NucEnvDB: A Database of Nuclear Envelope Proteins and Their Interactions
Abstract
1. Introduction
2. Materials and Methods
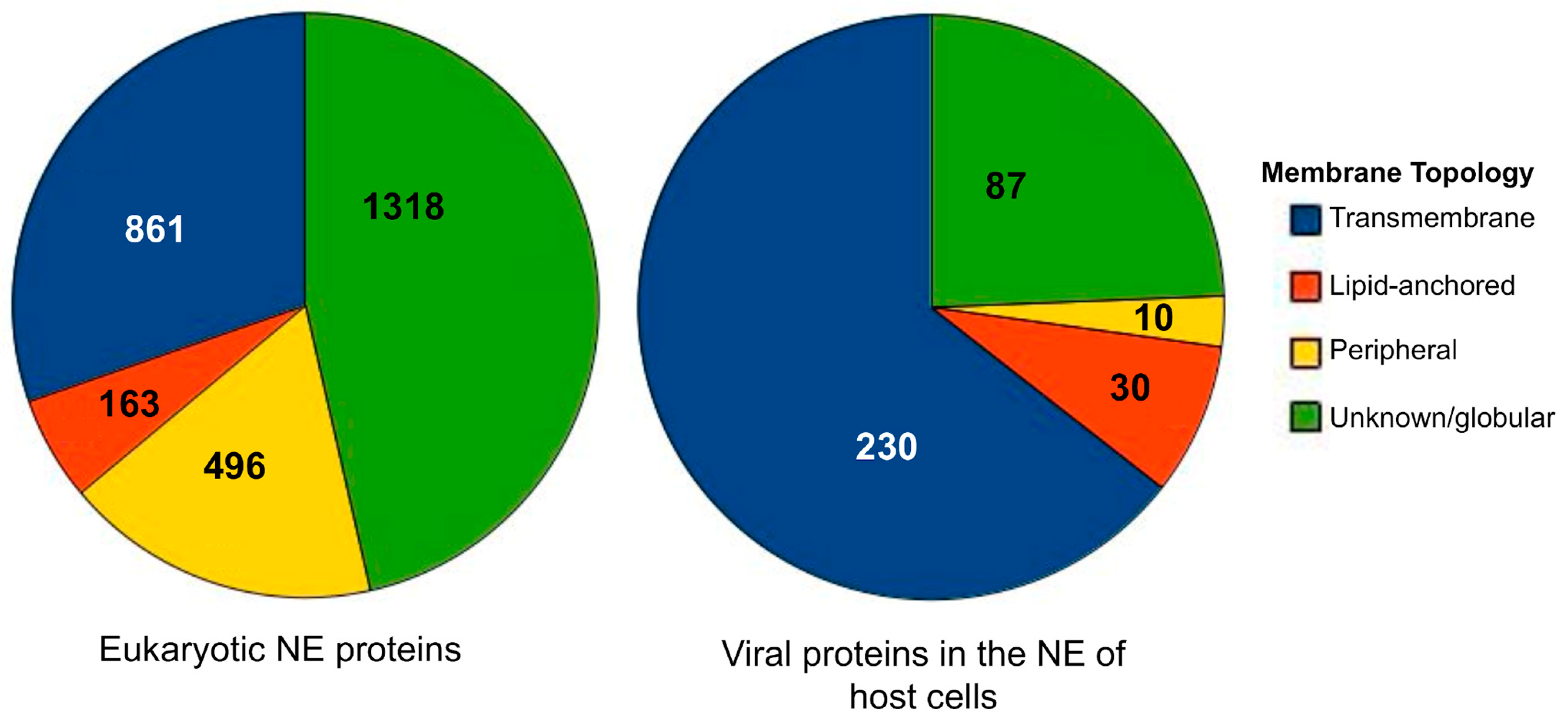
2.1. Data Collection, Annotation, and Classification
2.2. Database Organization and Implementation
2.3. Case Studies
3. Results and Discussion
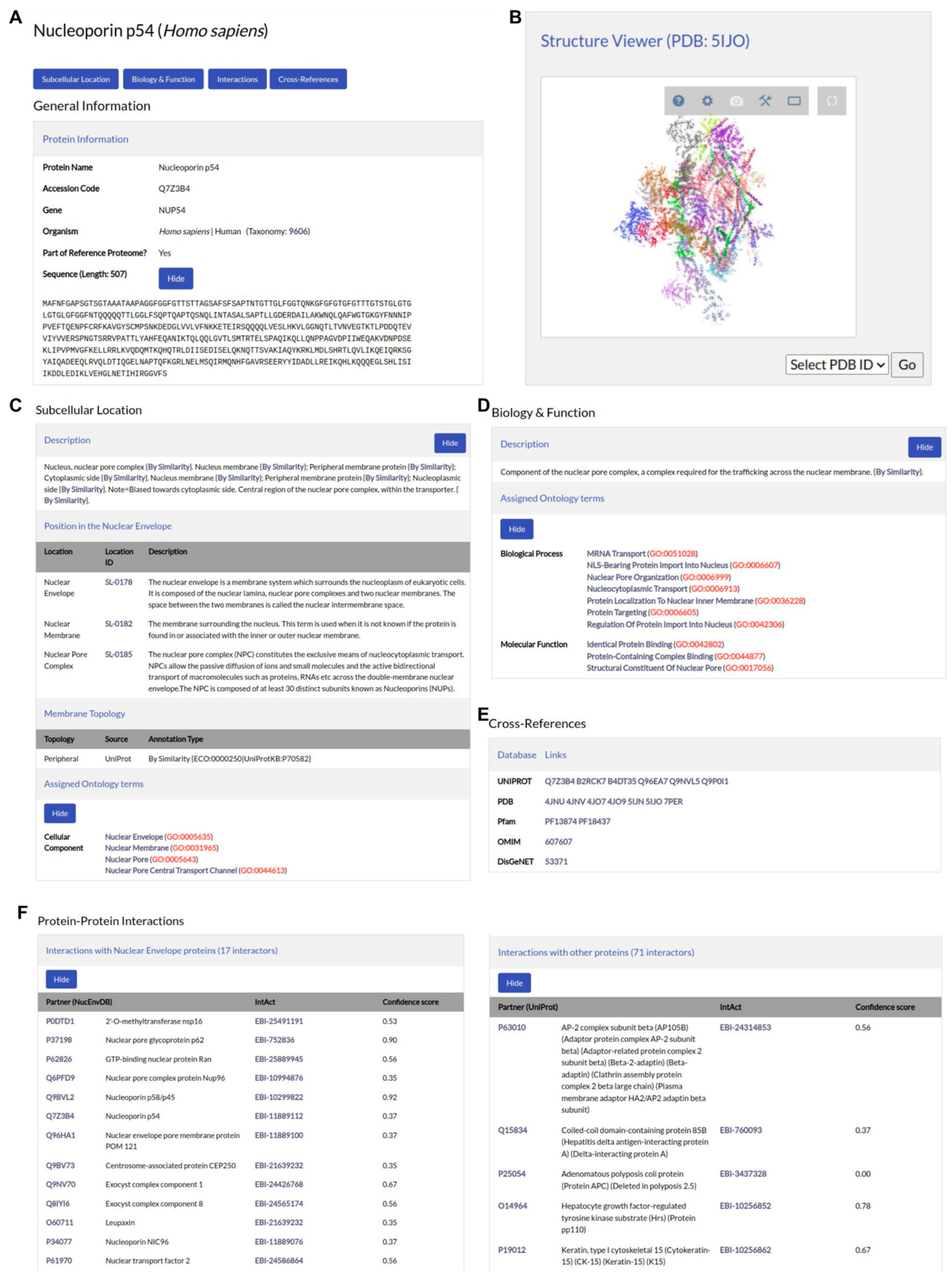
3.1. Database Components
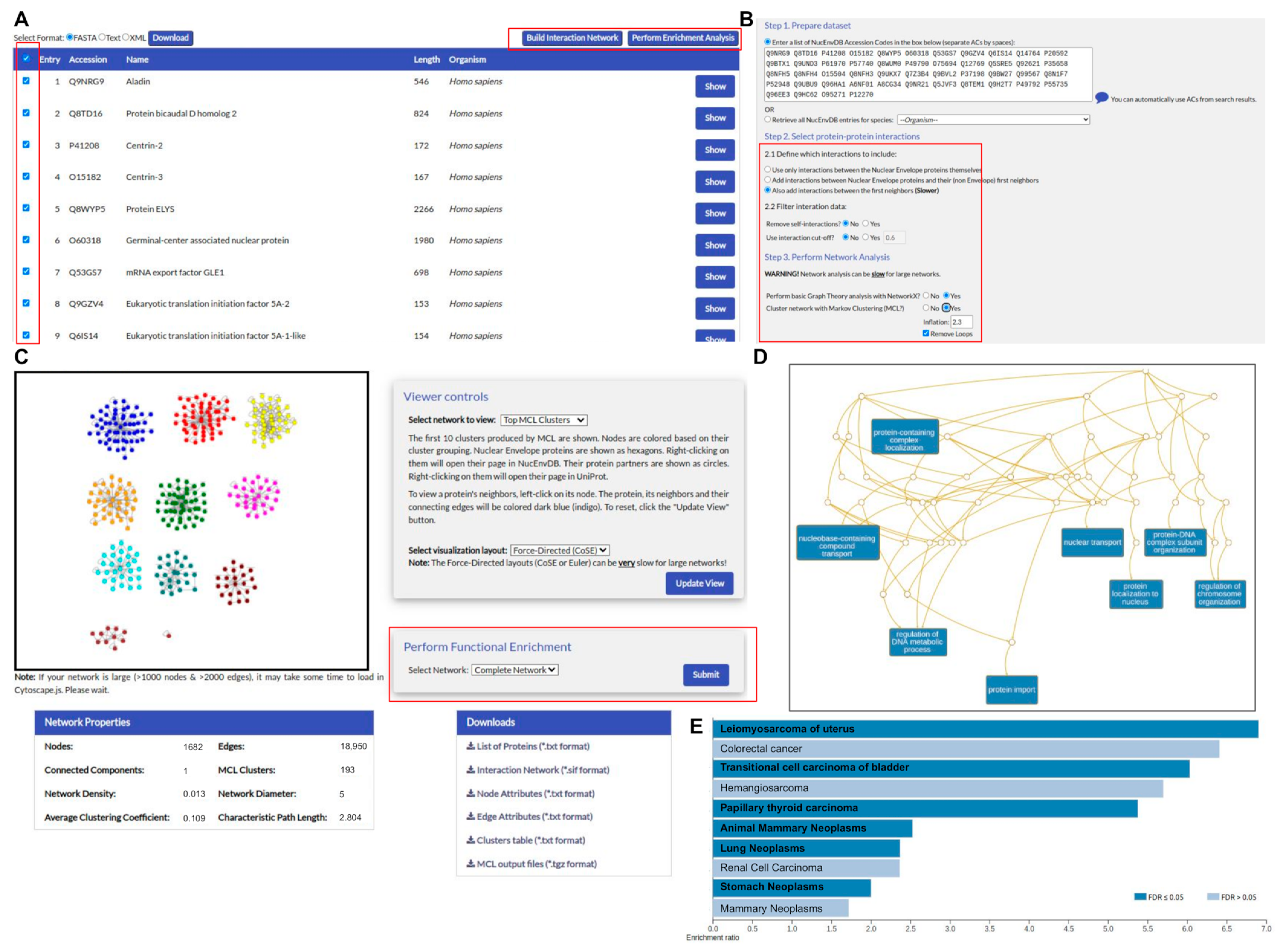
3.2. User Interface
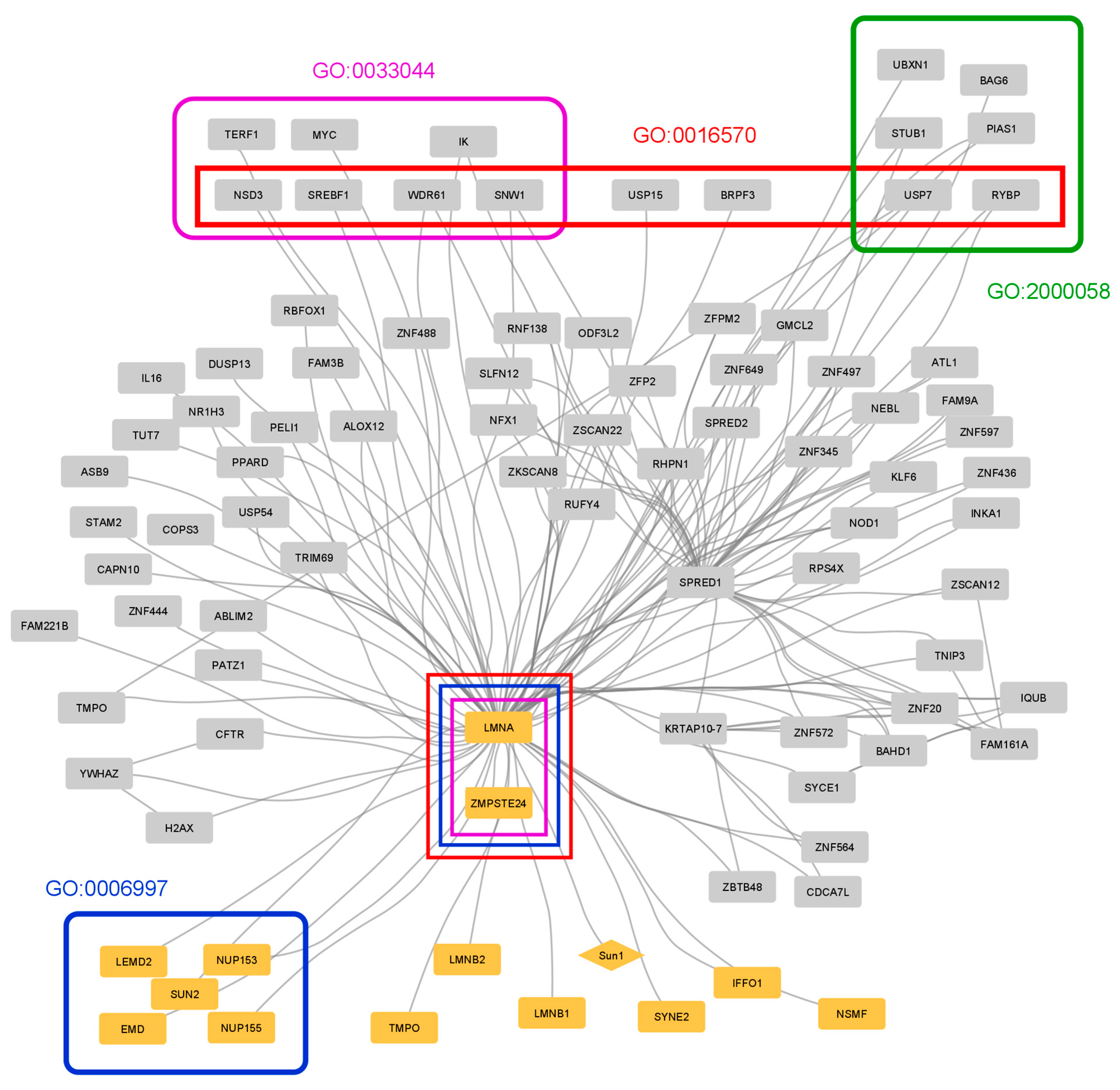
3.3. Case Study 1: Protein–Protein Interactions in Hutchinson–Gilford Progeria
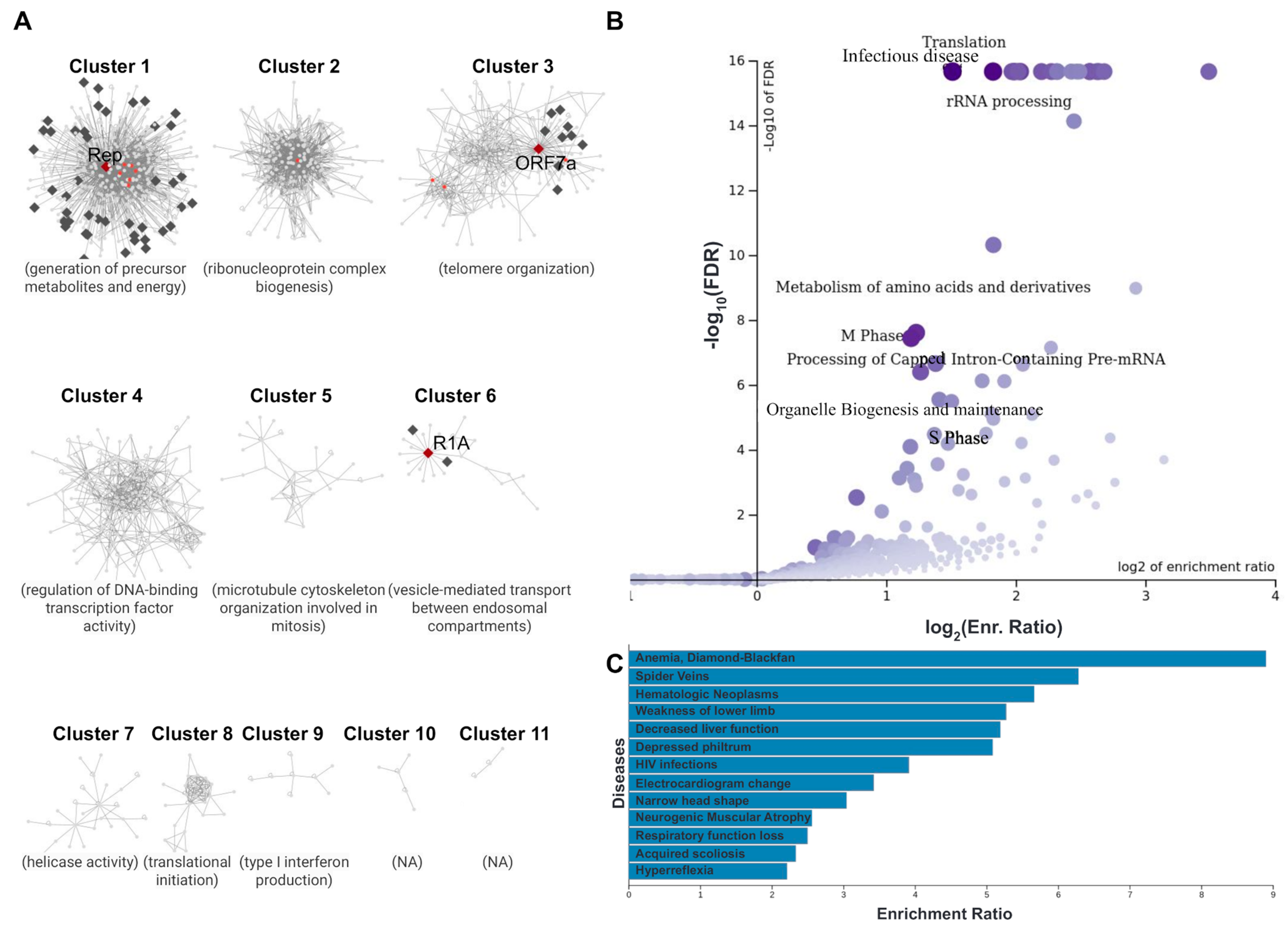
3.4. Case Study 2: Host–Pathogen Interactions at the Nuclear Envelope of Cells Infected with SARS-CoV-2
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wilson, K.L.; Berk, J.M. The Nuclear Envelope at a Glance. J. Cell. Sci. 2010, 123, 1973–1978. [Google Scholar] [CrossRef] [PubMed]
- Hetzer, M.W. The Nuclear Envelope. Cold Spring Harb. Perspect. Biol. 2010, 2, a000539. [Google Scholar] [CrossRef] [PubMed]
- Holmer, L.; Worman, H.J. Inner Nuclear Membrane Proteins: Functions and Targeting. Cell. Mol. Life Sci. 2001, 58, 1741–1747. [Google Scholar] [CrossRef] [PubMed]
- Katta, S.S.; Smoyer, C.J.; Jaspersen, S.L. Destination: Inner Nuclear Membrane. Trends Cell. Biol. 2014, 24, 221–229. [Google Scholar] [CrossRef]
- Lusk, C.P.; Blobel, G.; King, M.C. Highway to the Inner Nuclear Membrane: Rules for the Road. Nat. Rev. Mol. Cell. Biol. 2007, 8, 414–420. [Google Scholar] [CrossRef]
- Bouzid, T.; Kim, E.; Riehl, B.D.; Esfahani, A.M.; Rosenbohm, J.; Yang, R.; Duan, B.; Lim, J.Y. The LINC Complex, Mechanotransduction, and Mesenchymal Stem Cell Function and Fate. J. Biol. Eng. 2019, 13, 68. [Google Scholar] [CrossRef]
- Chen, I.-H.B.; Huber, M.; Guan, T.; Bubeck, A.; Gerace, L. Nuclear Envelope Transmembrane Proteins (NETs) That Are up-Regulated during Myogenesis. BMC Cell. Biol. 2006, 7, 38. [Google Scholar] [CrossRef]
- Wang, Y.-N.; Yamaguchi, H.; Hsu, J.-M.; Hung, M.-C. Nuclear Trafficking of the Epidermal Growth Factor Receptor Family Membrane Proteins. Oncogene 2010, 29, 3997–4006. [Google Scholar] [CrossRef]
- Wu, S.C.; O’Connell, T.D. Nuclear Compartmentalization of A1-Adrenergic Receptor Signaling in Adult Cardiac Myocytes. J. Cardiovasc. Pharm. 2015, 65, 91–100. [Google Scholar] [CrossRef]
- Vaniotis, G.; Del Duca, D.; Trieu, P.; Rohlicek, C.V.; Hébert, T.E.; Allen, B.G. Nuclear β-Adrenergic Receptors Modulate Gene Expression in Adult Rat Heart. Cell Signal. 2011, 23, 89–98. [Google Scholar] [CrossRef]
- Coucke, P.J.; Willaert, A.; Wessels, M.W.; Callewaert, B.; Zoppi, N.; De Backer, J.; Fox, J.E.; Mancini, G.M.S.; Kambouris, M.; Gardella, R.; et al. Mutations in the Facilitative Glucose Transporter GLUT10 Alter Angiogenesis and Cause Arterial Tortuosity Syndrome. Nat. Genet. 2006, 38, 452–457. [Google Scholar] [CrossRef]
- Rogers, S.; Macheda, M.L.; Docherty, S.E.; Carty, M.D.; Henderson, M.A.; Soeller, W.C.; Gibbs, E.M.; James, D.E.; Best, J.D. Identification of a Novel Glucose Transporter-like Protein-GLUT-12. Am. J. Physiol. Endocrinol. Metab. 2002, 282, E733–E738. [Google Scholar] [CrossRef]
- Burdine, R.D.; Preston, C.C.; Leonard, R.J.; Bradley, T.A.; Faustino, R.S. Nucleoporins in Cardiovascular Disease. J. Mol. Cell. Cardiol. 2020, 141, 43–52. [Google Scholar] [CrossRef]
- Ross, J.A.; Stroud, M.J. THE NUCLEUS: Mechanosensing in Cardiac Disease. Int. J. Biochem. Cell. Biol. 2021, 137, 106035. [Google Scholar] [CrossRef] [PubMed]
- Birks, S.; Uzer, G. At the Nuclear Envelope of Bone Mechanobiology. Bone 2021, 151, 116023. [Google Scholar] [CrossRef]
- Battey, E.; Stroud, M.J.; Ochala, J. Using Nuclear Envelope Mutations to Explore Age-Related Skeletal Muscle Weakness. Clin. Sci. 2020, 134, 2177–2187. [Google Scholar] [CrossRef]
- Hachiya, N.; Sochocka, M.; Brzecka, A.; Shimizu, T.; Gąsiorowski, K.; Szczechowiak, K.; Leszek, J. Nuclear Envelope and Nuclear Pore Complexes in Neurodegenerative Diseases-New Perspectives for Therapeutic Interventions. Mol. Neurobiol. 2021, 58, 983–995. [Google Scholar] [CrossRef]
- Mettenleiter, T.C. Breaching the Barrier—The Nuclear Envelope in Virus Infection. J. Mol. Biol. 2016, 428, 1949–1961. [Google Scholar] [CrossRef]
- Dellaire, G.; Farrall, R.; Bickmore, W.A. The Nuclear Protein Database (NPD): Sub-Nuclear Localisation and Functional Annotation of the Nuclear Proteome. Nucleic Acids Res. 2003, 31, 328–330. [Google Scholar] [CrossRef]
- Choura, M.; Rebaï, A. Application of Computational Approaches to Study Signalling Networks of Nuclear and Tyrosine Kinase Receptors. Biol. Direct. 2010, 5, 58. [Google Scholar] [CrossRef]
- The UniProt Consortium; Bateman, A.; Martin, M.-J.; Orchard, S.; Magrane, M.; Agivetova, R.; Ahmad, S.; Alpi, E.; Bowler-Barnett, E.H.; Britto, R.; et al. UniProt: The Universal Protein Knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a Reference Resource for Gene and Protein Annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef] [PubMed]
- Fabregat, A.; Jupe, S.; Matthews, L.; Sidiropoulos, K.; Gillespie, M.; Garapati, P.; Haw, R.; Jassal, B.; Korninger, F.; May, B.; et al. The Reactome Pathway Knowledgebase. Nucleic Acids Res. 2018, 46, D649–D655. [Google Scholar] [CrossRef] [PubMed]
- Pontén, F.; Jirström, K.; Uhlen, M. The Human Protein Atlas—A Tool for Pathology. J. Pathol. 2008, 216, 387–393. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the Unification of Biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- The Gene Ontology Consortium; Carbon, S.; Douglass, E.; Good, B.M.; Unni, D.R.; Harris, N.L.; Mungall, C.J.; Basu, S.; Chisholm, R.L.; Dodson, R.J.; et al. The Gene Ontology Resource: Enriching a GOld Mine. Nucleic Acids Res. 2021, 49, D325–D334. [Google Scholar] [CrossRef]
- Huntley, R.P.; Sawford, T.; Mutowo-Meullenet, P.; Shypitsyna, A.; Bonilla, C.; Martin, M.J.; O’Donovan, C. The GOA Database: Gene Ontology Annotation Updates for 2015. Nucleic Acids Res. 2015, 43, D1057–D1063. [Google Scholar] [CrossRef]
- Amberger, J.S.; Bocchini, C.A.; Scott, A.F.; Hamosh, A. OMIM.Org: Leveraging Knowledge across Phenotype-Gene Relationships. Nucleic Acids Res. 2019, 47, D1038–D1043. [Google Scholar] [CrossRef]
- Piñero, J.; Ramírez-Anguita, J.M.; Saüch-Pitarch, J.; Ronzano, F.; Centeno, E.; Sanz, F.; Furlong, L.I. The DisGeNET Knowledge Platform for Disease Genomics: 2019 Update. Nucleic Acids Res. 2020, 48, D845–D855. [Google Scholar] [CrossRef]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for Clustering the next-Generation Sequencing Data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- Del Toro, N.; Shrivastava, A.; Ragueneau, E.; Meldal, B.; Combe, C.; Barrera, E.; Perfetto, L.; How, K.; Ratan, P.; Shirodkar, G.; et al. The IntAct Database: Efficient Access to Fine-Grained Molecular Interaction Data. Nucleic Acids Res. 2022, 50, D648–D653. [Google Scholar] [CrossRef] [PubMed]
- Orchard, S.; Ammari, M.; Aranda, B.; Breuza, L.; Briganti, L.; Broackes-Carter, F.; Campbell, N.H.; Chavali, G.; Chen, C.; del-Toro, N.; et al. The MIntAct Project—IntAct as a Common Curation Platform for 11 Molecular Interaction Databases. Nucleic Acids Res. 2014, 42, D358–D363. [Google Scholar] [CrossRef] [PubMed]
- Sehnal, D.; Deshpande, M.; Vařeková, R.S.; Mir, S.; Berka, K.; Midlik, A.; Pravda, L.; Velankar, S.; Koča, J. LiteMol Suite: Interactive Web-Based Visualization of Large-Scale Macromolecular Structure Data. Nat. Methods 2017, 14, 1121–1122. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Eddy, S.R. Accelerated Profile HMM Searches. PLOS Comput. Biol. 2011, 7, e1002195. [Google Scholar] [CrossRef]
- Franz, M.; Lopes, C.T.; Huck, G.; Dong, Y.; Sumer, O.; Bader, G.D. Cytoscape.Js: A Graph Theory Library for Visualisation and Analysis. Bioinformatics 2016, 32, 309–311. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Hagberg, A.A.; Schult, D.A.; Swart, P.J. Exploring Network Structure, Dynamics, and Function Using NetworkX. In Proceedings of the 7th Python in Science Conference, Pasadena, CA, USA, 21 August 2008; Varoquaux, G., Vaught, T., Millman, J., Varoquaux, G., Vaught, T., Millman, J., Eds.; 2008; pp. 11–15. [Google Scholar]
- Enright, A.J. An Efficient Algorithm for Large-Scale Detection of Protein Families. Nucleic Acids Res. 2002, 30, 1575–1584. [Google Scholar] [CrossRef]
- Liao, Y.; Wang, J.; Jaehnig, E.J.; Shi, Z.; Zhang, B. WebGestalt 2019: Gene Set Analysis Toolkit with Revamped UIs and APIs. Nucleic Acids Res. 2019, 47, W199–W205. [Google Scholar] [CrossRef]
- Pollex, R.; Hegele, R. Hutchinson-Gilford Progeria Syndrome: Hutchinson-Gilford Progeria Syndrome. Clin. Genet. 2004, 66, 375–381. [Google Scholar] [CrossRef] [PubMed]
- Gonzalo, S.; Kreienkamp, R.; Askjaer, P. Hutchinson-Gilford Progeria Syndrome: A Premature Aging Disease Caused by LMNA Gene Mutations. Ageing Res. Rev. 2017, 33, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Porras, P.; Barrera, E.; Bridge, A.; del-Toro, N.; Cesareni, G.; Duesbury, M.; Hermjakob, H.; Iannuccelli, M.; Jurisica, I.; Kotlyar, M.; et al. Towards a Unified Open Access Dataset of Molecular Interactions. Nat. Commun. 2020, 11, 6144. [Google Scholar] [CrossRef] [PubMed]
- Assenov, Y.; Ramírez, F.; Schelhorn, S.-E.; Lengauer, T.; Albrecht, M. Computing Topological Parameters of Biological Networks. Bioinformatics 2008, 24, 282–284. [Google Scholar] [CrossRef]
- Morris, J.H.; Apeltsin, L.; Newman, A.M.; Baumbach, J.; Wittkop, T.; Su, G.; Bader, G.D.; Ferrin, T.E. ClusterMaker: A Multi-Algorithm Clustering Plugin for Cytoscape. BMC Bioinform. 2011, 12, 436. [Google Scholar] [CrossRef]
- Su, G.; Kuchinsky, A.; Morris, J.H.; States, D.J.; Meng, F. GLay: Community Structure Analysis of Biological Networks. Bioinformatics 2010, 26, 3135–3137. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The Protein Families Database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Goldman, R.D.; Shumaker, D.K.; Erdos, M.R.; Eriksson, M.; Goldman, A.E.; Gordon, L.B.; Gruenbaum, Y.; Khuon, S.; Mendez, M.; Varga, R.; et al. Accumulation of Mutant Lamin A Causes Progressive Changes in Nuclear Architecture in Hutchinson–Gilford Progeria Syndrome. Proc. Natl. Acad. Sci. USA 2004, 101, 8963–8968. [Google Scholar] [CrossRef]
- Yang, S.H.; Andres, D.A.; Spielmann, H.P.; Young, S.G.; Fong, L.G. Progerin Elicits Disease Phenotypes of Progeria in Mice Whether or Not It Is Farnesylated. J. Clin. Investig. 2008, 118, 3291–3300. [Google Scholar] [CrossRef]
- Schreiber, K.H.; Kennedy, B.K. When Lamins Go Bad: Nuclear Structure and Disease. Cell 2013, 152, 1365–1375. [Google Scholar] [CrossRef]
- Johne, C.; Matenia, D.; Li, X.; Timm, T.; Balusamy, K.; Mandelkow, E.-M. Spred1 and TESK1—Two New Interaction Partners of the Kinase MARKK/TAO1 That Link the Microtubule and Actin Cytoskeleton. MBoC 2008, 19, 1391–1403. [Google Scholar] [CrossRef] [PubMed]
- Stenvall, C.-G.A.; Nyström, J.H.; Butler-Hallissey, C.; Jansson, T.; Heikkilä, T.R.H.; Adam, S.A.; Foisner, R.; Goldman, R.D.; Ridge, K.M.; Toivola, D.M. Cytoplasmic Keratins Couple with and Maintain Nuclear Envelope Integrity in Colonic Epithelial Cells. MBoC 2022, 33, ar121. [Google Scholar] [CrossRef] [PubMed]
- Barrowman, J.; Wiley, P.A.; Hudon-Miller, S.E.; Hrycyna, C.A.; Michaelis, S. Human ZMPSTE24 Disease Mutations: Residual Proteolytic Activity Correlates with Disease Severity. Hum. Mol. Genet. 2012, 21, 4084–4093. [Google Scholar] [CrossRef] [PubMed]
- COVID-19 Data Explorer. Our World in Data. Available online: https://ourworldindata.org/explorers/coronavirus-data-explorer (accessed on 27 December 2022).
- Harrison, A.G.; Lin, T.; Wang, P. Mechanisms of SARS-CoV-2 Transmission and Pathogenesis. Trends Immunol. 2020, 41, 1100–1115. [Google Scholar] [CrossRef]
- Chen, M.; Ma, Y.; Chang, W. SARS-CoV-2 and the Nucleus. Int. J. Biol. Sci. 2022, 18, 4731–4743. [Google Scholar] [CrossRef] [PubMed]
- Uddin, M.H.; Zonder, J.A.; Azmi, A.S. Exportin 1 Inhibition as Antiviral Therapy. Drug Discov. Today 2020, 25, 1775–1781. [Google Scholar] [CrossRef]
- Burke, J.M.; St Clair, L.A.; Perera, R.; Parker, R. SARS-CoV-2 Infection Triggers Widespread Host MRNA Decay Leading to an MRNA Export Block. RNA 2021, 27, 1318–1329. [Google Scholar] [CrossRef]
- Li, J.; Guo, M.; Tian, X.; Wang, X.; Yang, X.; Wu, P.; Liu, C.; Xiao, Z.; Qu, Y.; Yin, Y.; et al. Virus-Host Interactome and Proteomic Survey Reveal Potential Virulence Factors Influencing SARS-CoV-2 Pathogenesis. Med 2021, 2, 99–112.e7. [Google Scholar] [CrossRef]
- Gordon, D.E.; Jang, G.M.; Bouhaddou, M.; Xu, J.; Obernier, K.; White, K.M.; O’Meara, M.J.; Rezelj, V.V.; Guo, J.Z.; Swaney, D.L.; et al. A SARS-CoV-2 Protein Interaction Map Reveals Targets for Drug Repurposing. Nature 2020, 583, 459–468. [Google Scholar] [CrossRef]
- Aslan, A.; Aslan, C.; Zolbanin, N.M.; Jafari, R. Acute Respiratory Distress Syndrome in COVID-19: Possible Mechanisms and Therapeutic Management. Pneumonia 2021, 13, 14. [Google Scholar] [CrossRef]
- Abdelrahman, M.M.; Abdel-baset, A.A.; Younis, M.A.; Mahmoud, M.G.; Shafik, N.S. Liver Function Test Abnormalities in COVID-19 Patients and Factors Affecting Them—A Retrospective Study. Clin. Exp. Hepatol. 2021, 7, 297–304. [Google Scholar] [CrossRef] [PubMed]
- Galyfos, G.; Sianou, A.; Frountzas, M.; Vasilios, K.; Vouros, D.; Theodoropoulos, C.; Michalopoulou, V.; Sigala, F.; Filis, K. Acute Limb Ischemia among Patients with COVID-19 Infection. J. Vasc. Surg. 2022, 75, 326–342. [Google Scholar] [CrossRef] [PubMed]
- Roy, N.B.A.; Telfer, P.; Eleftheriou, P.; de la Fuente, J.; Drasar, E.; Shah, F.; Roberts, D.; Atoyebi, W.; Trompeter, S.; Layton, D.M.; et al. Protecting Vulnerable Patients with Inherited Anaemias from Unnecessary Death during the COVID-19 Pandemic. Br. J. Haematol. 2020, 189, 635–639. [Google Scholar] [CrossRef] [PubMed]
- Leemans, W.; Antonis, S.; De Vooght, W.; Lemmens, R.; Van Damme, P. Neuromuscular Complications after COVID-19 Vaccination: A Series of Eight Patients. Acta Neurol. Belg. 2022, 122, 753–761. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, P.K.; Agrawal, C.; Blunden, G. Artemisia Extracts and Artemisinin-Based Antimalarials for COVID-19 Management: Could These Be Effective Antivirals for COVID-19 Treatment? Molecules 2022, 27, 3828. [Google Scholar] [CrossRef] [PubMed]
- Barh, D.; Tiwari, S.; Weener, M.E.; Azevedo, V.; Góes-Neto, A.; Gromiha, M.M.; Ghosh, P. Multi-Omics-Based Identification of SARS-CoV-2 Infection Biology and Candidate Drugs against COVID-19. Comput. Biol. Med. 2020, 126, 104051. [Google Scholar] [CrossRef]
- Rahnavard, A.; Mann, B.; Giri, A.; Chatterjee, R.; Crandall, K.A. Metabolite, Protein, and Tissue Dysfunction Associated with COVID-19 Disease Severity. Sci. Rep. 2022, 12, 12204. [Google Scholar] [CrossRef]
| Subcellular Location | SL-ID | Description | Proteins |
|---|---|---|---|
| Nuclear Envelope | SL-0178 | The complex membrane system that surrounds the nucleoplasm. It is composed of two membranes, the nuclear lamina, and nuclear pore complexes. The space between the two membranes is called the nuclear intermembrane space. | 1551 |
| Nuclear Membrane | SL-0182 | The membrane surrounding the nucleus. This term is used when it is not known if the protein is found in or associated with the inner or outer nuclear membrane. | 1037 |
| Nuclear Inner Membrane | SL-0179 | The inner membrane of the nucleus is the membrane which separates the nuclear matrix from the intermembrane space. | 211 |
| Nuclear Outer Membrane | SL-0183 | The outer membrane of the nucleus is the membrane facing the cytoplasm. | 89 |
| Nuclear Intermembrane Space | SL-0184 | The nuclear intermembrane space is the space between the inner and outer nuclear membranes. | 4 |
| Nuclear Pore Complex | SL-0185 | The nuclear pore complex (NPC) constitutes the exclusive means of nucleocytoplasmic transport. It is composed of at least 30 distinct subunits known as nucleoporins (NUPs). | 389 |
| Nuclear Lamina | SL-0180 | The nuclear lamina is a meshwork of intermediate filament proteins called lamins and lamin-binding proteins that are embedded in the inner nuclear membrane. | 32 |
| Perinuclear Region | SL-0198 | The perinuclear region is the cytoplasmic region just around the nucleus. | 1347 |
| Host Nuclear Envelope 1 | SL-0415 | Equivalent of SL-0178 for viral proteins that target the nuclear envelope of host cells. | 93 |
| Host Nuclear Membrane 1 | SL-0182 | Equivalent of SL-0178 for viral proteins that target the nuclear membrane of host cells. | 85 |
| Host Nuclear Inner Membrane 1 | SL-0419 | Equivalent of SL-0179 for viral proteins that target the inner nuclear membrane of host cells. | 57 |
| Host Nuclear Outer Membrane 1 | SL-0446 | Equivalent of SL-0183 for viral proteins that target the outer nuclear membrane of host cells. | 0 |
| