.; Cattinelli, I.; Neri, L.; Mari, F.; Martín-Guerrero, J.D.; Barbieri, C. Enhanced prediction of hemoglobin concentration in a very large cohort of hemodialysis patients by means of deep recurrent neural networks. Artif. Intell. Med. 2020, 107, 101898. [Google Scholar] [CrossRef]
- Lachover-Roth, I.; Peretz, S.; Zoabi, H.; Harel, E.; Livshits, L.; Filon, D.; Levin, C.; Koren, A. Support vector machine-based formula for detecting suspected α thalassemia carriers: A path toward universal screening. Int. J. Mol. Sci. 2024, 25, 6446. [Google Scholar] [CrossRef]
- Schipper, A.; Rutten, M.; van Gammeren, A.; Harteveld, C.L.; Urrechaga, E.; Weerkamp, F.; den Besten, G.; Krabbe, J.; Slomp, J.; Schoonen, L.; et al. Machine learning-based prediction of hemoglobinopathies using complete blood count data. Clin. Chem. 2024, 70, 1064–1075. [Google Scholar] [CrossRef]
- Wang, W.; Ye, R.; Tang, B.; Qi, Y. MultiThal-classifier, a machine learning-based multi-class model for thalassemia diagnosis and classification. Clin. Chim. Acta 2025, 567, 120025. [Google Scholar] [CrossRef]
- Christensen, F.; Kılıç, D.K.; Nielsen, I.E.; El-Galaly, T.C.; Glenthøj, A.; Helby, J.; Frederiksen, H.; Möller, S.; Fuglkjær, A.D. Classification of α-thalassemia data using machine learning models. Comput. Methods Programs Biomed. 2025, 260, 108581. [Google Scholar] [CrossRef]
- Feng, P.; Li, Y.; Liao, Z.; Yao, Z.; Lin, W.; Xie, S.; Hu, B.; Huang, C.; Liu, W.; Xu, H.; et al. An online alpha-thalassemia carrier discrimination model based on random forest and red blood cell parameters for low HbA2 cases. Clin. Chim. Acta 2022, 525, 1–5. [Google Scholar] [CrossRef]
- Alagu, S.; Ganesan, K.; Bagan, B. A novel deep learning approach for sickle cell anemia detection in human RBCs using an improved wrapper-based feature selection technique in microscopic blood smear images. Biomed. Eng. Biomed. Techol. 2023, 68, 175–185. [Google Scholar] [CrossRef]
- Douglass, P.M.; O’Connor, T.; Javidi, B. Automated sickle cell disease identification in human red blood cells using a lensless single random phase encoding biosensor and convolutional neural networks. Opt. Express 2022, 30, 35965. [Google Scholar] [CrossRef]
- Sani, A.; Tian, Y.; Shah, S.; Khan, M.I.; Abdurrahman, H.R.; Zha, G.; Zhang, Q.; Liu, W.; Abdullahi, I.L.; Wang, Y.; et al. Deep learning ResNet34 model-assisted diagnosis of sickle cell disease via microcolumn isoelectric focusing. Anal. Methods 2024, 16, 6517–6528. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, B.J.; Bucholz, E.M.; Desai, N.R.; Huang, C.; Curtis, J.P.; Masoudi, F.A.; Shaw, R.E.; Negahban, S.N.; Krumholz, H.M. Comparison of machine learning methods with national cardiovascular data registry models for prediction of risk of bleeding after percutaneous coronary intervention. JAMA Netw. Open 2019, 2, e196835. [Google Scholar] [CrossRef]
- Hamilton, D.E.; Albright, J.; Seth, M.; Painter, I.; Maynard, C.; Hira, R.S.; Sukul, D.; Gurm, H.S. Merging machine learning and patient preference: A novel tool for risk prediction of percutaneous coronary interventions. Eur. Heart J. 2024, 45, 601–609. [Google Scholar] [CrossRef] [PubMed]
- Rayfield, C.; Agasthi, P.; Mookadam, F.; Yang, E.H.; Venepally, N.R.; Ramakrishna, H.; Slomka, P.; Holmes, D.R., Jr.; Arsanjani, R. Machine learning on high-dimensional data to predict bleeding post percutaneous coronary intervention. J. Invasive Cardiol. 2020, 32, E122–E129. [Google Scholar] [CrossRef]
- Agarwal, S.; Sharma, S.; Kumar, M.; Venishetty, S.; Bhardwaj, A.; Kaushal, K.; Gopi, S.; Mohta, S.; Gunjan, D.; Saraya, A.; et al. Development of a machine learning model to predict bleed in esophageal varices in compensated advanced chronic liver disease: A proof of concept. J. Gastroenterol. Hepatol. 2021, 36, 2935–2942. [Google Scholar] [CrossRef]
- Wang, Y.; Hong, Y.; Wang, Y.; Zhou, X.; Gao, X.; Yu, C.; Lin, J.; Liu, L.; Gao, J.; Yin, M.; et al. Automated multimodal machine learning for esophageal variceal bleeding prediction based on endoscopy and structured data. J. Digit. Imaging 2022, 36, 326–338. [Google Scholar] [CrossRef]
- Hong, Y.; Yu, Q.; Mo, F.; Yin, M.; Xu, C.; Zhu, S.; Lin, J.; Xu, G.; Gao, J.; Liu, L.; et al. Deep learning to predict esophageal variceal bleeding based on endoscopic images. J. Int. Med. Res. 2023, 51, 03000605231200371. [Google Scholar] [CrossRef]
- Chen, D.; Wang, R.; Jiang, Y.; Xing, Z.; Sheng, Q.; Liu, X.; Wang, R.; Xie, H.; Zhao, L. Application of artificial neural network in daily prediction of bleeding in ICU patients treated with anti-thrombotic therapy. BMC Med. Inform. Decis. Mak. 2023, 23, 171. [Google Scholar] [CrossRef]
- Abbasi, A.; Li, C.; Dekle, M.; Bermudez, C.A.; Brodie, D.; Sellke, F.W.; Sodha, N.R.; Ventetuolo, C.E.; Eickhoff, C. Interpretable machine learning-based predictive modeling of patient outcomes following cardiac surgery. J. Thorac. Cardiovasc. Surg. 2025, 169, 114–123.e28. [Google Scholar] [CrossRef]
- Taggart, M.; Chapman, W.W.; Steinberg, B.A.; Ruckel, S.; Pregenzer-Wenzler, A.; Du, Y.; Ferraro, J.; Bucher, B.T.; Lloyd-Jones, D.M.; Rondina, M.T.; et al. Comparison of 2 natural language processing methods for identification of bleeding among critically ill patients. JAMA Netw. Open 2018, 1, e183451. [Google Scholar] [CrossRef]
- Shi, Y.; Zhang, G.; Ma, C.; Xu, J.; Xu, K.; Zhang, W.; Wu, J.; Xu, L. Machine learning algorithms to predict intraoperative hemorrhage in surgical patients: A modeling study of real-world data in Shanghai, China. BMC Med. Inform. Decis. Mak. 2023, 23, 156. [Google Scholar] [CrossRef] [PubMed]
- Lou, S.S.; Liu, H.; Lu, C.; Wildes, T.S.; Hall, B.L.; Kannampallil, T. Personalized surgical transfusion risk prediction using machine learning to guide preoperative type and screen orders. Anesthesiology 2022, 137, 55–66. [Google Scholar] [CrossRef] [PubMed]
- Zapf, M.A.C.; Fabbri, D.V.; Andrews, J.; Li, G.; Freundlich, R.E.; Al-Droubi, S.; Wanderer, J.P. Development of a machine learning model to predict intraoperative transfusion and guide type and screen ordering. J. Clin. Anesth. 2023, 91, 111272. [Google Scholar] [CrossRef] [PubMed]
- Kwon, D.; Mi Jung, Y.; Lee, H.C.; Kyong Kim, T.; Kim, K.; Lee, G.; Kim, D.; Lee, S.B.; Mi Lee, S. Non-invasive prediction of massive transfusion during surgery using intraoperative hemodynamic monitoring data. J. Biomed. Inform. 2024, 156, 104680. [Google Scholar] [CrossRef]
- Lee, S.L.S.; Chang, D.; Song, M.-H.; Kim, J.-Y.; Lee, S. Explainable machine learning based a packed red blood cell transfusion prediction and evaluation for major internal medical condition. J. Inf. Process. Syst. 2022, 18, 302–310. [Google Scholar] [CrossRef]
- Levi, R.; Carli, F.; Arévalo, A.R.; Altinel, Y.; Stein, D.J.; Naldini, M.M.; Grassi, F.; Zanoni, A.; Finkelstein, S.; Vieira, S.F.; et al. Artificial intelligence-based prediction of transfusion in the intensive care unit in patients with gastrointestinal bleeding. BMJ Health Care Inform. 2021, 28, e100245. [Google Scholar] [CrossRef]
- Shung, D.L.; Au, B.; Taylor, R.A.; Tay, J.K.; Laursen, S.B.; Stanley, A.J.; Dalton, H.R.; Ngu, J.; Schultz, M.; Laine, L. Validation of a machine learning model that outperforms clinical risk scoring systems for upper gastrointestinal bleeding. Gastroenterology 2020, 158, 160–167. [Google Scholar] [CrossRef]
- Bruun-Rasmussen, P.; Andersen, P.K.; Banasik, K.; Brunak, S.; Johansson, P.I. Estimating the effect of donor sex on red blood cell transfused patient mortality: A retrospective cohort study using a targeted learning and emulated trials-based approach. eClinicalMedicine 2022, 51, 101628. [Google Scholar] [CrossRef]
- Nguyen, M.; Pirracchio, R.; Kornblith, L.Z.; Callcut, R.; Fox, E.E.; Wade, C.E.; Schreiber, M.; Holcomb, J.B.; Coyle, J.; Cohen, M.; et al. Dynamic impact of transfusion ratios on outcomes in severely injured patients: Targeted machine learning analysis of the Pragmatic, Randomized Optimal Platelet and Plasma Ratios randomized clinical trial. J. Trauma Acute Care Surg. 2020, 89, 505–513. [Google Scholar] [CrossRef]
- Portela, G.T.; Ducrocq, G.; Bertolet, M.; Alexander, J.H.; Goodman, S.G.; Glynn, S.; Strom, J.B.; Swanson, S.A.; Lemesle, G.; Rao, S.V.; et al. Individualized transfusion decisions to minimize adverse cardiovascular outcomes in patients with acute myocardial infarction and anemia. Am. Heart J. 2025, 282, 146–155. [Google Scholar] [CrossRef]
- Chang, T.-C.; Yu, J.; Wang, Z.; Hankins, J.S.; Weiss, M.J.; Wu, G.; Westhoff, C.M.; Chou, S.T.; Zheng, Y. Machine learning to optimize automated RH genotyping using whole-exome sequencing data. Blood Adv. 2024, 8, 2651–2659. [Google Scholar] [CrossRef] [PubMed]
- Hyvärinen, K.; Haimila, K.; Moslemi, C.; Biobank, B.S.; Olsson, M.L.; Ostrowski, S.R.; Pedersen, O.B.; Erikstrup, C.; Partanen, J.; Ritari, J. A machine-learning method for biobank-scale genetic prediction of blood group antigens. PLoS Comput. Biol. 2024, 20, e1011977. [Google Scholar] [CrossRef]
- Wu, K.; Duan, S.; Wang, Y.; Wang, H.; Gao, X. Convolutional neural network-based automatic classification for incomplete antibody reaction intensity in solid phase anti-human globulin test image. Med. Biol. Eng. Comput. 2022, 60, 1211–1222. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Arnold, D.M.; Down, D.G.; Barty, R.; Blake, J.; Chiang, F.; Courtney, T.; Waito, M.; Trifunov, R.; Heddle, N.M. From demand forecasting to inventory ordering decisions for red blood cells through integrating machine learning, statistical modeling, and inventory optimization. Transfusion 2022, 62, 87–99. [Google Scholar] [CrossRef]
- Engelke, M.; Schmidt, C.S.; Baldini, G.; Parmar, V.; Hosch, R.; Borys, K.; Koitka, S.; Turki, A.T.; Haubold, J.; Horn, P.A.; et al. Optimizing platelet transfusion through a personalized deep learning risk assessment system for demand management. Blood 2023, 142, 2315–2326. [Google Scholar] [CrossRef]
- Sun, X.; Xu, Z.; Feng, Y.; Yang, Q.; Xie, Y.; Wang, D.; Yu, Y. RBC inventory-management system based on XGBoost model. Indian J. Hematol. Blood Transfus. 2021, 37, 126–133. [Google Scholar] [CrossRef]
- Ahmadimanesh, M.; Tavakoli, A.; Pooya, A.; Dehghanian, F. Designing an optimal inventory management model for the blood supply chain: Synthesis of reusable simulation and neural network. Medicine 2020, 99, e21208. [Google Scholar] [CrossRef]
- Xiang, R.F.; Quinn, J.G.; Watson, S.; Kumar-Misir, A.; Cheng, C. Application of unsupervised machine learning to identify areas of blood product wastage in transfusion medicine. Vox Sang. 2021, 116, 955–964. [Google Scholar] [CrossRef]
- Mora, D.; Mateo, J.; Nieto, J.A.; Bikdeli, B.; Yamashita, Y.; Barco, S.; Jimenez, D.; Demelo-Rodríguez, P.; Rosa, V.; Yoo, H.H.B.; et al. Machine learning to predict major bleeding during anticoagulation for venous thromboembolism: Possibilities and limitations. Br. J. Haematol. 2023, 201, 971–981. [Google Scholar] [CrossRef]
- Qian, Y.; Wanlin, L.; Maofeng, W. Machine learning derived model for the prediction of bleeding in dual antiplatelet therapy patients. Front. Cardiovasc. Med. 2024, 11, 1402672. [Google Scholar] [CrossRef]
- An, R.; Man, Y.; Iram, S.; Kucukal, E.; Hasan, M.N.; Huang, Y.; Goreke, U.; Bode, A.; Hill, A.; Cheng, K.; et al. Point-of-care microchip electrophoresis for integrated anemia and hemoglobin variant testing. Lab Chip 2021, 21, 3863–3875. [Google Scholar] [CrossRef]
- De Almeida, J.G.; Gudgin, E.; Besser, M.; Dunn, W.G.; Cooper, J.; Haferlach, T.; Vassiliou, G.S.; Gerstung, M. Computational analysis of peripheral blood smears detects disease-associated cytomorphologies. Nat. Commun. 2023, 14, 4378. [Google Scholar] [CrossRef] [PubMed]
- Shohat, N.; Ludwick, L.; Sherman, M.B.; Fillingham, Y.; Parvizi, J. Using machine learning to predict venous thromboembolism and major bleeding events following total joint arthroplasty. Sci. Rep. 2023, 13, 2197. [Google Scholar] [CrossRef] [PubMed]
- Acharya, S.; Swaminathan, D.; Das, S.; Kansara, K.; Chakraborty, S.; Kumar, R.D.; Francis, T.; Aatre, K.R. Non-invasive estimation of hemoglobin using a multi-model stacking regressor. IEEE J. Biomed. Health Inform. 2020, 24, 1717–1726. [Google Scholar] [CrossRef] [PubMed]
- Aghajanian, S.; Jafarabady, K.; Abbasi, M.; Mohammadifard, F.; Bakhshali Bakhtiari, M.; Shokouhi, N.; Saleh Gargari, S.; Bakhtiyari, M. Prediction of post-delivery hemoglobin levels with machine learning algorithms. Sci. Rep. 2024, 14, 13953. [Google Scholar] [CrossRef]
- AlAgha, A.S.; Faris, H.; Hammo, B.H.; Al-Zoubi, A.M. Identifying β-thalassemia carriers using a data mining approach: The case of the Gaza Strip, Palestine. Artif. Intell. Med. 2018, 88, 70–83. [Google Scholar] [CrossRef]
- Amendolia, S.R.; Brunetti, A.; Carta, P.; Cossu, G.; Ganadu, M.L.; Golosio, B.; Mura, G.M.; Pirastru, M.G. A real-time classification system of thalassemic pathologies based on artificial neural networks. Med. Decis. Mak. 2002, 22, 18–26. [Google Scholar] [CrossRef]
- Appiahene, P.; Arthur, E.J.; Korankye, S.; Afrifa, S.; Asare, J.W.; Donkoh, E.T. Detection of anemia using conjunctiva images: A smartphone application approach. Med. Nov. Technol. Devices 2023, 18, 100237. [Google Scholar] [CrossRef]
- Appiahene, P.; Asare, J.W.; Donkoh, E.T.; Dimauro, G.; Maglietta, R. Detection of iron deficiency anemia by medical images: A comparative study of machine learning algorithms. BioData Min. 2023, 16, 2. [Google Scholar] [CrossRef]
- Arjmand, M.; Kompany-Zareh, M.; Vasighi, M.; Parvizzadeh, N.; Zamani, Z.; Nazgooei, F. Nuclear magnetic resonance-based screening of thalassemia and quantification of some hematological parameters using chemometric methods. Talanta 2010, 81, 1229–1236. [Google Scholar] [CrossRef]
- Asare, J.W.; Brown-Acquaye, W.L.; Ujakpa, M.M.; Freeman, E.; Appiahene, P. Application of machine learning approach for iron deficiency anaemia detection in children using conjunctiva images. Inform. Med. Unlocked 2024, 45, 101451. [Google Scholar] [CrossRef]
- Ayyıldız, H.; Tuncer, S.A. Determination of the effect of red blood cell parameters in the discrimination of iron deficiency anemia and beta thalassemia via neighborhood component analysis feature selection-based machine learning. Chemom. Intell. Lab. Syst. 2020, 196, 103886. [Google Scholar] [CrossRef]
- Azarkhish, I.; Raoufy, M.R.; Gharibzadeh, S. Artificial intelligence models for predicting iron deficiency anemia and iron serum level based on accessible laboratory data. J. Med. Syst. 2012, 36, 2057–2061. [Google Scholar] [CrossRef] [PubMed]
- Barnhart-Magen, G.; Gotlib, V.; Marilus, R.; Einav, Y. Differential diagnostics of thalassemia minor by artificial neural networks model: New screening tool for thalassemia minor. J. Clin. Lab. Anal. 2013, 27, 481–486. [Google Scholar] [CrossRef]
- Chakraborty, S.; Kansara, K.; Kumar, R.D.; Swaminathan, D.; Aatre, K.; Acharya, S. Non-invasive estimation of clinical severity of anemia using hierarchical ensemble classifiers. J. Med. Biol. Eng. 2022, 42, 828–838. [Google Scholar] [CrossRef]
- Chen, Y.-M.; Miaou, S.-G.; Bian, H. Examining palpebral conjunctiva for anemia assessment with image processing methods. Comput. Methods Programs Biomed. 2016, 137, 125–135. [Google Scholar] [CrossRef]
- Chen, Z.; Mo, Y.; Ouyang, P.; Shen, H.; Li, D.; Zhao, R. Retinal vessel optical coherence tomography images for anemia screening. Med. Biol. Eng. Comput. 2019, 57, 953–966. [Google Scholar] [CrossRef]
- Chen, Y.; Zhong, K.; Zhu, Y.; Sun, Q. Two-stage hemoglobin prediction based on prior causality. Front. Public Health 2022, 10, 1079389. [Google Scholar] [CrossRef]
