Abstract
MicroRNAs (miRNAs) are key regulators of gene expression and play essential roles in physiological processes such as cell proliferation, apoptosis, and metabolism. They have emerged as promising biomarkers for the early diagnosis, prognosis, and treatment of Metabolic Syndrome (MetS). In recent years, exosome-derived miRNAs, known as “xeno-miRNAs”, which are derived from food, as well as circulating miRNAs, have emerged as areas of intense research due to their potential effects on metabolic disorders. miRNAs influence fasting blood glucose and insulin resistance through mechanisms such as β-cell differentiation, insulin gene transcription, and PI3K–AKT pathway activation. Additionally, miRNAs play important roles in regulating MetS components, as follows: obesity through adipogenesis and lipogenesis; hypertension through regulation of the renin–angiotensin system and vascular tone; and dyslipidemia by modulating lipid metabolism. Emerging evidence suggests that nutrients such as polyphenols and specific dietary patterns can alter miRNA expression, potentially impacting metabolic health. Understanding the interactions between diet and miRNA regulation offers novel insights into the prevention and treatment of MetS. This review explores the mechanisms by which miRNAs influence MetS components, and highlights the growing potential of nutrient-regulated miRNAs as therapeutic targets within the framework of precision nutrition and personalized metabolic disease management.
1. Introduction
Metabolic Syndrome (MetS) encompasses a cluster of interrelated metabolic disorders, including central obesity, hypertension, dyslipidemia, impaired glucose regulation, and type 2 diabetes mellitus (T2DM) [1]. The development of MetS is shaped by a combination of genetic predisposition, environmental influences, and lifestyle choices. Among these, diet significantly affects metabolic health by regulating gene expression through epigenetic modifications [2,3]. One notable group of epigenetic regulators, microRNAs (miRNAs), has attracted growing interest due to their regulatory influence on metabolic pathways linked to MetS [4]. These small, non-coding RNAs are increasingly recognized as valuable non-invasive biomarkers, with potential clinical applications in both the early detection and treatment of MetS [5].
miRNAs govern essential cellular functions such as proliferation, apoptosis, and cell differentiation by modulating gene expression at the post-transcriptional stage [6]. In MetS, distinct miRNA expression patterns significantly influence critical biochemical pathways, especially those related to insulin resistance, inflammation, blood pressure regulation, and lipid processing [7]. MiRNAs are suggested to modulate MetS through interconnected pathways that influence key metabolic processes, with the following proposed as major mechanisms in its pathophysiology [8]:
- i.
- Glucose homeostasis regulation—miRNAs influence neural signaling between the brain, muscle, liver, pancreas, and gastrointestinal tract. miR-132 regulates cAMP Response Element-Binding Protein (CREB) in the brain, involved in glucose homeostasis [9]. miR-9 and miR-124a also impact pancreatic beta-cell function, affecting insulin secretion by targeting transcription factors such as One Cut Homeobox 2 and Forkhead Box A2 [10]. miRNAs regulate insulin production, secretion, and sensitivity [11,12]. miR-375 influences beta-cell function and insulin exocytosis, while miR-124a and miR let-7b affect glucose regulation by repressing myotrophin [11]. Elevated glucose levels upregulate miR-30d, thereby enhancing insulin gene expression [12];
- ii.
- Adipogenesis and lipid metabolism—miRNAs regulate adipogenesis and lipid metabolism. miR-143 promotes adipocyte differentiation [13,14], while miR-103 enhances key markers like Glucose Transporter Type 4 (Glut4), Fatty Acid-Binding Protein 4 (Fabp4), and Peroxisome Proliferator-Activated Receptor Gamma 2 (Pparγ2) [15]. The miR-17-92 cluster promotes adipocyte differentiation by inhibiting Retinoblastoma 2/p130 [16]. In obesity, chronic inflammation alters miRNA expression, leading to a loss of miRNAs maintaining metabolically active adipocytes, contributing to obesity and metabolic disorders [8,17]. miR-30a-5p and miR-195 regulate hypothalamic brain-derived neurotrophic factor secretion, which influences appetite and energy balance.
- iii.
- Blood pressure mechanism—miRNAs play a central role in the development and regulation of hypertension. MiR-122 regulates hypertension by modulating the renin–angiotensin system (RAS), endothelial dysfunction, vascular fibrosis, autophagy, pulmonary vascular remodeling, and fibrogenesis through pathways like Nuclear Factor Kappa B (NF-κB), Transforming Growth Factor Beta (TGF-β), β-catenin, Phosphoinositide 3-Kinase/Protein Kinase B (PI3K-Akt), and cationic amino acid transporter [18,19,20,21,22]. MiR-204 inhibits the Src family kinases (SRC)/Signal Transducer and Activator of Transcription 3 (Stat3) module, regulating pulmonary hypertension [19]. The miR-143/145 cluster is a major player in the pathophysiology of hypoxia-induced pulmonary hypertension, which is prevented by miR-143 [20]. MiR-181 targets genes regulating blood pressure, contributing to hypertension, and miR-663 reduces blood pressure by controlling renin gene expression [21]. MiR-155 modulates the TGFβ signaling pathway, influencing cardiac hypertrophy and systemic hypertension [22].
Circulating miRNAs and dietary exosomal miRNAs (xeno-miRNAs) are increasingly acknowledged for their significant influence on metabolic pathways, as they mediate intercellular communication and help maintain metabolic balance [23,24]. Increasing evidence highlights the intricate interaction between circulating miRNAs, xeno-miRNAs, and MetS components, revealing their critical role in disease pathophysiology [5,7,23,25,26,27].
Circulating miRNAs, which are naturally produced and present in biological fluids like blood, saliva, and urine, are considered promising biomarkers for MetS owing to their tissue-specific expression patterns and correlation with important clinical and pathological indicators [27,28]. They regulate essential metabolic processes, including glucose metabolism, lipid homeostasis, and adipogenesis, making them valuable indicators for diagnosing and monitoring MetS progression [25]. The dysregulation of various microRNAs—including miR-33a/b, miR-21, miR-34a, miR-378, miR-107, miR-103, miR-146a, and miR-122, which are involved in regulating glucose metabolism, inflammatory signaling, and adipocyte differentiation—has been associated with the onset and progression of MetS [5,29,30]. Dysregulation of these miRNAs exacerbates hallmark features of MetS, including dyslipidaemia, insulin resistance, and impaired glucose metabolism, ultimately increasing the risk of T2DM and cardiovascular disease [27,31,32].
While endogenous miRNAs play essential roles in various physiological processes, recent studies have highlighted the existence of exogenous dietary miRNAs, known as xeno-miRNAs [33]. Xeno-miRNAs enter circulation via intestinal epithelial absorption mechanisms such as receptor-mediated endocytosis, transmembrane transporters, and phagocytosis [34,35]. Once absorbed, these miRNAs can modulate endogenous gene expression by targeting metabolic pathways related to insulin sensitivity, lipid metabolism, and inflammation [36]. Plant-derived miRNAs, including miR-156, miR-168a, and miR-172a, have been demonstrated to affect protein expression and metabolic processes, potentially contributing to metabolic health [37,38], while animal-derived miRNAs including elevated levels of miR-99a, miR-1, miR-206, and miR-133a-3p have been linked to the formation of lipid droplets in the livers of mice, along with disruptions in glucose and insulin metabolism, underscoring their important role in the progression of metabolic diseases [39].
The growing understanding of miRNA-mediated regulation highlights their potential as both biomarkers and therapeutic targets in MetS management [5,33]. Consequently, exosomal miRNAs emerge as promising candidates for diagnostic and therapeutic strategies aimed at mitigating metabolic disorders [24,40,41]. This study investigates the involvement of food-derived xeno-miRNAs and circulating miRNAs in the cluster of metabolic factors associated with MetS. We aim to investigate how dietary and endogenous miRNAs influence MetS pathophysiology, highlighting their biomarker potential and regulatory effects on metabolic processes.
2. Food Xeno-miRNAs and Circulating miRNAs in MetS
miRNAs are crucial regulators of gene expression, influencing metabolic disorders such as MetS through their role in key biological processes [42]. Recent research highlights the importance of both food-derived xeno-miRNAs and circulating miRNAs in modulating metabolic pathways, offering potential for novel diagnostic and therapeutic strategies [7,23,26].
2.1. Biogenesis of miRNAs
The biogenesis of miRNAs involves sequential nuclear and cytoplasmic processing [42]. In the nucleus, RNA polymerase II transcribes primary miRNAs (pri-miRNAs), which are subsequently processed into precursor miRNAs (pre-miRNAs) by the Drosha-DGCR8 complex [43,44,45,46]. Exportin 5 transports the pre-miRNAs to the cytoplasm, where the Dicer enzyme processes them into mature miRNAs. These mature miRNAs subsequently bind to the RNA-induced silencing complex (RISC), enabling the regulation of gene expression by promoting mRNA degradation or blocking translation [43,44]. While miRNAs primarily function in post-transcriptional gene regulation, they are also found in multiple cellular compartments, including the nucleus, mitochondria, and nucleolus [47].
Circulating miRNAs are secreted into biological fluids through exosomes, microvesicles, apoptotic bodies, RNA-binding proteins, and lipoproteins, facilitating intercellular communication and systemic metabolic regulation [48,49,50]. Due to their stability and resistance to enzymatic breakdown, these molecules represent promising biomarkers for metabolic disorders [7].
2.2. Food-Derived Xeno-miRNAs
Studies suggest that dietary miRNA levels in humans fluctuate in response to food intake [51,52,53,54]. Bioinformatic analyses suggest that animal-derived dietary miRNAs could participate in cancer-related pathways, whereas plant miRNAs demonstrate regulatory effects on metabolism [51]. Certain foods, such as poultry (miR-206), pork (miR-1), rapeseed (miR-156), wheat (miR-156a), barley (miR-168-5p), and corn (miR-319b), are particularly rich in miRNAs, whereas milk and cheese contain lower miRNA levels [55,56]. Moreover, specific miRNAs are found in other foods, such as gga-miR-92-3p, gga-miR-2188, and gga-miR-30c-5p in eggs [57], bta-miR-154c in beef [58], and miR-92, miR-125a, miR-223, miR-let-7c, miR-181, miR-21, miR-155-5p/miR-29b-5p, miR-17, and miR-148a-3p in milk [59,60,61,62,63]. Human milk contains miR-27b, miR-125b, miR-181a and b, and miR-155 [64,65]. Green and yellow vegetables, such as peas, green beans, broccoli, corn, and carrots, are rich in miR-156a [66], while bra-miR156g-5p (56), miR-168, and hvu-miR-168-3p are found in rice [67,68]. Ginger contains miR-168, and miR-167a, miR-1078, miR-34a, miR-143, and miR-145 [69,70,71]. Apple contains mdm-miR-7121d-h [72] and soybean contains miR-159a [73].
2.3. Stability and Absorption of Food Derived Xeno-miRNAs
The stability and absorption of dietary miRNAs remain areas of ongoing investigation, with numerous studies highlighting their unique biochemical properties and potential for gene regulation in host organisms [74]. Although dietary miRNAs have been detected in biological samples, research suggests that miRNAs can withstand processing and digestion [75].
A key factor supporting miRNA stability is their encapsulation within extracellular vesicles, such as exosomes, which protect them from enzymatic degradation and facilitate their transfer across biological barriers like the blood–brain barrier and placenta [76,77,78]. Breast milk extracellular vesicles, for instance, have been demonstrated to deliver miRNAs that play a role in immune development, suggesting that dietary miRNAs could exert functional effects when effectively delivered [79,80].
Recent evidence suggests that plant-derived miRNAs can resist degradation during gastrointestinal digestion. Their stability is enhanced by the food matrix and structural modifications such as 2′-O-methylation [81]. For instance, miRNAs in honey such as miR-30c-5p have demonstrated significant resilience under in vitro digestion conditions [82]. Furthermore, dietary miRNAs are taken up by intestinal epithelial cells via caveolin- and clathrin-mediated endocytosis, facilitated by specific membrane receptors [81]. However, circulating miRNAs are reported to remain stable under various storage conditions and are not significantly influenced by food intake [83]. These observations support the hypothesis that dietary miRNAs follow a metabolic absorption curve, although their physiological role post-absorption remains to be fully elucidated [84].
Some bioactive dietary compounds, such as resveratrol, have been found to influence miRNA expression, reducing miR-196b/miR-1290 in leukemia cells, suggesting potential antitumor properties [85,86]. However, the biological function of these absorbed miRNAs in mammalian systems requires further validation. Notably, distinctions have been proposed between endogenous miRNAs and absorbed dietary miRNAs, emphasizing the need to understand exosome secretion, transport, and target interactions [87].
2.4. Metabolic Effects of Food-Derived Xeno-miRNAs
The food-derived xeno-miRNAs have been considered to influence metabolic pathways, thereby playing a role in the modulation of various physiological processes. For instance, rice-derived miR-168 has been reported to decrease plasma low-density lipoprotein (LDL) clearance, potentially contributing to dyslipidaemia and increased cardiovascular risk [88,89]. Similarly, hvu-miR-168-3p, derived from rice aleurone, enhances glucose uptake by upregulating glucose transporter 1 (GLUT1), thereby lowering blood glucose levels [68]. Additionally, miR-22-3p inhibits the Wnt signaling pathway, preventing glucose intolerance and T2DM onset, while milk-derived miRNAs, like miR-148a and miR-30b, play a role in controlling lipid metabolism and obesity [90,91].
Rats fed freely for three months exhibited notable changes in miRNA levels depending on their diet. Specifically, miR-27, miR-122, and miR-451 were significantly reduced, while miR-200b, miR-429, and miR-200a were elevated in animals fed with a high-fat diet, a standard diet supplemented with high fructose, or a combination of high-fat and high-fructose diets. Furthermore, a marked decrease in miR-21 expression was observed exclusively in the groups consuming fructose-enriched diets [52].
The effects of plant- and animal-derived xeno-miRNAs on human gene expression and metabolism have been widely reported in the literature. For example, osa-miR168a, originating from rice, targets the human LDLRAP1 gene and has been shown to reduce LDL clearance in the liver, potentially influencing lipid metabolism [88]. Similarly, ath-miR156a, found in vegetables like spinach, lettuce, and cabbage, targets JAM-A in humans, and has been reported to reduce monocyte adhesion triggered by inflammatory stimuli [66]. From animal sources, bta-miR-148a, a bovine-derived miRNA, targets DNMT1 in humans, affecting epigenetic regulation and maintaining Th17/Treg cell balance [92]. Another notable example is miR2911 from honeysuckle, which possesses 28 binding sites in the SARS-CoV-2 genome and has been implicated in inhibiting viral replication [93]. Multiple plant miRNAs, including miR168 and miR156 from strawberry, bol-miR874 from cabbage, and osa-miR168 from rice, have been found to target the TLR3 gene in humans, thereby suppressing inflammatory responses [94]. Furthermore, gma-miR159 from soybean inhibits TCF7, which may suppress breast cancer cell proliferation [95], while gma-miR4995, also from soybean, targets MALAT1 and NEAT1, two long non-coding RNAs involved in tumor progression, thereby exerting anti-tumor effects [96].
2.5. Circulating miRNAs and Their Role in MetS
Circulating miRNAs exhibit cell- and tissue-specific expression profiles and play a crucial role in metabolic homeostasis [97]. These miRNAs mediate important events in the pathogenesis of MetS. They are actively released by viable cells or released into the bloodstream as a result of cell death [40].
Alterations in circulating miRNA profiles have been associated with factors that increase the risk of MetS, including insulin resistance, chronic inflammation, and dyslipidaemia [5,31,98]. For example, miR-122 plays a crucial role in hepatic lipid metabolism by regulating lipid synthesis and uptake [31]. miR-107 and miR-103 play a role in glucose homeostasis and insulin signaling [31], while miR-146a modulates NF-κB signaling, influencing inflammatory mechanisms and immune cell activation [99]. miR-146a also modulates glucose-stimulated insulin secretion and cell differentiation [99]. Various miRNAs, like miR-378/378, miR-27a/b, miR-34a, and miR-21, contribute to various aspects of lipid metabolism. Fatty liver disease and dyslipidemia have been linked to the dysregulation of these miRNAs [100].
Furthermore, miR-375, miR-16, miR-27a, miR-146a, miR-150, miR-320a, miR-155, miR-15a, and miR-30a-5p have been implicated in T2DM pathogenesis [5,101,102,103,104]. Various miRNAs, such as miR-7, miR-375, and miR-124a, are involved in the development of β-cell and insulin secretion [105,106]. miRNAs have also been shown to affect insulin resistance. For example, miR-320, miR-29, and miR-384-5p modulate components of the insulin signaling pathway, such as PI3K, Insulin Receptor Substrate (IRS) proteins, and the Insulin-like Growth Factor 1 Receptor (IGF-1R) [106]. A comprehensive analysis of miRNA expression in human β-cells and pancreatic islets revealed 384 unique miRNAs, of which 40 were predominantly expressed in islets [107]. Similarly, miR-199a-3p, miR-34a, miR-652-3p, miR-139, miR-223-3p, miR-30d, miR-16-5p, miR-18a-5p, miR-122, and miR-106a-5p have been linked to cardiovascular complications associated with MetS [108,109,110,111]. Elevated levels of miR-34a have been observed in patients with coronary artery disease, exhibiting a negative correlation with silent information regulator 1 (SIRT1) protein expression, thereby promoting endothelial senescence [108]. Numerous miRNAs have demonstrated inverse correlations with biomarkers associated with adverse clinical outcomes. In patients exhibiting the progression of heart failure within 48 h of hospitalization, several biomarkers were identified in association with specific miRNAs, miR-652-3p (soluble ST-2), miR-199a-3p (procalcitonin and galectin-3), miR-223-3p (growth differentiation factor 15), miR-18a-5p (procalcitonin), miR-106a-5p (creatinine), and miR-16-5p (C-reactive protein), implying their potential roles in the molecular pathways underlying cardiac dysfunction [109]. An elevated miR-122-5p/133b ratio has been significantly associated with poorer event-free survival in patients with ST-elevation myocardial infarction (STEMI), corresponding to an approximately ninefold increased risk of mortality or recurrent myocardial infarction and a fourfold greater likelihood of adverse cardiovascular outcomes, making it a potential prognostic biomarker for identifying high-risk patients after primary percutaneous coronary intervention [110]. Inhibiting miR-139-5p restores endothelial function by enhancing angiogenesis and blood perfusion. Its involvement in diabetic vascular complications is attributed to the inhibition of the c-Jun–vascular endothelial growth factor/platelet-derived growth factor-B signaling axis, leading to reduced endothelial cell viability and highlighting its potential as a therapeutic target [111]. The relationship between food-derived xeno-miRNAs, circulating miRNAs, and MetS factors is illustrated in Figure 1.
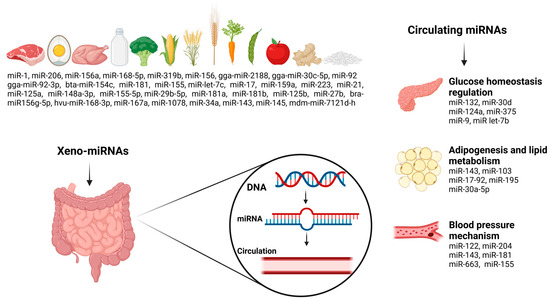
Figure 1.
Food xeno-miRNAs and circulating miRNAs in relation to metabolic factors in MetS.
Clinical studies further support the association between circulating miRNAs and metabolic diseases, with myocardial infarction, heart failure, and atherosclerosis being linked to altered circulating miRNA levels [112,113]. Research indicates that cardiac injuries, including myocardial infarction, result in increased circulating levels of heart-specific miRNAs, such as miR-499, miR-208, miR-1, and miR-133 [113,114]. In contrast, diseases linked to MetS, including coronary artery disease and T2DM, frequently exhibit reduced levels of endothelial-derived miRNAs like miR-126 [113]. Table 1 summarizes key miRNAs affecting components of MetS.

Table 1.
Key miRNAs affecting components of MetS.
3. miRNA in the MetS Hallmarks
3.1. Impaired Fasting Glucose
A major factor contributing to impaired fasting glucose in patients with MetS is insulin resistance. A breakdown in insulin signaling under metabolic stress can lead to persistent systemic hyperinsulinemia and insulin resistance, which is characterized as a reduction in the impact of insulin on important target tissues (including adipose tissue, liver, and skeletal muscle). The fundamental reasons behind insulin resistance linked to obesity are complex [115]. Obesity contributes to insulin resistance through multiple interrelated mechanisms. Increased free fatty acid levels can alter insulin receptor substrate-1 (IRS-1), which can disrupt insulin signaling. Adipokine imbalances—such as decreased adiponectin and increased leptin—further reduce insulin sensitivity. Ectopic fat accumulation in the liver, muscle, and heart disrupts glucose metabolism, while chronic inflammation driven by immune cell infiltration in visceral adipose tissue exacerbates insulin resistance. Endoplasmic reticulum stress, mitochondrial dysfunction, oxidative stress, and reduced hepatic insulin clearance are further contributing factors. Moreover, alterations in gut microbiota may lead to increased systemic inflammation via bacterial lipopolysaccharide translocation. Gene expression associated with metabolic dysfunction may also be regulated by epigenetic modifications, such as DNA methylation and histone modifications [115,116,117].