- Çil, B.; Ayyıldız, H.; Tuncer, T. Discrimination of β-thalassemia and iron deficiency anemia through extreme learning machine and regularized extreme learning machine based decision support system. Med. Hypotheses 2020, 138, 109611. [Google Scholar] [CrossRef]
- Çuvadar, B.; Yılmaz, H. Non-invasive hemoglobin estimation from conjunctival images using deep learning. Med. Eng. Phys. 2023, 120, 104038. [Google Scholar] [CrossRef]
- Darshan, B.S.D.; Sharma, P.; Chadaga, K.; Sampathila, N.; Bairy, G.M.; Belurkar, S.; Prabhu, S.; Swathi, K.S. An ensemble machine learning framework with explainable artificial intelligence for predicting haemoglobin anaemia considering haematological markers. Syst. Sci. Control Eng. 2024, 12, 2420927. [Google Scholar] [CrossRef]
- Darshan, B.S.D.; Sampathila, N.; Bairy, M.G.; Belurkar, S.; Prabhu, S.; Chadaga, K. Detection of anemic condition in patients from clinical markers and explainable artificial intelligence. Technol. Health Care 2024, 32, 2431–2444. [Google Scholar] [CrossRef] [PubMed]
- Darshan, B.S.D.; Sampathila, N.; Bairy, G.M.; Prabhu, S.; Belurkar, S.; Chadaga, K.; Nandish, S. Differential diagnosis of iron deficiency anemia from aplastic anemia using machine learning and explainable Artificial Intelligence utilizing blood attributes. Sci. Rep. 2025, 15, 505. [Google Scholar] [CrossRef] [PubMed]
- Das, R.; Saleh, S.; Nielsen, I.; Kaviraj, A.; Sharma, P.; Dey, K.; Saha, S. Performance analysis of machine learning algorithms and screening formulae for β–thalassemia trait screening of Indian antenatal women. Int. J. Med. Inf. 2022, 167, 104866. [Google Scholar] [CrossRef] [PubMed]
- Das, P.K.; Dash, A.; Meher, S. ACDSSNet: Atrous Convolution-Based Deep Semantic Segmentation Network for Efficient Detection of Sickle Cell Anemia. IEEE J. Biomed. Health Inform. 2024, 28, 5676–5684. [Google Scholar] [CrossRef]
- Dejene, B.E.; Abuhay, T.M.; Bogale, D.S. Predicting the level of anemia among Ethiopian pregnant women using homogeneous ensemble machine learning algorithm. BMC Med. Inform. Decis. Mak. 2022, 22, 247. [Google Scholar] [CrossRef]
- Diaz-del-Pino, S.; Trelles-Martinez, R.; González-Fernández, F.A.; Guil, N. Artificial intelligence to assist specialists in the detection of haematological diseases. Heliyon 2023, 9, e15940. [Google Scholar] [CrossRef]
- Dimauro, G.; Griseta, M.E.; Camporeale, M.G.; Clemente, F.; Guarini, A.; Maglietta, R. An intelligent non-invasive system for automated diagnosis of anemia exploiting a novel dataset. Artif. Intell. Med. 2023, 136, 102477. [Google Scholar] [CrossRef]
- Erten, M.; Tuncer, T. Automated differential diagnosis method for iron deficiency anemia and beta thalassemia trait based on iterative Chi2 feature selector. Int. J. Lab. Hematol. 2022, 44, 430–436. [Google Scholar] [CrossRef]
- Foy, B.H.; Stefely, J.A.; Bendapudi, P.K.; Hasserjian, R.P.; Al-Samkari, H.; Louissaint, A., Jr.; Fitzpatrick, M.J.; Hutchison, B.; Mow, C.; Collins, J.; et al. Computer vision quantitation of erythrocyte shape abnormalities provides diagnostic, prognostic, and mechanistic insight. Blood Adv. 2023, 7, 4621–4630. [Google Scholar] [CrossRef]
- Fu, H.; Tian, Y.; Zha, G.; Xiao, X.; Zhu, H.; Zhang, Q.; Yu, C.; Sun, W.; Li, C.M.; Wei, L.; et al. Microstrip isoelectric focusing with deep learning for simultaneous screening of diabetes, anemia, and thalassemia. Anal. Chim. Acta 2024, 1312, 342696. [Google Scholar] [CrossRef]
- Göl, M.; Aktürk, C.; Talan, T.; Vural, M.S.; Türkbeyler, İ.H. Predicting malnutrition-based anemia in geriatric patients using machine learning methods. J. Eval. Clin. Pract. 2025, 31, e14142. [Google Scholar] [CrossRef] [PubMed]
- Hennek, J.W.; Kumar, A.A.; Wiltschko, A.B.; Patton, M.R.; Lee, S.Y.R.; Brugnara, C.; Adams, R.P.; Whitesides, G.M. Diagnosis of iron deficiency anemia using density-based fractionation of red blood cells. Lab Chip 2016, 16, 3929–3939. [Google Scholar] [CrossRef] [PubMed]
- Jain, P.; Bauskar, S.; Gyanchandani, M. Neural network based non-invasive method to detect anemia from images of eye conjunctiva. Int. J. Imaging Syst. Technol. 2020, 30, 112–125. [Google Scholar] [CrossRef]
- Kabootarizadeh, L.; Jamshidnezhad, A.; Koohmareh, Z.; Ghamchili, A. Differential Diagnosis of Iron-Deficiency Anemia from beta-Thalassemia Trait Using an Intelligent Model in Comparison with Discriminant Indexes. Acta Inform. Medica 2019, 27, 78. [Google Scholar] [CrossRef]
- Kato, S.; Chagi, K.; Takagi, Y.; Hidaka, M.; Inoue, S.; Sekiguchi, M.; Adachi, N.; Sato, K.; Kawai, H.; Kato, M. Machine/deep learning-assisted hemoglobin level prediction using palpebral conjunctival images. Br. J. Haematol. 2024, 205, 1590–1598. [Google Scholar] [CrossRef]
- Kay, F.U.; Lumby, C.; Tanabe, Y.; Abbara, S.; Rajiah, P. Detection of Low Blood Hemoglobin Levels on Pulmonary CT Angiography: A Feasibility Study Combining Dual-Energy CT and Machine Learning. Tomography 2023, 9, 1538–1550. [Google Scholar] [CrossRef]
- Kim, G.; Jo, Y.; Cho, H.; Min, H.; Park, Y. Learning-based screening of hematologic disorders using quantitative phase imaging of individual red blood cells. Biosens. Bioelectron. 2019, 123, 69–76. [Google Scholar] [CrossRef]
- Kitaw, B.; Asefa, C.; Legese, F. Leveraging machine learning models for anemia severity detection among pregnant women following ANC: Ethiopian context. BMC Public Health 2024, 24, 3500. [Google Scholar] [CrossRef]
- Kurstjens, S.; De Bel, T.; Van Der Horst, A.; Kusters, R.; Krabbe, J.; Van Balveren, J. Automated prediction of low ferritin concentrations using a machine learning algorithm. Clin. Chem. Lab. Med. 2022, 60, 1921–1928. [Google Scholar] [CrossRef] [PubMed]
- Laengsri, V.; Shoombuatong, W.; Adirojananon, W.; Nantasenamat, C.; Prachayasittikul, V.; Nuchnoi, P. ThalPred: A web-based prediction tool for discriminating thalassemia trait and iron deficiency anemia. BMC Med. Inform. Decis. Mak. 2019, 19, 212. [Google Scholar] [CrossRef]
- Lin, E.-T.; Lu, S.C.; Liu, A.S.; Ko, C.H.; Huang, C.H.; Tsai, C.L.; Fu, L.C. Deep learning-based model for non-invasive hemoglobin estimation via body parts images: A retrospective analysis and a prospective emergency department study. J. Imaging Inform. Med. 2024, 38, 775–792. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Martínez, J.M.; Escandell-Montero, P.; Barbieri, C.; Soria-Olivas, E.; Mari, F.; Martínez-Sober, M.; Amato, C.; Serrano López, A.J.; Bassi, M.; Magdalena-Benedito, R.; et al. Prediction of the hemoglobin level in hemodialysis patients using machine learning techniques. Comput. Methods Programs Biomed. 2014, 117, 208–217. [Google Scholar] [CrossRef]
- Masala, G.L.; Golosio, B.; Cutzu, R.; Pola, R. A two-layered classifier based on the radial basis function for the screening of thalassaemia. Comput. Biol. Med. 2013, 43, 1724–1731. [Google Scholar] [CrossRef]
- Matović, V.; Jeftić, B.; Trbojević-Stanković, J.; Matija, L. Predicting anemia using NIR spectrum of spent dialysis fluid in hemodialysis patients. Sci. Rep. 2021, 11, 10549. [Google Scholar] [CrossRef]
- Meitei, A.J.; Saini, A.; Mohapatra, B.B.; Singh, K.J. Predicting child anaemia in the North-Eastern states of India: A machine learning approach. Int. J. Syst. Assur. Eng. Manag. 2022, 13, 2949–2962. [Google Scholar] [CrossRef]
- Memmolo, P.; Aprea, G.; Bianco, V.; Russo, R.; Andolfo, I.; Mugnano, M.; Merola, F.; Miccio, L.; Iolascon, A.; Ferraro, P.; et al. Differential diagnosis of hereditary anemias from a fraction of blood drop by digital holography and hierarchical machine learning. Biosens. Bioelectron. 2022, 201, 113945. [Google Scholar] [CrossRef]
- Mo, D.; Zheng, Q.; Xiao, B.; Li, L. Predicting thalassemia using deep neural network based on red blood cell indices. Clin. Chim. Acta 2023, 543, 117329. [Google Scholar] [CrossRef]
- Moreno, G.; Camargo, A.; Ayala, L.; Zimic, M.; Del Carpio, C. An algorithm for the estimation of hemoglobin level from digital images of palpebral conjunctiva based in digital image processing and artificial intelligence. Int. J. Online Biomed. Eng. 2024, 20, 33–46. [Google Scholar] [CrossRef]
- Muljono; Wulandari, S.A.; Azies, H.A.; Naufal, M.; Prasetyanto, W.A.; Zahra, F.A. Breaking boundaries in diagnosis: Non-invasive anemia detection empowered by AI. IEEE Access 2024, 12, 9292–9307. [Google Scholar] [CrossRef]
- Muthalagu, R.; Bai, V.T.; Gracias, D.; John, S. Developmental screening tool: Accuracy and feasibility of non-invasive anaemia estimation. Technol. Health Care 2018, 26, 723–727. [Google Scholar] [CrossRef] [PubMed]
- Muyama, L.; Neuraz, A.; Coulet, A. Deep reinforcement learning for personalized diagnostic decision pathways using electronic health records: A comparative study on anemia and systemic lupus erythematosus. Artif. Intell. Med. 2024, 157, 102994. [Google Scholar] [CrossRef] [PubMed]
- Navya, K.T.; Akshatha, K.R.; Prasad, K.; Singh, B.M.K. An empirical study of object detection models for the detection of iron deficiency anemia using peripheral blood smear images. Biomed. Phys. Eng. Express 2025, 11, 015024. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Du, R.; Han, X.; Zhu, W.; Peng, D.; Tu, Y.; Han, J.; Bao, Y.; Yu, H. Machine learning prediction of iron deficiency anemia in Chinese premenopausal women 12 months after sleeve gastrectomy. Nutrients 2023, 15, 3385. [Google Scholar] [CrossRef]
- Petrović, N.; Moyà-Alcover, G.; Jaume-i-Capó, A.; González-Hidalgo, M. Sickle-cell disease diagnosis support selecting the most appropriate machine learning method: Towards a general and interpretable approach for cell morphology analysis from microscopy images. Comput. Biol. Med. 2020, 126, 104027. [Google Scholar] [CrossRef]
- Pullakhandam, S.; McRoy, S. Classification and explanation of iron deficiency anemia from complete blood count data using machine learning. BioMedInformatics 2024, 4, 661–672. [Google Scholar] [CrossRef]
- Qadah, T.; Munshi, A. Prediction of anemia from multi-data attribute co-existence. IEEE Access 2024, 12, 182923–182942. [Google Scholar] [CrossRef]
- Ranganathan, H.; Gunasekaran, N. Simple method for estimation of hemoglobin in human blood using color analysis. IEEE Trans. Inf. Technol. Biomed. 2006, 10, 657–662. [Google Scholar] [CrossRef]
- Rizzuto, V.; Mencattini, A.; Álvarez-González, B.; Di Giuseppe, D.; Martinelli, E.; Beneitez-Pastor, D.; Mañú-Pereira, M.M.; López-Martínez, M.J.; Samitier, J. Combining microfluidics with machine learning algorithms for RBC classification in rare hereditary hemolytic anemia. Sci. Rep. 2021, 11, 13553. [Google Scholar] [CrossRef]
- Rustam, F.; Ashraf, I.; Jabbar, S.; Tutusaus, K.; Mazas, C.; Barrera, A.E.P.; de la Torre Diez, I. Prediction of β-thalassemia carriers using complete blood count features. Sci. Rep. 2022, 12, 19999. [Google Scholar] [CrossRef]
- Salma, N.; Ali, M.K.M. Evaluating the factors and forecasting childhood anemia through machine learning algorithms. Malays. J. Fundam. Appl. Sci. 2025, 21, 1529–1541. [Google Scholar] [CrossRef]
- Saputra, D.C.E.; Sunat, K.; Ratnaningsih, T. A new artificial intelligence approach using extreme learning machine as the potentially effective model to predict and analyze the diagnosis of anemia. Healthcare 2023, 11, 697. [Google Scholar] [CrossRef]
- Saputra, D.C.E.; Sunat, K.; Ratnaningsih, T. SMOTE-MRS: A novel SMOTE-multiresolution sampling technique for imbalanced distribution to improve prediction of anemia. IEEE Access 2024, 12, 154675–154699. [Google Scholar] [CrossRef]
- Sarsam, S.M.; Al-Samarraie, H.; Alzahrani, A.I.; Shibghatullah, A.S. A non-invasive machine learning mechanism for early disease recognition on Twitter: The case of anemia. Artif. Intell. Med. 2022, 134, 102428. [Google Scholar] [CrossRef] [PubMed]
- Su, Z.; Nong, Z.; Huang, F.; Zhou, C.; Yu, C. Anemia risk prediction model for osteosarcoma patients post-chemotherapy using artificial intelligence. Cancer Med. 2024, 13, e70427. [Google Scholar] [CrossRef]
- Terzi, E.; Sarıbacak, B.; Sağlam, F.; Cengiz, M.A. A novel expert system for diagnosis of iron deficiency anemia. Comput. Math. Methods Med. 2022, 2022, 7352096. [Google Scholar] [CrossRef]
- Tesfaye, S.H.; Seboka, B.T.; Sisay, D. Application of machine learning methods for predicting childhood anaemia: Analysis of Ethiopian Demographic Health Survey of 2016. PLoS ONE 2024, 19, e0300172. [Google Scholar] [CrossRef]
- Uçucu, S.; Azik, F. Artificial intelligence-driven diagnosis of β-thalassemia minor & iron deficiency anemia using machine learning models. J. Med. Biochem. 2024, 43, 11–18. [Google Scholar] [CrossRef]
- Vohra, R.; Hussain, A.; Dudyala, A.K.; Pahareeya, J.; Khan, W. Multi-class classification algorithms for the diagnosis of anemia in an outpatient clinical setting. PLoS ONE 2022, 17, e0269685. [Google Scholar] [CrossRef]
- Yagmur, N.; Dag, I.; Temurtas, H. A new computer-aided diagnostic method for classifying anaemia disease: Hybrid use of Tree Bagger and metaheuristics. Expert Syst. 2024, 41, e13528. [Google Scholar] [CrossRef]
- Yılmaz, Z.; Bozkurt, M.R. Determination of women iron deficiency anemia using neural networks. J. Med. Syst. 2012, 36, 2941–2945. [Google Scholar] [CrossRef]
- Zemariam, A.B.; Yimer, A.; Kibret Abebe, G.; Wondie, W.T.; Abate, B.B.; Alamaw, A.W.; Yilak, G.; Melaku, T.M.; Ngusie, H.S. Employing supervised machine learning algorithms for classification and prediction of anemia among youth girls in Ethiopia. Sci. Rep. 2024, 14, 9080. [Google Scholar] [CrossRef]
- Zhang, A.; Lou, J.; Pan, Z.; Luo, J.; Zhang, X.; Zhang, H.; Li, J.; Wang, L.; Cui, X.; Ji, B.; et al. Prediction of anemia using facial images and deep learning technology in the emergency department. Front. Public Health 2022, 10, 964385. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Yang, J.; Wang, Y.; Cai, M.; Ouyang, J.; Li, J. TT@MHA: A machine learning-based webpage tool for discriminating thalassemia trait from microcytic hypochromic anemia patients. Clin. Chim. Acta 2023, 545, 117368. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chun, Y.; Fu, H.; Jiao, W.; Bao, J.; Jiang, T.; Cui, L.; Hu, X.; Cui, J.; Qiu, X.; et al. A risk warning model for anemia based on facial visible light reflectance spectroscopy: Cross-sectional study. JMIR Med. Inform. 2025, 13, e64204. [Google Scholar] [CrossRef] [PubMed]
- Barbieri, C.; Molina, M.; Ponce, P.; Tothova, M.; Cattinelli, I.; Titapiccolo, J.I.; Mari, F.; Amato, C.; Leipold, F.; Wehmeyer, W.; et al. A new machine learning approach for predicting the response to anemia treatment in a large cohort of end stage renal disease patients undergoing dialysis. Comput. Biol. Med. 2015, 61, 56–61. [Google Scholar] [CrossRef]
- Barbieri, C.; Molina, M.; Ponce, P.; Tothova, M.; Cattinelli, I.; Titapiccolo, J.I.; Mari, F.; Amato, C.; Leipold, F.; Wehmeyer, W.; et al. An international observational study suggests that artificial intelligence for clinical decision support optimizes anemia management in hemodialysis patients. Kidney Int. 2016, 90, 422–429. [Google Scholar] [CrossRef]
- Barbieri, C.; Bolzoni, E.; Mari, F.; Cattinelli, I.; Bellocchio, F.; Martin, J.D.; Amato, C.; Stopper, A.; Gatti, E.; Macdougall, I.C.; et al. Performance of a predictive model for long-term hemoglobin response to darbepoetin and iron administration in a large cohort of hemodialysis patients. PLoS ONE 2016, 11, e0148938. [Google Scholar] [CrossRef]
- Brier, M.E.; Gaweda, A.E. Predictive modeling for improved anemia management in dialysis patients. Curr. Opin. Nephrol. Hypertens. 2011, 20, 573–576. [Google Scholar] [CrossRef]
- Bucalo, M.L.; Barbieri, C.; Roca, S.; Titapiccolo, J.I.; Romero, M.S.; Ramos, R.; Albaladejo, M.; Manzano, D.; Mari, F.; Molina, M. The anaemia control model: Does it help nephrologists in therapeutic decision-making in the management of anaemia? Nefrología 2018, 38, 491–502. [Google Scholar] [CrossRef]
- Gabutti, L.; Lötscher, N.; Bianda, J.; Marone, C.; Mombelli, G.; Burnier, M. Would artificial neural networks implemented in clinical wards help nephrologists in predicting epoetin responsiveness? BMC Nephrol. 2006, 7, 13. [Google Scholar] [CrossRef][Green Version]
- Gaweda, A.E.; Muezzinoglu, M.K.; Aronoff, G.R.; Jacobs, A.A.; Zurada, J.M.; Brier, M.E. Individualization of pharmacological anemia management using reinforcement learning. Neural Netw. 2005, 18, 826–834. [Google Scholar] [CrossRef]
- Gaweda, A.E.; Jacobs, A.A.; Brier, M.E. Application of fuzzy logic to predicting erythropoietic response in hemodialysis patients. Int. J. Artif. Organs 2008, 31, 1035–1042. [Google Scholar] [CrossRef]
- Gaweda, A.E.; Jacobs, A.A.; Aronoff, G.R.; Brier, M.E. Model predictive control of erythropoietin administration in the anemia of ESRD. Am. J. Kidney Dis. 2008, 51, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Guerrero, J.D.M.; Olivas, E.S.; Valls, G.C.; López, A.J.S.; Ruixo, J.J.P.; Torres, N.V.J. Use of neural networks for dosage individualisation of erythropoietin in patients with secondary anemia to chronic renal failure. Comput. Biol. Med. 2003, 33, 361–373. [Google Scholar] [CrossRef] [PubMed]
- Kang, C.; Han, J.; Son, S.; Lee, S.; Baek, H.; Hwang, D.D.; Park, J.I. Optimizing anemia management using artificial intelligence for patients undergoing hemodialysis. Sci. Rep. 2024, 14, 26739. [Google Scholar] [CrossRef] [PubMed]
- Martín-Guerrero, J.D.; Camps-Valls, G.; Soria-Olivas, E.; Serrano-López, A.J.; Pérez-Ruixo, J.J.; Jiménez-Torres, N.V. Dosage individualization of erythropoietin using a profile-dependent support vector regression. IEEE Trans. Biomed. Eng. 2003, 50, 1136–1142. [Google Scholar] [CrossRef]
- Yun, H.-R.; Lee, G.; Jeon, M.J.; Kim, H.W.; Joo, Y.S.; Kim, H.; Chang, T.I.; Park, J.T.; Han, S.H.; Kang, S.W.; et al. Erythropoiesis stimulating agent recommendation model using recurrent neural networks for patient with kidney failure with replacement therapy. Comput. Biol. Med. 2021, 137, 104718. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, Z. Haemoglobin response modelling under erythropoietin treatment: Physiological model-informed machine learning method. Can. J. Chem. Eng. 2023, 101, 4307–4319. [Google Scholar] [CrossRef]
- Alsayegh, F.; Alkhamis, M.A.; Ali, F.; Attur, S.; Fountain-Jones, N.M.; Zubaid, M. Anemia or other comorbidities? Using machine learning to reveal deeper insights into the drivers of acute coronary syndromes in hospital admitted patients. PLoS ONE 2022, 17, e0262997. [Google Scholar] [CrossRef]
- D’Ascenzo, F.; De Filippo, O.; Gallone, G.; Mittone, G.; Deriu, M.A.; Iannaccone, M.; Ariza-Solé, A.; Liebetrau, C.; Manzano-Fernández, S.; Quadri, G.; et al. Machine learning-based prediction of adverse events following an acute coronary syndrome (PRAISE): A modelling study of pooled datasets. Lancet 2021, 397, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Kou, Y.; Ye, S.; Tian, Y.; Yang, K.; Qin, L.; Huang, Z.; Luo, B.; Ha, Y.; Zhan, L.; Ye, R.; et al. Risk factors for gastrointestinal bleeding in patients with acute myocardial infarction: Multicenter retrospective cohort study. J. Med. Internet Res. 2025, 27, e67346. [Google Scholar] [CrossRef] [PubMed]
- Niimi, N.; Shiraishi, Y.; Sawano, M.; Ikemura, N.; Inohara, T.; Ueda, I.; Fukuda, K.; Kohsaka, S. Machine learning models for prediction of adverse events after percutaneous coronary intervention. Sci. Rep. 2022, 12, 6262. [Google Scholar] [CrossRef] [PubMed]
- Ahmadzadeh, E.; Kim, J.; Lee, J.; Kwon, N.; Kim, H.W.; Park, J.; Hong, J.H. Early prediction of intraventricular hemorrhage in very low birth weight infants using deep neural networks with attention in low-resource settings. Sci. Rep. 2025, 15, 10102. [Google Scholar] [CrossRef]
- Dong, J.; Jin, Z.; Li, C.; Yang, J.; Jiang, Y.; Li, Z.; Chen, C.; Zhang, B.; Ye, Z.; Hu, Y.; et al. Machine learning models with prognostic implications for predicting gastrointestinal bleeding after coronary artery bypass grafting and guiding personalized medicine: Multicenter cohort study. J. Med. Internet Res. 2025, 27, e68509. [Google Scholar] [CrossRef]
- Gao, Y.; Liu, X.; Wang, L.; Wang, S.; Yu, Y.; Ding, Y.; Wang, J.; Ao, H. Machine learning algorithms to predict major bleeding after isolated coronary artery bypass grafting. Front. Cardiovasc. Med. 2022, 9, 881881. [Google Scholar] [CrossRef]
- Harm, T.; Fu, X.; Frey, M.; Dittrich, K.; Brun, A.; Castor, T.; Borst, O.; Müller, K.A.L.; Geisler, T.; Rath, D. Machine learning insights into thrombo-ischemic risks and bleeding events through platelet lysophospholipids and acylcarnitine species. Sci. Rep. 2024, 14, 6089. [Google Scholar] [CrossRef]
- Hu, Y.; Zhang, X.; Wei, M.; Yang, T.; Chen, J.; Wu, X.; Zhu, Y.; Chen, X.; Lou, S.; Zhu, J. Using machine learning to predict the bleeding risk for patients with cardiac valve replacement treated with warfarin in hospitalized. Pharmacoepidemiol. Drug Saf. 2024, 33, e5756. [Google Scholar] [CrossRef]
- Huang, R.S.; Nedelcu, E.; Bai, Y.; Wahed, A.; Klein, K.; Tint, H.; Gregoric, I.; Patel, M.; Kar, B.; Loyalka, P. Post-operative bleeding risk stratification in cardiac pulmonary bypass patients using artificial neural network. Ann. Clin. Lab. Sci. 2015, 45, 181–186. [Google Scholar]
- Hui, V.; Litton, E.; Edibam, C.; Geldenhuys, A.; Hahn, R.; Larbalestier, R.; Wright, B.; Pavey, W. Using machine learning to predict bleeding after cardiac surgery. Eur. J. Cardiothorac. Surg. 2023, 64, ezad297. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Jang, I. Predictors of bleeding event among elderly patients with mechanical valve replacement using random forest model: A retrospective study. Medicine 2021, 100, e25875. [Google Scholar] [CrossRef] [PubMed]
- Meyer, A.; Zverinski, D.; Pfahringer, B.; Kempfert, J.; Kuehne, T.; Sündermann, S.H.; Stamm, C.; Hofmann, T.; Falk, V.; Eickhoff, C. Machine learning for real-time prediction of complications in critical care: A retrospective study. Lancet Respir. Med. 2018, 6, 905–914. [Google Scholar] [CrossRef]
- Perduca, V.; Bouaziz, O.; Zannis, K.; Beaussier, M.; Untereiner, O. Can machine learning provide preoperative predictions of biological hemostasis after extracorporeal circulation for cardiac surgery? J. Thorac. Cardiovasc. Surg. 2024, 168, 1120–1129.e9. [Google Scholar] [CrossRef]
- Yang, Y.; Yan, Y.; Zhou, Z.; Zhang, J.; Han, H.; Zhang, W.; Wang, X.; Chen, C.; Ge, W.; Pan, J.; et al. Accurate prediction of bleeding risk after coronary artery bypass grafting with dual antiplatelet therapy: A machine learning model vs. the PRECISE-DAPT score. Int. J. Cardiol. 2025, 421, 132925. [Google Scholar] [CrossRef]
- Zhao, X.; Wang, J.; Yang, J.; Chen, T.; Song, Y.; Li, X.; Xie, G.; Gao, X.; Xu, H.; Gao, R.; et al. Machine learning for prediction of bleeding in acute myocardial infarction patients after percutaneous coronary intervention. Ther. Adv. Chronic Dis. 2023, 14, 20406223231158561. [Google Scholar] [CrossRef]
- Abbasi, A.; Karasu, Y.; Li, C.; Sodha, N.R.; Eickhoff, C.; Ventetuolo, C.E. Machine learning to predict hemorrhage and thrombosis during extracorporeal membrane oxygenation. Crit. Care 2020, 24, 689. [Google Scholar] [CrossRef]
- Ge, Y.; Chang, W.; Xie, L.; Gao, Y.; Xu, Y.; Zhu, H. A predictive model for secondary posttonsillectomy hemorrhage in pediatric patients: An 8-year retrospective study. Laryngoscope Investig. Otolaryngol. 2025, 10, e70080. [Google Scholar] [CrossRef]
- Pizzi, N.J. Bleeding predisposition assessments in tonsillectomy/adenoidectomy patients using fuzzy interquartile encoded neural networks. Artif. Intell. Med. 2001, 21, 65–90. [Google Scholar] [CrossRef]
- Herrin, J.; Abraham, N.S.; Yao, X.; Noseworthy, P.A.; Inselman, J.; Shah, N.D.; Ngufor, C. Comparative effectiveness of machine learning approaches for predicting gastrointestinal bleeds in patients receiving antithrombotic treatment. JAMA Netw. Open 2021, 4, e2110703. [Google Scholar] [CrossRef]
- Hou, Y.; Yu, H.; Zhang, Q.; Yang, Y.; Liu, X.; Wang, X.; Jiang, Y. Machine learning-based model for predicting the esophagogastric variceal bleeding risk in liver cirrhosis patients. Diagn. Pathol. 2023, 18, 29. [Google Scholar] [CrossRef]
- Larkin, J.W.; Lama, S.; Chaudhuri, S.; Willetts, J.; Winter, A.C.; Jiao, Y.; Stauss-Grabo, M.; Usvyat, L.A.; Hymes, J.L.; Maddux, F.W.; et al. Prediction of gastrointestinal bleeding hospitalization risk in hemodialysis using machine learning. BMC Nephrol. 2024, 25, 366. [Google Scholar] [CrossRef]
- Liu, H.; Sun, J.; Liu, G.; Liu, X.; Zhou, Q.; Zhou, J. Establishment of a non-invasive prediction model for the risk of oesophageal variceal bleeding using radiomics based on CT. Clin. Radiol. 2022, 77, 368–376. [Google Scholar] [CrossRef]
- Na, J.E.; Lee, Y.C.; Kim, T.J.; Lee, H.; Won, H.H.; Min, Y.W.; Min, B.H.; Lee, J.H.; Rhee, P.L.; Kim, J.J. Utility of a deep learning model and a clinical model for predicting bleeding after endoscopic submucosal dissection in patients with early gastric cancer. World J. Gastroenterol. 2022, 28, 2721–2732. [Google Scholar] [CrossRef]
- Peng, Y.-J.; Liu, X.; Liu, Y.; Tang, X.; Zhao, Q.-P.; Du, Y. Computed tomography-based multi-organ radiomics nomogram model for predicting the risk of esophagogastric variceal bleeding in cirrhosis. World J. Gastroenterol. 2024, 30, 4044–4056. [Google Scholar] [CrossRef]
- Seo, D.-W.; Yi, H.; Park, B.; Kim, Y.J.; Jung, D.H.; Woo, I.; Sohn, C.H.; Ko, B.S.; Kim, N.; Kim, W.Y. Prediction of adverse events in stable non-variceal gastrointestinal bleeding using machine learning. J. Clin. Med. 2020, 9, 2603. [Google Scholar] [CrossRef]
- Wong, G.L.; Ma, A.J.; Deng, H.; Ching, J.Y.; Wong, V.W.; Tse, Y.K.; Yip, T.C.; Lau, L.H.; Liu, H.H.; Leung, C.M.; et al. Machine learning model to predict recurrent ulcer bleeding in patients with history of idiopathic gastroduodenal ulcer bleeding. Aliment. Pharmacol. Ther. 2019, 49, 912–918. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Li, Y.; Fan, C.; Zhang, Y.; Zhang, S.; Wang, Z.; Huang, T.; Ding, Z.; Hu, K.; Li, L.; et al. A novel machine learning-based radiomic model for diagnosing high bleeding risk esophageal varices in cirrhotic patients. Hepatol. Int. 2022, 16, 423–432. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Zhang, X.; Huang, B.; Shi, X.; Xiao, L.; Li, Z. Application of machine learning methods for predicting esophageal variceal bleeding in patients with cirrhosis. Eur. Radiol. 2024, 35, 1440–1450. [Google Scholar] [CrossRef]
- Zhuang, Y.; Xia, S.; Chen, J.; Ke, J.; Lin, S.; Lin, Q.; Tang, X.; Huang, H.; Zheng, N.; Wang, Y.; et al. Construction of a prediction model for rebleeding in patients with acute upper gastrointestinal bleeding. Eur. J. Med. Res. 2023, 28, 351. [Google Scholar] [CrossRef]
- Chen, D.; Afzal, N.; Sohn, S.; Habermann, E.B.; Naessens, J.M.; Larson, D.W.; Liu, H. Postoperative bleeding risk prediction for patients undergoing colorectal surgery. Surgery 2018, 164, 1209–1216. [Google Scholar] [CrossRef] [PubMed]
- Hsu, J.L.; Chen, K.A.; Butler, L.R.; Bahraini, A.; Kapadia, M.R.; Gomez, S.M.; Farrell, T.M. Application of machine learning to predict postoperative gastrointestinal bleed in bariatric surgery. Surg. Endosc. 2023, 37, 7121–7127. [Google Scholar] [CrossRef] [PubMed]
- Ikuta, S.; Fujikawa, M.; Nakajima, T.; Kasai, M.; Aihara, T.; Yamanaka, N. Machine learning approach to predict postpancreatectomy hemorrhage following pancreaticoduodenectomy: A retrospective study. Langenbecks Arch. Surg. 2024, 409, 29. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Jia, Y.M.; Zhang, Z.L.; Liu, C.Y.; Jiang, Z.W.; Hao, Z.W.; Peng, L. Development and validation of a machine learning-based early prediction model for massive intraoperative bleeding in patients with primary hepatic malignancies. World J. Gastrointest. Oncol. 2024, 16, 90–101. [Google Scholar] [CrossRef]
- Liu, J.; Cao, B.; Luo, Y.; Chen, X.; Han, H.; Li, L.; Zeng, J. Risk factors of major bleeding detected by machine learning method in patients undergoing liver resection with controlled low central venous pressure technique. Postgrad. Med. J. 2023, 99, 1280–1286. [Google Scholar] [CrossRef]
- Loftus, T.J.; Brakenridge, S.C.; Croft, C.A.; Smith, R.S.; Efron, P.A.; Moore, F.A.; Mohr, A.M.; Jordan, J.R. Neural network prediction of severe lower intestinal bleeding and the need for surgical intervention. J. Surg. Res. 2017, 212, 42–47. [Google Scholar] [CrossRef]
- Park, S.; Park, K.; Lee, J.G.; Choi, T.Y.; Heo, S.; Koo, B.N.; Chae, D. Development of machine learning models predicting estimated blood loss during liver transplant surgery. J. Pers. Med. 2022, 12, 1028. [Google Scholar] [CrossRef]
- Wakiya, T.; Ishido, K.; Kimura, N.; Nagase, H.; Kubota, S.; Fujita, H.; Hagiwara, Y.; Kanda, T.; Matsuzaka, M.; Sasaki, Y.; et al. Prediction of massive bleeding in pancreatic surgery based on preoperative patient characteristics using a decision tree. PLoS ONE 2021, 16, e0259682. [Google Scholar] [CrossRef]
- Weller, G.B.; Lovely, J.; Larson, D.W.; Earnshaw, B.A.; Huebner, M. Leveraging electronic health records for predictive modeling of post-surgical complications. Stat. Methods Med. Res. 2018, 27, 3271–3285. [Google Scholar] [CrossRef]
- Xue, Q.; Zhu, Y.; Yang, L.; Duan, W.; Li, Z.; Ji, M.; Tong, J.; Yang, J.J.; Zhou, C.M. Predicting intraoperative bleeding in patients undergoing a hepatectomy using multiple machine learning and deep learning techniques. J. Clin. Anesth. 2021, 74, 110444. [Google Scholar] [CrossRef]
- An, Z.Y.; Wu, Y.J.; Hou, Y.; Mei, H.; Nong, W.X.; Li, W.Q.; Zhou, H.; Feng, R.; Shen, J.P.; Peng, J.; et al. A life-threatening bleeding prediction model for immune thrombocytopenia based on personalized machine learning: A nationwide prospective cohort study. Sci. Bull. 2023, 68, 2106–2114. [Google Scholar] [CrossRef] [PubMed]
- Chu, Q.; Wei, W.; Lao, H.; Li, Y.; Tan, Y.; Wei, X.; Huang, B.; Qin, C.; Tang, Y. Machine learning algorithms for integrating clinical features to predict intracranial hemorrhage in patients with acute leukemia. Int. J. Neurosci. 2023, 133, 977–986. [Google Scholar] [CrossRef] [PubMed]
- Kang, K.W.; Choi, Y.; Lim, H.J.; Kwak, K.; Choi, Y.S.; Park, Y.; Kim, B.S.; Lee, K.S.; Ahn, K.H. Impact of platelet transfusion and bleeding risk stratification in patients with immune thrombocytopenia before procedures. Sci. Rep. 2025, 15, 5174. [Google Scholar] [CrossRef] [PubMed]
- Sidonio, R.F.; Lu, A.; Hale, S.; Caicedo, J.; Bullano, M.; Xing, S. Early diagnosis of persons with von Willebrand disease using a machine learning algorithm and real-world data. Expert Rev. Hematol. 2024, 17, 261–268. [Google Scholar] [CrossRef]
- Bernardini, A.; Bindini, L.; Antonucci, E.; Berteotti, M.; Giusti, B.; Testa, S.; Palareti, G.; Poli, D.; Frasconi, P.; Marcucci, R. Machine learning approach for prediction of outcomes in anticoagulated patients with atrial fibrillation. Int. J. Cardiol. 2024, 407, 132088. [Google Scholar] [CrossRef]
- Chen, C.; Yin, C.; Wang, Y.; Zeng, J.; Wang, S.; Bao, Y.; Xu, Y.; Liu, T.; Fan, J.; Liu, X. XGBoost-based machine learning test improves the accuracy of hemorrhage prediction among geriatric patients with long-term administration of rivaroxaban. BMC Geriatr. 2023, 23, 418. [Google Scholar] [CrossRef]
- Chen, T.; Lei, W.; Wang, M. Predictive model of internal bleeding in elderly aspirin users using XGBoost machine learning. Risk Manag. Healthc. Policy 2024, 17, 2255–2269. [Google Scholar] [CrossRef]
- Fard, S.S.; Perkins, T.J.; Wells, P.S. Machine learning analysis of bleeding status in venous thromboembolism patients. Res. Pract. Thromb. Haemost. 2024, 8, 102403. [Google Scholar] [CrossRef]
- Fard, S.S.; Perkins, T.J.; Wells, P.S. A deep-learning approach to predict bleeding risk over time in patients on extended anticoagulation therapy. J. Thromb. Haemost. 2024, 22, 1997–2008. [Google Scholar] [CrossRef]
- Watanabe, E.; Noyama, S.; Kiyono, K.; Inoue, H.; Atarashi, H.; Okumura, K.; Yamashita, T.; Lip, G.Y.H.; Kodani, E.; Origasa, H. Comparison among random forest, logistic regression, and existing clinical risk scores for predicting outcomes in patients with atrial fibrillation: A report from the J-RHYTHM registry. Clin. Cardiol. 2021, 44, 1305–1315. [Google Scholar] [CrossRef]
- Cui, S.; Song, H.; Ren, H.; Wang, X.; Xie, Z.; Wen, H.; Li, Y. Prediction of hemorrhagic complication after thrombolytic therapy based on multimodal data from multiple centers: An approach to machine learning and system implementation. J. Pers. Med. 2022, 12, 2052. [Google Scholar] [CrossRef]
- Dharmasaroja, P.; Dharmasaroja, P.A. Prediction of intracerebral hemorrhage following thrombolytic therapy for acute ischemic stroke using multiple artificial neural networks. Neurol. Res. 2012, 34, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Heo, J.; Sim, Y.; Kim, B.M.; Kim, D.J.; Kim, Y.D.; Nam, H.S.; Choi, Y.S.; Lee, S.K.; Kim, E.Y.; Sohn, B. Radiomics using non-contrast CT to predict hemorrhagic transformation risk in stroke patients undergoing revascularization. Eur. Radiol. 2024, 34, 6005–6015. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Liu, J.; Hong, J.; Chen, Y.; Wang, Z.; Hu, J.; Gai, S.; Yu, X.; Fu, J. Machine learning, clinical-radiomics approach with HIM for hemorrhagic transformation prediction after thrombectomy and treatment. Front. Neurol. 2025, 16, 1471274. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Xu, C.; Shang, C.; Wang, Y.; Xu, J.; Zhou, Q. Machine learning predicts the risk of hemorrhagic transformation of acute cerebral infarction and in-hospital death. Comput. Methods Programs Biomed. 2023, 237, 107582. [Google Scholar] [CrossRef]
- Li, X.; Jones, P.; Zhao, M. Identifying potential (re)hemorrhage among sporadic cerebral cavernous malformations using machine learning. Sci. Rep. 2024, 14, 11022. [Google Scholar] [CrossRef]
- Lin, Y.; Li, Y.; Luo, Y.; Han, J. Development and validation of an explainable machine learning prediction model of hemorrhagic transformation after intravenous thrombolysis in stroke. Front. Neurol. 2025, 15, 1446250. [Google Scholar] [CrossRef]
- Raman, T.; Vasquez-Montes, D.; Varlotta, C.; Passias, P.G.; Errico, T.J. Decision tree-based modelling for identification of predictors of blood loss and transfusion requirement after adult spinal deformity surgery. Int. J. Spine Surg. 2020, 14, 87–95. [Google Scholar] [CrossRef]
- Ren, H.; Song, H.; Cui, S.; Xiong, H.; Long, B.; Li, Y. Deep learning of noncontrast CT for fast prediction of hemorrhagic transformation of acute ischemic stroke: A multicenter study. Eur. Radiol. Exp. 2025, 9, 8. [Google Scholar] [CrossRef]
- Ru, X.; Zhao, S.; Chen, W.; Wu, J.; Yu, R.; Wang, D.; Dong, M.; Wu, Q.; Peng, D.; Song, Y. A weakly supervised deep learning model integrating noncontrasted computed tomography images and clinical factors facilitates haemorrhagic transformation prediction after intravenous thrombolysis in acute ischaemic stroke patients. Biomed. Eng. OnLine 2023, 22, 129. [Google Scholar] [CrossRef]
- Saeedi, E.; Mashhadinejad, M.; Tavallaii, A. Development of a machine learning model for prediction of intraventricular hemorrhage in premature neonates. Childs Nerv. Syst. 2025, 41, 51. [Google Scholar] [CrossRef]
- Saggi, S.; Winkler, E.A.; Ammanuel, S.G.; Morshed, R.A.; Garcia, J.H.; Young, J.S.; Semonche, A.; Fullerton, H.J.; Kim, H.; Cooke, D.L.; et al. Machine learning for predicting hemorrhage in pediatric patients with brain arteriovenous malformation. J. Neurosurg. Pediatr. 2022, 30, 203–209. [Google Scholar] [CrossRef]
- Shi, X.; Cui, Y.; Wang, S.; Pan, Y.; Wang, B.; Lei, M. Development and validation of a web-based artificial intelligence prediction model to assess massive intraoperative blood loss for metastatic spinal disease using machine learning techniques. Spine J. 2024, 24, 146–160. [Google Scholar] [CrossRef] [PubMed]
- Turcato, G.; Cipriano, A.; Park, N.; Zaboli, A.; Ricci, G.; Riccardi, A.; Barbieri, G.; Gianpaoli, S.; Guiddo, G.; Santini, M.; et al. Decision tree analysis for assessing the risk of post-traumatic haemorrhage after mild traumatic brain injury in patients on oral anticoagulant therapy. BMC Emerg. Med. 2022, 22, 47. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Gao, L.; Wang, X.; Wei, J.; Xia, B.; Liu, X.; Zheng, P. Application of supervised machine learning algorithms to predict the risk of hidden blood loss during the perioperative period in thoracolumbar burst fracture patients complicated with neurological compromise. Front. Public Health 2022, 10, 969919. [Google Scholar] [CrossRef]
- Zernikow, B.; Holtmannspoetter, K.; Michel, E.; Theilhaber, M.; Pielemeier, W.; Hennecke, K. Artificial neural network for predicting intracranial haemorrhage in preterm neonates. Acta Paediatr. 1998, 87, 969–975. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.; Yang, Y.; Tang, J.; Xiong, T. Machine learning for predicting intraventricular hemorrhage in preterm infants. J. Evid.-Based Med. 2024, 17, 7–9. [Google Scholar] [CrossRef]
- Nopp, S.; Spielvogel, C.P.; Schmaldienst, S.; Klauser-Braun, R.; Lorenz, M.; Bauer, B.N.; Pabinger, I.; Säemann, M.; Königsbrügge, O.; Ay, C. Bleeding risk assessment in end-stage kidney disease: Validation of existing risk scores and evaluation of a machine learning-based approach. Thromb. Haemost. 2022, 122, 1558–1566. [Google Scholar] [CrossRef]
- Ahmadzia, H.K.; Dzienny, A.C.; Bopf, M.; Phillips, J.M.; Federspiel, J.J.; Amdur, R.; Rice, M.M.; Rodriguez, L. Machine learning models for prediction of maternal hemorrhage and transfusion: Model development study. JMIR Bioinform. Biotechnol. 2024, 5, e52059. [Google Scholar] [CrossRef]
- Akazawa, M.; Hashimoto, K.; Katsuhiko, N.; Kaname, Y. Machine learning approach for the prediction of postpartum hemorrhage in vaginal birth. Sci. Rep. 2021, 11, 22620. [Google Scholar] [CrossRef]
- Akazawa, M.; Hashimoto, K. A multimodal deep learning model for predicting severe hemorrhage in placenta previa. Sci. Rep. 2023, 13, 17320. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, H.; Guo, D.; Yang, S.; Liu, B.; Hao, Y.; Liu, Q.; Zhang, T.; Meng, F.; Sun, L.; et al. Risk of intraoperative hemorrhage during cesarean scar ectopic pregnancy surgery: Development and validation of an interpretable machine learning prediction model. eClinicalMedicine 2024, 78, 102969. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.; Park, J.; Kim, H.; Lee, Y.J.; Lee, Y.; Choi, Y.S.; Yeo, S.G.; Kang, J.; Rahmati, M.; Lee, H.; et al. Artificial intelligence models predicting abnormal uterine bleeding after COVID-19 vaccination. Sci. Rep. 2025, 15, 7081. [Google Scholar] [CrossRef] [PubMed]
- Futterman, I.D.; Sher, O.; Saroff, C.; Cohen, A.; Doulaveris, G.; Dar, P.; Griffin, M.M.; Limaye, M.; Owens, T.; Brustman, L.; et al. Machine learning for the prediction of surgical morbidity in placenta accreta spectrum. Am. J. Perinatol. 2025, 42, 281–292. [Google Scholar] [CrossRef] [PubMed]
- Guedalia, J.; Lipschuetz, M.; Daoud-Sabag, L.; Cohen, S.M.; Novoselsky-Persky, M.; Yagel, S.; Unger, R.; Karavani, G. Prediction of neonatal subgaleal hemorrhage using first stage of labor data: A machine-learning based model. J. Gynecol. Obstet. Hum. Reprod. 2022, 51, 102320. [Google Scholar] [CrossRef]
- Holcroft, S.; Karangwa, I.; Little, F.; Behoor, J.; Bazirete, O. Predictive modelling of postpartum haemorrhage using early risk factors: A comparative analysis of statistical and machine learning models. Int. J. Environ. Res. Public Health 2024, 21, 600. [Google Scholar] [CrossRef]
- Krishnamoorthy, S.; Liu, Y.; Liu, K. A novel oppositional binary crow search algorithm with optimal machine learning based postpartum hemorrhage prediction model. BMC Pregnancy Childbirth 2022, 22, 560. [Google Scholar] [CrossRef]
- Lengerich, B.J.; Caruana, R.; Painter, I.; Weeks, W.B.; Sitcov, K.; Souter, V. Interpretable machine learning predicts postpartum hemorrhage with severe maternal morbidity in a lower-risk laboring obstetric population. Am. J. Obstet. Gynecol. MFM 2024, 6, 101391. [Google Scholar] [CrossRef]
- Liu, J.; Wang, C.; Yan, R.; Lu, Y.; Bai, J.; Wang, H.; Li, R. Machine learning-based prediction of postpartum hemorrhage after vaginal delivery: Combining bleeding high risk factors and uterine contraction curve. Arch. Gynecol. Obstet. 2022, 306, 1015–1025. [Google Scholar] [CrossRef]
- Mehrnoush, V.; Ranjbar, A.; Farashah, M.V.; Darsareh, F.; Shekari, M.; Jahromi, M.S. Prediction of postpartum hemorrhage using traditional statistical analysis and a machine learning approach. AJOG Glob. Rep. 2023, 3, 100185. [Google Scholar] [CrossRef]
- Meyer, S.R.; Carver, A.; Joo, H.; Venkatesh, K.K.; Jelovsek, J.E.; Klumpner, T.T.; Singh, K. External validation of postpartum hemorrhage prediction models using electronic health record data. Am. J. Perinatol. 2024, 41, 598–605. [Google Scholar] [CrossRef]
- Shazly, S.A.; Hortu, I.; Shih, J.C.; Melekoglu, R.; Fan, S.; Ahmed, F.U.A.; Karaman, E.; Fatkullin, I.; Pinto, P.V.; Irianti, S.; et al. Prediction of clinical outcomes in women with placenta accreta spectrum using machine learning models: An international multicenter study. J. Matern. Fetal Neonatal Med. 2024, 35, 6644–6653. [Google Scholar] [CrossRef]
- Susanu, C.; Hărăbor, A.; Vasilache, I.-A.; Harabor, V.; Călin, A.-M. Predicting intra- and postpartum hemorrhage through artificial intelligence. Medicina 2024, 60, 1604. [Google Scholar] [CrossRef] [PubMed]
- Walczak, S.; Mikhail, E. Predicting estimated blood loss and transfusions in gynecologic surgery using artificial neural networks. Int. J. Healthc. Inf. Syst. Inform. 2021, 16, 1–15. [Google Scholar] [CrossRef]
- Wang, M.; Yi, G.; Zhang, Y.; Li, M.; Zhang, J. Quantitative prediction of postpartum hemorrhage in cesarean section on machine learning. BMC Med. Inform. Decis. Mak. 2024, 24, 166. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Liao, C.; Zhang, H.; Hu, Y. Postpartum haemorrhage risk prediction model developed by machine learning algorithms: A single-centre retrospective analysis of clinical data. Clin. Exp. Obstet. Gynecol. 2024, 51, 60. [Google Scholar] [CrossRef]
- Westcott, J.M.; Hughes, F.; Liu, W.; Grivainis, M.; Hoskins, I.; Fenyo, D. Prediction of maternal hemorrhage using machine learning: Retrospective cohort study. J. Med. Internet Res. 2022, 24, e34108. [Google Scholar] [CrossRef]
- Zheng, C.; Yue, P.; Cao, K.; Wang, Y.; Zhang, C.; Zhong, J.; Xu, X.; Lin, C.; Liu, Q.; Zou, Y.; et al. Predicting intraoperative blood loss during cesarean sections based on multi-modal information: A two-center study. Abdom. Radiol. 2024, 49, 2325–2339. [Google Scholar] [CrossRef]
- Zheutlin, A.B.; Vieira, L.; Shewcraft, R.A.; Li, S.; Wang, Z.; Schadt, E.; Gross, S.; Dolan, S.M.; Stone, J.; Li, L.; et al. Improving postpartum hemorrhage risk prediction using longitudinal electronic medical records. J. Am. Med. Inform. Assoc. 2022, 29, 296–305. [Google Scholar] [CrossRef]
- Grdinic, A.G.; Radovanovic, S.; Gleditsch, J.; Jørgensen, C.T.; Asady, E.; Pettersen, H.H.; Delibasic, B.; Ghanima, W. Developing a machine learning model for bleeding prediction in patients with cancer-associated thrombosis receiving anticoagulation therapy. J. Thromb. Haemost. 2024, 22, 1094–1104. [Google Scholar] [CrossRef]
- Muñoz Martín, A.J.; Lecumberri, R.; Souto, J.C.; Obispo, B.; Sanchez, A.; Aparicio, J.; Aguayo, C.; Gutierrez, D.; García Palomo, A.; Benavent, D.; et al. Prediction model for major bleeding in anticoagulated patients with cancer-associated venous thromboembolism using machine learning and natural language processing. Clin. Transl. Oncol. 2024, 27, 1816–1825. [Google Scholar] [CrossRef]
- Truong, B.; Zheng, J.; Hornsby, L.; Fox, B.; Chou, C.; Qian, J. Development and validation of machine learning algorithms to predict 1-year ischemic stroke and bleeding events in patients with atrial fibrillation and cancer. Cardiovasc. Toxicol. 2024, 24, 365–374. [Google Scholar] [CrossRef]
- Valiente Fernández, M.; García Fuentes, C.; Delgado Moya, F.P.; Marcos Morales, A.; Fernández Hervás, H.; Barea Mendoza, J.A.; Mudarra Reche, C.; Bermejo Aznárez, S.; Muñoz Calahorro, R.; López García, L.; et al. Could machine learning algorithms help us predict massive bleeding at prehospital level? Med. Intensiv. 2023, 47, 681–690. [Google Scholar] [CrossRef]
- Guo, C.; Gong, M.; Ji, L.; Pan, F.; Han, H.; Li, C.; Li, T. A prediction model for massive hemorrhage in trauma: A retrospective observational study. BMC Emerg. Med. 2022, 22, 180. [Google Scholar] [CrossRef]
- Lee, S.W.; Kung, H.C.; Huang, J.F.; Hsu, C.P.; Wang, C.C.; Wu, Y.T.; Wen, M.S.; Cheng, C.T.; Liao, C.H. The clinical application of machine learning-based models for early prediction of hemorrhage in trauma intensive care units. J. Pers. Med. 2022, 12, 1901. [Google Scholar] [CrossRef]
- Li, K.; Wu, H.; Pan, F.; Chen, L.; Feng, C.; Liu, Y.; Hui, H.; Cai, X.; Che, H.; Ma, Y.; et al. A machine learning–based model to predict acute traumatic coagulopathy in trauma patients upon emergency hospitalization. Clin. Appl. Thromb. 2020, 26, 1076029619897827. [Google Scholar] [CrossRef] [PubMed]
- Perkins, Z.B.; Yet, B.; Marsden, M.; Glasgow, S.; Marsh, W.; Davenport, R.; Brohi, K.; Tai, N.R.M. Early identification of trauma-induced coagulopathy: Development and validation of a multivariable risk prediction model. Ann. Surg. 2021, 274, e1119–e1128. [Google Scholar] [CrossRef] [PubMed]
- Richards, J.E.; Yang, S.; Kozar, R.A.; Scalea, T.M.; Hu, P. A machine learning–based coagulation risk index predicts acute traumatic coagulopathy in bleeding trauma patients. J. Trauma Acute Care Surg. 2025, 98, 614–620. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Peng, C.; Peng, L.; Wang, J.; Li, Y.; Li, W. A machine learning approach for the prediction of traumatic brain injury–induced coagulopathy. Front. Med. 2021, 8, 792689. [Google Scholar] [CrossRef]
- Xiong, X.; Fu, H.; Xu, B.; Wei, W.; Zhou, M.; Hu, P.; Ren, Y.; Mao, Q. Ten machine learning models for predicting preoperative and postoperative coagulopathy in patients with trauma: Multicenter cohort study. J. Med. Internet Res. 2025, 27, e66612. [Google Scholar] [CrossRef]
- Meng, R.; Wang, W.; Zhai, Z.; Zuo, C. Machine learning algorithm to predict postoperative bleeding complications after lateral decubitus percutaneous nephrolithotomy. Medicine 2024, 103, e37050. [Google Scholar] [CrossRef]
- Laursen, M.S.; Pedersen, J.S.; Hansen, R.S.; Savarimuthu, T.R.; Lynggaard, R.B.; Vinholt, P.J. Doctors identify hemorrhage better during chart review when assisted by artificial intelligence. Appl. Clin. Inform. 2023, 14, 743–751. [Google Scholar] [CrossRef] [PubMed]
- Mitra, A.; Rawat, B.P.S.; McManus, D.; Yu, H. Bleeding entity recognition in electronic health records: A comprehensive analysis of end-to-end systems. AMIA Annu. Symp. Proc. 2021, 2020, 860–869. [Google Scholar] [PubMed]
- Mittman, B.G.; Sheehan, M.; Kojima, L.; Casacchia, N.J.; Lisheba, O.; Hu, B.; Pappas, M.A.; Rothberg, M.B. Development and internal validation of the Cleveland Clinic Bleeding Model to predict major bleeding risk at admission in medical inpatients. J. Thromb. Haemost. 2024, 22, 2855–2863. [Google Scholar] [CrossRef] [PubMed]
- Nwanosike, E.M.; Merchant, H.A.; Sunter, W.; Ansari, M.A.; Conway, B.R.; Hasan, S.S. Direct oral anticoagulants and the risk of adverse clinical outcomes among patients with different body weight categories: A large hospital-based study. Eur. J. Clin. Pharmacol. 2024, 80, 163–173. [Google Scholar] [CrossRef]
- Pedersen, J.S.; Laursen, M.S.; Rajeeth Savarimuthu, T.; Hansen, R.S.; Alnor, A.B.; Bjerre, K.V.; Kjær, I.M.; Gils, C.; Thorsen, A.F.; Andersen, E.S.; et al. Deep learning detects and visualizes bleeding events in electronic health records. Res. Pract. Thromb. Haemost. 2021, 5, e12505. [Google Scholar] [CrossRef]
- Rickards, C.A.; Vyas, N.; Ryan, K.L.; Ward, K.R.; Andre, D.; Hurst, G.M.; Barrera, C.R.; Convertino, V.A. Are you bleeding? Validation of a machine-learning algorithm for determination of blood volume status: Application to remote triage. J. Appl. Physiol. 2014, 116, 486–494. [Google Scholar] [CrossRef][Green Version]
- Yoon, D.; Yoo, M.; Kim, B.S.; Kim, Y.G.; Lee, J.H.; Lee, E.; Min, G.H.; Hwang, D.Y.; Baek, C.; Cho, M.; et al. Automated deep learning model for estimating intraoperative blood loss using gauze images. Sci. Rep. 2024, 14, 2597. [Google Scholar] [CrossRef]
- Che, J.; Yang, B.; Xie, Y.; Wang, L.; Chang, Y.; Han, J.; Zhang, H. A precise blood transfusion evaluation model for aortic surgery: A single-center retrospective study. J. Clin. Monit. Comput. 2024, 38, 691–699. [Google Scholar] [CrossRef]
- Chen, C.; Wang, Y.; Yang, X.; Zhang, M.; He, J.; Yang, L.; Qin, L.; Chen, B.; Chen, B.; Wang, Q. Machine learning–based prediction of intraoperative red blood cell transfusion in aortic valve replacement surgery. Clin. Lab. 2024, 70. [Google Scholar] [CrossRef]
- Cunha, C.B.C.D.; Lima, T.A.; Ferraz, D.L.M.; Silva, I.T.C.; Santiago, M.K.D.; Sena, G.R.; Monteiro, V.S.; Andrade, L.B. Predicting the need for blood transfusions in cardiac surgery: A comparison between machine learning algorithms and established risk scores in the Brazilian population. Braz. J. Cardiovasc. Surg. 2024, 39, e20230212. [Google Scholar] [CrossRef]
- Hur, S.; Yoo, J.; Min, J.Y.; Jeon, Y.J.; Cho, J.H.; Seo, J.Y.; Cho, D.; Kim, K.; Lee, Y.; Cha, W.C. Development, validation, and usability evaluation of machine learning algorithms for predicting personalized red blood cell demand among thoracic surgery patients. Int. J. Med. Inf. 2024, 191, 105543. [Google Scholar] [CrossRef]
- Li, Q.; Lv, H.; Chen, Y.; Shen, J.; Shi, J.; Zhou, C.; Yan, F. Development and validation of a machine learning prediction model for perioperative red blood cell transfusions in cardiac surgery. Int. J. Med. Inf. 2024, 184, 105343. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Xue, B.; Kannampallil, T.; Lu, C.; Abraham, J. A novel generative multi-task representation learning approach for predicting postoperative complications in cardiac surgery patients. J. Am. Med. Inform. Assoc. 2025, 32, 459–469. [Google Scholar] [CrossRef]
- Sun, Z.; Cui, Z.; Xie, Y.; Wang, L.; Li, Z.; Yang, X.; Zhang, X.; Wang, J. Evaluation of the factors influencing blood transfusion during minimally invasive direct coronary artery bypass surgery. Cardiology 2025, 150, 98–110. [Google Scholar] [CrossRef] [PubMed]
- Tschoellitsch, T.; Böck, C.; Mahečić, T.T.; Hofmann, A.; Meier, J. Machine learning–based prediction of massive perioperative allogeneic blood transfusion in cardiac surgery. Eur. J. Anaesthesiol. 2022, 39, 766–773. [Google Scholar] [CrossRef]
- Wang, Z.; Zhe, S.; Zimmerman, J.; Morrisey, C.; Tonna, J.E.; Sharma, V.; Metcalf, R.A. Development and validation of a machine learning method to predict intraoperative red blood cell transfusions in cardiothoracic surgery. Sci. Rep. 2022, 12, 1355. [Google Scholar] [CrossRef]
- Zhou, R.; Li, Z.; Liu, J.; Qian, D.; Meng, X.; Guan, L.; Sun, X.; Li, H.; Yu, M. Prediction of intraoperative red blood cell transfusion in valve replacement surgery: Machine learning algorithm development based on non-anemic cohort. Front. Cardiovasc. Med. 2024, 11, 1344170. [Google Scholar] [CrossRef]
- Sheikhalishahi, S.; Goss, S.; Seidlmayer, L.K.; Zaghdoudi, S.; Hinske, L.C.; Kaspar, M. Predicting blood transfusion demand in intensive care patients after surgery by comparative analysis of temporally extended data selection. BMC Med. Inform. Decis. Mak. 2024, 24, 397. [Google Scholar] [CrossRef]
- Shung, D.; Huang, J.; Castro, E.; Tay, J.K.; Simonov, M.; Laine, L.; Batra, R.; Krishnaswamy, S. Neural network predicts need for red blood cell transfusion for patients with acute gastrointestinal bleeding admitted to the intensive care unit. Sci. Rep. 2021, 11, 8827. [Google Scholar] [CrossRef]
- Chen, S.; Liu, L.P.; Wang, Y.J.; Zhou, X.H.; Dong, H.; Chen, Z.W.; Wu, J.; Gui, R.; Zhao, Q.Y. Advancing prediction of risk of intraoperative massive blood transfusion in liver transplantation with machine learning models: A multicenter retrospective study. Front. Neuroinform. 2022, 16, 893452. [Google Scholar] [CrossRef]
- Liu, L.P.; Zhao, Q.Y.; Wu, J.; Luo, Y.W.; Dong, H.; Chen, Z.W.; Gui, R.; Wang, Y.J. Machine learning for the prediction of red blood cell transfusion in patients during or after liver transplantation surgery. Front. Med. 2021, 8, 632210. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Jiang, L.; Zhu, X. A machine learning–modified novel nomogram to predict perioperative blood transfusion of total gastrectomy for gastric cancer. Front. Oncol. 2022, 12, 826760. [Google Scholar] [CrossRef] [PubMed]
- Jalali, A.; Lonsdale, H.; Zamora, L.V.; Ahumada, L.; Nguyen, A.T.H.; Rehman, M.; Fackler, J.; Stricker, P.A.; Fernandez, A.M. Machine learning applied to registry data: Development of a patient-specific prediction model for blood transfusion requirements during craniofacial surgery using the pediatric craniofacial perioperative registry dataset. Anesth. Analg. 2021, 132, 160–171. [Google Scholar] [CrossRef]
- Puladi, B.; Ooms, M.; Rieg, A.; Taubert, M.; Rashad, A.; Hölzle, F.; Röhrig, R.; Modabber, A. Development of machine learning and multivariable models for predicting blood transfusion in head and neck microvascular reconstruction for risk-stratified patient blood management. Head Neck 2023, 45, 1389–1405. [Google Scholar] [CrossRef]
- Stehrer, R.; Hingsammer, L.; Staudigl, C.; Hunger, S.; Malek, M.; Jacob, M.; Meier, J. Machine learning–based prediction of perioperative blood loss in orthognathic surgery. J. Cranio-Maxillofac. Surg. 2019, 47, 1676–1681. [Google Scholar] [CrossRef] [PubMed]
- Cabrera, A.; Bouterse, A.; Nelson, M.; Razzouk, J.; Ramos, O.; Chung, D.; Cheng, W.; Danisa, O. Use of random forest machine learning algorithm to predict short-term outcomes following posterior cervical decompression with instrumented fusion. J. Clin. Neurosci. 2023, 107, 167–171. [Google Scholar] [CrossRef]
- Durand, W.M.; DePasse, J.M.; Daniels, A.H. Predictive modeling for blood transfusion after adult spinal deformity surgery: A tree-based machine learning approach. Spine 2018, 43, 1058–1066. [Google Scholar] [CrossRef]
- Dong, S.; Li, W.; Tang, Z.R.; Wang, H.; Pei, H.; Yuan, B. Development and validation of a novel predictive model and web calculator for evaluating transfusion risk after spinal fusion for spinal tuberculosis: A retrospective cohort study. BMC Musculoskelet. Disord. 2021, 22, 825. [Google Scholar] [CrossRef]
- Fatima, N.; Zheng, H.; Massaad, E.; Hadzipasic, M.; Shankar, G.M.; Shin, J.H. Development and validation of machine learning algorithms for predicting adverse events after surgery for lumbar degenerative spondylolisthesis. World Neurosurg. 2020, 140, 627–641. [Google Scholar] [CrossRef]
- Lang, F.-F.; Liu, L.-Y.; Wang, S.-W. Predictive modeling of perioperative blood transfusion in lumbar posterior interbody fusion using machine learning. Front. Physiol. 2023, 14, 1306453. [Google Scholar] [CrossRef]
- De La Garza Ramos, R.; Hamad, M.K.; Ryvlin, J.; Krol, O.; Passias, P.G.; Fourman, M.S.; Shin, J.H.; Yanamadala, V.; Gelfand, Y.; Murthy, S.; et al. An artificial neural network model for the prediction of perioperative blood transfusion in adult spinal deformity surgery. J. Clin. Med. 2022, 11, 4436. [Google Scholar] [CrossRef]
- Tunthanathip, T.; Sae-heng, S.; Oearsakul, T.; Kaewborisutsakul, A.; Taweesomboonyat, C. Development and deployment of web application using machine learning for predicting intraoperative transfusions in neurosurgical operations. Iran. J. Neurosurg. 2022, 8, 25. [Google Scholar] [CrossRef]
- Xiao, S.; Liu, F.; Yu, L.; Li, X.; Ye, X.; Gong, X. Development and validation of a nomogram for blood transfusion during intracranial aneurysm clamping surgery: A retrospective analysis. BMC Med. Inform. Decis. Mak. 2023, 23, 71. [Google Scholar] [CrossRef]
- Xiao, S.; Jiang, F.; Chen, Y.; Gong, X. Development and validation of a prediction tool for intraoperative blood transfusion in brain tumor resection surgery: A retrospective analysis. Sci. Rep. 2023, 13, 17428. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Cao, B.; Yang, J.; Ren, H.; Xia, X.; Zhang, X.; Yan, W.; Liang, X.; Li, C. Construction and effect evaluation of prediction model for red blood cell transfusion requirement in cesarean section based on artificial intelligence. BMC Med. Inform. Decis. Mak. 2023, 23, 213. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-W.; Park, B.; Seo, J.; Lee, S.; Sim, J.-H. Development of a machine learning approach for prediction of red blood cell transfusion in patients undergoing cesarean section at a single institution. Sci. Rep. 2024, 14, 16628. [Google Scholar] [CrossRef]
- Raz, N.R.; Anoushirvani, A.A.; Rahimian, N.; Ghoerishi, M.; Alibeik, N.; Rad, M.S. Nano fuzzy alarming system for blood transfusion requirement detection in cancer using deep learning. Sci. Rep. 2024, 14, 15958. [Google Scholar] [CrossRef]
- Laios, A.; Kalampokis, E.; Mamalis, M.E.; Thangavelu, A.; Tan, Y.S.; Hutson, R.; Munot, S.; Broadhead, T.; Nugent, D.; Theophilou, G.; et al. Explaining the elusive nature of a well-defined threshold for blood transfusion in advanced epithelial ovarian cancer cytoreductive surgery. Diagnostics 2023, 14, 94. [Google Scholar] [CrossRef]
- Buddhiraju, A.; Shimizu, M.R.; Subih, M.A.; Chen, T.L.-W.; Seo, H.H.; Kwon, Y.-M. Validation of machine learning model performance in predicting blood transfusion after primary and revision total hip arthroplasty. J. Arthroplasty 2023, 38, 1959–1966. [Google Scholar] [CrossRef]
- Cavazos, D.R.; Sayeed, Z.; Court, T.; Chen, C.; Little, B.E.; Darwiche, H.F. Predicting factors for blood transfusion in primary total knee arthroplasty using a machine learning method. J. Am. Acad. Orthop. Surg. 2023, 31, e845–e858. [Google Scholar] [CrossRef]
- Chen, X.; Pan, J.; Li, Y.; Tang, R. Application of machine learning model in predicting the likelihood of blood transfusion after hip fracture surgery. Aging Clin. Exp. Res. 2023, 35, 2643–2656. [Google Scholar] [CrossRef]
- Chen, Y.; Cai, X.; Cao, Z.; Lin, J.; Huang, W.; Zhuang, Y.; Xiao, L.; Guan, X.; Wang, Y.; Xia, X.; et al. Prediction of red blood cell transfusion after orthopedic surgery using an interpretable machine learning framework. Front. Surg. 2023, 10, 1047558. [Google Scholar] [CrossRef]
- Cohen-Levy, W.B.; Klemt, C.; Tirumala, V.; Burns, J.C.; Barghi, A.; Habibi, Y.; Kwon, Y.M. Artificial neural networks for the prediction of transfusion rates in primary total hip arthroplasty. Arch. Orthop. Trauma Surg. 2022, 143, 1643–1650. [Google Scholar] [CrossRef]
- Deng, J.; Zhou, C.; Xiao, F.; Chen, J.; Li, C.; Xie, Y. Construction of a predictive model for blood transfusion in patients undergoing total hip arthroplasty and identification of clinical heterogeneity. Sci. Rep. 2024, 14, 724. [Google Scholar] [CrossRef] [PubMed]
- Dreizin, D.; Zhou, Y.; Chen, T.; Li, G.; Yuille, A.L.; McLenithan, A.; Morrison, J.J. Deep learning-based quantitative visualization and measurement of extraperitoneal hematoma volumes in patients with pelvic fractures: Potential role in personalized forecasting and decision support. J. Trauma Acute Care Surg. 2020, 88, 425–433. [Google Scholar] [CrossRef]
- Faure, N.; Knecht, S.; Tran, P.; Tamine, L.; Orban, J.C.; Bronsard, N.; Gonzalez, J.F.; Micicoi, G. Prediction of transfusion risk after total knee arthroplasty: Use of a machine learning algorithm. Orthop. Traumatol. Surg. Res. 2025, 111, 103985. [Google Scholar] [CrossRef] [PubMed]
- Gowd, A.K.; Agarwalla, A.; Amin, N.H.; Romeo, A.A.; Nicholson, G.P.; Verma, N.N.; Liu, J.N. Construct validation of machine learning in the prediction of short-term postoperative complications following total shoulder arthroplasty. J. Shoulder Elb. Surg. 2019, 28, e410–e421. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; He, Q.; Li, Y. Development and validation of machine learning models to predict perioperative transfusion risk for hip fractures in the elderly. Ann. Med. 2024, 56, 2357225. [Google Scholar] [CrossRef]
- Huang, X.; Wang, Y.; Chen, B.; Huang, Y.; Wang, X.; Chen, L.; Gui, R.; Ma, X. Ability of a machine learning algorithm to predict the need for perioperative red blood cells transfusion in pelvic fracture patients: A multicenter cohort study in China. Front. Med. 2021, 8, 694733. [Google Scholar] [CrossRef]
- Huang, Z.; Martin, J.; Huang, Q.; Ma, J.; Pei, F.; Huang, C. Predicting postoperative transfusion in elective total hip and knee arthroplasty: Comparison of different machine learning models of a case-control study. Int. J. Surg. 2021, 96, 106183. [Google Scholar] [CrossRef]
- Jo, C.; Ko, S.; Shin, W.C.; Han, H.S.; Lee, M.C.; Ko, T.; Ro, D.H. Transfusion after total knee arthroplasty can be predicted using the machine learning algorithm. Knee Surg. Sports Traumatol. Arthrosc. 2020, 28, 1757–1764. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, H.; Huang, Y.; Memtsoudis, S.; Parks, M.; Huang, Y.; Ma, Y. Utilization of machine learning methods for predicting surgical outcomes after total knee arthroplasty. PLoS ONE 2022, 17, e0263897. [Google Scholar] [CrossRef] [PubMed]
- Seong, H.; Lee, K.S.; Choi, Y.; Na, D.; Kim, J.; Shin, H.J.; Ahn, K.H. Explainable artificial intelligence for predicting red blood cell transfusion in geriatric patients undergoing hip arthroplasty: Machine learning analysis using national health insurance data. Medicine 2024, 103, e36909. [Google Scholar] [CrossRef] [PubMed]
- Zang, H.; Hu, A.; Xu, X.; Ren, H.; Xu, L. Development of machine learning models to predict perioperative blood transfusion in hip surgery. BMC Med. Inform. Decis. Mak. 2024, 24, 158. [Google Scholar] [CrossRef]
- Zhou, Y.; Wang, S.; Wu, Z.; Chen, W.; Yang, D.; Chen, C.; Zhao, G.; Hong, Q. An explainable and supervised machine learning model for prediction of red blood cell transfusion in patients during hip fracture surgery. BMC Anesthesiol. 2024, 24, 467. [Google Scholar] [CrossRef]
- Zhu, J.; Xu, C.; Jiang, Y.; Zhu, J.; Tu, M.; Yan, X.; Shen, Z.; Lou, Z. Development and validation of a machine learning algorithm to predict the risk of blood transfusion after total hip replacement in patients with femoral neck fractures: A multicenter retrospective cohort study. Orthop. Surg. 2024, 16, 2066–2080. [Google Scholar] [CrossRef]
- Liu, Y.; Tao, Y.; Cai, T.N.; Yang, Y.; Mao, H.M.; Zhong, S.J.; Guo, W.L. Development of a simplified model and nomogram for the prediction of pulmonary hemorrhage in respiratory distress syndrome in extremely preterm infants. BMC Pediatr. 2024, 24, 760. [Google Scholar] [CrossRef]
- El-Menyar, A.; Naduvilekandy, M.; Asim, M.; Rizoli, S.; Al-Thani, H. Machine learning models predict triage levels, massive transfusion protocol activation, and mortality in trauma utilizing patients’ hemodynamics on admission. Comput. Biol. Med. 2024, 179, 108880. [Google Scholar] [CrossRef]
- Feng, Y.-N.; Xu, Z.-H.; Liu, J.-T.; Sun, X.-L.; Wang, D.-Q.; Yu, Y. Intelligent prediction of RBC demand in trauma patients using decision tree methods. Mil. Med. Res. 2021, 8, 33. [Google Scholar] [CrossRef]
- Hodgman, E.; Cripps, M.W.; Mina, M.J.; Bulger, E.M.; Schreiber, M.A.; Brasel, K.J.; Cohen, M.J.; Muskat, P.; Myers, J.G.; Alarcon, L.H.; et al. External validation of a smartphone app model to predict the need for massive transfusion using five different definitions. J. Trauma Acute Care Surg. 2018, 84, 397–402. [Google Scholar] [CrossRef]
- Lammers, D.; Marenco, C.; Morte, K.; Conner, J.; Williams, J.; Bax, T.; Martin, M.; Eckert, M.; Bingham, J. Machine learning for military trauma: Novel massive transfusion predictive models in combat zones. J. Surg. Res. 2022, 270, 369–375. [Google Scholar] [CrossRef]
- Nederpelt, C.J.; Mokhtari, A.K.; Alser, O.; Tsiligkaridis, T.; Roberts, J.; Cha, M.; Fawley, J.A.; Parks, J.J.; Mendoza, A.E.; Fagenholz, P.J.; et al. Development of a field artificial intelligence triage tool: Confidence in the prediction of shock, transfusion, and definitive surgical therapy in patients with truncal gunshot wounds. J. Trauma Acute Care Surg. 2021, 90, 1054–1060. [Google Scholar] [CrossRef] [PubMed]
- Nikouline, A.; Feng, J.; Rudzicz, F.; Nathens, A.; Nolan, B. Machine learning in the prediction of massive transfusion in trauma: A retrospective analysis as a proof-of-concept. Eur. J. Trauma Emerg. Surg. 2024, 50, 1073–1081. [Google Scholar] [CrossRef]
- Shahi, N.; Shahi, A.K.; Phillips, R.; Shirek, G.; Bensard, D.; Moulton, S.L. Decision-making in pediatric blunt solid organ injury: A deep learning approach to predict massive transfusion, need for operative management, and mortality risk. J. Pediatr. Surg. 2021, 56, 379–384. [Google Scholar] [CrossRef] [PubMed]
- Fernández, M.V. Letter to the editor: Assessment of machine learning methods to predict massive blood transfusion in trauma. World J. Surg. 2023, 47, 3445–3446. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, T.M.; Milestone, Z.P.; Tempel, P.E.; Gao, S.; Burd, R.S. Development and validation of a Bayesian belief network predicting the probability of blood transfusion after pediatric injury. J. Trauma Acute Care Surg. 2023, 94, 304–311. [Google Scholar] [CrossRef]
- Walczak, S. Artificial neural network medical decision support tool: Predicting transfusion requirements of ER patients. IEEE Trans. Inf. Technol. Biomed. 2005, 9, 468–474. [Google Scholar] [CrossRef]
- Engel, D.; Beilstein, C.M.; Jerney, P.; Furrer, M.A.; Burkhard, F.C.; Löffel, L.M.; Wuethrich, P.Y. Predictors for perioperative blood transfusion in patients undergoing open cystectomy and urinary diversion and development of a nomogram: An observational cohort study. J. Clin. Med. 2021, 10, 2797. [Google Scholar] [CrossRef]
- Li, B.; Verma, R.; Beaton, D.; Tamim, H.; Hussain, M.A.; Hoballah, J.J.; Lee, D.S.; Wijeysundera, D.N.; de Mestral, C.; Mamdani, M.; et al. Predicting outcomes following lower extremity open revascularization using machine learning. Sci. Rep. 2024, 14, 2899. [Google Scholar] [CrossRef]
- Hayn, D.; Kreiner, K.; Ebner, H.; Kastner, P.; Breznik, N.; Rzepka, A.; Hofmann, A.; Gombotz, H.; Schreier, G. Development of multivariable models to predict and benchmark transfusion in elective surgery supporting patient blood management. Appl. Clin. Inform. 2017, 8, 617–631. [Google Scholar] [CrossRef]
- Lee, S.M.; Lee, G.; Kim, T.K.; Le, T.; Hao, J.; Jung, Y.M.; Park, C.W.; Park, J.S.; Jun, J.K.; Lee, H.C.; et al. Development and validation of a prediction model for need for massive transfusion during surgery using intraoperative hemodynamic monitoring data. JAMA Netw. Open 2022, 5, e2246637. [Google Scholar] [CrossRef] [PubMed]
- Mitterecker, A.; Hofmann, A.; Trentino, K.M.; Lloyd, A.; Leahy, M.F.; Schwarzbauer, K.; Tschoellitsch, T.; Böck, C.; Hochreiter, S.; Meier, J. Machine learning–based prediction of transfusion. Transfusion 2020, 60, 1977–1986. [Google Scholar] [CrossRef]
- Park, I.; Park, J.H.; Yoon, J.; Koo, C.H.; Oh, A.Y.; Kim, J.H.; Ryu, J.H. Assessment of machine learning classifiers for predicting intraoperative blood transfusion in non-cardiac surgery. Transfus. Clin. Biol. 2025, 32, 1–8. [Google Scholar] [CrossRef]
- Walczak, S.; Velanovich, V. Prediction of perioperative transfusions using an artificial neural network. PLoS ONE 2020, 15, e0229450. [Google Scholar] [CrossRef]
- Yao, Y.; Cifuentes, J.; Zheng, B.; Yan, M. Computer algorithm can match physicians’ decisions about blood transfusions. J. Transl. Med. 2019, 17, 340. [Google Scholar] [CrossRef]
- Epah, J.; Gülec, I.; Winter, S.; Dörr, J.; Geisen, C.; Haecker, E.; Link, D.; Schwab, M.; Seifried, E.; Schäfer, R. From unit to dose: A machine learning approach for precise prediction of hemoglobin and iron content in individual packed red blood cell units. Adv. Sci. 2022, 9, 2204077. [Google Scholar] [CrossRef]
- Borgmann, D.M.; Mayr, S.; Polin, H.; Schaller, S.; Dorfer, V.; Obritzberger, L.; Endmayr, T.; Gabriel, C.; Winkler, S.M.; Jacak, J. Single molecule fluorescence microscopy and machine learning for Rhesus D antigen classification. Sci. Rep. 2016, 6, 32317. [Google Scholar] [CrossRef]
- Ferraz, A.; Brito, J.H.; Carvalho, V.; Machado, J. Blood type classification using computer vision and machine learning. Neural Comput. Appl. 2017, 28, 2029–2040. [Google Scholar] [CrossRef]
- Fung, M.K.; AuBuchon, J.P.; Stephens, L.D. Classification of posttransfusion adverse events using a publicly available artificial intelligence system. Transfusion 2024, 64, 590–596. [Google Scholar] [CrossRef]
- Larpant, N.; Niamsi, W.; Noiphung, J.; Chanakiat, W.; Sakuldamrongpanich, T.; Kittichai, V.; Tongloy, T.; Chuwongin, S.; Boonsang, S.; Laiwattanapaisal, W. Simultaneous phenotyping of five Rh red blood cell antigens on a paper-based analytical device combined with deep learning for rapid and accurate interpretation. Anal. Chim. Acta 2022, 1207, 339807. [Google Scholar] [CrossRef]
- Lai, M.; De Stefano, V.; Landolfi, R. Autoimmune hemolytic anemia with gel-based immunohematology tests: Neural network analysis. Immunol. Res. 2014, 58, 70–74. [Google Scholar] [CrossRef]
- Mahmud, S.; Donmez, T.B.; Mansour, M.; Kutlu, M.; Freeman, C. Anemia detection through non-invasive analysis of lip mucosa images. Front. Big Data 2023, 6, 1241899. [Google Scholar] [CrossRef] [PubMed]
- Moslemi, C.; Sækmose, S.; Larsen, R.; Brodersen, T.; Bay, J.T.; Didriksen, M.; Nielsen, K.R.; Bruun, M.T.; Dowsett, J.; Dinh, K.M.; et al. A deep learning approach to prediction of blood group antigens from genomic data. Transfusion 2024, 64, 2179–2195. [Google Scholar] [CrossRef] [PubMed]
- Ngufor, C.; Murphree, D.; Upadhyaya, S.; Madde, N.; Kor, D.; Pathak, J. Effects of plasma transfusion on perioperative bleeding complications: A machine learning approach. Stud. Health Technol. Inform. 2015, 216, 721–725. [Google Scholar]
- Ngufor, C.; Warner, M.A.; Murphree, D.H.; Liu, H.; Carter, R.; Storlie, C.B.; Kor, D.J. Identification of clinically meaningful plasma transfusion subgroups using unsupervised random forest clustering. AMIA Annu. Symp. Proc. 2018, 2017, 1332–1341. [Google Scholar] [PubMed]
- Whitaker, B.; Pizarro, J.; Deady, M.; Williams, A.; Ezzeldin, H.; Belov, A.; Kanderian, S.; Billings, D.; Cook, K.; Hettinger, A.Z.; et al. Detection of allergic transfusion-related adverse events from electronic medical records. Transfusion 2022, 62, 2029–2038. [Google Scholar] [CrossRef]
- Zhu, S.; Zhou, L.; Feng, Y.; Zhu, J.; Shu, Q.; Li, H. Understanding the risk factors for adverse events during exchange transfusion in neonatal hyperbilirubinemia using explainable artificial intelligence. BMC Pediatr. 2022, 22, 567. [Google Scholar] [CrossRef]
- Cifuentes, J.; Yao, Y.; Yan, M.; Zheng, B. Blood transfusion prediction using restricted Boltzmann machines. Comput. Methods Biomech. Biomed. Engin. 2020, 23, 510–517. [Google Scholar] [CrossRef]
- Etchells, T.A.; Harrison, M.J. Orthogonal search-based rule extraction for modelling the decision to transfuse. Anaesthesia 2006, 61, 335–338. [Google Scholar] [CrossRef]
- Fanoodi, B.; Malmir, B.; Jahantigh, F.F. Reducing demand uncertainty in the platelet supply chain through artificial neural networks and ARIMA models. Comput. Biol. Med. 2019, 113, 103415. [Google Scholar] [CrossRef]
- Feng, Y.; Xu, Z.; Sun, X.; Wang, D.; Yu, Y. Machine learning for predicting preoperative red blood cell demand. Transfus. Med. 2021, 31, 262–270. [Google Scholar] [CrossRef]
- Miri-Moghaddam, E.; Bizhaem, S.K.; Moezzifar, Z.; Salmani, F. Long-term prediction of Iranian blood product supply using LSTM: A 5-year forecast. BMC Med. Inform. Decis. Mak. 2024, 24, 213. [Google Scholar] [CrossRef]
- Motamedi, M.; Dawson, J.; Li, N.; Down, D.G.; Heddle, N.M. Demand forecasting for platelet usage: From univariate time series to multivariable models. PLoS ONE 2024, 19, e0297391. [Google Scholar] [CrossRef]
- Sarvestani, S.E.; Hatam, N.; Seif, M.; Kasraian, L.; Lari, F.S.; Bayati, M. Forecasting blood demand for different blood groups in Shiraz using autoregressive integrated moving average (ARIMA) and artificial neural network (ANN) and a hybrid approaches. Sci. Rep. 2022, 12, 22031. [Google Scholar] [CrossRef] [PubMed]
- Shih, H.; Rajendran, S. Comparison of time series methods and machine learning algorithms for forecasting Taiwan Blood Services Foundation’s blood supply. J. Healthc. Eng. 2019, 2019, 6123745. [Google Scholar] [CrossRef] [PubMed]
- Afonso, J.; Saraiva, M.M.; Ferreira, J.P.S.; Ribeiro, T.; Cardoso, H.; Macedo, G. Performance of a convolutional neural network for automatic detection of blood and hematic residues in small bowel lumen. Dig. Liver Dis. Off. J. Ital. Soc. Gastroenterol. Ital. Assoc. Study Liver 2021, 53, 654–657. [Google Scholar] [CrossRef] [PubMed]
- Aiwale, S.; Kolte, M.; Harpale, V.; Bendre, V.; Khurge, D.; Bhandari, S.U.; Kadam, S.; Mulani, A. Non-invasive Anemia Detection and Prediagnosis. J. Pharmacol. Pharmacother. 2024, 15, 408–416. [Google Scholar] [CrossRef]
- Akazawa, M.; Hashimoto, K. Prediction of hemorrhage in placenta previa: Radiomics analysis of pelvic MRI images. Eur. J. Obstet. Gynecol. Reprod. Biol. 2024, 299, 37–42. [Google Scholar] [CrossRef]
- Alnor, A.B.; Lynggaard, R.B.; Laursen, M.S.; Vinholt, P.J. Natural language processing for identifying major bleeding risk in hospitalised medical patients. Comput. Biol. Med. 2025, 190, 110093. [Google Scholar] [CrossRef]
- Aminsharifi, A.; Irani, D.; Pooyesh, S.; Parvin, H.; Dehghani, S.; Yousofi, K.; Fazel, E.; Zibaie, F. Artificial Neural Network System to Predict the Postoperative Outcome of Percutaneous Nephrolithotomy. J. Endourol. 2017, 31, 461–467. [Google Scholar] [CrossRef]
- Aoki, T.; Yamada, A.; Kato, Y.; Saito, H.; Tsuboi, A.; Nakada, A.; Niikura, R.; Fujishiro, M.; Oka, S.; Ishihara, S.; et al. Automatic detection of blood content in capsule endoscopy images based on a deep convolutional neural network. J. Gastroenterol. Hepatol. 2020, 35, 1196–1200. [Google Scholar] [CrossRef] [PubMed]
- Asadi, H.; Dowling, R.; Yan, B.; Mitchell, P. Machine learning for outcome prediction of acute ischemic stroke post intra-arterial therapy. PLoS ONE 2014, 9, e88225. [Google Scholar] [CrossRef] [PubMed]
- Asadi, H.; Kok, H.K.; Looby, S.; Brennan, P.; O’Hare, A.; Thornton, J. Outcomes and Complications After Endovascular Treatment of Brain Arteriovenous Malformations: A Prognostication Attempt Using Artificial Intelligence. World Neurosurg. 2016, 96, 562–569.e1. [Google Scholar] [CrossRef]
- Ayling, R.M.; Lewis, S.J.; Cotter, F. Potential roles of artificial intelligence learning and faecal immunochemical testing for prioritisation of colonoscopy in anaemia. Br. J. Haematol. 2019, 185, 311–316. [Google Scholar] [CrossRef]
- Khameneh, N.B.; Arabalibeik, H.; Salehian, P.; Setayeshi, S. Abnormal red blood cells detection using adaptive neuro-fuzzy system. Stud. Health Technol. Inform. 2012, 173, 30–34. [Google Scholar]
- Bahar, B.; Gehrie, E.A.; Mo, Y.D.; Jacquot, C.; Delaney, M. Using trends and outliers in managing delayed transfusions. Transfus. Med. Oxf. Engl. 2023, 33, 263–267. [Google Scholar] [CrossRef]
- Baiardi, P.; Piazza, V.; Montagna, G.; Mazzoleni, M.C. Use of statistical classifiers as support tools for the diagnosis of iron-deficiency anemia in patients on chronic hemodialysis. Stud. Health Technol. Inform. 1997, 43 Pt B, 666–670. [Google Scholar]
- Barbieri, C.; Neri, L.; Chermisi, M.; Bolzoni, E.; Cattinelli, I.; Decker, W.; Stuard, S.; Martín-Guerrero, J.D.; Mari, F. How to assess the risks associated with the usage of a medical device based on predictive modeling: The case of an anemia control model certified as medical device. Expert Rev. Med. Devices 2021, 18, 1117–1121. [Google Scholar] [CrossRef]
- Battles, J.B.; Kaplan, H.S.; Van der Schaaf, T.W.; Shea, C.E. The attributes of medical event-reporting systems: Experience with a prototype medical event-reporting system for transfusion medicine. Arch. Pathol. Lab. Med. 1998, 122, 231–238. [Google Scholar]
- Beck, J.R.; O’Donnell, J.F.; Hirai, F.; Simmons, J.J.; Healy, J.C.; Lyon, H.C. Computer-based exercises in anemia diagnosis (PlanAlyzer). Methods Inf. Med. 1989, 28, 364–369. [Google Scholar] [CrossRef] [PubMed]
- Bensalah, K. Perioperative outcomes of off-clamp vs complete hilar control laparoscopic partial nephrectomy. BJU Int. 2013, 111 Pt B, E242. [Google Scholar] [CrossRef] [PubMed]
- Birndorf, N.I.; Pentecost, J.O.; Coakley, J.R.; Spackman, K.A. An expert system to diagnose anemia and report results directly on hematology forms. Comput. Biomed. Res. Int. J. 1996, 29, 16–26. [Google Scholar] [CrossRef] [PubMed]
- Bonatti, J.; Schachner, T.; Wiedemann, D.; Weidinger, F.; Kolbitsch, C.; Knotzer, H.; Kon, Z.N.; Bonaros, N. Factors influencing blood transfusion requirements in robotic totally endoscopic coronary artery bypass grafting on the arrested heart. Eur. J. Cardio-Thorac. Surg. Off. J. Eur. Assoc. Cardio-Thorac. Surg. 2011, 39, 262–267. [Google Scholar] [CrossRef][Green Version]
- Causer, M.B.; Findlay, G.A.; Hawes, C.R.; Boswell, D.R. Assessment of a computerized system for the diagnosis of iron deficiency. Pathology 1994, 26, 37–39. [Google Scholar] [CrossRef]
- Chen, L.; Xia, S.; Lin, Y.; Chen, Y.; Xian, L.; Yang, Y.; Qiu, X.; Xu, L.; Xingshu, Z.; Chen, D.; et al. The role of coagulopathy and subdural hematoma thickness at admission in predicting the prognoses of patients with severe traumatic brain injury: A multicenter retrospective cohort study from China. Int. J. Surg. Lond. Engl. 2024, 110, 5545–5562. [Google Scholar] [CrossRef]
- Christie, S.A.; Conroy, A.S.; Callcut, R.A.; Hubbard, A.E.; Cohen, M.J. Dynamic multi-outcome prediction after injury: Applying adaptive machine learning for precision medicine in trauma. PLoS ONE 2019, 14, e0213836. [Google Scholar] [CrossRef]
- Chu, J.; Dong, W.; He, K.; Duan, H.; Huang, Z. Using neural attention networks to detect adverse medical events from electronic health records. J. Biomed. Inform. 2018, 87, 118–130. [Google Scholar] [CrossRef]
- Chu, Y.; Huang, F.; Gao, M.; Zou, D.W.; Zhong, J.; Wu, W.; Wang, Q.; Shen, X.N.; Gong, T.T.; Li, Y.Y.; et al. Convolutional neural network-based segmentation network applied to image recognition of angiodysplasias lesion under capsule endoscopy. World J. Gastroenterol. 2023, 29, 879–889. [Google Scholar] [CrossRef]
- Convertino, V.A.; Howard, J.T.; Hinojosa-Laborde, C.; Cardin, S.; Batchelder, P.; Mulligan, J.; Grudic, G.Z.; Moulton, S.L.; MacLeod, D.B. Individual-Specific, Beat-to-beat Trending of Significant Human Blood Loss: The Compensatory Reserve. Shock Augusta Ga 2015, 44 (Suppl. S1), 27–32. [Google Scholar] [CrossRef]
- Convertino, V.A.; Koons, N.J. The compensatory reserve: Potential for accurate individualized goal-directed whole blood resuscitation. Transfusion 2020, 60 (Suppl. S3), S150–S157. [Google Scholar] [CrossRef]
- Convertino, V.A.; Techentin, R.W.; Poole, R.J.; Dacy, A.C.; Carlson, A.N.; Cardin, S.; Haider, C.R.; Holmes, D.R., III; Wiggins, C.C.; Joyner, M.J.; et al. AI-Enabled Advanced Development for Assessing Low Circulating Blood Volume for Emergency Medical Care: Comparison of Compensatory Reserve Machine-Learning Algorithms. Sensors 2022, 22, 2642. [Google Scholar] [CrossRef]
- Darrin, M.; Samudre, A.; Sahun, M.; Atwell, S.; Badens, C.; Charrier, A.; Helfer, E.; Viallat, A.; Cohen-Addad, V.; Giffard-Roisin, S. Classification of red cell dynamics with convolutional and recurrent neural networks: A sickle cell disease case study. Sci. Rep. 2023, 13, 745. [Google Scholar] [CrossRef]
- Das, D.K.; Chakraborty, C.; Mitra, B.; Maiti, A.K.; Ray, A.K. Quantitative microscopy approach for shape-based erythrocytes characterization in anaemia. J. Microsc. 2013, 249, 136–149. [Google Scholar] [CrossRef]
- Dengler, N.F.; Madai, V.I.; Unteroberdörster, M.; Zihni, E.; Brune, S.C.; Hilbert, A.; Livne, M.; Wolf, S.; Vajkoczy, P.; Frey, D. Outcome prediction in aneurysmal subarachnoid hemorrhage: A comparison of machine learning methods and established clinico-radiological scores. Neurosurg. Rev. 2021, 44, 2837–2846. [Google Scholar] [CrossRef]
- Deshmukh, F.; Merchant, S.S. Explainable Machine Learning Model for Predicting GI Bleed Mortality in the Intensive Care Unit. Am. J. Gastroenterol. 2020, 115, 1657–1668. [Google Scholar] [CrossRef] [PubMed]
- Dong, T.S.; Kalani, A.; Aby, E.S.; Le, L.; Luu, K.; Hauer, M.; Kamath, R.; Lindor, K.D.; Tabibian, J.H. Machine Learning-based Development and Validation of a Scoring System for Screening High-Risk Esophageal Varices. Clin. Gastroenterol. Hepatol. Off. Clin. Pract. J. Am. Gastroenterol. Assoc. 2019, 17, 1894–1901.e1. [Google Scholar] [CrossRef] [PubMed]
- Dumont, T.M.; Rughani, A.I.; Tranmer, B.I. Prediction of symptomatic cerebral vasospasm after aneurysmal subarachnoid hemorrhage with an artificial neural network: Feasibility and comparison with logistic regression models. World Neurosurg. 2011, 75, 57–63; discussion 25–28. [Google Scholar] [CrossRef] [PubMed]
- Erler, B.S.; Vitagliano, P.; Lee, S. Superiority of neural networks over discriminant functions for thalassemia minor screening of red blood cell microcytosis. Arch. Pathol. Lab. Med. 1995, 119, 350–354. [Google Scholar]
- Escandell-Montero, P.; Chermisi, M.; Martínez-Martínez, J.M.; Gómez-Sanchis, J.; Barbieri, C.; Soria-Olivas, E.; Mari, F.; Vila-Francés, J.; Stopper, A.; Gatti, E.; et al. Optimization of anemia treatment in hemodialysis patients via reinforcement learning. Artif. Intell. Med. 2014, 62, 47–60. [Google Scholar] [CrossRef]
- Farooq, M.S.; Muhammad, M.H.G.; Ali, O.; Zeeshan, Z.; Saleem, M.; Ahmad, M.; Abbas, S.; Khan, M.A.; Ghazal, T.M. Developing a Transparent Anaemia Prediction Model Empowered with Explainable Artificial Intelligence. IEEE Access 2025, 13, 1307–1318. [Google Scholar] [CrossRef]
- Ferret, L.; Luyckx, M.; Merlin, B.; Ficheur, G.; Chazard, E.; Beuscart, R. Evaluation of a computerized tool allowing retrospective detection of potential vitamin K antagonist overdoses in complex contexts. Stud. Health Technol. Inform. 2013, 192, 553–556. [Google Scholar]
- Formeister, E.J.; Baum, R.; Knott, P.D.; Seth, R.; Ha, P.; Ryan, W.; El-Sayed, I.; George, J.; Larson, A.; Plonowska, K.; et al. Machine Learning for Predicting Complications in Head and Neck Microvascular Free Tissue Transfer. Laryngoscope 2020, 130, E843–E849. [Google Scholar] [CrossRef]
- Ge, Y.; Rosendahl, P.; Duran, C.; Topfner, N.; Ciucci, S.; Guck, J.; Cannistraci, C.V. Cell Mechanics Based Computational Classification of Red Blood Cells Via Machine Intelligence Applied to Morpho-Rheological Markers. IEEE/ACM Trans. Comput. Biol. Bioinform. 2021, 18, 1405–1415. [Google Scholar] [CrossRef] [PubMed]
- Ghobadi, Z.; Esmaili, S.S. Use of a neuro-fuzzy technique to predict complete Rockall score in patients with upper gastrointestinal bleeding. Int. J. Med. Eng. Inform. 2024, 16, 503–515. [Google Scholar] [CrossRef]
- Gómez, J.G.; Urueta, C.P.; Álvarez, D.S.; Riaño, V.H.; Ramirez-Gonzalez, G. Anemia Classification System Using Machine Learning. Informatics 2025, 12, 19. [Google Scholar] [CrossRef]
- Graham, J.E. Transfusion e-learning for junior doctors: The educational role of ‘LearnBloodTransfusion’. Transfus. Med. Oxf. Engl. 2015, 25, 144–150. [Google Scholar] [CrossRef]
- Guo, P.; Zhang, C.; Liu, D.; Sun, Z.; He, J.; Wang, J. Evaluation of artificial intelligence-assisted morphological analysis for platelet count estimation. Int. J. Lab. Hematol. 2024, 46, 1012–1020. [Google Scholar] [CrossRef]
- Haghani, S.; Sedehi, M.; Kheiri, S. Artificial Neural Network to Modeling Zero-inflated Count Data: Application to Predicting Number of Return to Blood Donation. J. Res. Health Sci. 2017, 17, e00392. [Google Scholar]
- Hahn-Klimroth, M.; Loick, P.; Kim-Wanner, S.-Z.; Seifried, E.; Bonig, H. Generation and validation of a formula to calculate hemoglobin loss on a cohort of healthy adults subjected to controlled blood loss. J. Transl. Med. 2021, 19, 116. [Google Scholar] [CrossRef]
- Hurley, N.C.; Schroeder, K.M.; Hess, A.S. Would doctors dream of electric blood bankers? Large language model-based artificial intelligence performs well in many aspects of transfusion medicine. Transfusion 2023, 63, 1833–1840. [Google Scholar] [CrossRef] [PubMed]
- Imbert, M.; Priolet, G.; Dadi, W.; Sultan, C. An expert system applied to the diagnosis of anemia with special reference to myelodysplastic syndromes. Blood Cells 1989, 15, 563–570; discussion 570–571. [Google Scholar] [PubMed]
- Isiksacan, Z.; D’Alessandro, A.; Wolf, S.M.; McKenna, D.H.; Tessier, S.N.; Kucukal, E.; Gokaltun, A.A.; William, N.; Sandlin, R.D.; Bischof, J.; et al. Assessment of stored red blood cells through lab-on-a-chip technologies for precision transfusion medicine. Proc. Natl. Acad. Sci. USA 2023, 120, e2115616120. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Liu, L.; Wang, Y.; Ji, H.; Ma, X.; Wu, J.; Huang, Y.; Wang, X.; Gui, R.; Zhao, Q.; et al. Machine Learning for the Prediction of Complications in Patients After Mitral Valve Surgery. Front. Cardiovasc. Med. 2021, 8, 771246. [Google Scholar] [CrossRef]
- Jørgensen, C.C.; Michelsen, C.; Petersen, T.; Kehlet, H. Preoperative hemoglobin thresholds for increased risk of ‘medical’ complications in fast-track total hip and knee arthroplasty, a secondary analysis of a machine-learning algorithm. Transfusion 2024, 64, 438–442. [Google Scholar] [CrossRef]
- Kang, I.K.; Shin, T.Y.; Kim, J.H. Observation-informed modeling of artificial neural networks to predict flow and bleeding of cement-based materials. Constr. Build. Mater. 2023, 409, 133811. [Google Scholar] [CrossRef]
- Karkouti, K.; Cohen, M.M.; McCluskey, S.A.; Sher, G.D. A multivariable model for predicting the need for blood transfusion in patients undergoing first-time elective coronary bypass graft surgery. Transfusion 2001, 41, 1193–1203. [Google Scholar] [CrossRef]
- Kassaw, A.K.; Yimer, A.; Abey, W.; Molla, T.L.; Zemariam, A.B. The application of machine learning approaches to determine the predictors of anemia among under five children in Ethiopia. Sci. Rep. 2023, 13, 22919. [Google Scholar] [CrossRef]
- Kimura, K.; Tabe, Y.; Ai, T.; Takehara, I.; Fukuda, H.; Takahashi, H.; Naito, T.; Komatsu, N.; Uchihashi, K.; Ohsaka, A. A novel automated image analysis system using deep convolutional neural networks can assist to differentiate MDS and AA. Sci. Rep. 2019, 9, 13385. [Google Scholar] [CrossRef]
- Kneifati-Hayek, J.; Fleischman, W.; Bernstein, L.H.; Riccioli, A.; Bellevue, R. A model for automated screening of thalassemia in hematology (math study). Lab. Hematol. Off. Publ. Int. Soc. Lab. Hematol. 2007, 13, 119–123. [Google Scholar]
- Knowlton, S.M.; Sencan, I.; Aytar, Y.; Khoory, J.; Heeney, M.M.; Ghiran, I.C.; Tasoglu, S. Sickle cell detection using a smartphone. Sci. Rep. 2015, 5, 15022. [Google Scholar] [CrossRef]
- Kolin, D.A.; Lyman, S.; Della Valle, A.G.; Ast, M.P.; Landy, D.C.; Chalmers, B.P. Predicting Postoperative Anemia and Blood Transfusion Following Total Knee Arthroplasty. J. Arthroplast. 2023, 38, 1262–1266.e2. [Google Scholar] [CrossRef] [PubMed]
- Koons, N.J.; Owens, G.A.; Parsons, D.L.; Schauer, S.G.; Buller, J.L.; Convertino, V.A. Combat medic testing of a novel monitoring capability for early detection of hemorrhage. J. Trauma Acute Care Surg. 2020, 89 (Suppl. S2), S146–S152. [Google Scholar] [CrossRef] [PubMed]
- Kordzadeh, A.; Hanif, M.A.; Ramirez, M.J.; Railton, N.; Prionidis, I.; Browne, T. Prediction, pattern recognition and modelling of complications post-endovascular infra renal aneurysm repair by artificial intelligence. Vascular 2021, 29, 171–182. [Google Scholar] [CrossRef]
- Kunze, K.N.; Karhade, A.V.; Polce, E.M.; Schwab, J.H.; Levine, B.R. Development and internal validation of machine learning algorithms for predicting complications after primary total hip arthroplasty. Arch. Orthop. Trauma Surg. 2022, 143, 2181–2188. [Google Scholar] [CrossRef]
- Kurmanaliyev, A.; Sutiene, K.; Braukylienė, R.; Aldujeli, A.; Jurenas, M.; Kregzdyte, R.; Braukyla, L.; Zhumagaliyev, R.; Aitaliyev, S.; Zhanabayev, N.; et al. An Integrative Machine Learning Model for Predicting Early Safety Outcomes in Patients Undergoing Transcatheter Aortic Valve Implantation. Med. Kaunas Lith. 2025, 61, 374. [Google Scholar] [CrossRef]
- Lee, K.C.; Lin, T.C.; Chiang, H.F.; Horng, G.J.; Hsu, C.C.; Wu, N.C.; Su, H.C.; Chen, K.T. Predicting outcomes after trauma: Prognostic model development based on admission features through machine learning. Medicine 2021, 100, e27753. [Google Scholar] [CrossRef]
- Li, X.; Han, S.; Zhao, L.; Gong, C.; Liu, X. New Dandelion Algorithm Optimizes Extreme Learning Machine for Biomedical Classification Problems. Comput. Intell. Neurosci. 2017, 2017, 4523754. [Google Scholar] [CrossRef]
- Liu, Z.W.; Olweny, E.O.; Yin, G.; Faddegon, S.; Tan, Y.K.; Han, W.K.; Cadeddu, J.A. Prediction of perioperative outcomes following minimally invasive partial nephrectomy: Role of the R.E.N.A.L nephrometry score. World J. Urol. 2013, 31, 1183–1189. [Google Scholar] [CrossRef]
- Lo, B.W.Y.; Macdonald, R.L.; Baker, A.; Levine, M.A.H. Clinical outcome prediction in aneurysmal subarachnoid hemorrhage using Bayesian neural networks with fuzzy logic inferences. Comput. Math. Methods Med. 2013, 2013, 904860. [Google Scholar] [CrossRef]
- Luo, Z.; Qin, L.; Xu, S.; Yang, X.; Peng, Z.; Huang, C. Impact of fresh frozen plasma transfusion on mortality in extracorporeal membrane oxygenation. Perfusion 2024, 39, 294–303. [Google Scholar] [CrossRef]
- Das Mahapatra, P.P.; Roy, C.; Agarwal, K.; Sharma, S.D.; Chowdhury, S.R.; Sharma, S.; Rajagopal, H.; Meher, D. Non-invasive hemoglobin screening device: A promising digital method for reducing anemia prevalence through routine screening and timely intervention. Hematol. Amst. Neth. 2024, 29, 2365078. [Google Scholar] [CrossRef]
- Mascarella, M.A.; Muthukrishnan, N.; Maleki, F.; Kergoat, M.J.; Richardson, K.; Mlynarek, A.; Forest, V.I.; Reinhold, C.; Martin, D.R.; Hier, M.; et al. Above and Beyond Age: Prediction of Major Postoperative Adverse Events in Head and Neck Surgery. Ann. Otol. Rhinol. Laryngol. 2022, 131, 697–703. [Google Scholar] [CrossRef] [PubMed]
- Merath, K.; Hyer, J.M.; Mehta, R.; Farooq, A.; Bagante, F.; Sahara, K.; Tsilimigras, D.I.; Beal, E.; Paredes, A.Z.; Wu, L.; et al. Use of Machine Learning for Prediction of Patient Risk of Postoperative Complications After Liver, Pancreatic, and Colorectal Surgery. J. Gastrointest. Surg. Off. J. Soc. Surg. Aliment. Tract. 2020, 24, 1843–1851. [Google Scholar] [CrossRef] [PubMed]
- Merrill, R.K.; Ferrandino, R.M.; Hoffman, R.; Shaffer, G.W.; Ndu, A. Machine Learning Accurately Predicts Short-Term Outcomes Following Open Reduction and Internal Fixation of Ankle Fractures. J. Foot Ankle Surg. Off. Publ. Am. Coll. Foot Ankle Surg. 2019, 58, 410–416. [Google Scholar] [CrossRef] [PubMed]
- Meyer, C.H.; Nguyen, J.; ElHabr, A.; Venkatayogi, N.; Steed, T.; Gichoya, J.; Sciarretta, J.D.; Sikora, J.; Dente, C.; Lyons, J.; et al. TiME OUT: Time-specific machine-learning evaluation to optimize ultramassive transfusion. J. Trauma Acute Care Surg. 2024, 96, 443–454. [Google Scholar] [CrossRef]
- Mikić, M.; Milutinović, D.; Aranđelović, B.; Stojaković, N.; Obradović, M.; Plećaš-Đurić, A.; Rašović, P.; Vranješ, M. The advantages of using tranexamic acid in anterior cruciate ligament reconstruction: A randomized controlled trial. Cir. Cir. 2024, 92, 525–531. [Google Scholar] [CrossRef]
- Miyagi, Y.; Tada, K.; Yasuhi, I.; Maekawa, Y.; Okura, N.; Kawakami, K.; Yamaguchi, K.; Ogawa, M.; Kodama, T.; Nomiyama, M.; et al. New method for determining fibrinogen and FDP threshold criteria by artificial intelligence in cases of massive hemorrhage during delivery. J. Obstet. Gynaecol. Res. 2020, 46, 256–265. [Google Scholar] [CrossRef]
- Miyagi, Y.; Tada, K.; Yasuhi, I.; Tsumura, K.; Maegawa, Y.; Tanaka, N.; Mizunoe, T.; Emoto, I.; Maeda, K.; Kawakami, K.; et al. A Novel Method for Determining Fibrin/Fibrinogen Degradation Products and Fibrinogen Threshold Criteria via Artificial Intelligence in Massive Hemorrhage during Delivery with Hematuria. J. Clin. Med. 2024, 13, 1826. [Google Scholar] [CrossRef]
- Moreillon, B.; Krumm, B.; Saugy, J.J.; Saugy, M.; Botrè, F.; Vesin, J.M.; Faiss, R. Prediction of plasma volume and total hemoglobin mass with machine learning. Physiol. Rep. 2023, 11, e15834. [Google Scholar] [CrossRef]
- Moura, P.L.; Dobbe, J.G.G.; Streekstra, G.J.; Rab, M.A.E.; Veldthuis, M.; Fermo, E.; van Wijk, R.; van Zwieten, R.; Bianchi, P.; Toye, A.M.; et al. Rapid diagnosis of hereditary haemolytic anaemias using automated rheoscopy and supervised machine learning. Br. J. Haematol. 2020, 190, e250–e255. [Google Scholar] [CrossRef] [PubMed]
- Narula, I.S.; Diaper, D. Third World continuing medical education with hypertext: The Liverpool Anaemia Guide System. Ergonomics 1991, 34, 1147–1159. [Google Scholar] [CrossRef] [PubMed]
- Nazarian, S.; Lo, F.P.W.; Qiu, J.; Patel, N.; Lo, B.; Ayaru, L. Development and validation of machine learning models to predict the need for haemostatic therapy in acute upper gastrointestinal bleeding. Ther. Adv. Gastrointest. Endosc. 2024, 17, 26317745241246899. [Google Scholar] [CrossRef] [PubMed]
- Nemeth, C.; Amos-Binks, A.; Rule, G.; Laufersweiler, D.; Keeney, N.; Flint, I.; Pinevich, Y.; Herasevich, V. TCCC Decision Support with Machine Learning Prediction of Hemorrhage Risk, Shock Probability. Mil. Med. 2023, 188 (Suppl. S6), 659–665. [Google Scholar] [CrossRef]
- Nguyen, A.N.; Hartwell, E.A.; Milam, J.D. A rule-based expert system for laboratory diagnosis of hemoglobin disorders. Arch. Pathol. Lab. Med. 1996, 120, 817–827. [Google Scholar]
- O’Connor, M.L.; McKinney, T. The diagnosis of microcytic anemia by a rule-based expert system using VP-Expert. Arch. Pathol. Lab. Med. 1989, 113, 985–988. [Google Scholar]
- Ohara, T.; Ikeda, H.; Sugitani, Y.; Suito, H.; Huynh, V.Q.H.; Kinomura, M.; Haraguchi, S.; Sakurama, K. Artificial intelligence supported anemia control system (AISACS) to prevent anemia in maintenance hemodialysis patients. Int. J. Med. Sci. 2021, 18, 1831–1839. [Google Scholar] [CrossRef]
- Orlando, J.I.; Prokofyeva, E.; Del Fresno, M.; Blaschko, M.B. An ensemble deep learning based approach for red lesion detection in fundus images. Comput. Methods Programs Biomed. 2018, 153, 115–127. [Google Scholar] [CrossRef]
- Padrão, E.M.H.; Bustos, B.; Mahesh, A.; Fonseca, G.H.H.; Taniguchi, L.U. Phenotypes of sickle cell intensive care admissions: An unsupervised machine learning approach in a single-center retrospective cohort. Ann. Hematol. 2022, 101, 1951–1957. [Google Scholar] [CrossRef]
- Paterakis, G.S.; Terzoglou, G.; Vasilioy, E. The performance characteristics of an expert system for the ‘on-line’ assessment of thalassemia trait and iron deficiency—Micro Hema Screen. Blood Cells 1989, 15, 541–560; discussion 560–561. [Google Scholar]
- Pollei, T.R.; Hinni, M.L.; Moore, E.J.; Hayden, R.E.; Olsen, K.D.; Casler, J.D.; Walter, L.C. Analysis of postoperative bleeding and risk factors in transoral surgery of the oropharynx. JAMA Otolaryngol. Head Neck Surg. 2013, 139, 1212–1218. [Google Scholar] [CrossRef]
- Qasrawi, R.; Badrasawi, M.; Al-Halawa, D.A.; Polo, S.V.; Khader, R.A.; Al-Taweel, H.; Alwafa, R.A.; Zahdeh, R.; Hahn, A.; Schuchardt, J.P. Identification and prediction of association patterns between nutrient intake and anemia using machine learning techniques: Results from a cross-sectional study with university female students from Palestine. Eur. J. Nutr. 2024, 63, 1635–1649. [Google Scholar] [CrossRef] [PubMed]
- Ramzan, M.; Sheng, J.; Saeed, M.U.; Wang, B.; Duraihem, F.Z. Revolutionizing anemia detection: Integrative machine learning models and advanced attention mechanisms. Vis. Comput. Ind. Biomed. Art 2024, 7, 18. [Google Scholar] [CrossRef] [PubMed]
- Rangaraj, S.; Islam, M.; Vs, V.; Wijethilake, N.; Uppal, U.; See, A.A.Q.; Chan, J.; James, M.L.; King, N.K.K.; Ren, H. Identifying risk factors of intracerebral hemorrhage stability using explainable attention model. Med. Biol. Eng. Comput. 2022, 60, 337–348. [Google Scholar] [CrossRef] [PubMed]
- Rao, B.N.; Mohanty, S.; Sen, K.; Acharya, U.R.; Cheong, K.H.; Sabut, S. Deep Transfer Learning for Automatic Prediction of Hemorrhagic Stroke on CT Images. Comput. Math. Methods Med. 2022, 2022, 3560507. [Google Scholar] [CrossRef]
- Rashiq, S.; Shah, M.; Chow, A.K.; O’Connor, P.J.; Finegan, B.A. Predicting allogeneic blood transfusion use in total joint arthroplasty. Anesth. Analg. 2004, 99, 1239–1244. [Google Scholar] [CrossRef]
- Rashedi, N.; Sun, Y.; Vaze, V.; Shah, P.; Halter, R.; Elliott, J.T.; Paradis, N.A. Prediction of Occult Hemorrhage in the Lower Body Negative Pressure Model: Initial Validation of Machine Learning Approaches. Mil. Med. 2024, 189, e1629–e1636. [Google Scholar] [CrossRef]
- Roubinian, N.H.; Chowdhury, D.; Hendrickson, J.E.; Triulzi, D.J.; Gottschall, J.L.; Looney, M.R.; Matthay, M.A.; Kor, D.J.; Brambilla, D.; Kleinman, S.H.; et al. NT-proBNP levels in the identification and classification of pulmonary transfusion reactions. Transfusion 2020, 60, 2548–2556. [Google Scholar] [CrossRef]
- Rubbert, C.; Patil, K.R.; Beseoglu, K.; Mathys, C.; May, R.; Kaschner, M.G.; Sigl, B.; Teichert, N.A.; Boos, J.; Turowski, B.; et al. Prediction of outcome after aneurysmal subarachnoid haemorrhage using data from patient admission. Eur. Radiol. 2018, 28, 4949–4958. [Google Scholar] [CrossRef]
- Santipas, B.; Suvithayasiri, S.; Trathitephun, W.; Wilartratsami, S.; Luksanapruksa, P. Developmental and Validation of Machine Learning Model for Prediction Complication After Cervical Spine Metastases Surgery. Clin. Spine Surg. 2025, 38, E81–E88. [Google Scholar] [CrossRef]
- Shah, A.A.; Devana, S.K.; Lee, C.; Olson, T.E.; Upfill-Brown, A.; Sheppard, W.L.; Lord, E.L.; Shamie, A.N.; van der Schaar, M.; SooHoo, N.F.; et al. Development and External Validation of a Risk Calculator for Prediction of Major Complications and Readmission After Anterior Cervical Discectomy and Fusion. Spine 2023, 48, 460–467. [Google Scholar] [CrossRef] [PubMed]
- Sharma, G.; Shah, M.; Ahluwalia, P.; Dasgupta, P.; Challacombe, B.J.; Bhandari, M.; Ahlawat, R.; Rawal, S.; Buffi, N.M.; Sivaraman, A.; et al. Development and Validation of a Nomogram Predicting Intraoperative Adverse Events During Robot-assisted Partial Nephrectomy. Eur. Urol. Focus 2023, 9, 345–351. [Google Scholar] [CrossRef] [PubMed]
- Shi, B.; Wang, H.Y.; Liu, J.; Cai, Z.; Song, C.; Yin, D.; Wang, H.; Dong, Q.; Song, W.; Dou, K.F. Prognostic Value of Machine-Learning-Based PRAISE Score for Ischemic and Bleeding Events in Patients with Acute Coronary Syndrome Undergoing Percutaneous Coronary Intervention. J. Am. Heart Assoc. 2023, 12, e025812. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, P.; Lohse, H.; Bhatla, C.; McCartney, H.; Alzaki, A.; Sandhu, N.; Oli, P.K.; Chaudhary, S.; Amid, A.; Onell, R.; et al. Evaluation of low-cost techniques to detect sickle cell disease and β-thalassemia: An open-label, international, multicentre study. Lancet Reg. Health Southeast Asia 2025, 35, 100571. [Google Scholar] [CrossRef]
- Shung, D.; Tsay, C.; Laine, L.; Chang, D.; Li, F.; Thomas, P.; Partridge, C.; Simonov, M.; Hsiao, A.; Tay, J.K.; et al. Early identification of patients with acute gastrointestinal bleeding using natural language processing and decision rules. J. Gastroenterol. Hepatol. 2021, 36, 1590–1597. [Google Scholar] [CrossRef]
- Shung, D.L.; Chan, C.E.; You, K.; Nakamura, S.; Saarinen, T.; Zheng, N.S.; Simonov, M.; Li, D.K.; Tsay, C.; Kawamura, Y.; et al. Validation of an Electronic Health Record-Based Machine Learning Model Compared with Clinical Risk Scores for Gastrointestinal Bleeding. Gastroenterology 2024, 167, 1198–1212. [Google Scholar] [CrossRef]
- Schweta, N.; Pande, S.D. Prediction of Anemia using various Ensemble Learning and Boosting Techniques. EAI Endorsed Trans. Pervasive Health Technol. 2023, 9. [Google Scholar] [CrossRef]
- Sielaff, B.H.; Connelly, D.P.; Scott, E.P. ESPRE: A knowledge-based system to support platelet transfusion decisions. IEEE Trans. Biomed. Eng. 1989, 36, 541–546. [Google Scholar] [CrossRef]
- Sielaff, B.H.; Scott, E.P.; Connelly, D.P. Design and preliminary evaluation of an expert system for platelet request evaluation. Transfusion 1991, 31, 600–606. [Google Scholar] [CrossRef]
- Sofo, L.; Caprino, P.; Schena, C.A.; Sacchetti, F.; Potenza, A.E.; Ciociola, A. New perspectives in the prediction of postoperative complications for high-risk ulcerative colitis patients: Machine learning preliminary approach. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 12781–12787. [Google Scholar] [CrossRef]
- Sorace, J.M.; Carnahan, G.E.; Moore, G.W.; Berman, J.J. Automated review of blood donor screening test patterns at a regional blood center. Am. J. Clin. Pathol. 1992, 98, 334–344. [Google Scholar] [CrossRef] [PubMed]
- Stewart, C.L.; Nawn, C.D.; Mulligan, J.; Grudic, G.; Moulton, S.L.; Convertino, V.A. Compensatory Reserve for Early and Accurate Prediction of Hemodynamic Compromise: Case Studies for Clinical Utility in Acute Care and Physical Performance. J. Spec. Oper. Med. Peer Rev. J. SOF Med. Prof. 2016, 16, 6–13. [Google Scholar] [CrossRef]
- Sultan, C.; Imbert, M.; Priolet, G. Decision-making system (DMS) applied to hematology. Diagnosis of 180 cases of anemia secondary to a variety of hematologic disorders. Hematol. Pathol. 1988, 2, 221–228. [Google Scholar]
- Susič, D.; Tavčar, L.B.; Lučovnik, M.; Hrobat, H.; Gornik, L.; Gradišek, A. Wellbeing Forecasting in Postpartum Anemia Patients. Healthcare 2023, 11, 1694. [Google Scholar] [CrossRef]
- Tonetti, M.S.; Deng, K.; Christiansen, A.; Bogetti, K.; Nicora, C.; Thurnay, S.; Cortellini, P. Self-reported bleeding on brushing as a predictor of bleeding on probing: Early observations from the deployment of an internet of things network of intelligent power-driven toothbrushes in a supportive periodontal care population. J. Clin. Periodontol. 2020, 47, 1219–1226. [Google Scholar] [CrossRef]
- Tsai, S.C.; Lin, C.H.; Chu, C.J.; Lo, H.Y.; Ng, C.J.; Hsu, C.C.; Chen, S.Y. Machine Learning Models for Predicting Mortality in Patients with Cirrhosis and Acute Upper Gastrointestinal Bleeding at an Emergency Department: A Retrospective Cohort Study. Diagnostics 2024, 14, 1919. [Google Scholar] [CrossRef]
- Turgut, A.; Yalçin, Ö. Applications of deep learning to the assessment of red blood cell deformability. Biorheology 2021, 58, 51–60. [Google Scholar] [CrossRef]
- Turna, A.; Mercan, C.A.; Bedirhan, M.A. Prediction of Morbidity After Lung Resection in Patients with Lung Cancer Using Fuzzy Logic. Thorac. Cardiovasc. Surg. 2005, 53, 368–374. [Google Scholar] [CrossRef]
- Valafar, H.; Valafar, F.; Darvill, A.; Albersheim, P.; Kutlar, A.; Woods, K.F.; Hardin, J. Predicting the effectiveness of hydroxyurea in individual sickle cell anemia patients. Artif. Intell. Med. 2000, 18, 133–148. [Google Scholar] [CrossRef]
- Vicent, M.; Simon, K.; Yonasi, S. An algorithm to detect overlapping red blood cells for sickle cell disease diagnosis. IET Image Process. 2022, 16, 1669–1677. [Google Scholar] [CrossRef]
- Vittoraki, A.G.; Fylaktou, A.; Tarassi, K.; Tsinaris, Z.; Petasis, G.C.; Gerogiannis, D.; Kheav, V.D.; Carmagnat, M.; Lehmann, C.; Doxiadis, I.; et al. Patterns of 1748 Unique Human Alloimmune Responses Seen by Simple Machine Learning Algorithms. Front. Immunol. 2020, 11, 1667. [Google Scholar] [CrossRef] [PubMed]
- Waisberg, E.; Ong, J.; Zaman, N.; Kamran, S.A.; Lee, A.G.; Tavakkoli, A. A non-invasive approach to monitor anemia during long-duration spaceflight with retinal fundus images and deep learning. Life Sci. Space Res. 2022, 33, 69–71. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.-L.; Jin, G.-L.; Yuan, Z.-G. Artificial neural network predicts hemorrhagic contusions following decompressive craniotomy in traumatic brain injury. J. Neurosurg. Sci. 2021, 65, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, Z.; Chen, M.; Xiao, Y.; Chen, S.; Wu, L.; Yao, L.; Jiang, X.; Li, J.; Xu, M.; et al. An interpretable artificial intelligence system for detecting risk factors of gastroesophageal variceal bleeding. NPJ Digit. Med. 2022, 5, 183. [Google Scholar] [CrossRef]
- Welsby, I.; Crow, J.; Bandarenko, N.; Lappas, G.; Phillips-Bute, B.; Stafford-Smith, M. A clinical prediction tool to estimate the number of units of red blood cells needed in primary elective coronary artery bypass surgery. Transfusion 2010, 50, 2337–2343. [Google Scholar] [CrossRef]
- Wu, X.; Hu, J.; Zhang, J. Machine learning-based model for predicting major adverse cardiovascular and cerebrovascular events in patients aged 65 years and older undergoing noncardiac surgery. BMC Geriatr. 2023, 23, 819. [Google Scholar] [CrossRef]
- Xu, M.; Papageorgiou, D.P.; Abidi, S.Z.; Dao, M.; Zhao, H.; Karniadakis, G.E. A deep convolutional neural network for classification of red blood cells in sickle cell anemia. PLoS Comput. Biol. 2017, 13, e1005746. [Google Scholar] [CrossRef]
- Xu, S.; Guan, L.-J.; Shi, B.-Q.; Tan, Y.-S.; Zhang, X.-J. Recurrent Hemoptysis After Bronchial Artery Embolization: Prediction Using a Nomogram and Artificial Neural Network Model. AJR Am. J. Roentgenol. 2020, 215, 1490–1498. [Google Scholar] [CrossRef]
- Yamakuni, R.; Sekino, H.; Saito, M.; Kakamu, T.; Takahashi, K.; Hara, J.; Suenaga, H.; Ishii, S.; Fukushima, K.; Ito, H. Prediction of Anemia From Cerebral Venous Sinus Attenuation on Deep-Learning Reconstructed Brain Computed Tomography Images. J. Comput. Assist. Tomogr. 2023, 47, 796–805. [Google Scholar] [CrossRef]
- Yang, S.; Mackenzie, C.F.; Rock, P.; Lin, C.; Floccare, D.; Scalea, T.; Stumpf, F.; Winans, C.; Galvagno, S.; Miller, C.; et al. Comparison of massive and emergency transfusion prediction scoring systems after trauma with a new Bleeding Risk Index score applied in-flight. J. Trauma Acute Care Surg. 2021, 90, 268–273. [Google Scholar] [CrossRef]
- Yang, Y.; He, H.; Wang, J.; Chen, L.; Xu, Y.; Ge, C.; Li, S. Blood quality evaluation via on-chip classification of cell morphology using a deep learning algorithm. Lab. Chip 2023, 23, 2113–2121. [Google Scholar] [CrossRef]
- Zador, Z.; Huang, W.; Sperrin, M.; Lawton, M.T. Multivariable and Bayesian Network Analysis of Outcome Predictors in Acute Aneurysmal Subarachnoid Hemorrhage: Review of a Pure Surgical Series in the Post-International Subarachnoid Aneurysm Trial Era. Oper. Neurosurg. 2018, 14, 603–610. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Ge, J.; Gong, Z.; Chen, J.; Wang, C.; Sun, Y. Evaluation of machine learning-driven automated Kleihauer-Betke counting: A method comparison study. Int. J. Lab. Hematol. 2021, 43, 372–377. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, L.; Li, Y.; Zeng, Z. Machine learning model-based risk prediction of severe complications after off-pump coronary artery bypass grafting. Adv. Clin. Exp. Med. Off. Organ Wroclaw Med. Univ. 2023, 32, 185–194. [Google Scholar] [CrossRef]
- Zhang, F.; Zhan, J.; Wang, Y.; Cheng, J.; Wang, M.; Chen, P.; Ouyang, J.; Li, J. Enhancing thalassemia gene carrier identification in non-anemic populations using artificial intelligence erythrocyte morphology analysis and machine learning. Eur. J. Haematol. 2024, 112, 692–700. [Google Scholar] [CrossRef]
- Zhou, F.; Su, Z.; Chai, X.; Chen, L. Detection of foreign matter in transfusion solution based on Gaussian background modeling and an optimized BP neural network. Sensors 2014, 14, 19945–19962. [Google Scholar] [CrossRef]
- Zhu, F.; Pan, Z.; Tang, Y.; Fu, P.; Cheng, S.; Hou, W.; Zhang, Q.; Huang, H.; Sun, Y. Machine learning models predict coagulopathy in spontaneous intracerebral hemorrhage patients in ER. CNS Neurosci. Ther. 2021, 27, 92–100. [Google Scholar] [CrossRef]
- Zhu, D.Q.; Chen, Q.; Xiang, Y.L.; Zhan, C.Y.; Zhang, M.Y.; Chen, C.; Zhuge, Q.C.; Chen, W.J.; Yang, X.M.; Yang, Y.J. Predicting intraventricular hemorrhage growth with a machine learning-based, radiomics-clinical model. Aging 2021, 13, 12833–12848. [Google Scholar] [CrossRef]
- Zhu, G.; Ozkara, B.B.; Chen, H.; Zhou, B.; Jiang, B.; Ding, V.Y.; Wintermark, M. Enhancing hospital course and outcome prediction in patients with traumatic brain injury: A machine learning study. Neuroradiol. J. 2024, 37, 74–83. [Google Scholar] [CrossRef]
- Zürn, C.; Höhn, R.; Hübner, D.; Umhau, M.; Kroll, J.; Kari, F.A.; Humburger, F.; Maier, S.; Stiller, B. Risk Assessment of Red Cell Transfusion in Congenital Heart Disease. Thorac. Cardiovasc. Surg. 2022, 70, e15–e20. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).