Recent genomics research has demonstrated that miRNAs control gene expression and have a role in the development of insulin resistance and diabetes in MetS [106,118]. miRNAs participate in controlling the synthesis, secretion, and signaling pathways of insulin-related proteins. There is evidence that miRNAs may be important regulators and biomarkers in the early phases of the development and management of insulin resistance [119]. It has been discovered that prediabetic or type 2 diabetic patients have decreased levels of circulating miR-126, miR-15a, miR-223 and miR-29b, which may be useful in predicting the onset of diabetes [120]. Another study discovered that miRNAs (miR-let-7c, miR-30a, miR-122, miR-139 and miR-126) were shown to have elevated plasma levels in patients with MetS and insulin resistance and/or elevated fasting plasma glucose [25]. Also, it was stated that the miRNAs potentially linked to insulin resistance in MetS include miR-375, miR-320b, miR-148a-3p, miR-197-3p, miR-122-5p, miR-19b-3p, miR-342-3p, miR-103, miR-155-5p, and miR-130a [121], and insulin resistance can be managed by using these miRNAs as biomarkers. The majority of miRNAs target several genes, which supports both their usage as biomarkers and their potential use as therapeutics [122]. Findings indicate that adding miRNAs to the ROC curve in addition to traditional indicators like HbA1c and fasting blood glucose can improve diagnosis, help track treatment progress, and be used to evaluate treatment efficacy [123]. In a related study that established a multi-parameter diagnostic model for the early identification of type 2 diabetes using miR-130a, miR-148b, miR-19a and miR-223, the validation set’s AUC was 0.88, and the sensitivity and specificity were 78.36% and 87.63%, respectively [124].
miRNAs may contribute to insulin resistance in MetS by influencing insulin production, mature β-cell function, and β-cell differentiation. The overexpression of islet-specific miR-375 inhibits the insulin gene transcription and glucose-stimulated insulin production [117]. miR-375 modifies secretion via a mechanism that is directly related to insulin exocytosis and unrelated to changes in intracellular Ca2+ signaling or glucose metabolism. It was confirmed that miR-375 targets myotrophin (Mtpn) [125]. Due to its potential role in β-cell function regulation, miR-30d may be associated with insulin resistance. A study investigating the circulating levels of specific miRNAs in Indian type 2 diabetic patients found that miR-30d was significantly higher in patients compared to healthy controls [126]. In β cells, glucose-induced insulin production has been shown to be negatively modulated by another miRNA, miR-7. It has been demonstrated that miR-7a2 affects insulin granule fusion with the plasma membrane by targeting the genes involved in insulin synthesis and release [122]. The Mir-200 miRNA family—miR-200c, miR-200a and miR-200b—and miR-141 and miR-429 have been linked to both β-cell dedifferentiation and protection against β-cell apoptosis. The ablation of all miRNAs in this family led to increased protection against β-cell apoptosis, but the deletion of miR-200c, one of the most abundant miRNAs, provides some protection against β-cell apoptosis following oxidative stress [127]. A considerable number of genes are controlled, either directly or indirectly, by miR-34a, which also suppresses the production of the anti-apoptotic protein Bcl-II, resulting in decreased insulin secretion and cell apoptosis [128]. miR-29a is highly expressed in the β cells that secrete insulin. Although miR-29a is necessary for regular exocytosis in pancreatic β cells, elevated levels are linked to impaired β-cell activity [129]. In addition, miR-184, miR-124a2, miR-185-5p, miR-24 and miR-204 have also been connected to the management of insulin production and β-cell proliferation during insulin resistance [117]. Insulin secretion is remarkably influenced by miR-184. It has been found that this miRNA directly targets the mitochondrial glutamate carrier Slc25a22. Slc25a22 suppression reduces the insulin level secreted by pancreatic β cells [122]. The upregulation of miR-204 suppresses the synthesis of insulin by adversely affecting MafA, a transcription factor necessary for insulin transcription [127].
The presence of chronic low-grade inflammation and insulin resistance in MetS is influenced by a variety of microRNAs that facilitate the polarization of macrophages from an anti-inflammatory M2 phenotype to a proinflammatory M1 phenotype [130]. miR-30 suppression increases pro-inflammatory cytokine production and M1 macrophage polarization. The upregulation of miR-1249-3p also reduced pro-inflammatory cytokine molecule expressions, such as IL-6, IL-1β and TNF-α [117]. Insulin resistance and systemic inflammatory incidence are positively correlated with elevated levels of miR-34a and miR-122 and reduced levels of miR-26a and miR-126-3p expression [131,132]. A decreased expression of miR-1934 raises the levels of TNF-α, IL-1β and IL-6. Similarly, the reduced expression of miR-146-a stimulates the inflammatory genes’ expressions after an increase in NF-kB [131]. In reaction to obesity, adipose tissue macrophages release more miR-155, which targets PPARγ and inhibits insulin signaling in muscle cells, hepatocytes, and adipocytes. These findings point to a mechanism by which insulin resistance and inflammation development are impacted by obesity-mediated elevations in miR-155 [133]. Similarly, by blocking insulin signaling, extracellular miR-27a elevation in response to obesity causes insulin resistance. Obesity-induced extracellular miR-27a inhibits the expression of the transcription factor peroxisome proliferator-activated receptor γ (PPARγ), which inhibits the production of inflammatory cytokines [133,134].
Following its synthesis and release from the pancreas, insulin attaches itself to the insulin receptor (INSR) and causes the target tissues to uptake glucose. Tyrosine residues of the insulin receptor substrate IRS-1 or -2 become phosphorylated when insulin binds to its receptor and starts a downstream signaling cascade. Phosphoinositide-dependent kinase-1 (Akt-1 and -2), ribosomal protein S6 kinase beta 1 (S6K1), phosphatidylinositol 3-kinase (PI3K), protein kinase C (PKC), and mammalian target of rapamycin (mTOR) are the next in line. Following this, there will be a greater translocation of glucose to the membrane via glucose transporter 4 (GLUT4) [128]. By controlling the INSR and IRS, several miRNAs have been shown to contribute to insulin resistance. IRS-1 is targeted by miR-96 and miR-126, miR-144 and INSR, and IRS-2 by miR-33, miR-7, miR-27 and miR-13a [128,135]. miR-135 also inhibits the insulin-induced phosphorylation of PI3K/AKT and adversely regulates IRS-2 expression [135]. miR-378 inhibits the production of p110α, the catalytic subunit of phosphoinositide 3-kinase (PI3K). Insulin resistance is caused by the persistent overexpression of miR-378 because PI3K is a crucial transcriptional regulator of insulin [136]. In addition, insulin resistance results from the inhibition of regulatory component p85α by miR-320, which degrades insulin-PI3K signaling [128]. Through its targeting of the oxysterol-binding protein-related protein (ORP)-8, obesity-induced overexpression of miR-143 also contributes to insulin resistance. This causes the suppression of insulin-induced PI3K–AKT pathway activation [136]. The exaggerated expression of miR-29 and miR-153 can also cause insulin resistance by disrupting the AKT phosphorylation pathway [128].
The skeletal muscle, adipose tissue and liver are among the target tissues where miRNA dysregulation also results in insulin resistance. miR-122 controls both the insulin signaling pathway and glucose metabolism in hepatic insulin resistance. By dephosphorylating tyrosine residues in IRS and INSR, a decrease in miR-122 inhibits the action of hepatic insulin [137]. Nguyen et al. demonstrated the crucial role that saturated fatty acid inducible miR-183-5p plays in the pathophysiology of hepatic insulin resistance and the control of the insulin signaling pathway [138]. The overexpression of miR-27a/b inhibits hepatic glucose production and increases hyperglycaemia via targeting fork head box protein O1 (FOXO1) [139]. The overexpression of miR-144, miR-33, miR-148a and miR-128-1, and the inhibition of miR-30c, miR-224, and miR-520d, are associated with an increase in low-density and reduction in high-density lipoprotein (HDL) production and LDL clearance in hepatocytes [117]. Due to the inhibition of insulin sensitivity and glucose uptake in hepatocytes, an obesity-induced reduction in miR-141-3p can result in insulin resistance [133]. In addition, alterations in miR-125a-5p, miR-21, miR-802, miR-592, miR-29 and miR-543 induce hepatic insulin resistance through various mechanisms [137]. In adipose tissue insulin resistance, it was found that increased levels of a miR-29 family (a, b, and c) negatively affect glucose uptake via lowering GLUT4 expression. Caveolin-1 is the target of miR-107 and miR-103, and it has been discovered that two miRNAs play a critical role in controlling insulin sensitivity. Insulin sensitivity and signaling were improved, fat cell size was reduced, and caveolin-1 expression increased when these two miRNAs were inhibited. Decreased miR-330-5p expression is connected to the emergence of insulin resistance, as this miRNA has an effect on T cell immunoglobulin expression [137]. By inhibiting KLF-15, miR-133a-1, miR-133a-2, and miR-133b reduce the uptake of glucose mediated by insulin in skeletal muscle and adipocytes. Consequently, this implies that miR-133 regulates the pathophysiology of insulin resistance [128]. Decreased miR-182 expression affects enzyme activation and phosphorylation, while miR-16 overexpression plays a role in reducing insulin sensitivity by reducing protein synthesis in muscles. Lastly, it was found that Rock-1 expression levels are inhibited by the overexpression of miR-135, miR-214, and miR-202, which leads to insulin signaling and insulin resistance [137].
The gut microbiota–miR crosstalk is also important in maintaining the insulin signaling pathway and glucose homeostasis. It was demonstrated that the expression of miR-122-5p was adversely connected with specific gut bacteria, such as Phascolarctobacterium faecium and Bacteroides uniformis. Since miR-122-5p is essential for controlling glucose metabolism, these results imply that alterations in the gut microbiome may influence glucose homeostasis and insulin resistance through modulation of this microRNA [140]. Moreover, oral miR-10a-5p therapy lowers insulin resistance brought on by a high-fat diet via modifying the circadian cycle of intestinal Lachnospiraceae and its metabolite butyric acid [141]. The effects of microRNAs on insulin resistance in MetS are given Figure 2.
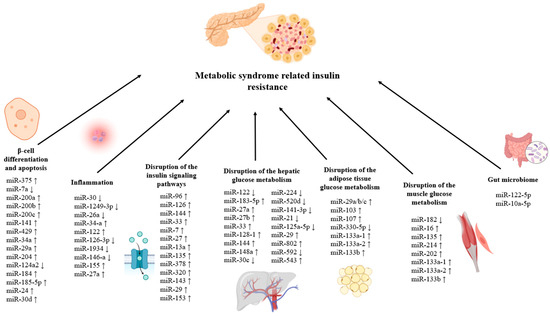
Figure 2.
Effects of microRNAs on insulin resistance in MetS. (↓) shows that the expression of the relevant miRNA decreases, (↑) shows that the expression of the relevant miRNA increases.
In conclusion, circulating miRNAs play a crucial role in the regulation of insulin signaling and glucose metabolism, making them key contributors to the development and progression of insulin resistance. They can change gene expression in tissues that are susceptible to insulin, including muscle, adipose tissue, and the liver. However, current research is still lacking in several key aspects, despite increased interest in circulating miRNAs as possible biomarkers and therapeutic targets for insulin resistance in MetS. Future investigations should focus on larger, well-characterized studies with standardized protocols, investigate the mechanistic involvement of circulating miRNAs in insulin signaling pathways, and evaluate how therapeutic interventions affect the dynamic changes in circulating miRNA profiles.
Xeno-miRNAs could enter circulatory systems and organs, where they may influence health and diseases. It was indicated that food intake modifies the inflammatory conditions and redox homeostasis that underline pathological processes in MetS and MetS-related symptoms via delivering xeno-miRNAs. Nonetheless, their absorption, stability, availability, and epigenetic functions are fascinating and debated topics nowadays, but there is scarce information on the physiological and possible adverse consequences of xeno-miRNAs [36]. miRNA-148a in bovine milk appears to encourage β-cell dedifferentiation to the immature mTORC1-high/AMPK-low phenotype, which is characterized by functional deficiencies in insulin production, increased endoplasmic reticulum stress caused by mTORC1, decreased early β-cell apoptosis, and autophagy. Boiling, ultra-heat treatment, and bacterial fermentation deactivate milk’s miRNAs. This raises the suspicion that the continuous consumption of pasteurized milk may cause diabetes via miR-148a [63]. Similarly, milk-derived miR-21 disrupts the homeostasis of α-synuclein, leading to its overexpression and aggregation. This can have harmful consequences on pancreatic β-cells, perhaps contributing to the pathophysiology of T2DM [36]. miRNAs are a significant component of breast milk. The presence of stem cells and breast milk exosome-derived miRNAs is responsible for the highest concentration of miRNAs in breast milk. Second, breast milk miRNAs are extremely resilient to extreme circumstances like UV light, RNase digestion, low pH, high temperatures, repeated freeze/thaw cycles and pasteurization. Thirdly, only a minor portion of miRNA comes from the circulation of maternal blood; the majority comes from mammary epithelial cells. As a result, a variety of organ-derived miRNAs, including pancreatic, liver and hematopoietic-derived miRNAs, are given to breastfed infants. Additionally, xeno-miRNAs have been discovered in breast milk, proving that miRNAs are not just produced endogenously (by the lactation glands or by the mother’s blood circulation) [142]. Some miRNAs abundant in breast milk are essential for immune system-related pathways: miR-155 regulates the maturation of T and B cells and the innate immune response, miR-92s are responsible for T- and B-cell and monocyte development, miR-181a and miR-181b regulate B-cell differentiation, miR-17 and miR-125b inhibit tumor necrosis factor-alpha production, miR-146b negatively regulates innate immunity, and miR-223 regulates the activation and proliferation of neutrophils [143].
When evaluated in terms of plant-based xeno-miRNAs, it was noted that rice aleurone-derived hvu-miR-168-3p decreased blood glucose levels and enhanced glucose transporter 1 (GLUT1) expression [36]. In another study, it was shown that carrot-derived miRNAs target genes’ regulating pathways (insulin signaling pathway, AMPK signaling pathway, MAPK signaling pathway, TGF-beta signaling pathway, etc.) associated with MetS. It was concluded that the management of obesity and MetS could benefit from the development of curative and preventative methods based on foods that are enriched with these miRNAs [144]. Diez-Sainz et al. have shown that miR162, miR159, and miR156e were found in edible plants in high abundance, and miR159’s projected target genes were associated with the insulin signaling system, the negative control of insulin production involved in the physiological response to glucose stimulation, and the positive regulation of fatty acid beta-oxidation and glucose metabolism. However, these miRNAs were not detected in serum, and their interactions with enterocytes and the gut microbiome are assumed to be the cause of their potential effects [34]. Another study showed that miR-482c-5p and miR-482f from plant-based foods such as fruits and vegetables, grains, legumes, and oils can increase the expression of anti-inflammatory genes in macrophages [145]. Anti-inflammatory activity is crucial in preventing the development and progression of many metabolic conditions. However, although these miRNAs are resistant to destruction during the digestive process, they cannot be absorbed. For this reason, they were not detected in serum and are only effective at the intestinal level [145]. Similarly, it has been shown that dry nut-derived miR159a and miR156c can attenuate the Tnfrsf1a protein and TNF-α signaling pathway in adipose cells, thus effectively suppressing inflammation. It has been noted that these xeno-miRNAs could be crucial in the management of metabolic conditions associated with inflammation, such as insulin resistance [146]. It is also anticipated that the target gene of miR-5781 derived from soybeans is IL-17A, that of miR-4996 is IL-10, and that of miR-5671a is IL-33; the target of miR-164a derived from hami melon is IL-16; that of miR-398b derived from oranges is IL-1α; that of miR-1078 derived from ginger is IL-6; and that of miR-4995 derived from tomatoes is IL-5 [69]. However, new approaches and nanocarriers are needed to extend the bioavailability of plant-derived xeno-miRNAs beyond the intestine [34].
miRNAs can have their tissue expression and plasma levels altered by food, nutrition, diet, or non-nutrient substances. Although studies on xeno-miRNAs are limited [34,63,143,144,145,146], the impact of nutrition on the expression of miRNAs has been studied more broadly. When the effects of foods on miRNAs were evaluated, it was stated that milk consumption causes a rise in the levels of miR-200c and miR-29b, which are linked to insulin resistance [23]. According to clinical data from a cohort of women with overweight and insulin resistance, blood orange juice consumption (500 mL per day) for four weeks resulted in decreased let-7f-5p and miR-126-3p levels in peripheral blood mononuclear cells and increased miR-144-3p, miR-424-5p, miR-144-3p, and miR-130b-3p. The increase in the mentioned miRNAs resulted in the decreased expression of NF-κB and IL-6 [147]. In another study, healthy participants who had 500 mL of orange juice showed an immediately enhanced expression of miR-375 and decreased hyperglycemia brought on by a high-calorie meal [148]. In randomized crossover research including 49 prediabetic patients, Hernández-Alonso et al. found that two weeks of pistachio consumption (57 g/day) decreased miR-375 and miR-192 expressions [149]. One more study by Rodriguez-Mateos et al. found that eating wild blueberries affected the expression of miRNAs linked to the insulin signaling pathway in healthy males. The researchers demonstrated that eating wild blueberries 11 g twice a day decreased miR-30c-5p and miR-126-5p expressions [150]. Mir-let7a was also lower in patients with MetS who drank at least one sweetened beverage, ate at least three pieces of fruit each day, and ate less red meat than white meat [151].
When the effects of nutrients on mRNAs were evaluated, palmitic acid facilitated the development of insulin resistance in hepatic cells by activating the PI3K-AKT pathway [36]. By boosting the expression of miR-96-5p and miR-27a-3p, quercetin and catechin epigallocatechin gallate reduced insulin resistance by lowering the transcription of gluconeogenic enzymes and glucose synthesis [36]. Polyunsaturated fatty acid (PUFA) therapy also resulted in the downregulation of miRNAs linked to inflammation, including miR-21, miR-125a, miR-155, miR-146a and miR-146b [36]. The HDL fraction had a large amount of miR-155, and smoking increased miR-155 expression. The miR-155 level in the HDL3 fraction was reduced by 92% in the smoking group, following 8 weeks of vitamin C supplementation (1250 mg/day). This suggests that a high dose of vitamin C can enhance the anti-inflammatory effect by reducing inflammatory miRNA [152]. However, in 21 prediabetic participants, taking 2000 units of cholecalciferol (vitamin D) every day for four months decreased the plasma levels of miR-7 and miR-192 while raising levels of miR-152 [153]. When the impacts of diets on the expressions of miRNA were assessed in a study, the effects of two different dietary strategies for weight loss on miRNA expression in MetS were evaluated. After 8 weeks, it was discovered that 49 miRNAs have varied expression, with 35 associated with the control diet (based on American Heart Association recommendations) and 14 with the RESMENA diet (Mediterranean diet with high meal frequency). Notably, significant expression changes were observed in miR-214, miR-410, miR-190, and miR-637 as a result of the weight-loss intervention [154]. A pilot study reported a decrease in two plasma miRNAs (hsa-miR-26a-5p and hsa-miR-126-3p) that have been shown to be associated with diabetes in a traditional Korean diet group characterized by low red meat, moderate to high fish and legumes, and high fiber and vegetables levels [155]. The regulation of several miRNAs linked to MetS may also be influenced by the Mediterranean diet [156]. It was found that there is a correlation between increased serum levels of miR-590 and Mediterranean diet adherence [151].
In conclusion, xeno-miRNAs can affect fasting glucose and insulin resistance through a range of pathways. Nonetheless, their absorption, stability, availability, and epigenetic functions are fascinating and debated topics nowadays, but there is scarce information on the physiological and possible adverse consequences of xeno-miRNAs [37]. Standardized techniques are required to identify and measure the xeno-miRNAs in target tissues and circulation. Also, studies on this subject are quite limited. Further research is required to completely clarify the mechanisms by which miRNAs affect impaired fasting glucose, to examine their clinical applications in diagnosis and treatment, and to investigate the nutritional sources, metabolism, and mechanisms of xeno-miRNAs.
3.2. Abdominal Obesity
Being a major component of MetS, obesity is a complex health problem that occurs as a result of an abnormal increase in fat mass in the body, causing endocrine, metabolic, and behavioral changes [157]. Environmental, microbiological, epigenetic, and genetic risk factors all contribute to obesity. Obesity and associated metabolic disorders arise when epigenetic mechanisms—such as DNA methylation, histone changes, and miRNA-mediated processes—are disrupted [158]. A well-known pathogenic physiological state that involves modifications in different miRNA expressions is obesity. The abnormal expression of miRNAs, which are effective in regulating adipose tissue metabolism, contributes to the emergence of obesity [159]. Exosomal miRNAs in the circulation are mostly derived from adipose tissue, and different fat depots release different exosomal miRNAs into the blood [160]. miRNA genes are associated with brown fat cell differentiation, which plays a role in obesity [161]. Some miRNAs that promote or inhibit adipogenesis are shown in Figure 3 [162,163].
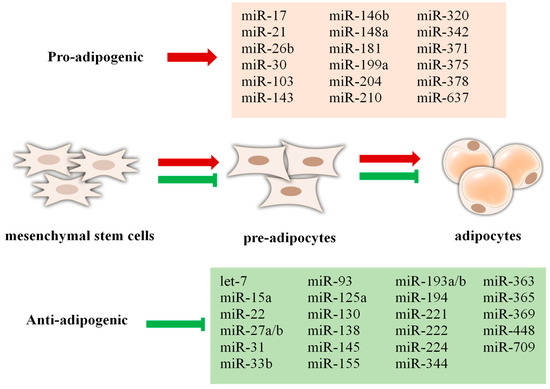
Figure 3.
Some miRNAs that promote or inhibit adipogenesis weight increase. Red arrow indicates promote, green arrow indicates inhibit.
The development of obesity involves the interplay of multiple biological pathways. miRNAs have been shown to modulate essential metabolic pathways, including adipogenesis, lipogenesis, glycolysis, gluconeogenesis, thermogenesis, and cytokine signaling [164]. A diet with high fat is a major factor contributing to the development of obesity, leading to increased lipogenesis, fat accumulation, and overall body weight gain. miRNA-143 expression plays a role in the regulation of lipogenesis by increasing the glycoprotein nonmetastatic melanoma protein B (GPNMB), which the liver secretes, and this encourages white adipose tissue to produce lipogenesis [164,165]. In a study, miR-145 was shown to attenuate lipolytic activity in white adipose tissue by directly suppressing the expression of key lipolysis-promoting factors, including FoxO1 and abhydrolase domain containing 5 (ABHD5/Cgi58) [166]. It has been shown that miR-345-5p, which is negatively associated with obesity, is decreased when focusing on vascular endothelial growth factors during adipogenic differentiation. miR-345-5p upregulation contributes to decreased fat storage in adipose tissue, and plays a key role in controlling gene expression related to lipid metabolism, including lipogenesis, fatty acid biosynthesis, and transport mechanisms [167].
Adipogenesis can be defined as the process of increased adipocyte formation and differentiation into mature adipocytes [168]. In one study, miR-27a-3p was shown to be a critical regulator of human adipogenesis. This miRNA promoted fat cell formation by increasing the expression of genes that promote adipocyte differentiation. These results imply a potential regulatory function of miR-27a-3p in the adipogenic differentiation process [169]. In a study conducted in bone marrow mesenchymal stem cells, miR-422a and miR-483-5p were shown to promote the adipogenesis process. These miRNAs suppressed the expression of the Methyl-CpG binding protein 2 gene, leading to an increase in adipocyte markers (adiponectin, leptin, FABP4). In addition, miR-422a was found to inhibit osteogenesis markers and was specifically increased during adipogenesis. miR-422a appears to exert a significant influence on the molecular pathways governing adipocyte formation [170].
Brown adipose tissue, known for its involvement in body fat regulation and energy homeostasis, contributes to reduced adiposity primarily through the enhancement of thermogenic activity. By altering thermogenic energy expenditure and brown adipogenesis, some miRNAs contribute to obesity [171,172]. Brown adipocyte differentiation is negatively regulated by miR-133, but brown adipogenesis is positively regulated by miR-196a. The overexpression of miR-196a in adipose tissue prevents obesity by increasing brown adipose tissue activity and energy expenditure [173].
The first miRNA linked to metabolic disorders in humans is miR-122, which is found in large quantities in the liver. It contributes to the control of lipid metabolism and cholesterol [174]. Increased miRNA-122 levels reduce brown adipose tissue activity [173]. Furthermore, it has been demonstrated that there is a positive correlation between BMI and the level of miR-122 in the blood [174]. A prospective population-based study identified elevated circulating levels of miR-122 as a significant predictor of the development of MetS in the general population [175]. Circulating miR-122 expression was shown to be considerably higher in patients with MetS than in healthy controls in a study of those with the diagnosis, and there was a positive association between miR-122 levels and BMI. These results imply that miR-122 could be a viable biomarker candidate for MetS diagnosis [176].
Numerous circulating microRNAs, such as miR-101a, miR-16, miR-186, miR-200c, miR-21, miR-299, miR-30c-2, miR-467b, and miR-877, have been linked to the regulation of adiposity [23]. In a study exploring the associations between circulating miRNAs and parameters such as obesity, the distribution of adipose tissue, and total fat accumulation, twelve miRNAs were identified as significantly correlated with these phenotypes. Among them, eight miRNAs—including miR-149-3p, miR-193a-5p, miR-4478, miR-7107-5p, miR-6088, miR-6799-5p, miR-6803-5p, and miR-6821-5p—showed positive associations, whereas four miRNAs—miR-4433b-5p, miR-345-5p, miR-145-5p, and miR-3937—were inversely related to adiposity measures [177].
Heianza et al. reported that adipose tissue-related miRNA-99/100s control PPARγ expression, adipose tissue inflammation, lipid storage, and other critical metabolic factors, hence modulating adiposity, weight gain, and liver steatosis. They reported that reductions in the expression of miR-99-5p/100-5p in circulation induced by dietary therapy and physical activity intervention were linked to both ectopic fat formation and better body fat distribution [178].
Adipocyte-derived adipokines are key regulators of various metabolic processes, including energy homeostasis, inflammatory responses, and the pathophysiology of obesity. There is an interaction between adipokines and miRNAs in maintaining metabolic homeostasis [171]. miRNAs are key regulators of adipokine production and release in adipose tissue. Conversely, certain adipokines have also been demonstrated to affect particular miRNAs’ expression profiles, indicating a bidirectional regulatory relationship between miRNAs and adipokine signaling [30]. The elevated expression of miR-27b and miR-145 in obese patients has been associated with increased leptin receptor expression and activity, implying a possible regulatory role in leptin-mediated metabolic processes [179]. A study found that leptin and resistin significantly reduced miR-143 expression in human preadipocytes [180]. In a study conducted on morbidly obese adolescents, reduced levels of miR-15a, miR-146a, miR-423-5p, miR-520c-3p, and miR-532-5p and elevated levels of miR-130, miR-140-5p, miR-142-3p, miR-143, and miR-222 were detected. These circulating miRNAs were strongly associated with waist–height ratio, BMI, adiponectin, leptin, leptin-adiponectin ratio, fasting blood glucose, C-peptide, insulin, HOMA-IR, and circulating plasma lipids (triglyceride, LDL-C, and HDL-C). It has been demonstrated that adipokines and factors related to MetS may change through the regulation of miRNA expression [174].
Dietary habits and endogenous miRNA regulation are tightly associated. In particular, the energy content of the diet and fat intake have been implicated as key factors affecting miRNA levels [53]. In a systematic review, energy intake has been reported to be an important regulating factor in the human miRNA profile. Changes in endogenous miRNA levels are caused by variations in dietary energy. The length of the dietary intervention and the nutritional circumstances may affect these results. In addition, it has been suggested that these miRNAs have an effect on metabolic pathways and alter the development process of metabolic diseases. It has been stated that miR-22 controls metabolic homeostasis, and treatment for metabolic disorders like obesity and hepatic steatosis may benefit from suppressing this miRNA [53]. A meta-analysis examining extracellular miRNAs in the context of weight loss revealed that circulating levels of miR-223, miR-126, and miR-26a significantly increased post-weight reduction, whereas miR-142 levels showed a marked decrease. In contrast, miR-122, miR-140, miR-146, and miR-221 exhibited no notable changes [181]. After the energy-restricted diet, miR-193a-5p and miR-122-5p levels decreased with weight loss, while miR-126a-3p and miR-222-3p levels increased [182].
The Mediterranean diet, which possesses antioxidant, anti-inflammatory, and anti-atherosclerotic properties, is effective in regulating various miRNAs associated with MetS. In a study conducted in morbidly obese people, higher serum miR-590 levels were were linked to increased Mediterranean diet adherence. A healthier miRNA profile in circulation was linked to greater adherence to the Mediterranean diet, while high-calorie diet consumption was linked to elevated levels of miRNAs involved in the onset and progression of obesity [151].
The inclusion of specific polyphenols or plant-derived bioactive compounds in the diet leads to notable alterations in peripheral tissue miRNA expression, suggesting a mechanistic link between dietary components and gene regulation. In particular, these compounds affected the miRNA dysregulation that would lead to obesity, permitting weight loss and adipogenesis inhibition [183]. In an in vivo study with curcumin supplementation, curcumin inhibited adipogenic differentiation by suppressing miR-17-5p expression in white adipose tissue [184]. In a study including people with MetS who received daily resveratrol (150 mg/day resveratrol and 250 mg/day δ-tocotrienol) supplement as a mixture for six months, improvements were observed in MetS characteristics, including hypertension, dyslipidemia, impaired fasting glucose, and central obesity. Via the examination of serum miRNA levels of individuals, it was shown that resveratrol supplementation significantly reduced the expression of miR-122 and, statistically, considerably decreased the expression of miR-130b and miR-221-5p. Among the miRNAs whose expressions changed, miR130b contributed to the prevention of central obesity in particular by suppressing PPAR-γ, which contributes to the development of adipocytes and adipogenesis in adipose tissue [185]. Green tea, rich in flavonoids and polyphenolic constituents, exerts various health-promoting effects, including the enhancement of redox balance, the attenuation of inflammatory responses, and improved glucose and lipid metabolism [186]. One study evaluated the potential of green tea supplementation given to obese women to alter miRNA levels by reducing markers of oxidative stress and inflammation induced by a high-fat and high-saturated-fat meal. Green tea reduced the levels of 62 miRNAs that are activated after a fatty meal and prevented these molecules from affecting cellular signaling pathways such as TGF-beta, methyltransferase 1, S6 kinase, and bone morphogenetic proteins [187]. Selenium protects against oxidative stress through selenoproteins, and is therefore important in reducing the occurrence of chronic diseases. In a study conducted to determine the effects of Brazil nut intake on circulating miRNAs in obese women with and without MetS, one Brazil nut per day (approximately 1261 μg Selenium) was given for 2 months. Circulating expression levels of miR-454-3p and miR-584-5p rose in obese women at the end of the research, whereas miR-375 levels dramatically dropped in women with MetS. Furthermore, a relationship has been shown between calcium homeostasis, vitamin D metabolism, and selenium intake [188].
It has been stated that miRNAs can respond to various nutritional interventions and can be regulated by changes in diet and lifestyle factors, thus correcting metabolic disorders [163]. Table 2 summarizes the changes in miRNAs according to nutritional interventions, diet, and lifestyle factors.

Table 2.
The changes in miRNAs according to nutritional interventions, diet and lifestyle factors.
According to recent research, dietary intake containing xeno-miRNAs modulates individual miRNA profiles and alters inflammation and redox homeostasis, which are the foundations of diseases like insulin resistance, type 2 diabetes, cancer, and MetS [195,196]. Despite growing interest, the role of food-derived xeno-miRNAs in human health and disease has not yet been clearly elucidated, and their clinical implications remain largely uncertain [36].
One of the important sources of miRNAs is breast milk [197]. miR-22-3p, miR-200a/c-3p, miR-148a-3p, miR-30a/d-5p, miR-146b-5p, and members of the let-7 family have been identified in the cellular, lipid, and skim milk components of human breast milk, which represents the initial source of nutrition in the neonatal period [198]. There are some factors that affect these miRNA expressions, both maternal and milk-related. Factors related to the mother include pregnancy nutrition, pregnancy body weight, stress status during pregnancy, time of birth of the baby, delivery method, mother’s nutrition, mother’s body weight and chronic disease conditions of the mother, while factors related to milk include breast milk storage conditions and heat application to breast milk [199]. A study on breast milk sought to determine how maternal obesity affected specific miRNAs (miR-148a, miR-30b, miR-29a, miR-29b, miR-let-7a, and miR-32) involved in glucose metabolism and adipogenesis, as well as how these miRNAs related to measurements of infant body composition during the initial six months after birth. In this study by Shah et al., it was reported that miRNA-30b and miRNA-148a were abundant in breast milk exosomes, and the amounts of these miRNAs were affected by the mother’s body weight and breastfeeding duration. At one month, miR-30b was favorably correlated with infant body weight, body fat percentage, and fat mass, but miR-148a was negatively correlated with infant body weight, fat mass, and lean mass. A certain miRNA content in human milk was inversely correlated with maternal obesity. It has been reported that miRNA 148a and 30b may be significant indicators of early infancy fat accumulation and baby growth [197]. Milk-derived miR-148a and miR-30b have been associated with adipogenic effects, suggesting a relationship between the prevalence of obesity and milk consumption [91].
As a result, miRNAs play important roles in the pathophysiology of obesity, and the potential roles of these molecules as both therapeutic agents and diagnostic biomarkers have been revealed. miRNAs function in many biological processes associated with obesity, such as energy metabolism, adipose tissue homeostasis, inflammation, insulin resistance, and cellular stress by regulating gene expression at the post-transcriptional level. In this context, miRNAs are considered promising targets in the early diagnosis of obesity-related metabolic complications and in individualized treatment approaches. However, changes in lifestyle, like improving diet, exercising more, and losing weight, can alter miRNA expression profiles, and we still do not fully understand the long-term effects of these changes. The lack of longitudinal studies examining the permanent effects of lifestyle changes applied in obesity treatment on miRNAs limits the sustainability of these molecules as biomarkers and their stability as therapeutic targets. Therefore, future research should focus on prospective studies to evaluate the effects of lifestyle interventions on these molecules in the long term. Thus, the clinical use of miRNAs in obesity diagnosis, follow-up, and treatment will be based on more solid scientific foundations.
3.3. High Blood Pressure
One of the main components of the MetS is hypertension, and its complications cause significant increases in morbidity and mortality. The renin–angiotensin–aldosterone system (RAAS), insulin resistance, obesity, catecholamines, oxidative stress, inflammatory mediators, and high fructose and salt intake are all MetS-related factors that lead to the development of hypertension [200]. miRNAs control hypertension through intricate and varied molecular processes [18]. Some miRNAs have been reported to affect blood pressure and the development of hypertension through vascular, renal, and other physiological mechanisms [201]. In total, 30 miRNAs have been identified that are abundant in human microvascular endothelial cells, and are associated with target genes known to be involved in blood pressure regulation or hypertension [202]. Some miRNAs that act on the kidney to regulate hypertension have been shown to include miR-214-3p, miR-195-5p, miR-192, miR-181a, and miR-133a. miR-214-3p reduces arterial pressure by suppressing endothelial nitric oxide synthase. miRNA-195-5p reduces hypertension by inhibiting Na-K-2Cl cotransporter 2 isoform A (NKCC2A) along the thick ascending limb of the loop of Henle. miR-192 targets Na-K-ATPase β1, a transporter; miR-133a acts by targeting angiotensinogen in the proximal tubule. miR-181a regulates blood pressure by affecting the renin–angiotensin–aldosterone system [201].
It has been proposed in recent years that miRNAs may serve as possible biomarkers for numerous pathological conditions, including hypertension [203]. The combination of miR-122-5p, miR-199a-3p, miR 223-3p, and miR-208a-3p has been suggested as a potential marker for the diagnosis of hypertension, and miRNA dysregulation raises the risk of developing hypertension [204].
The expression of miRNA is changed in the context of hypertension. A study has shown that miR-126-3p and miR-182-5p expressions are significantly higher in hypertensive individuals than in normotensive individuals. It has also been emphasized that these miRNAs can be used for the diagnosis of hypertension, and are important for the development of miRNA-based treatments of hypertension [205].
Inflammatory processes, including endothelial dysfunction, vascular inflammation, and reactive oxygen species (ROS) production, are effective in the formation of hypertension [206]. These inflammatory processes are mediated by miRNAs [207]. Exosomal miRNAs affect the balance between proantioxidant and antioxidant components, including superoxide dismutase (SOD) and NAD(P)H oxidase (NOX) enzymes. miR-21 downregulates SOD, which protects against ROS with antioxidants. It has been reported that miR-21 directly binds to SOD3 in the 3′-UTR region to regulate ROS levels, and indirectly stimulates SOD2 expression by suppressing tumor necrosis factor-alpha (TNF-α). In addition, miR-21 reduces ROS levels by increasing cytochrome b expression in mitochondria. It is thought that the ROS balance disrupted in hypertension may be corrected in this way [208]. Likewise, many miRNAs, including miR-155, miR-212, miR-21, miR-19a/b, and miR-20b, mediate inflammatory processes that lead to hypertension [209].
miRNAs have been shown to regulate the expression of blood pressure-associated genes within the RAAS, thereby contributing to the molecular mechanisms underlying the development of hypertension. The RAAS functions as a critical neurohormonal system that maintains blood pressure homeostasis and electrolyte balance through tightly regulated endocrine signaling pathways [209]. It has been shown that exosomal miRNAs can regulate the RAAS pathways, such as the angiotensin-converting enzyme (ACE)/angiotensin II pathway and the ACE2/Mas pathway. It has been demonstrated that the overexpression of miR-155-5p lowers blood pressure and vascular proliferation by directly lowering ACE gene and angiotensin II peptide levels [210]. In one study, miRNA-143/145 was found to reduce ACE1 expression via adenosine monophosphate-activated protein kinase α2 (AMPKα2), thus slowing down the conversion of Angiotensin I to vasoconstrictive Angiotensin II and showing atheroprotective and antihypertensive effects in endothelial cells [211]. Another miRNA with antihypertensive effects, miR-181a, directly reduces renin levels. The excessive activation of the sympathetic nervous system, which is associated with high blood pressure, inhibits miR-181a expression, and thus increases renin activity, leading to increased blood pressure [209,212].
miR-122 has been implicated in the regulation of multiple target genes associated with the renin–angiotensin system, thereby influencing key pathological processes such as hypertension and cardiovascular fibrosis. Furthermore, sirtuin 6 (SIRT6), elabela, growth differentiation factor 15 (GDF15), porimin, and connective tissue growth factor (CTGF) are among the factors that miR-122 controls in relation to cardiovascular function. In addition, miR-122 causes angiotensin II-induced endothelial dysfunction and vascular fibrosis by targeting sirtuin 6 (SIRT6), a negative regulator of the renin-angiotensin system [18]. Angiotensin II plays a crucial part in the pathophysiological increase in blood pressure by stimulating the proliferation of vascular smooth muscle cells through its vasoconstrictive effects [213].
Environmental and genetic factors cause dysbiosis by inducing pathogenic bacteria to multiply and disrupt the intestinal microbiota. It is suggested that the changing microbiome composition with intestinal dysbiosis affects the pathogenesis of essential hypertension [214]. miRNA expression may be impacted downstream by dietary disruptions in the gut–liver axis, as evidenced by the study of nutritional epigenetics. Both macronutrients and micronutrients have been reported to influence regulatory miRNAs, thereby altering multiple cellular processes that contribute to hypertension and its comorbidities [215]. The effects of altered miRNA expressions in response to diet on the development of high blood pressure are shown in Figure 4.
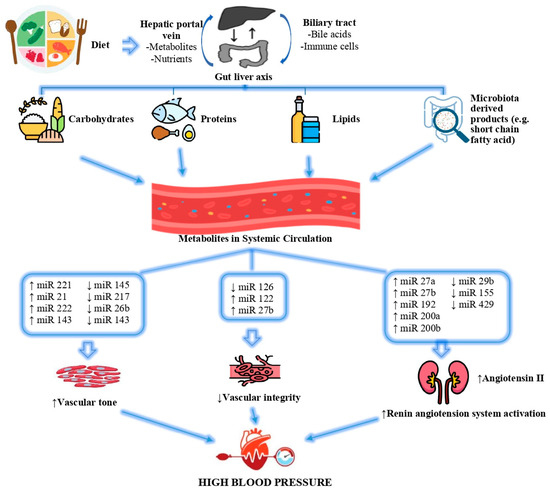
Figure 4.
Effects of altered miRNA expressions in response to diet on the development of high blood pressure [215]. (↓) shows that the expression of the relevant miRNA decreases, (↑) shows that the expression of the relevant miRNA increases.
Excessive dietary sodium consumption is a major environmental determinant of blood pressure regulation, and is strongly associated with an increased risk of cardiovascular disease. Although numerous complex and interrelated physiological pathways have been implicated in the salt–blood pressure relationship, many of these underlying mechanisms remain incompletely understood [216,217]. Studies have shown that changes in miRNA expression play a role in the pathological events underlying hypertension [218,219]. In one study, reducing dietary sodium intake increased circulating levels of miR-143-3p. The increased expression of miR-143-3p has been shown to reduce systolic blood pressure and improve arterial stiffness and skin capillary density [220]. TNF-α, produced by kidney epithelial cells and known for its proinflammatory effects, has been shown to reduce increases in blood pressure in response to high sodium intake by inhibiting NKCC2A [221]. In fact, another study by the same authors determined that TNF-α, which increases with high sodium intake, induces miRNA-195a-5p, and miRNA-195a-5p, and in turn achieves this reduction in blood pressure through a mechanism that regulates NKCC2A mRNA [222].
Unlike the well-known effects of excessive sodium intake, insufficient potassium consumption also contributes to elevated blood pressure, and is recognized as an independent risk factor. Blood pressure regulation exhibits sensitivity not only to sodium but also to potassium, with this trait influenced by both genetic and environmental factors [223]. In a dietary intervention study examining sodium and potassium intake, researchers identified notable associations between genetic variations in specific miRNAs and individuals’ blood pressure responses to these dietary changes. For instance, polymorphisms such as rs115254818 in miR-26b-3p, rs11191676 and rs2292807 in miR-1307-5p, and rs6601178 in miR-4638-3p were linked to increased blood pressure following high sodium intake. Meanwhile, some of these same variants—particularly in miR-26b-3p and miR-1307-5p—were associated with blood pressure reduction after potassium supplementation. Moreover, the study tracked participants over a 14-year period, and found that miRNA-related genetic polymorphisms were significantly correlated with both long-term blood pressure trajectories and the risk of developing hypertension [224].
Fructose, an independent risk factor for hypertension, affects the expressions of some miRNAs that play a role in the pathogenesis of hypertension. While the expressions of miR-19b and miR-101a, which have anti-atherogenic properties, are suppressed after a high-fructose diet, the expression of miR-145a is increased. In addition, high fructose consumption changes blood pressure by affecting miRNAs that affect the renin–angiotensin–aldosterone system [215].
In conclusion, it has been demonstrated that miRNAs may influence the onset of hypertension by controlling the renin–angiotensin–aldosterone system’s constituent parts, peripheral resistance-inducing processes like inflammation, and modifications to the structural and functional characteristics of vascular smooth muscle and endothelial cells. miRNA expression is altered in hypertension. Excessive dietary salt consumption, a modifiable factor, alters blood pressure by affecting circulating miRNA expressions. More comprehensive, prospective human studies are needed to determine how miRNA expression changes in patients with hypertension, and to clarify the long-term effects of miRNAs on hypertension and their potential use in clinical applications.
3.4. Dyslipidaemia
Lipid transport, fat cell development, and cholesterol synthesis are all regulated by miRNAs, which in turn can impact lipid metabolism [100,225,226,227]. Because miRNAs affect the expressions of genes that control the metabolism of HDL, LDL, and very-low-density lipoprotein (VLDL), they play a role in maintaining cholesterol homeostasis and lipoprotein metabolism [228]. These miRNAs are emerging as new biomarkers for dyslipidaemia, and play significant roles in lipid metabolism by controlling the expressions of genes related to lipid metabolism, such as ATP-binding cassette subfamily A member 1 (ABCA1) and LDL receptor (LDLR) [229,230]. Some miRNAs associated with lipid metabolism, the genes they regulate, their mechanisms, and target organs are shown in Figure 5.
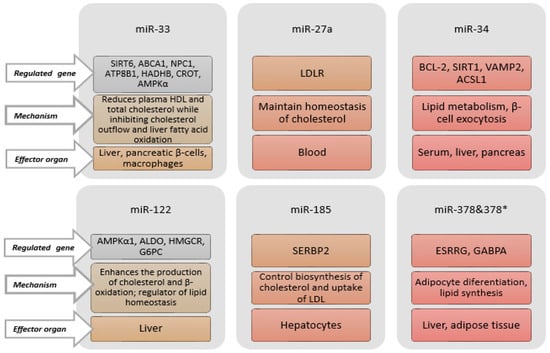
Figure 5.
Some miRNAs related to lipid metabolism, regulated genes, mechanisms, and effector organs. ABCA1: ATP-binding cassette subfamily A member 1. ACSL1: Long-chain acyl-CoA synthetase 1. ALDO: Aldolaz. AMPKα: AMP-activated protein kinase alpha. ATP8B1: ATPase phospholipid transporting 8B1. BCL2: B-cell lymphoma gene-2. CROT: Carnitine o-octanoyltransferase. ESRRG: Estrogen Receptor-Related Receptor. GABPA: GA-Binding Protein Transcription Factor Subunit Alpha. G6PC: Glucose-6-Phosphatase Catalytic. HADHB: Hydroxyacyl-Coa Dehydrogenase Trifunctional Multienzyme Complex Subunit Beta. HDL: High-density lipoprotein. HMGCR: 3-Hydroxy-3-Methylglutaryl-CoA Reductase. LDL: Low-density lipoprotein. LDLR: Low-density lipoprotein receptor. NPC1: Niemann–Pick disease, type C1. SERBP2: Sterol Regulatory Element Binding Protein 2. SIRT1: Sirtuin 1. SIRT6: Sirtuin 6. VAMP2: Vesicle-Associated Membrane Protein 2.
Among these, miR-122, miR33, and miR-27a attract attention due to their role in regulating lipid homeostasis genes [231,232,233]. miR-27a controls the metabolism of cholesterol [229], and miR-33a is a possible biomarker for treatment targets due to its responses to dietary interventions [234]. In patients with dyslipidaemia, synbiotic supplementation has been shown to significantly reduce the expression levels of miR-27a and miR-33a [235]. By enhancing reverse cholesterol transport, the downregulation of miR-33a via elevated ABCA1 and ABCG1 gene expression may raise circulating HDL-C levels [236]. Additionally, Simionescu et al. found that miR-33a expression had an important positive connection with total cholesterol (TC), TAG, LDL-C, and apoB-100, and an inverse correlation (albeit not statistically significant) with HDL-C [237]. It has also been shown that the inhibition of miR-27a can increase the expression of the LDLR (a primary pathway for clearance of LDL-C from circulation) gene, and reduce LDLR degradation by decreasing proprotein convertase subtilisin/kexin type 9 (PCSK9) levels [238].
It has been shown that miRNAs in HDL are different in normal and familial hypercholesterolemic individuals, and have potential for use as biomarkers [239]. The best-studied and best-characterized miRNAs among those involved in HDL-C metabolism are members of the miR-33 family, specifically miR-33a and miR-33b. The intronic miRNA known as the miR-33 family is encoded by the sterol regulatory element-binding proteins (SREBP)-2 and SREBP-1 genes, which are found in intron-16 of two protein-coding genes for SREBP [240]. Together with their host genes SREBP-2 and SREBP-1, miR-33a and miR-33b regulate cholesterol efflux and fatty acid metabolism by preserving the equilibrium between the transcriptional induction and transcriptional repression of genes involved in lipid metabolism [241,242]. The main way that miR-33 controls the metabolism of cholesterol is by directly blocking the ABCA1 and ABCG1 transporters [240,241,242]. While the antagonism of miR-33 with antisense oligonucleotides (ASOs) is sufficient to upregulate hepatic ABCA1 and ABCG1 and boost circulating HDL-C, the overexpression of miR-33 downregulates ABCA1 and ABCG1, lowering HDL-C levels in plasma [241,242]. Patients with MetS have higher levels of VLDL and lower levels of serum HDL-C due to elevated levels of miR-33b and the host gene SREBP1C in the liver. MiR-33a/b inhibition raises HDL-C and lowers VLDL plasma levels [243]. Price et al. demonstrated that AMP-activated kinase (Ampkα1), which governs lipid metabolism, is expressed in response to miR-33a. When miR-33a inhibits AMPKα1, intracellular levels of fatty acids and cholesterol may rise. Thus, by promoting the breakdown of fatty acids, the endogenous suppression of miR-33a in human hepatic cells lowers cholesterol levels [244]. Additionally, one study demonstrated that the suppression of miR-33a and miR-33b resulted in higher HDL-C and lower VLDL-C plasma levels. These findings imply that creating miR-33 antagonists could be a viable method of treating dyslipidemia linked to MetS and cardiometabolic diseases [31,243].
The first miRNA linked to metabolic control, miR-122, is positively correlated with LDL-C, and exhibits increased expression in those with MetS [245]. miR-122 contributes to the protection of liver functions by playing a role in maintaining liver cholesterol balance and regulating fatty acid metabolism. It shows these effects by regulating gene expression and affecting regulatory enzymes involved in cholesterol biosynthesis. As a result, LDL, HDL, TC and apolipoprotein B levels decrease [29,233]. In patients with type 2 diabetes, miR-122 has also been found to exhibit a positive and important connection with LDL-C. [233]. Similarly, in T2DM patients with and without coronary artery disease, circulating miR-122 was found to be significantly correlated with LDL-C, TG and TC by Rashed et al. [246]. When comparing individuals with hyperlipidemia to the control group, Gao et al. discovered a positive correlation between miR-122 expression and LDL-C, TG and TC [247]. The significant down-regulation of genes involved in de novo cholesterol synthesis, including squalene epoxidase (SQLE), seven dehydrocholesterol reductase (DHCR7), 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR), and 3-hydroxy-3-methylglutaryl-CoA synthase 1 (HMGCS1), was observed upon the suppression of miR-122 [248]. In miR-122 suppression settings, a 25–35% reduction in total plasma cholesterol was noted, which is consistent with these findings [245]. Fatty acid and cholesterol synthesis is decreased when miR-122 expression levels for a number of genes, including SREBP, are downregulated, whereas obesity results from overexpression, according to Novák et al. [249]. According to one study, dyslipidemia and elevated levels of miR-122 in the blood were associated positively [175]. In addition, miR-122 has been shown to play an important role in regulating TC and TG levels. It shows this effect by controlling cholesterol biosynthesis in the liver and leading to the release of VLDL. The inhibition of miR-122 has been shown to reduce cholesterol and lipid accumulation in the liver by decreasing the expressions of various genes involved in regulating lipid biosynthesis [244]. A study by Rottiers and Näär showed that one of the significant metabolic roles of miR-122 and miR-33 is maintaining cholesterol levels and lipid biosynthesis [31]. Although miR-33 and 122 still require studies to be fully integrated into clinical practice, applying these two miRNAs in treating lipid disorders is important and promising for the future [250].
By controlling hepatic LDLR expression and VLDL generation, several miRNAs, including miR-148a, miR-128-1, miR-483, miR-520d, miR-224, miR-30c, and miR-122, regulate plasma LDL-C and VLDL-C levels [251]. By raising hepatic LDL receptor and ABCA1 expression, the inhibition of miR-148a lowers LDL-C and raises HDL-C levels (in vivo) [252]. In a systematic review examining the links between MetS components and circulating miRNA, lower levels of miR-363, miR-375, miR-486 and miR-16 were observed in the entire cohort (primarily in men when considered by gender) in association with lower HDL concentrations [25]. These miRNAs have been implicated in bile acid production and secretion, cholesterol efflux, hepatic uptake, and HDL-C biogenesis [253]. In a systematic review, it was reported that the levels of sixteen circulating miRNAs (miR-629-5p, miR-33, miR-375, miR-454-3p, miR-320b, miR-25-5p, miR-1180, miR-103, miR-339-5p, miR-628-3p, miR-29a-3p, miR-122-5p, miR-29b-2-5p, miR-185-5p, miR-19a-3p, miR-886-5p) were associated with HDL-C (p < 0.05) [121]. In another review, it was reported that miR-302, miR-27, miR-148a, miR-34a, miR-10b, miR-128-1, miR-26, miR-758, miR-19b and miR-144 regulate plasma HDL-C levels by directly affecting ABCA1 expression in the liver or macrophages [228]. miR-34a is known to be one of the primary regulators of hepatic lipid homeostasis [31].
Overweight/obese persons with hypertension have higher levels of miR-605 and miR-623; there have been reports of negative correlations between miR-623 and HDL-c levels and positive correlations between miR-605 and TC and LDL-C levels [254]. According to one study, dyslipidemia is linked to plasma markers that indicate diabetes dyslipidemia, and miRNAs including miR-143, miR-122, miR-132, miR-218, miR-21, and miR-155 are involved in both the up- and the down-regulation of these indicators [255]. Additionally, there is a positive correlation between plasma LDL-C and triglyceride levels and let-7 family miRNA levels [256]. Furthermore, it has been documented that people with insulin resistance and/or hyperlipidemia express more miR-33b, miR-146a, miR-10a, miR-125a, miR-33a and miR-21. Further, it has been stated that these microRNAs show a positive correlation with TC, TG and LDL-C levels in individuals with hyperglycemia and hyperlipidemia [5,237]. Table 3 summarizes the correlation between specific miRNAs and lipid biomarkers in populations with MetS components.

Table 3.
Association between miRNAs and lipid-related biomarkers in diverse populations with MetS components.
The discovery of exogenous plant miR-168a, which has been proposed as a new biomarker for its likely functional role in the metabolism of LDL-C, enables the identification of food-derived xeno-miRs in disease etiology [88,261]. Studies have shown that xeno-miRNAs can perform cross-kingdom roles [88,262,263]. For instance, it has been demonstrated that rice’s miR-168a controls the expression of genes in mammals upon dietary intake [88,263]. Following rice consumption, in vitro and in vivo studies showed that miR168a may bind to the mRNA of the human/mouse low-density lipoprotein receptor adaptor protein 1 (LDLRAP1), suppress the liver’s expression of LDLRAP1, and thus reduce the elimination of LDL from mouse plasma [88,89]. Compared to control mice, rice-fed mice showed higher levels of circulating LDL-C and a greater buildup of miR168a in the liver. Consequently, it was shown that miR168a can decrease LDL clearance in the bloodstream by directly targeting LDLRAP1 in the liver. Exogenous plant miRNAs from a variety of sources have been shown to accumulate in particular tissues, enter the circulatory system from the gastrointestinal tract, and have significant biological effects [88]. In contrast, Dickinson and colleagues reported that they could not detect miR168a in the serum of mice after rice consumption, and did not observe any changes in LDLRAP1 expression levels in the liver. However, they stated that this result may have been due to the experimental methods and applications employed [264].
Because of their structural resemblance to cholesterol, phytosterols work in a similar way. The absorption of cholesterol is decreased when consumed alongside plant-based diets [265]. The daily consumption of a nutraceutical combination including 400 mg phytosterols, 100 mg bergamot extract, 20 mg olive extract, and 52 μM vitamin K2 for up to 12 weeks did not change the serum lipid profile or inflammation-associated microRNAs and biomarkers, according to a study on hypercholesterolemia [266]. Diet-induced dyslipidemia can be avoided by downregulating miRNA-96 and consuming a high-fat, high-carb meal together with foods that include a blend of polyphenolic flavonoids, such as orange juice and grape seed extract [267].
In conclusion, by controlling important genes involved in lipid synthesis, transport, and degradation, miRNAs may have an impact on lipid metabolism. Circulating miRNAs may be potential biomarkers that can be used to assess changes in lipid profiles and monitor responses. Some dietary xeno-miRNAs may also affect lipid metabolism, but studies investigating the effects of dietary xeno-miRNAs on dyslipidemia are quite limited, and current findings are contradictory. Further studies are needed to elucidate the dietary sources and mechanisms of xeno-miRNAs in lipid homeostasis.
4. Conclusions and Future Perspectives
miRNAs and diet-derived xeno-miRNAs have received increasing attention for their potential effects on MetS and related pathological processes. As we know, a single miRNA can affect the expressions of several genes, and multiple miRNAs can affect the expression of a single gene. MiRNAs may, therefore, be crucial biomarkers for the prompt diagnosis, prognosis, and management of metabolic disorders. This review offers a current, thorough analysis of the important and contentious role that circulating miRNA and xeno-miRNA activities play in regulating chronic processes linked to MetS and its constituent parts.
miRNAs have been shown to affect T2DM by influencing β-cell differentiation, GLUT4 expression, and insulin sensitivity; obesity by affecting adipogenesis, lipolysis, and energy metabolism; hypertension through their regulatory effects on the renin–angiotensin system and vascular tone; and dyslipidemia by modulating cholesterol and lipoprotein metabolism (HDL, LDL, and VLDL levels). With our current review, it can be concluded that there are significant relationships between miRNA levels and abdominal obesity, insulin resistance, hypertension, and plasma high LDL-C and low HDL-C concentrations. With our current review, it is possible to say that there are important relationships between miRNA levels and abdominal obesity, insulin resistance, hypertension, and plasma high LDL-C and low HDL-C concentrations.
Nutrition and diet change redox homeostasis and inflammatory conditions, which affect the pathological processes in MetS and related symptoms, thus altering individual miRNA profiles. The relevance of dietary miRNAs in the diet is highlighted by research on their functions, which also provides fresh perspectives on the possible advantages of the foods we eat on a regular basis. These miRNAs can enter the bloodstream and circulate throughout the body, potentially influencing various physiological processes and contributing to the regulation of MetS and its components by modulating gene expression and cellular functions.
Research suggests that dietary patterns such as breast milk, polyphenols such as curcumin, resveratrol and EGCG, and food and dietary components such as rice, orange juice, and the Mediterranean diet may influence miRNA expression and contribute to the prevention of MetS, indicating their potential use as therapeutic targets in supporting metabolic health.
In this context, understanding the complexities of diet–gene interactions and determining lifestyle and dietary habits that affect miRNA expression will enable us to exploit the potential of precision nutrition more effectively in preventing and treating chronic diseases such as MetS. This approach, which combines an understanding of diet and gene interactions with advances in molecular nutrition, may allow for personalized solutions that consider individual differences rather than generic interventions.
Although the clinical use of these molecules as biomarkers and their relationship with nutrition-based interventions are promising, further research is needed on their bioavailability, metabolism, and mechanisms of action. In the future, developing new technologies to increase the bioavailability of dietary miRNAs may enable the use of these molecules as personalized nutrition strategies in the prevention and treatment of metabolic diseases. Future research should focus on increasing our understanding of how different nutrients modulate miRNA profiles, and how individual genetic and environmental factors contribute to these effects. In addition, the development of miRNA-based treatment approaches and the investigation of the therapeutic potential of these molecules in clinical nutrition are important research areas that need to be focused on in the future. In particular, their effects on lipid metabolism, inflammation, and insulin sensitivity may open a new window for disease-specific intervention strategies. Also, personalized nutrition applications based on the analysis of miRNA expression profiles according to the genetic background, metabolic status, and environmental factors of individuals (precision nutrition) may be an important area of research in the future. With this approach, it will be possible to develop more effective nutrition strategies to prevent and manage metabolic diseases such as obesity, type 2 diabetes, cardiovascular diseases, and MetS.
Author Contributions
Conceptualization, D.A.; project administration, D.A.; writing—original draft preparation, M.N.Ç., B.D.G., B.A., B.S. and M.A.I.; writing—review and editing, D.A. and F.B.; visualization, M.N.Ç., B.D.G., B.A., B.S. and M.A.I.; supervision, D.A. and F.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Giangregorio, F.; Mosconi, E.; Debellis, M.G.; Provini, S.; Esposito, C.; Garolfi, M.; Oraka, S.; Kaloudi, O.; Mustafazade, G.; Marín-Baselga, R.; et al. A systematic review of metabolic syndrome: Key correlated pathologies and non-invasive diagnostic approaches. J. Clin. Med. 2024, 13, 5880. [Google Scholar] [CrossRef] [PubMed]
- Alemany, M. The metabolic syndrome, a human disease. Int. J. Mol. Sci. 2024, 25, 2251. [Google Scholar] [PubMed]
- Silva-Ochoa, A.D.; Velasteguí, E.; Falconí, I.B.; García-Solorzano, V.I.; Rendón-Riofrio, A.; Sanguna-Soliz, G.A.; Berghe, W.V.; Orellana-Manzano, A. Metabolic syndrome: Nutri-epigenetic cause or consequence? Heliyon 2023, 9, e21106. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Lei, Y.; Luo, S.; Huang, Z.; Chen, C.; Wang, K.; Yang, P.; Huang, X. MicroRNA expression profile and identification of novel microRNA biomarkers for metabolic syndrome. Bioengineered 2021, 12, 3864–3872. [Google Scholar] [CrossRef]
- Brandao-Lima, P.N.; de Carvalho, G.B.; Payolla, T.B.; Sarti, F.M.; Rogero, M.M. Circulating microRNA related to cardiometabolic risk factors for metabolic syndrome: A systematic review. Metabolites 2022, 12, 1044. [Google Scholar] [CrossRef]
- Plawgo, K.; Raczynska, K.D. Context-dependent regulation of gene expression by non-canonical small RNAs. Non-Coding RNA 2022, 8, 29. [Google Scholar] [CrossRef]
- Ramzan, F.; Vickers, M.H.; Mithen, R.F. Epigenetics, microRNA and metabolic syndrome: A comprehensive review. Int. J. Mol. Sci. 2021, 22, 5047. [Google Scholar] [CrossRef]
- Heneghan, H.; Miller, N.; Kerin, M. Role of microRNAs in obesity and the metabolic syndrome. Obes. Rev. 2010, 11, 354–361. [Google Scholar] [CrossRef]
- Mayr, B.; Montminy, M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat. Rev. Mol. Cell Biol. 2001, 2, 599–609. [Google Scholar] [CrossRef]
- Plaisance, V.; Abderrahmani, A.; Perret-Menoud, V.; Jacquemin, P.; Lemaigre, F.; Regazzi, R. MicroRNA-9 controls the expression of Granuphilin/Slp4 and the secretory response of insulin-producing cells. J. Biol. Chem. 2006, 281, 26932–26942. [Google Scholar] [CrossRef]
- Krek, A.; Grün, D.; Poy, M.N.; Wolf, R.; Rosenberg, L.; Epstein, E.J.; MacMenamin, P.; Da Piedade, I.; Gunsalus, K.C.; Stoffel, M.; et al. Combinatorial microRNA target predictions. Nat. Genet. 2005, 37, 495–500. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Muniappan, L.; Tang, G.; Özcan, S. Identification of glucose-regulated miRNAs from pancreatic β cells reveals a role for miR-30d in insulin transcription. RNA 2009, 15, 287–293. [Google Scholar] [CrossRef] [PubMed]
- Takanabe, R.; Ono, K.; Abe, Y.; Takaya, T.; Horie, T.; Wada, H.; Kita, T.; Satoh, N.; Shimatsu, A.; Hasegawa, K. Up-regulated expression of microRNA-143 in association with obesity in adipose tissue of mice fed high-fat diet. Biochem. Biophys. Res. Commun. 2008, 376, 728–732. [Google Scholar] [CrossRef] [PubMed]
- Esau, C.; Kang, X.; Peralta, E.; Hanson, E.; Marcusson, E.G.; Ravichandran, L.V.; Sun, Y.; Koo, S.; Perera, R.J.; Jain, R.; et al. MicroRNA-143 regulates adipocyte differentiation. J. Biol. Chem. 2004, 279, 52361–52365. [Google Scholar] [CrossRef]
- Xie, H.; Lim, B.; Lodish, H.F. MicroRNAs induced during adipogenesis that accelerate fat cell development are downregulated in obesity. Diabetes 2009, 58, 1050–1057. [Google Scholar] [CrossRef]
- Wang, Q.; Li, Y.C.; Wang, J.; Kong, J.; Qi, Y.; Quigg, R.J.; Li, X. miR-17-92 cluster accelerates adipocyte differentiation by negatively regulating tumor-suppressor Rb2/p130. Proc. Natl. Acad. Sci. USA 2008, 105, 2889–2894. [Google Scholar] [CrossRef]
- Mellios, N.; Huang, H.-S.; Grigorenko, A.; Rogaev, E.; Akbarian, S. A set of differentially expressed miRNAs, including miR-30a-5p, act as post-transcriptional inhibitors of BDNF in prefrontal cortex. Hum. Mol. Genet. 2008, 17, 3030–3042. [Google Scholar] [CrossRef]
- Shaheen, N.; Shaheen, A.; Diab, R.A.; Desouki, M.T. MicroRNAs (miRNAs) role in hypertension: Pathogenesis and promising therapeutics. Ann. Med. Surg. 2024, 86, 319–328. [Google Scholar] [CrossRef]
- Paulin, R.; Meloche, J.; Jacob, M.H.; Bisserier, M.; Courboulin, A.; Bonnet, S. Dehydroepiandrosterone inhibits the Src/STAT3 constitutive activation in pulmonary arterial hypertension. Am. J. Physiol.-Heart Circ. Physiol. 2011, 301, H1798–H1809. [Google Scholar] [CrossRef]
- Deng, L.; Blanco, F.J.; Stevens, H.; Lu, R.; Caudrillier, A.; McBride, M.; McClure, J.D.; Grant, J.; Thomas, M.; Frid, M.; et al. MicroRNA-143 activation regulates smooth muscle and endothelial cell crosstalk in pulmonary arterial hypertension. Circ. Res. 2015, 117, 870–883. [Google Scholar] [CrossRef]
- Improta-Caria, A.C.; Aras, M.G.; Nascimento, L.; De Sousa, R.A.L.; Aras-Júnior, R.; Souza, B.S.d.F. MicroRNAs regulating renin–angiotensin–aldosterone system, sympathetic nervous system and left ventricular hypertrophy in systemic arterial hypertension. Biomolecules 2021, 11, 1771. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Liu, Y.; Han, Q. Puerarin attenuates cardiac hypertrophy partly through increasing mir-15b/195 expression and suppressing non-canonical transforming growth factor beta (Tgfβ) signal pathway. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2016, 22, 1516. [Google Scholar] [CrossRef] [PubMed]
- Deveci, G.; Capasso, R.; Ağagündüz, D. Xeno-miRs and Circulating miRNAs as Novel Biomarkers in Certain Diseases. Biologics 2022, 3, 1–10. [Google Scholar] [CrossRef]
- Vukelić, I.; Šuša, B.; Klobučar, S.; Buljević, S.; Liberati Pršo, A.-M.; Belančić, A.; Rahelić, D.; Detel, D. Exosome-Derived microRNAs: Bridging the Gap Between Obesity and Type 2 Diabetes in Diagnosis and Treatment. Diabetology 2024, 5, 706–724. [Google Scholar] [CrossRef]
- Brandão-Lima, P.N.; de Carvalho, G.B.; Payolla, T.B.; Sarti, F.M.; Fisberg, R.M.; Malcomson, F.C.; Mathers, J.C.; Rogero, M.M. Circulating microRNAs Showed Specific Responses according to Metabolic Syndrome Components and Sex of Adults from a Population-Based Study. Metabolites 2023, 13, 2. [Google Scholar] [CrossRef]
- Barbiera, A.; Pelosi, L.; Sica, G.; Scicchitano, B.M. Nutrition and microRNAs: Novel insights to fight sarcopenia. Antioxidants 2020, 9, 951. [Google Scholar] [CrossRef]
- Włodarski, A.; Strycharz, J.; Wróblewski, A.; Kasznicki, J.; Drzewoski, J.; Śliwińska, A. The role of microRNAs in metabolic syndrome-related oxidative stress. Int. J. Mol. Sci. 2020, 21, 6902. [Google Scholar] [CrossRef]
- Pozniak, T.; Shcharbin, D.; Bryszewska, M. Circulating microRNAs in medicine. Int. J. Mol. Sci. 2022, 23, 3996. [Google Scholar] [CrossRef]
- Fernández-Hernando, C.; Ramírez, C.M.; Goedeke, L.; Suárez, Y. MicroRNAs in metabolic disease. Arterioscler. Thromb. Vasc. Biol. 2013, 33, 178–185. [Google Scholar] [CrossRef]
- Kurylowicz, A. microRNAs in human adipose tissue physiology and dysfunction. Cells 2021, 10, 3342. [Google Scholar] [CrossRef]
- Rottiers, V.; Näär, A.M. MicroRNAs in metabolism and metabolic disorders. Nat. Rev. Mol. Cell Biol. 2012, 13, 239–250. [Google Scholar] [CrossRef] [PubMed]
- Suksangrat, T.; Phannasil, P.; Jitrapakdee, S. miRNA regulation of glucose and lipid metabolism in relation to diabetes and non-alcoholic fatty liver disease. In Reviews on Biomarker Studies of Metabolic and Metabolism-Related Disorders; Springer: Berlin/Heidelberg, Germany, 2019; pp. 129–148. [Google Scholar]
- Witwer, K.W. XenomiRs and miRNA homeostasis in health and disease: Evidence that diet and dietary miRNAs directly and indirectly influence circulating miRNA profiles. RNA Biol. 2012, 9, 1147–1154. [Google Scholar] [CrossRef] [PubMed]
- Díez-Sainz, E.; Milagro, F.I.; Aranaz, P.; Riezu-Boj, J.I.; Lorente-Cebrián, S. MicroRNAs from edible plants reach the human gastrointestinal tract and may act as potential regulators of gene expression. J. Physiol. Biochem. 2024, 80, 655–670. [Google Scholar] [CrossRef] [PubMed]
- Gai, C.; Pomatto, M.A.C.; Deregibus, M.C.; Dieci, M.; Piga, A.; Camussi, G. Edible plant-derived extracellular vesicles for oral mRNA vaccine delivery. Vaccines 2024, 12, 200. [Google Scholar] [CrossRef]
- Martino, E.; D’Onofrio, N.; Balestrieri, A.; Colloca, A.; Anastasio, C.; Sardu, C.; Marfella, R.; Campanile, G.; Balestrieri, M.L. Dietary epigenetic modulators: Unravelling the still-controversial benefits of miRNAs in nutrition and disease. Nutrients 2024, 16, 160. [Google Scholar] [CrossRef]
- Zhu, W.-J.; Liu, Y.; Cao, Y.-N.; Peng, L.-X.; Yan, Z.-Y.; Zhao, G. Insights into health-promoting effects of plant MicroRNAs: A review. J. Agric. Food Chem. 2021, 69, 14372–14386. [Google Scholar] [CrossRef]
- Lukasik, A.; Zielenkiewicz, P. Plant microRNAs—Novel players in natural medicine? Int. J. Mol. Sci. 2016, 18, 9. [Google Scholar] [CrossRef]
- Shen, L.; Ma, J.; Yang, Y.; Liao, T.; Wang, J.; Chen, L.; Zhang, S.; Zhao, Y.; Niu, L.; Hao, X.; et al. Cooked pork-derived exosome nanovesicles mediate metabolic disorder—microRNA could be the culprit. J. Nanobiotechnol. 2023, 21, 83. [Google Scholar] [CrossRef]
- Cunha e Rocha, K.; Ying, W.; Olefsky, J.M. Exosome-Mediated Impact on Systemic Metabolism. Annu. Rev. Physiol. 2024, 86, 225–253. [Google Scholar] [CrossRef]
- Kiani, A.K.; Bonetti, G.; Donato, K.; Kaftalli, J.; Herbst, K.L.; Stuppia, L.; Fioretti, F.; Nodari, S.; Perrone, M.; Chiurazzi, P.; et al. Polymorphisms, diet and nutrigenomics. J. Prev. Med. Hyg. 2022, 63, E125–E141. [Google Scholar]
- Hill, M.; Tran, N. miRNA interplay: Mechanisms and consequences in cancer. Dis. Models Mech. 2021, 14, dmm047662. [Google Scholar] [CrossRef] [PubMed]
- Du, T.; Zamore, P.D. Beginning to understand microRNA function. Cell Res. 2007, 17, 661–663. [Google Scholar] [CrossRef] [PubMed]
- Cullen, B.R. Five questions about viruses and microRNAs. PLoS Pathog. 2010, 6, e1000787. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Lee, Y.; Yeom, K.-H.; Kim, Y.-K.; Jin, H.; Kim, V.N. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004, 18, 3016–3027. [Google Scholar] [CrossRef]
- Landthaler, M.; Yalcin, A.; Tuschl, T. The human DiGeorge syndrome critical region gene 8 and Its D. melanogaster homolog are required for miRNA biogenesis. Curr. Biol. 2004, 14, 2162–2167. [Google Scholar] [CrossRef]
- Makarova, J.A.; Shkurnikov, M.U.; Wicklein, D.; Lange, T.; Samatov, T.R.; Turchinovich, A.A.; Tonevitsky, A.G. Intracellular and extracellular microRNA: An update on localization and biological role. Prog. Histochem. Cytochem. 2016, 51, 33–49. [Google Scholar] [CrossRef]
- Bayraktar, R.; Van Roosbroeck, K.; Calin, G.A. Cell-to-cell communication: MicroRNAs as hormones. Mol. Oncol. 2017, 11, 1673–1686. [Google Scholar] [CrossRef]
- Xu, L.; Yang, B.f.; Ai, J. MicroRNA transport: A new way in cell communication. J. Cell. Physiol. 2013, 228, 1713–1719. [Google Scholar] [CrossRef]
- Vickers, K.C.; Remaley, A.T. Lipid-based carriers of microRNAs and intercellular communication. Curr. Opin. Lipidol. 2012, 23, 91–97. [Google Scholar] [CrossRef]
- Pirim, D.; Dogan, B. In silico identification of putative roles of food-derived xeno-mirs on diet-associated cancer. Nutr. Cancer 2020, 72, 481–488. [Google Scholar] [CrossRef]
- Alisi, A.; Da Sacco, L.; Bruscalupi, G.; Piemonte, F.; Panera, N.; De Vito, R.; Leoni, S.; Bottazzo, G.F.; Masotti, A.; Nobili, V. Mirnome analysis reveals novel molecular determinants in the pathogenesis of diet-induced nonalcoholic fatty liver disease. Lab. Investig. 2011, 91, 283–293. [Google Scholar] [CrossRef] [PubMed]
- DeLucas, M.; Sánchez, J.; Palou, A.; Serra, F. The impact of diet on miRNA regulation and its implications for health: A systematic review. Nutrients 2024, 16, 770. [Google Scholar] [CrossRef] [PubMed]
- Ferrero, G.; Carpi, S.; Polini, B.; Pardini, B.; Nieri, P.; Impeduglia, A.; Grioni, S.; Tarallo, S.; Naccarati, A. Intake of natural compounds and circulating microRNA expression levels: Their relationship investigated in healthy subjects with different dietary habits. Front. Pharmacol. 2021, 11, 619200. [Google Scholar] [CrossRef] [PubMed]
- Wagner, A.E.; Piegholdt, S.; Ferraro, M.; Pallauf, K.; Rimbach, G. Food derived microRNAs. Food Funct. 2015, 6, 714–718. [Google Scholar] [CrossRef]
- Link, J.; Thon, C.; Schanze, D.; Steponaitiene, R.; Kupcinskas, J.; Zenker, M.; Canbay, A.; Malfertheiner, P.; Link, A. Food-derived Xeno-microRNAs: Influence of diet and detectability in gastrointestinal tract—Proof-of-principle study. Mol. Nutr. Food Res. 2019, 63, 1800076. [Google Scholar] [CrossRef]
- Wade, B.; Cummins, M.; Keyburn, A.; Crowley, T.M. Isolation and detection of microRNA from the egg of chickens. BMC Res. Notes 2016, 9, 283. [Google Scholar] [CrossRef]
- Pieri, M.; Theori, E.; Dweep, H.; Flourentzou, M.; Kalampalika, F.; Maniori, M.A.; Papagregoriou, G.; Papaneophytou, C.; Felekkis, K. A bovine miRNA, bta-miR-154c, withstands in vitro human digestion but does not affect cell viability of colorectal human cell lines after transfection. FEBS Open Bio 2022, 12, 925–936. [Google Scholar] [CrossRef]
- Shome, S.; Jernigan, R.; Beitz, D.; Clark, S.; Testroet, E. Non-coding RNA in raw and commercially processed milk and putative targets related to growth and immune-response. BMC Genom. 2021, 22, 749. [Google Scholar] [CrossRef]
- Melnik, B.C.; Stadler, R.; Weiskirchen, R.; Leitzmann, C.; Schmitz, G. Potential pathogenic impact of cow’s milk consumption and bovine milk-derived exosomal MicroRNAs in diffuse large B-Cell lymphoma. Int. J. Mol. Sci. 2023, 24, 6102. [Google Scholar] [CrossRef]
- García-Martínez, J.; Pérez-Castillo, Í.M.; Salto, R.; López-Pedrosa, J.M.; Rueda, R.; Girón, M.D. Beneficial effects of bovine milk exosomes in metabolic interorgan cross-talk. Nutrients 2022, 14, 1442. [Google Scholar] [CrossRef]
- Chen, Z.; Xie, Y.; Luo, J.; Chen, T.; Xi, Q.; Zhang, Y.; Sun, J. Milk exosome-derived miRNAs from water buffalo are implicated in immune response and metabolism process. BMC Vet. Res. 2020, 16, 123. [Google Scholar] [CrossRef] [PubMed]
- Melnik, B.C. Milk exosomal miRNAs: Potential drivers of AMPK-to-mTORC1 switching in β-cell de-differentiation of type 2 diabetes mellitus. Nutr. Metab. 2019, 16, 85. [Google Scholar] [CrossRef] [PubMed]
- Lönnerdal, B. Human milk microRNAs/exosomes: Composition and biological effects. Nestle Nutr. Inst. Workshop Ser. 2019, 90, 83–92. [Google Scholar] [PubMed]
- Martino, E.; Balestrieri, A.; Mele, L.; Sardu, C.; Marfella, R.; D’Onofrio, N.; Campanile, G.; Balestrieri, M.L. Milk exosomal miR-27b worsen endoplasmic reticulum stress mediated colorectal cancer cell death. Nutrients 2022, 14, 5081. [Google Scholar] [CrossRef]
- Hou, D.; He, F.; Ma, L.; Cao, M.; Zhou, Z.; Wei, Z.; Xue, Y.; Sang, X.; Chong, H.; Tian, C.; et al. The potential atheroprotective role of plant MIR156a as a repressor of monocyte recruitment on inflamed human endothelial cells. J. Nutr. Biochem. 2018, 57, 197–205. [Google Scholar] [CrossRef]
- Xiao, X.; Sticht, C.; Yin, L.; Liu, L.; Karakhanova, S.; Yin, Y.; Georgikou, C.; Gladkich, J.; Gross, W.; Gretz, N.; et al. Novel plant microRNAs from broccoletti sprouts do not show cross-kingdom regulation of pancreatic cancer. Oncotarget 2020, 11, 1203. [Google Scholar] [CrossRef][Green Version]
- Akao, Y.; Kuranaga, Y.; Heishima, K.; Sugito, N.; Morikawa, K.; Ito, Y.; Soga, T.; Ito, T. Plant hvu-MIR168-3p enhances expression of glucose transporter 1 (SLC2A1) in human cells by silencing genes related to mitochondrial electron transport chain complex I. J. Nutr. Biochem. 2022, 101, 108922. [Google Scholar] [CrossRef]
- Xiao, J.; Feng, S.; Wang, X.; Long, K.; Luo, Y.; Wang, Y.; Ma, J.; Tang, Q.; Jin, L.; Li, X.; et al. Identification of exosome-like nanoparticle-derived microRNAs from 11 edible fruits and vegetables. PeerJ 2018, 6, e5186. [Google Scholar] [CrossRef]
- Manzaneque-López, M.C.; Sánchez-López, C.M.; Pérez-Bermúdez, P.; Soler, C.; Marcilla, A. Dietary-derived exosome-like nanoparticles as bacterial modulators: Beyond microRNAs. Nutrients 2023, 15, 1265. [Google Scholar] [CrossRef]
- Mlotshwa, S.; Pruss, G.J.; MacArthur, J.L.; Endres, M.W.; Davis, C.; Hofseth, L.J.; Peña, M.M.; Vance, V. A novel chemopreventive strategy based on therapeutic microRNAs produced in plants. Cell Res. 2015, 25, 521–524. [Google Scholar] [CrossRef]
- Komori, H.; Fujita, D.; Shirasaki, Y.; Zhu, Q.; Iwamoto, Y.; Nakanishi, T.; Nakajima, M.; Tamai, I. MicroRNAs in Apple-Derived Nanoparticles Modulate Intestinal Expression of Organic Anion–Transporting Peptide 2B1/SLCO2B1 in Caco-2 Cells. Drug Metab. Dispos. 2021, 49, 803–809. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, F.; Weng, Z.; Sui, X.; Fang, Y.; Tang, X.; Shen, X. Soybean-derived miRNAs specifically inhibit proliferation and stimulate apoptosis of human colonic Caco-2 cancer cells but not normal mucosal cells in culture. Genomics 2020, 112, 2949–2958. [Google Scholar] [CrossRef] [PubMed]
- López de Las Hazas, M.-C.; del Pozo-Acebo, L.; Hansen, M.S.; Gil-Zamorano, J.; Mantilla-Escalante, D.C.; Gómez-Coronado, D.; Marín, F.; Garcia-Ruiz, A.; Rasmussen, J.T.; Dávalos, A. Dietary bovine milk miRNAs transported in extracellular vesicles are partially stable during GI digestion, are bioavailable and reach target tissues but need a minimum dose to impact on gene expression. Eur. J. Nutr. 2022, 61, 1043–1056. [Google Scholar] [CrossRef] [PubMed]
- Philip, A.; Ferro, V.A.; Tate, R.J. Determination of the potential bioavailability of plant microRNAs using a simulated human digestion process. Mol. Nutr. Food Res. 2015, 59, 1962–1972. [Google Scholar] [CrossRef]
- Vashisht, M.; Rani, P.; Onteru, S.K.; Singh, D. Curcumin encapsulated in milk exosomes resists human digestion and possesses enhanced intestinal permeability in vitro. Appl. Biochem. Biotechnol. 2017, 183, 993–1007. [Google Scholar] [CrossRef]
- Benmoussa, A.; Lee, C.H.C.; Laffont, B.; Savard, P.; Laugier, J.; Boilard, E.; Gilbert, C.; Fliss, I.; Provost, P. Commercial dairy cow milk microRNAs resist digestion under simulated gastrointestinal tract conditions. J. Nutr. 2016, 146, 2206–2215. [Google Scholar] [CrossRef]
- Colombo, M.; Raposo, G.; Théry, C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef]
- Tome-Carneiro, J.; Fernandez-Alonso, N.; Tomas-Zapico, C.; Visioli, F.; Iglesias-Gutierrez, E.; Davalos, A. Breast milk microRNAs harsh journey towards potential effects in infant development and maturation. Lipid encapsulation can help. Pharmacol. Res. 2018, 132, 21–32. [Google Scholar] [CrossRef]
- Dávalos, A.; Pinilla, L.; de Las Hazas, M.-C.L.; Pinto-Hernández, P.; Barbé, F.; Iglesias-Gutiérrez, E.; de Gonzalo-Calvo, D. Dietary microRNAs and cancer: A new therapeutic approach? Semin. Cancer Biol. 2021, 73, 19–29. [Google Scholar] [CrossRef]
- Wang, X.; Ren, X.; Ning, L.; Wang, P.; Xu, K. Stability and absorption mechanism of typical plant miRNAs in an in vitro gastrointestinal environment: Basis for their cross-kingdom nutritional effects. J. Nutr. Biochem. 2020, 81, 108376. [Google Scholar] [CrossRef]
- Abrego-Guandique, D.M.; Ilori, O.A.; Caroleo, M.C.; Cannataro, R.; Cione, E.; Tucci, P. Differential Digestive Stability of Food-Derived microRNAs: The Case of miR-30c-5p and miR-92a-3p in Polyfloral Honey. Curr. Issues Mol. Biol. 2024, 46, 7473–7485. [Google Scholar] [CrossRef] [PubMed]
- Kupec, T.; Bleilevens, A.; Iborra, S.; Najjari, L.; Wittenborn, J.; Maurer, J.; Stickeler, E. Stability of circulating microRNAs in serum. PLoS ONE 2022, 17, e0268958. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Lei, L.; Ye, F.; Zhou, Y.; Chang, H.; Zhao, G. Nutritive implications of dietary microRNAs: Facts, controversies, and perspectives. Food Funct. 2019, 10, 3044–3056. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Wang, S.; Ying, Y.; Zhou, R.; Mao, P. miR-196b/miR-1290 participate in the antitumor effect of resveratrol via regulation of IGFBP3 expression in acute lymphoblastic leukemia. Oncol. Rep. 2017, 37, 1075–1083. [Google Scholar] [CrossRef]
- Özyalçin, B.; Sanlier, N. The effect of diet components on cancer with epigenetic mechanisms. Trends Food Sci. Technol. 2020, 102, 138–145. [Google Scholar] [CrossRef]
- Witwer, K.W.; Zhang, C.-Y. Diet-derived microRNAs: Unicorn or silver bullet? Genes Nutr. 2017, 12, 15. [Google Scholar] [CrossRef]
- Zhang, L.; Hou, D.; Chen, X.; Li, D.; Zhu, L.; Zhang, Y.; Li, J.; Bian, Z.; Liang, X.; Cai, X.; et al. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: Evidence of cross-kingdom regulation by microRNA. Cell Res. 2012, 22, 107–126. [Google Scholar] [CrossRef]
- Rabuma, T.; Gupta, O.P.; Chhokar, V. Recent advances and potential applications of cross-kingdom movement of miRNAs in modulating plant’s disease response. RNA Biol. 2022, 19, 519–532. [Google Scholar] [CrossRef]
- Kaur, K.; Vig, S.; Srivastava, R.; Mishra, A.; Singh, V.P.; Srivastava, A.K.; Datta, M. Elevated hepatic miR-22-3p expression impairs gluconeogenesis by silencing the Wnt-responsive transcription factor Tcf7. Diabetes 2015, 64, 3659–3669. [Google Scholar] [CrossRef]
- Abbas, M.A.; Al-Saigh, N.N.; Saqallah, F.G. Regulation of adipogenesis by exosomal milk miRNA. Rev. Endocr. Metab. Disord. 2023, 24, 297–316. [Google Scholar] [CrossRef]
- Melnik, B.C.; Schmitz, G. MicroRNAs: Milk’s epigenetic regulators. Best Pract. Res. Clin. Endocrinol. Metab. 2017, 31, 427–442. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.-K.; Zhou, Z.; Jiang, X.-M.; Zheng, Y.; Chen, X.; Fu, Z.; Xiao, G.; Zhang, C.-Y.; Zhang, L.-K.; Yi, Y. Absorbed plant MIR2911 in honeysuckle decoction inhibits SARS-CoV-2 replication and accelerates the negative conversion of infected patients. Cell Discov. 2020, 6, 54. [Google Scholar] [CrossRef] [PubMed]
- Cavalieri, D.; Rizzetto, L.; Tocci, N.; Rivero, D.; Asquini, E.; Si-Ammour, A.; Bonechi, E.; Ballerini, C.; Viola, R. Plant microRNAs as novel immunomodulatory agents. Sci. Rep. 2016, 6, 25761. [Google Scholar] [CrossRef] [PubMed]
- Chin, A.R.; Fong, M.Y.; Somlo, G.; Wu, J.; Swiderski, P.; Wu, X.; Wang, S.E. Cross-kingdom inhibition of breast cancer growth by plant miR159. Cell Res. 2016, 26, 217–228. [Google Scholar] [CrossRef]
- Marzano, F.; Caratozzolo, M.F.; Consiglio, A.; Licciulli, F.; Liuni, S.; Sbisà, E.; D’Elia, D.; Tullo, A.; Catalano, D. Plant miRNAs reduce cancer cell proliferation by targeting MALAT1 and NEAT1: A beneficial cross-kingdom interaction. Front. Genet. 2020, 11, 552490. [Google Scholar] [CrossRef]
- Shaikh, F.S.; Siegel, R.J.; Srivastava, A.; Fox, D.A.; Ahmed, S. Challenges and promise of targeting miRNA in rheumatic diseases: A computational approach to identify miRNA association with cell types, cytokines, and disease mechanisms. Front. Immunol. 2024, 14, 1322806. [Google Scholar] [CrossRef]
- Huang, Y.; Yan, Y.; Xv, W.; Qian, G.; Li, C.; Zou, H.; Li, Y. A new insight into the roles of MiRNAs in metabolic syndrome. BioMed Res. Int. 2018, 2018, 7372636. [Google Scholar] [CrossRef]
- Huszar, J.; Payne, C. miR-146a Influences energy metabolism, cell differentiation and innate immunity. Metabolomics 2013, 3, 119. [Google Scholar]
- Yang, Z.; Cappello, T.; Wang, L. Emerging role of microRNAs in lipid metabolism. Acta Pharm. Sin. B 2015, 5, 145–150. [Google Scholar] [CrossRef]
- Jiménez-Lucena, R.; Camargo, A.; Alcalá-Diaz, J.F.; Romero-Baldonado, C.; Luque, R.M.; Van Ommen, B.; Delgado-Lista, J.; Ordovás, J.M.; Pérez-Martínez, P.; Rangel-Zúñiga, O.A.; et al. A plasma circulating miRNAs profile predicts type 2 diabetes mellitus and prediabetes: From the CORDIOPREV study. Exp. Mol. Med. 2018, 50, 1–12. [Google Scholar] [CrossRef]
- Corral-Fernández, N.; Salgado-Bustamante, M.; Martínez-Leija, M.; Cortez-Espinosa, N.; García-Hernández, M.; Reynaga-Hernández, E.; Quezada-Calvillo, R.; Portales-Pérez, D. Dysregulated miR-155 expression in peripheral blood mononuclear cells from patients with type 2 diabetes. Exp. Clin. Endocrinol. Diabetes 2013, 121, 347–353. [Google Scholar] [CrossRef] [PubMed]
- Mensà, E.; Giuliani, A.; Matacchione, G.; Gurău, F.; Bonfigli, A.R.; Romagnoli, F.; De Luca, M.; Sabbatinelli, J.; Olivieri, F. Circulating miR-146a in healthy aging and type 2 diabetes: Age-and gender-specific trajectories. Mech. Ageing Dev. 2019, 180, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Karolina, D.S.; Tavintharan, S.; Armugam, A.; Sepramaniam, S.; Pek, S.L.T.; Wong, M.T.; Lim, S.C.; Sum, C.F.; Jeyaseelan, K. Circulating miRNA profiles in patients with metabolic syndrome. J. Clin. Endocrinol. Metab. 2012, 97, E2271–E2276. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Valverde, S.L.; Taft, R.J.; Mattick, J.S. MicroRNAs in β-cell biology, insulin resistance, diabetes and its complications. Diabetes 2011, 60, 1825. [Google Scholar] [CrossRef]
- Chakraborty, C.; Doss, C.G.P.; Bandyopadhyay, S.; Agoramoorthy, G. Influence of miRNA in insulin signaling pathway and insulin resistance: Micro-molecules with a major role in type-2 diabetes. Wiley Interdiscip. Rev. RNA 2014, 5, 697–712. [Google Scholar] [CrossRef]
- Van de Bunt, M.; Gaulton, K.J.; Parts, L.; Moran, I.; Johnson, P.R.; Lindgren, C.M.; Ferrer, J.; Gloyn, A.L.; McCarthy, M.I. The miRNA profile of human pancreatic islets and beta-cells and relationship to type 2 diabetes pathogenesis. PLoS ONE 2013, 8, e55272. [Google Scholar] [CrossRef]
- Tabuchi, T.; Satoh, M.; Itoh, T.; Nakamura, M. MicroRNA-34a regulates the longevity-associated protein SIRT1 in coronary artery disease: Effect of statins on SIRT1 and microRNA-34a expression. Clin. Sci. 2012, 123, 161–171. [Google Scholar] [CrossRef]
- Vegter, E.L.; Schmitter, D.; Hagemeijer, Y.; Ovchinnikova, E.S.; van der Harst, P.; Teerlink, J.R.; O’Connor, C.M.; Metra, M.; Davison, B.A.; Bloomfield, D.; et al. Use of biomarkers to establish potential role and function of circulating microRNAs in acute heart failure. Int. J. Cardiol. 2016, 224, 231–239. [Google Scholar] [CrossRef]
- Cortez-Dias, N.; Costa, M.C.; Carrilho-Ferreira, P.; Silva, D.; Jorge, C.; Calisto, C.; Pessoa, T.; Martins, S.R.; de Sousa, J.C.; da Silva, P.C.; et al. Circulating miR-122-5p/miR-133b ratio is a specific early prognostic biomarker in acute myocardial infarction. Circ. J. 2016, 80, 2183–2191. [Google Scholar] [CrossRef]
- Luo, Y.-F.; Wan, X.-X.; Zhao, L.-L.; Guo, Z.; Shen, R.-T.; Zeng, P.-Y.; Wang, L.-H.; Yuan, J.-J.; Yang, W.-J.; Yue, C.; et al. MicroRNA-139-5p upregulation is associated with diabetic endothelial cell dysfunction by targeting c-jun. Aging 2020, 13, 1186. [Google Scholar] [CrossRef]
- Creemers, E.E.; Tijsen, A.J.; Pinto, Y.M. Circulating microRNAs: Novel biomarkers and extracellular communicators in cardiovascular disease? Circ. Res. 2012, 110, 483–495. [Google Scholar] [CrossRef] [PubMed]
- Fichtlscherer, S.; Zeiher, A.M.; Dimmeler, S. Circulating microRNAs: Biomarkers or mediators of cardiovascular diseases? Arterioscler. Thromb. Vasc. Biol. 2011, 31, 2383–2390. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Lü, J.; Liang, Z.; Liu, D.; Yao, Q. Overview of 8 circulating microRNAs and their functions as major biomarkers for cardiovascular diseases. Clin. Pract. Rev. Meta-Anal. 2020, 7, e924530. [Google Scholar] [CrossRef]
- Tong, Y.; Xu, S.; Huang, L.; Chen, C. Obesity and insulin resistance: Pathophysiology and treatment. Drug Discov. Today 2022, 27, 822–830. [Google Scholar] [CrossRef]
- Zhao, X.; An, X.; Yang, C.; Sun, W.; Ji, H.; Lian, F. The crucial role and mechanism of insulin resistance in metabolic disease. Front. Endocrinol. 2023, 14, 1149239. [Google Scholar] [CrossRef]
- Li, M.; Chi, X.; Wang, Y.; Setrerrahmane, S.; Xie, W.; Xu, H. Trends in insulin resistance: Insights into mechanisms and therapeutic strategy. Signal Transduct. Target. Ther. 2022, 7, 216. [Google Scholar] [CrossRef]
- Mi, S.; Zhang, J.; Zhang, W.; Huang, R.S. Circulating microRNAs as biomarkers for inflammatory diseases. MicroRNA 2013, 2, 64–72. [Google Scholar] [CrossRef]
- Capetini, V.C.; Quintanilha, B.J.; Garcia, B.R.E.V.; Rogero, M.M. Dietary modulation of microRNAs in insulin resistance and type 2 diabetes. J. Nutr. Biochem. 2024, 133, 109714. [Google Scholar] [CrossRef]
- Zampetaki, A.; Kiechl, S.; Drozdov, I.; Willeit, P.; Mayr, U.; Prokopi, M.; Mayr, A.; Weger, S.; Oberhollenzer, F.; Bonora, E.; et al. Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ. Res. 2010, 107, 810–817. [Google Scholar] [CrossRef]
- Solís-Toro, D.; Mosquera Escudero, M.; García-Perdomo, H.A. Association between circulating microRNAs and the metabolic syndrome in adult populations: A systematic review. Diabetes Metab. Syndr. Clin. Res. Rev. 2022, 16, 102376. [Google Scholar] [CrossRef]
- Kaur, P.; Kotru, S.; Singh, S.; Behera, B.S.; Munshi, A. Role of miRNAs in the pathogenesis of T2DM, insulin secretion, insulin resistance, and β cell dysfunction: The story so far. J. Physiol. Biochem. 2020, 76, 485–502. [Google Scholar] [CrossRef] [PubMed]
- Sou, Y.L.; Chilian, W.M.; Ratnam, W.; Zain, S.M.; Syed Abdul Kadir, S.Z.; Pan, Y.; Pung, Y.F. Exosomal miRNAs and isomiRs: Potential biomarkers for type 2 diabetes mellitus. Precis. Clin. Med. 2024, 7, pbae021. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.N.; Zhang, X.; Xu, F.; Fan, Y.Y.; Ge, B.; Guo, H.; Li, Z.L. Four-microRNA signature for detection of type 2 diabetes. World J. Clin. Cases 2020, 8, 1923–1931. [Google Scholar] [CrossRef] [PubMed]
- Poy, M.N.; Eliasson, L.; Krutzfeldt, J.; Kuwajima, S.; Ma, X.; Macdonald, P.E.; Pfeffer, S.; Tuschl, T.; Rajewsky, N.; Rorsman, P.; et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature 2004, 432, 226–230. [Google Scholar] [CrossRef]
- Sucharita, S.; Ashwini, V.; Prabhu, J.; Avadhany, S.; Ayyar, V.; Bantwal, G. The role of circulating microRNA in the regulation of beta cell function and insulin resistance among Indians with type 2 diabetes. Indian J. Endocrinol. Metab. 2018, 22, 770–773. [Google Scholar] [CrossRef]
- Formichi, C.; Nigi, L.; Grieco, G.E.; Maccora, C.; Fignani, D.; Brusco, N.; Licata, G.; Sebastiani, G.; Dotta, F. Non-Coding RNAs: Novel Players in Insulin Resistance and Related Diseases. Int. J. Mol. Sci. 2021, 22, 7716. [Google Scholar] [CrossRef]
- Mendonca, A.; Thandapani, P.; Nagarajan, P.; Venkatesh, S.; Sundaresan, S. Role of microRNAs in regulation of insulin secretion and insulin signaling involved in type 2 diabetes mellitus. J. Biosci. 2022, 47, 58. [Google Scholar] [CrossRef]
- Dalgaard, L.T.; Sørensen, A.E.; Hardikar, A.A.; Joglekar, M.V. The microRNA-29 family: Role in metabolism and metabolic disease. Am. J. Physiol.-Cell Physiol. 2022, 323, C367–C377. [Google Scholar] [CrossRef]
- Cruz, K.J.C.; de Oliveira, A.R.S.; Morais, J.B.S.; Severo, J.S.; do Nascimento Marreiro, D. Role of microRNAs on adipogenesis, chronic low-grade inflammation, and insulin resistance in obesity. Nutrition 2017, 35, 28–35. [Google Scholar] [CrossRef]
- Klimczak, S.; Śliwińska, A. Epigenetic regulation of inflammation in insulin resistance. Semin. Cell Dev. Biol. 2024, 154, 185–192. [Google Scholar] [CrossRef]
- Zeinali, F.; Aghaei Zarch, S.M.; Jahan-Mihan, A.; Kalantar, S.M.; Vahidi Mehrjardi, M.Y.; Fallahzadeh, H.; Hosseinzadeh, M.; Rahmanian, M.; Mozaffari-Khosravi, H. Circulating microRNA-122, microRNA-126-3p and microRNA-146a are associated with inflammation in patients with pre-diabetes and type 2 diabetes mellitus: A case control study. PLoS ONE 2021, 16, e0251697. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Kim, O.-K. Potential Roles of Adipocyte Extracellular Vesicle–Derived miRNAs in Obesity-Mediated Insulin Resistance. Adv. Nutr. 2021, 12, 566–574. [Google Scholar] [CrossRef] [PubMed]
- Martin, H. Role of PPAR-gamma in inflammation. Prospects for therapeutic intervention by food components. Mutat. Res. 2010, 690, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Pérez-García, A.; Torrecilla-Parra, M.; Fernández-de Frutos, M.; Martín-Martín, Y.; Pardo-Marqués, V.; Ramírez, C.M. Posttranscriptional Regulation of Insulin Resistance: Implications for Metabolic Diseases. Biomolecules 2022, 12, 208. [Google Scholar] [CrossRef]
- Agbu, P.; Carthew, R.W. MicroRNA-mediated regulation of glucose and lipid metabolism. Nat. Rev. Mol. Cell Biol. 2021, 22, 425–438. [Google Scholar] [CrossRef]
- Abdel Mageed, S.S.; Doghish, A.S.; Ismail, A.; El-Husseiny, A.A.; Fawzi, S.F.; Mahmoud, A.M.A.; El-Mahdy, H.A. The role of miRNAs in insulin resistance and diabetic macrovascular complications—A review. Int. J. Biol. Macromol. 2023, 230, 123189. [Google Scholar] [CrossRef]
- Nguyen, M.T.; Min, K.-H.; Lee, W. MiR-183-5p induced by saturated fatty acids hinders insulin signaling by downregulating IRS-1 in hepatocytes. Int. J. Mol. Sci. 2022, 23, 2979. [Google Scholar] [CrossRef]
- Wang, S.; Ai, H.; Liu, L.; Zhang, X.; Gao, F.; Zheng, L.; Yi, J.; Sun, L.; Yu, C.; Zhao, H.; et al. Micro-RNA-27a/b negatively regulates hepatic gluconeogenesis by targeting FOXO1. Am. J. Physiol.-Endocrinol. Metab. 2019, 317, E911–E924. [Google Scholar] [CrossRef]
- Li, L.; Li, C.; Lv, M.; Hu, Q.; Guo, L.; Xiong, D. Correlation between alterations of gut microbiota and miR-122-5p expression in patients with type 2 diabetes mellitus. Ann. Transl. Med. 2020, 8, 1481. [Google Scholar] [CrossRef]
- Guo, Y.; Zhu, X.; Zeng, S.; He, M.; Xing, X.; Wang, C. miRNA-10a-5p Alleviates Insulin Resistance and Maintains Diurnal Patterns of Triglycerides and Gut Microbiota in High-Fat Diet-Fed Mice. Mediat. Inflamm. 2020, 2020, 8192187. [Google Scholar] [CrossRef]
- Hatmal, M.m.M.; Al-Hatamleh, M.A.I.; Olaimat, A.N.; Alshaer, W.; Hasan, H.; Albakri, K.A.; Alkhafaji, E.; Issa, N.N.; Al-Holy, M.A.; Abderrahman, S.M.; et al. Immunomodulatory Properties of Human Breast Milk: MicroRNA Contents and Potential Epigenetic Effects. Biomedicines 2022, 10, 1219. [Google Scholar] [CrossRef] [PubMed]
- Khurana, P.; Gupta, A.; Varshney, R. Diet-Derived Exogenous miRNAs as Functional Food Components: Facts and New Perspectives. Preprints 2021. [Google Scholar] [CrossRef]
- Reza-Zaldívar, E.E.; Jacobo-Velázquez, D.A. Targeting Metabolic Syndrome Pathways: Carrot microRNAs As Potential Modulators. ACS Omega 2024, 9, 21891–21903. [Google Scholar] [CrossRef] [PubMed]
- Díez-Sainz, E.; Lorente-Cebrian, S.; Aranaz, P.; Amri, E.-Z.; Riezu-Boj, J.I.; Milagro, F.I. miRf and miRc-p from edible plant-derived foods inhibit the expression of pro-inflammatory genes in human THP-macrophages. In Genome-Based Nutrition Strategies for Preventing Diet-related Chronic Diseases: Where Genes, Diet, and Food Culture Meet; Panduro, A., Ramos-Lopez, O., Ojeda-Granados, C., Eds.; Frontiers Media SA: Lausanne, Switzerland, 2024. [Google Scholar]
- Aquilano, K.; Ceci, V.; Gismondi, A.; De Stefano, S.; Iacovelli, F.; Faraonio, R.; Di Marco, G.; Poerio, N.; Minutolo, A.; Minopoli, G.; et al. Adipocyte metabolism is improved by TNF receptor-targeting small RNAs identified from dried nuts. Commun. Biol. 2019, 2, 317. [Google Scholar] [CrossRef]
- Capetini, V.C.; Quintanilha, B.J.; de Oliveira, D.C.; Nishioka, A.H.; de Matos, L.A.; Ferreira, L.R.P.; Ferreira, F.M.; Sampaio, G.R.; Hassimotto, N.M.A.; Lajolo, F.M.; et al. Blood orange juice intake modulates plasma and PBMC microRNA expression in overweight and insulin-resistant women: Impact on MAPK and NFκB signaling pathways. J. Nutr. Biochem. 2023, 112, 109240. [Google Scholar] [CrossRef]
- Quintanilha, B.J.; Chaves, D.F.; Brasili, E.; Corrêa, T.A.; Capetini, V.C.; Ferreira, F.M.; Castro, I.A.; Hassimotto, N.M.; Rogero, M.M.; Lajolo, F.M. Ingestion of orange juice prevents hyperglycemia and increases plasma miR-375 expression. Clin. Nutr. ESPEN 2022, 47, 240–245. [Google Scholar] [CrossRef]
- Hernández-Alonso, P.; Giardina, S.; Salas-Salvadó, J.; Arcelin, P.; Bulló, M. Chronic pistachio intake modulates circulating microRNAs related to glucose metabolism and insulin resistance in prediabetic subjects. Eur. J. Nutr. 2017, 56, 2181–2191. [Google Scholar] [CrossRef]
- Rodriguez-Mateos, A.; Istas, G.; Boschek, L.; Feliciano, R.P.; Mills, C.E.; Boby, C.; Gomez-Alonso, S.; Milenkovic, D.; Heiss, C. Circulating anthocyanin metabolites mediate vascular benefits of blueberries: Insights from randomized controlled trials, metabolomics, and nutrigenomics. J. Gerontol. Ser. A 2019, 74, 967–976. [Google Scholar] [CrossRef]
- Fontalba-Romero, M.I.; Lopez-Enriquez, S.; Lago-Sampedro, A.; García-Escobar, E.; Pastori, R.L.; Domínguez-Bendala, J.; Álvarez-Cubela, S.; Valdes, S.; Rojo, G.; Garcia-Fuentes, E.; et al. Association between the Mediterranean Diet and Metabolic Syndrome with Serum Levels of miRNA in Morbid Obesity. Nutrients 2021, 13, 436. [Google Scholar] [CrossRef]
- Kim, S.-M.; Lim, S.-M.; Yoo, J.-A.; Woo, M.-J.; Cho, K.-H. Consumption of high-dose vitamin C (1250 mg per day) enhances functional and structural properties of serum lipoprotein to improve anti-oxidant, anti-atherosclerotic, and anti-aging effects via regulation of anti-inflammatory microRNA. Food Funct. 2015, 6, 3604–3612. [Google Scholar] [CrossRef]
- Lopez, Y.O.N.; Pittas, A.G.; Pratley, R.E.; Seyhan, A.A. Circulating levels of miR-7, miR-152 and miR-192 respond to vitamin D supplementation in adults with prediabetes and correlate with improvements in glycemic control. J. Nutr. Biochem. 2017, 49, 117–122. [Google Scholar] [CrossRef] [PubMed]
- Marsetti, P.S.; Milagro, F.I.; Zulet, M.Á.; Martínez, J.A.; Lorente-Cebrián, S. Changes in miRNA expression with two weight-loss dietary strategies in a population with metabolic syndrome. Nutrition 2021, 83, 111085. [Google Scholar] [CrossRef] [PubMed]
- Shin, P.-K.; Kim, M.S.; Park, S.-J.; Kwon, D.Y.; Kim, M.J.; Yang, H.J.; Kim, S.-H.; Kim, K.; Chun, S.; Lee, H.-J.; et al. A Traditional Korean Diet Alters the Expression of Circulating MicroRNAs Linked to Diabetes Mellitus in a Pilot Trial. Nutrients 2020, 12, 2558. [Google Scholar] [CrossRef] [PubMed]
- Cannataro, R.; Fazio, A.; La Torre, C.; Caroleo, M.C.; Cione, E. Polyphenols in the Mediterranean diet: From dietary sources to microRNA modulation. Antioxidants 2021, 10, 328. [Google Scholar] [CrossRef]
- Akman, K.; Öney, B.; Bilgin, M.G. Fiziksel Aktivitenin Metabolik Sendrom Üzerine Etkisi. Haliç Üniversitesi Sağlık Bilimleri Dergisi 2023, 6, 1–9. [Google Scholar] [CrossRef]
- Wu, F.-Y.; Yin, R.-X. Recent progress in epigenetics of obesity. Diabetol. Metab. Syndr. 2022, 14, 171. [Google Scholar] [CrossRef]
- Landrier, J.-F.; Derghal, A.; Mounien, L. MicroRNAs in obesity and related metabolic disorders. Cells 2019, 8, 859. [Google Scholar] [CrossRef]
- Thomou, T.; Mori, M.A.; Dreyfuss, J.M.; Konishi, M.; Sakaguchi, M.; Wolfrum, C.; Rao, T.N.; Winnay, J.N.; Garcia-Martin, R.; Grinspoon, S.K.; et al. Adipose-derived circulating miRNAs regulate gene expression in other tissues. Nature 2017, 542, 450–455. [Google Scholar] [CrossRef]
- Fujii, Y.R. Food XenomiRNA Arise: MicroRNA Storm and Space. In The MicroRNA 2000 Transformer: Quantum Computing and Artificial Intelligence for Health; Springer Nature Singapore: Singapore, 2023; pp. 49–81. [Google Scholar]
- Kornmueller, K.; Amri, E.-Z.; Scheideler, M.; Prassl, R. Delivery of miRNAs to the adipose organ for metabolic health. Adv. Drug Deliv. Rev. 2022, 181, 114110. [Google Scholar] [CrossRef]
- Iacomino, G.; Siani, A. Role of microRNAs in obesity and obesity-related diseases. Genes Nutr. 2017, 12, 23. [Google Scholar] [CrossRef]
- Benavides-Aguilar, J.A.; Torres-Copado, A.; Isidoro-Sánchez, J.; Pathak, S.; Duttaroy, A.K.; Banerjee, A.; Paul, S. The Regulatory Role of MicroRNAs in Obesity and Obesity-Derived Ailments. Genes 2023, 14, 2070. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, H.; Zeng, D.; Xiong, J.; Luo, J.; Chen, X.; Chen, T.; Xi, Q.; Sun, J.; Ren, X.; et al. The novel importance of miR-143 in obesity regulation. Int. J. Obes. 2023, 47, 100–108. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-Y.; Chou, C.-F.; Giovarelli, M.; Briata, P.; Gherzi, R.; Chen, C.-Y. KSRP and MicroRNA 145 are negative regulators of lipolysis in white adipose tissue. Mol. Cell. Biol. 2014, 34, 2339–2349. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; He, Y.; Feng, Z.; Sheng, J.; Dong, A.; Zhang, M.; Cao, L. miR-345-5p regulates adipogenesis via targeting VEGF-B. Aging 2020, 12, 17114. [Google Scholar] [CrossRef]
- Yandım, M.K. Obezitede Hücre Sinyal İletimi. Turk. Klin. Intern. Med.-Spec. Top. 2022, 2, 1–6. [Google Scholar]
- Wu, H.; Pula, T.; Tews, D.; Amri, E.-Z.; Debatin, K.-M.; Wabitsch, M.; Fischer-Posovszky, P.; Roos, J. microRNA-27a-3p but not-5p is a crucial mediator of human adipogenesis. Cells 2021, 10, 3205. [Google Scholar] [CrossRef]
- Giuliani, A.; Sabbatinelli, J.; Amatori, S.; Graciotti, L.; Silvestrini, A.; Matacchione, G.; Ramini, D.; Mensà, E.; Prattichizzo, F.; Babini, L.; et al. MiR-422a promotes adipogenesis via MeCP2 downregulation in human bone marrow mesenchymal stem cells. Cell. Mol. Life Sci. 2023, 80, 75. [Google Scholar] [CrossRef]
- Ma, L.; Gilani, A.; Yi, Q.; Tang, L. MicroRNAs as mediators of adipose thermogenesis and potential therapeutic targets for obesity. Biology 2022, 11, 1657. [Google Scholar] [CrossRef]
- Shamsi, F.; Zhang, H.; Tseng, Y.-H. MicroRNA regulation of brown adipogenesis and thermogenic energy expenditure. Front. Endocrinol. 2017, 8, 205. [Google Scholar] [CrossRef]
- Okamatsu-Ogura, Y.; Matsushita, M.; Bariuan, J.V.; Nagaya, K.; Tsubota, A.; Saito, M. Association of circulating exosomal miR-122 levels with BAT activity in healthy humans. Sci. Rep. 2019, 9, 13243. [Google Scholar] [CrossRef]
- Al-Rawaf, H.A. Circulating microRNAs and adipokines as markers of metabolic syndrome in adolescents with obesity. Clin. Nutr. 2019, 38, 2231–2238. [Google Scholar] [CrossRef] [PubMed]
- Willeit, P.; Skroblin, P.; Moschen, A.R.; Yin, X.; Kaudewitz, D.; Zampetaki, A.; Barwari, T.; Whitehead, M.; Ramírez, C.M.; Goedeke, L.; et al. Circulating microRNA-122 is associated with the risk of new-onset metabolic syndrome and type 2 diabetes. Diabetes 2017, 66, 347–357. [Google Scholar] [CrossRef] [PubMed]
- Refeat, M.M.; Hassan, N.A.-M.; Ahmad, I.H.; Mostafa, E.R.M.; Amr, K.S. Correlation of circulating miRNA-33a and miRNA-122 with lipid metabolism among Egyptian patients with metabolic syndrome. J. Genet. Eng. Biotechnol. 2021, 19, 147. [Google Scholar] [CrossRef] [PubMed]
- Abozaid, Y.J.; Zhang, X.; Mens, M.M.; Ahmadizar, F.; Limpens, M.; Ikram, M.A.; Rivadeneira, F.; Voortman, T.; Kavousi, M.; Ghanbari, M. Plasma circulating microRNAs associated with obesity, body fat distribution, and fat mass: The Rotterdam study. Int. J. Obes. 2022, 46, 2137–2144. [Google Scholar] [CrossRef]
- Heianza, Y.; Krohn, K.; Xue, Q.; Meir, A.Y.; Ziesche, S.; Ceglarek, U.; Blüher, M.; Keller, M.; Kovacs, P.; Shai, I.; et al. Changes in circulating microRNAs-99/100 and reductions of visceral and ectopic fat depots in response to lifestyle interventions: The CENTRAL trial. Am. J. Clin. Nutr. 2022, 116, 165–172. [Google Scholar] [CrossRef]
- Viesti, A.; Collares, R.; Salgado, W., Jr.; Pretti da Cunha Tirapelli, D.; dos Santos, J.S. The expression of LEP, LEPR, IGF1 and IL10 in obesity and the relationship with microRNAs. PLoS ONE 2014, 9, e93512. [Google Scholar]
- Zhu, L.; Shi, C.; Ji, C.; Xu, G.; Chen, L.; Yang, L.; Fu, Z.; Cui, X.; Lu, Y.; Guo, X. FFAs and adipokine-mediated regulation of hsa-miR-143 expression in human adipocytes. Mol. Biol. Rep. 2013, 40, 5669–5675. [Google Scholar] [CrossRef]
- Veie, C.H.; Nielsen, I.M.; Frisk, N.L.; Dalgaard, L.T. Extracellular microRNAs in Relation to Weight Loss—A Systematic Review and Meta-Analysis. Non-Coding RNA 2023, 9, 53. [Google Scholar] [CrossRef]
- Hess, A.L.; Larsen, L.H.; Udesen, P.B.; Sanz, Y.; Larsen, T.M.; Dalgaard, L.T. Levels of circulating miR-122 are associated with weight loss and metabolic syndrome. Obesity 2020, 28, 493–501. [Google Scholar] [CrossRef]
- Filardi, T.; Sabato, C.; Lubrano, C.; Santangelo, C.; Morano, S.; Lenzi, A.; Migliaccio, S.; Ferretti, E.; Catanzaro, G. MicroRNA modulation by dietary supplements in obesity. Biomedicines 2020, 8, 545. [Google Scholar] [CrossRef]
- Tian, L.; Song, Z.; Shao, W.; Du, W.W.; Zhao, L.R.; Zeng, K.; Yang, B.B.; Jin, T. Curcumin represses mouse 3T3-L1 cell adipogenic differentiation via inhibiting miR-17-5p and stimulating the Wnt signalling pathway effector Tcf7l2. Cell Death Dis. 2018, 8, e2559. [Google Scholar] [CrossRef] [PubMed]
- Fatima, S.; Khan, D.A.; Fatima, F.; Aamir, M.; Ijaz, A.; Hafeez, A. Role of δ-tocotrienol and resveratrol supplementation in the regulation of micro RNAs in patients with metabolic syndrome: A randomized controlled trial. Complement. Ther. Med. 2023, 74, 102950. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, M.; Silva, D.; de Morais, A., Jr.; Mota, J.; Botelho, P. Therapeutic potential of green tea on risk factors for type 2 diabetes in obese adults–a review. Obes. Rev. 2016, 17, 1316–1328. [Google Scholar] [CrossRef] [PubMed]
- Bastos, R.V.; Dorna, M.S.; Chiuso-Minicucci, F.; Felix, T.F.; Fernandes, A.A.; Azevedo, P.S.; Franco, E.T.; Polegato, B.F.; Rogero, M.M.; Mota, G.A.; et al. Acute green tea intake attenuates circulating microRNA expression induced by a high-fat, high-saturated meal in obese women: A randomized crossover study. J. Nutr. Biochem. 2023, 112, 109203. [Google Scholar] [CrossRef]
- Reis, B.Z.; Duarte, G.B.S.; Vargas-Mendez, E.; Ferreira, L.R.P.; Barbosa, F., Jr.; Cercato, C.; Rogero, M.M.; Cozzolino, S.M.F. Brazil nut intake increases circulating miR-454-3p and miR-584-5p in obese women. Nutr. Res. 2019, 67, 40–52. [Google Scholar] [CrossRef]
- Assmann, T.S.; Riezu-Boj, J.I.; Milagro, F.I.; Martínez, J.A. Circulating adiposity-related microRNAs as predictors of the response to a low-fat diet in subjects with obesity. J. Cell. Mol. Med. 2020, 24, 2956–2967. [Google Scholar] [CrossRef]
- Duggan, C.; Tapsoba, J.d.D.; Scheel, J.; Wang, C.-Y.; McTiernan, A. Weight loss reduces circulating micro-RNA related to obesity and breast cancer in postmenopausal women. Epigenetics 2022, 17, 2082–2095. [Google Scholar] [CrossRef]
- Ojeda-Rodríguez, A.; Assmann, T.S.; Alonso-Pedrero, L.; Azcona-Sanjulian, M.C.; Milagro, F.I.; Marti, A. Circulating miRNAs in girls with abdominal obesity: miR-221-3p as a biomarker of response to weight loss interventions. Pediatr. Obes. 2022, 17, e12910. [Google Scholar] [CrossRef]
- Manning, P.; Munasinghe, P.E.; Bellae Papannarao, J.; Gray, A.R.; Sutherland, W.; Katare, R. Acute weight loss restores dysregulated circulating microRNAs in individuals who are obese. J. Clin. Endocrinol. Metab. 2019, 104, 1239–1248. [Google Scholar] [CrossRef]
- Heianza, Y.; Xue, Q.; Rood, J.; Bray, G.A.; Sacks, F.M.; Qi, L. Circulating thrifty microRNA is related to insulin sensitivity, adiposity, and energy metabolism in adults with overweight and obesity: The POUNDS Lost trial. Am. J. Clin. Nutr. 2023, 117, 121–129. [Google Scholar] [CrossRef]
- Hernández-Gómez, K.G.; Velázquez-Villegas, L.A.; Granados-Portillo, O.; Avila-Nava, A.; González-Salazar, L.E.; Serralde-Zúñiga, A.E.; Palacios-González, B.; Pichardo-Ontiveros, E.; Guizar-Heredia, R.; López-Barradas, A.M.; et al. Acute Effects of Dietary Protein Consumption on the Postprandial Metabolic Response, Amino Acid Levels and Circulating MicroRNAs in Patients with Obesity and Insulin Resistance. Int. J. Mol. Sci. 2024, 25, 7716. [Google Scholar] [CrossRef] [PubMed]
- Javaid, A.; Zahra, D.; Rashid, F.; Mashraqi, M.; Alzamami, A.; Khurshid, M.; Ashfaq, U.A. Regulation of micro-RNA, epigenetic factor by natural products for the treatment of cancers: Mechanistic insight and translational association. Saudi J. Biol. Sci. 2022, 29, 103255. [Google Scholar] [CrossRef] [PubMed]
- Jia, M.; He, J.; Bai, W.; Lin, Q.; Deng, J.; Li, W.; Bai, J.; Fu, D.; Ma, Y.; Ren, J.; et al. Cross-kingdom regulation by dietary plant miRNAs: An evidence-based review with recent updates. Food Funct. 2021, 12, 9549–9562. [Google Scholar] [CrossRef] [PubMed]
- Shah, K.B.; Chernausek, S.D.; Garman, L.D.; Pezant, N.P.; Plows, J.F.; Kharoud, H.K.; Demerath, E.W.; Fields, D.A. Human milk exosomal microRNA: Associations with maternal overweight/obesity and infant body composition at 1 month of life. Nutrients 2021, 13, 1091. [Google Scholar] [CrossRef]
- Tingö, L.; Ahlberg, E.; Johansson, L.; Pedersen, S.A.; Chawla, K.; Sætrom, P.; Cione, E.; Simpson, M.R. Non-coding RNAs in human breast milk: A systematic review. Front. Immunol. 2021, 12, 725323. [Google Scholar] [CrossRef]
- Çelik, E.; Cemali, Ö.; Şahin, T.Ö.; Deveci, G.; Biçer, N.Ç.; Hirfanoğlu, İ.M.; Ağagündüz, D.; Budán, F. Human breast milk exosomes: Affecting factors, their possible health outcomes, and future directions in dietetics. Nutrients 2024, 16, 3519. [Google Scholar] [CrossRef]
- Soleimani, M.; Barone, S.; Luo, H.; Zahedi, K. Pathogenesis of hypertension in metabolic syndrome: The role of fructose and salt. Int. J. Mol. Sci. 2023, 24, 4294. [Google Scholar] [CrossRef]
- He, L.; Liu, Y.; Widlansky, M.E.; Kriegel, A.J.; Qiu, Q.; Liang, M. microRNA and Hypertension. Hypertension 2025, 82, 181–184. [Google Scholar] [CrossRef]
- Kriegel, A.J.; Baker, M.A.; Liu, Y.; Liu, P.; Cowley, A.W., Jr.; Liang, M. Endogenous microRNAs in human microvascular endothelial cells regulate mRNAs encoded by hypertension-related genes. Hypertension 2015, 66, 793–799. [Google Scholar] [CrossRef]
- Kostiniuk, D.; Marttila, S.; Raitoharju, E. Circulatory miRNAs in essential hypertension. Atherosclerosis 2025, 401, 119069. [Google Scholar] [CrossRef]
- Altıntaş, N.; Tonk, O.; Yılmaz, Ö.S. Effects of microRNAs in hypertension disease. Eur. Res. J. 2022, 8, 131–138. [Google Scholar] [CrossRef]
- Matshazi, D.M.; Weale, C.J.; Erasmus, R.T.; Kengne, A.P.; Davids, S.F.; Raghubeer, S.; Davison, G.M.; Matsha, T.E. Circulating levels of MicroRNAs associated with hypertension: A cross-sectional study in male and female South African participants. Front. Genet. 2021, 12, 710438. [Google Scholar] [CrossRef] [PubMed]
- Wojtasińska, A.; Frąk, W.; Lisińska, W.; Sapeda, N.; Młynarska, E.; Rysz, J.; Franczyk, B. Novel insights into the molecular mechanisms of atherosclerosis. Int. J. Mol. Sci. 2023, 24, 13434. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Luo, Y.; Gao, X.; Liang, Y.; Yang, F.; Wu, J.; Fang, D.; Luo, M. MicroRNA regulation of phenotypic transformations in vascular smooth muscle: Relevance to vascular remodeling. Cell. Mol. Life Sci. 2023, 80, 144. [Google Scholar] [CrossRef]
- Zhang, X.; Ng, W.-L.; Wang, P.; Tian, L.; Werner, E.; Wang, H.; Doetsch, P.; Wang, Y. MicroRNA-21 modulates the levels of reactive oxygen species by targeting SOD3 and TNF α. Cancer Res. 2012, 72, 4707–4713. [Google Scholar] [CrossRef]
- Adamcova, M.; Kawano, I.; Simko, F. The impact of microRNAs in renin–angiotensin-system-induced cardiac remodelling. Int. J. Mol. Sci. 2021, 22, 4762. [Google Scholar] [CrossRef]
- Ren, X.-S.; Tong, Y.; Qiu, Y.; Ye, C.; Wu, N.; Xiong, X.-Q.; Wang, J.-J.; Han, Y.; Zhou, Y.-B.; Zhang, F.; et al. MiR155-5p in adventitial fibroblasts-derived extracellular vesicles inhibits vascular smooth muscle cell proliferation via suppressing angiotensin-converting enzyme expression. J. Extracell. Vesicles 2020, 9, 1698795. [Google Scholar] [CrossRef]
- Kohlstedt, K.; Trouvain, C.; Boettger, T.; Shi, L.; Fisslthaler, B.; Fleming, I. AMP-activated protein kinase regulates endothelial cell angiotensin-converting enzyme expression via p53 and the post-transcriptional regulation of microRNA-143/145. Circ. Res. 2013, 112, 1150–1158. [Google Scholar] [CrossRef]
- Yaacoub, S.; Boudaka, A.; AlKhatib, A.; Pintus, G.; Sahebkar, A.; Kobeissy, F.; Eid, A.H. The pharmaco-epigenetics of hypertension: A focus on microRNA. Mol. Cell. Biochem. 2024, 479, 3255–3271. [Google Scholar] [CrossRef]
- Sun, Y.; Shang, Q. Research hotspots and trends regarding microRNAs in hypertension: A bibliometric analysis. Clin. Exp. Hypertens. 2024, 46, 2304017. [Google Scholar] [CrossRef]
- Li, J.; Zhao, F.; Wang, Y.; Chen, J.; Tao, J.; Tian, G.; Wu, S.; Liu, W.; Cui, Q.; Geng, B.; et al. Gut microbiota dysbiosis contributes to the development of hypertension. Microbiome 2017, 5, 14. [Google Scholar] [CrossRef] [PubMed]
- Golonka, R.M.; Cooper, J.K.; Issa, R.; Devarasetty, P.P.; Gokula, V.; Busken, J.; Zubcevic, J.; Hill, J.; Vijay-Kumar, M.; Menon, B.; et al. Impact of nutritional epigenetics in essential hypertension: Targeting microRNAs in the gut-liver axis. Curr. Hypertens. Rep. 2021, 23, 28. [Google Scholar] [CrossRef] [PubMed]
- Covic, A.; Bilha, S.C.; Scripcariu, D.; Kanbay, M. Salt and Hypertension. J. Hypertens. Res. 2019, 5, 65–75. [Google Scholar]
- He, F.J.; Tan, M.; Ma, Y.; MacGregor, G.A. Salt reduction to prevent hypertension and cardiovascular disease: JACC state-of-the-art review. J. Am. Coll. Cardiol. 2020, 75, 632–647. [Google Scholar] [CrossRef]
- Nemecz, M.; Alexandru, N.; Tanko, G.; Georgescu, A. Role of microRNA in endothelial dysfunction and hypertension. Curr. Hypertens. Rep. 2016, 18, 87. [Google Scholar] [CrossRef]
- Guo, T.-S.; Zhang, J.; Mu, J.-J.; Liu, F.-Q.; Yuan, Z.-Y.; Ren, K.-Y.; Wang, D. High-salt intake suppressed microRNA-133a expression in Dahl SS rat myocardium. Int. J. Mol. Sci. 2014, 15, 10794–10805. [Google Scholar] [CrossRef]
- Chen, L.; He, F.J.; Dong, Y.; Huang, Y.; Harshfield, G.A.; Zhu, H. Sodium reduction, miRNA profiling and CVD risk in untreated hypertensives: A randomized, double-blind, placebo-controlled trial. Sci. Rep. 2018, 8, 12729. [Google Scholar] [CrossRef]
- Hao, S.; Hao, M.; Ferreri, N.R. Renal-specific silencing of TNF (tumor necrosis factor) unmasks salt-dependent increases in blood pressure via an NKCC2A (Na+-K+-2Cl− cotransporter isoform A)-dependent mechanism. Hypertension 2018, 71, 1117–1125. [Google Scholar] [CrossRef]
- Hao, S.; Zhao, H.; Hao, D.H.; Ferreri, N.R. MicroRNA-195a-5p regulates blood pressure by inhibiting NKCC2A. Hypertension 2023, 80, 426–439. [Google Scholar] [CrossRef]
- Nierenberg, J.L.; Li, C.; He, J.; Gu, D.; Chen, J.; Lu, X.; Li, J.; Wu, X.; Gu, C.C.; Hixson, J.E.; et al. Blood pressure genetic risk score predicts blood pressure responses to dietary sodium and potassium: The GenSalt study (genetic epidemiology network of salt sensitivity). Hypertension 2017, 70, 1106–1112. [Google Scholar] [CrossRef]
- Zhang, X.; Yao, S.; Wang, Y.; Chu, C.; Du, M.; Mu, J. Associations of microRNA Gene Polymorphisms with Salt Sensitivity, Longitudinal Blood Pressure Changes, and Hypertension Incidence in the Chinese Population. J. Clin. Hypertens. 2025, 27, e70019. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Mao, L.; Zuo, M.-L.; Song, G.-L.; Tan, L.-M.; Yang, Z.-B. The role of microRNAs in hyperlipidemia: From pathogenesis to therapeutical application. Mediat. Inflamm. 2022, 2022, 3101900. [Google Scholar] [CrossRef] [PubMed]
- Párrizas, M.; Novials, A. Circulating microRNAs as biomarkers for metabolic disease. Best Pract. Res. Clin. Endocrinol. Metab. 2016, 30, 591–601. [Google Scholar] [CrossRef] [PubMed]
- Silva, D.C.P.d.; Carneiro, F.D.; Almeida, K.C.d.; Fernandes-Santos, C. Role of miRNAs on the Pathophysiology of Cardiovascular Diseases. Arq. Bras. Cardiol. 2018, 111, 738–746. [Google Scholar] [CrossRef]
- Fernández-Tussy, P.; Ruz-Maldonado, I.; Fernández-Hernando, C. MicroRNAs and circular RNAs in lipoprotein metabolism. Curr. Atheroscler. Rep. 2021, 23, 33. [Google Scholar] [CrossRef]
- Khan, A.A.; Agarwal, H.; Reddy, S.S.; Arige, V.; Natarajan, B.; Gupta, V.; Kalyani, A.; Barthwal, M.K.; Mahapatra, N.R. MicroRNA 27a is a key modulator of cholesterol biosynthesis. Mol. Cell. Biol. 2020, 40, e00470-19. [Google Scholar] [CrossRef]
- Ono, K. Functions of microRNA-33a/b and microRNA therapeutics. J. Cardiol. 2016, 67, 28–33. [Google Scholar] [CrossRef]
- Zhang, X.; Price, N.L.; Fernández-Hernando, C. Non-coding RNAs in lipid metabolism. Vasc. Pharmacol. 2019, 114, 93–102. [Google Scholar] [CrossRef]
- Singh, A.K.; Aryal, B.; Zhang, X.; Fan, Y.; Price, N.L.; Suárez, Y.; Fernández-Hernando, C. Posttranscriptional regulation of lipid metabolism by non-coding RNAs and RNA binding proteins. Semin. Cell Dev. Biol. 2018, 81, 129–140. [Google Scholar] [CrossRef]
- Mokhtari Ardekani, A.; Mohammadzadehsaliani, S.; Behrouj, H.; Moridi, H.; Moradi, M.N.; Ghasemi, H. miR-122 dysregulation is associated with type 2 diabetes mellitus-induced dyslipidemia and hyperglycemia independently of its rs17669 variant. Mol. Biol. Rep. 2023, 50, 4217–4224. [Google Scholar] [CrossRef]
- Flowers, E.; Froelicher, E.S.; Aouizerat, B.E. MicroRNA regulation of lipid metabolism. Metabolism 2013, 62, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Salamat, S.; Jahan-Mihan, A.; Tabandeh, M.R.; Mansoori, A. Effects of Multi-species Synbiotic Supplementation on Circulating miR-27a, miR-33a Levels, and Lipid Parameters in Adult Men with Dyslipidemia; A Randomized, Double-Blind, Placebo-Controlled Clinical Trial. Br. J. Nutr. 2024, 133, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Kim, G.J.; Umemura, T.; Lee, S.G.; Cho, K.J. Aberrant expression of plasma microRNA-33a in an atherosclerosis-risk group. Mol. Biol. Rep. 2017, 44, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Simionescu, N.; Niculescu, L.S.; Sanda, G.M.; Margina, D.; Sima, A.V. Analysis of circulating microRNAs that are specifically increased in hyperlipidemic and/or hyperglycemic sera. Mol. Biol. Rep. 2014, 41, 5765–5773. [Google Scholar] [CrossRef]
- Alvarez, M.L.; Khosroheidari, M.; Eddy, E.; Done, S.C. MicroRNA-27a decreases the level and efficiency of the LDL receptor and contributes to the dysregulation of cholesterol homeostasis. Atherosclerosis 2015, 242, 595–604. [Google Scholar] [CrossRef]
- Vickers, K.C.; Palmisano, B.T.; Shoucri, B.M.; Shamburek, R.D.; Remaley, A.T. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 2011, 13, 423–433. [Google Scholar] [CrossRef]
- Marquart, T.J.; Allen, R.M.; Ory, D.S.; Baldán, Á. miR-33 links SREBP-2 induction to repression of sterol transporters. Proc. Natl. Acad. Sci. USA 2010, 107, 12228–12232. [Google Scholar] [CrossRef]
- Najafi-Shoushtari, S.H.; Kristo, F.; Li, Y.; Shioda, T.; Cohen, D.E.; Gerszten, R.E.; Näär, A.M. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science 2010, 328, 1566–1569. [Google Scholar] [CrossRef]
- Rayner, K.J.; Suárez, Y.; Dávalos, A.; Parathath, S.; Fitzgerald, M.L.; Tamehiro, N.; Fisher, E.A.; Moore, K.J.; Fernández-Hernando, C. MiR-33 contributes to the regulation of cholesterol homeostasis. Science 2010, 328, 1570–1573. [Google Scholar] [CrossRef]
- Rayner, K.J.; Esau, C.C.; Hussain, F.N.; McDaniel, A.L.; Marshall, S.M.; van Gils, J.M.; Ray, T.D.; Sheedy, F.J.; Goedeke, L.; Liu, X.; et al. Inhibition of miR-33a/b in non-human primates raises plasma HDL and lowers VLDL triglycerides. Nature 2011, 478, 404–407. [Google Scholar] [CrossRef]
- Price, N.L.; Singh, A.K.; Rotllan, N.; Goedeke, L.; Wing, A.; Canfrán-Duque, A.; Diaz-Ruiz, A.; Araldi, E.; Baldán, Á.; Camporez, J.-P.; et al. Genetic ablation of miR-33 increases food intake, enhances adipose tissue expansion, and promotes obesity and insulin resistance. Cell Rep. 2018, 22, 2133–2145. [Google Scholar] [CrossRef] [PubMed]
- Esau, C.; Davis, S.; Murray, S.F.; Yu, X.X.; Pandey, S.K.; Pear, M.; Watts, L.; Booten, S.L.; Graham, M.; McKay, R.; et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006, 3, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Rashad, N.M.; Ezzat, T.M.; Allam, R.M.; Soliman, M.H.; Yousef, M.S. The expression level of microRNA-122 in patients with type 2 diabetes mellitus in correlation with risk and severity of coronary artery disease. Egypt. J. Intern. Med. 2019, 31, 593–601. [Google Scholar] [CrossRef]
- Gao, W.; He, H.-W.; Wang, Z.-M.; Zhao, H.; Lian, X.-Q.; Wang, Y.-S.; Zhu, J.; Yan, J.-J.; Zhang, D.-G.; Yang, Z.-J.; et al. Plasma levels of lipometabolism-related miR-122 and miR-370 are increased in patients with hyperlipidemia and associated with coronary artery disease. Lipids Health Dis. 2012, 11, 55. [Google Scholar] [CrossRef]
- Krützfeldt, J.; Rajewsky, N.; Braich, R.; Rajeev, K.G.; Tuschl, T.; Manoharan, M.; Stoffel, M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature 2005, 438, 685–689. [Google Scholar] [CrossRef]
- Novák, J.; Bienertová-Vašků, J.; Kára, T.; Novák, M. MicroRNAs involved in the lipid metabolism and their possible implications for atherosclerosis development and treatment. Mediat. Inflamm. 2014, 2014, 275867. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, X.; Li, H.; Yu, J.; Ren, X. The role of miRNA-29 family in cancer. Eur. J. Cell Biol. 2013, 92, 123–128. [Google Scholar] [CrossRef]
- Goedeke, L.; Wagschal, A.; Fernández-Hernando, C.; Näär, A.M. miRNA regulation of LDL-cholesterol metabolism. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2016, 1861, 2047–2052. [Google Scholar] [CrossRef]
- Goedeke, L.; Rotllan, N.; Canfrán-Duque, A.; Aranda, J.F.; Ramírez, C.M.; Araldi, E.; Lin, C.-S.; Anderson, N.N.; Wagschal, A.; De Cabo, R.; et al. MicroRNA-148a regulates LDL receptor and ABCA1 expression to control circulating lipoprotein levels. Nat. Med. 2015, 21, 1280–1289. [Google Scholar] [CrossRef]
- Canfrán-Duque, A.; Lin, C.-S.; Goedeke, L.; Suárez, Y.; Fernández-Hernando, C. Micro-RNAs and high-density lipoprotein metabolism. Arterioscler. Thromb. Vasc. Biol. 2016, 36, 1076–1084. [Google Scholar] [CrossRef]
- Badawy, H.K.; Abo-Elmatty, D.M.; Mesbah, N.M. Association between serum microRNA-605 and microRNA-623 expression and essential hypertension in Egyptian patients. Meta Gene 2018, 16, 62–65. [Google Scholar] [CrossRef]
- Nemecz, M.; Stefan, D.S.; Comarița, I.K.; Constantin, A.; Tanko, G.; Guja, C.; Georgescu, A. Microvesicle-associated and circulating microRNAs in diabetic dyslipidemia: miR-218, miR-132, miR-143, and miR-21, miR-122, miR-155 have biomarker potential. Cardiovasc. Diabetol. 2023, 22, 260. [Google Scholar] [CrossRef] [PubMed]
- Giardina, S.; Hernández-Alonso, P.; Díaz-López, A.; Salas-Huetos, A.; Salas-Salvadó, J.; Bulló, M. Changes in circulating miRNAs in healthy overweight and obese subjects: Effect of diet composition and weight loss. Clin. Nutr. 2019, 38, 438–443. [Google Scholar] [CrossRef] [PubMed]
- De Candia, P.; Spinetti, G.; Specchia, C.; Sangalli, E.; La Sala, L.; Uccellatore, A.; Lupini, S.; Genovese, S.; Matarese, G.; Ceriello, A. A unique plasma microRNA profile defines type 2 diabetes progression. PLoS ONE 2017, 12, e0188980. [Google Scholar] [CrossRef]
- Wang, R.; Hong, J.; Cao, Y.; Shi, J.; Gu, W.; Ning, G.; Zhang, Y.; Wang, W. Elevated circulating microRNA-122 is associated with obesity and insulin resistance in young adults. Eur. J. Endocrinol. 2015, 172, 291–300. [Google Scholar] [CrossRef]
- Krause, B.J.; Carrasco-Wong, I.; Dominguez, A.; Arnaiz, P.; Farias, M.; Barja, S.; Mardones, F.; Casanello, P. Micro-RNAs Let7e and 126 in plasma as markers of metabolic dysfunction in 10 to 12 years old children. PLoS ONE 2015, 10, e0128140. [Google Scholar] [CrossRef]
- Prats-Puig, A.; Ortega, F.J.; Mercader, J.M.; Moreno-Navarrete, J.M.; Moreno, M.; Bonet, N.; Ricart, W.; López-Bermejo, A.; Fernández-Real, J.M. Changes in circulating microRNAs are associated with childhood obesity. J. Clin. Endocrinol. Metab. 2013, 98, E1655–E1660. [Google Scholar] [CrossRef]
- Zempleni, J.; Baier, S.R.; Howard, K.M.; Cui, J. Gene regulation by dietary microRNAs. Can. J. Physiol. Pharmacol. 2015, 93, 1097–1102. [Google Scholar] [CrossRef]
- Sanchita; Trivedi, R.; Asif, M.H.; Trivedi, P.K. Dietary plant miRNAs as an augmented therapy: Cross-kingdom gene regulation. RNA Biol. 2018, 15, 1433–1439. [Google Scholar] [CrossRef]
- Zhao, Q.; Liu, Y.; Zhang, N.; Hu, M.; Zhang, H.; Joshi, T.; Xu, D. Correction: Evidence for plant-derived xenomiRs based on a large-scale analysis of public small RNA sequencing data from human samples. PLoS ONE 2020, 15, e0230146. [Google Scholar] [CrossRef]
- Dickinson, B.; Zhang, Y.; Petrick, J.S.; Heck, G.; Ivashuta, S.; Marshall, W.S. Lack of detectable oral bioavailability of plant microRNAs after feeding in mice. Nat. Biotechnol. 2013, 31, 965–967. [Google Scholar] [CrossRef] [PubMed]
- Smet, E.D.; Mensink, R.P.; Plat, J. Effects of plant sterols and stanols on intestinal cholesterol metabolism: Suggested mechanisms from past to present. Mol. Nutr. Food Res. 2012, 56, 1058–1072. [Google Scholar] [CrossRef] [PubMed]
- Protic, O.; Di Pillo, R.; Montesanto, A.; Galeazzi, R.; Matacchione, G.; Giuliani, A.; Sabbatinelli, J.; Gurău, F.; Silvestrini, A.; Olivieri, F.; et al. Randomized, Double-Blind, placebo-controlled trial to test the effects of a Nutraceutical Combination Monacolin K-Free on the lipid and Inflammatory Profile of subjects with hypercholesterolemia. Nutrients 2022, 14, 2812. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Jia, M.; Xu, L.; Fang, Z.; Wu, W.; Zhang, Q.; Chung, P.; Lin, Y.; Wang, S.; Zhang, Y. miR-96 and autophagy are involved in the beneficial effect of grape seed proanthocyanidins against high-fat-diet-induced dyslipidemia in mice. Phytother. Res. 2019, 33, 1222–1232. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).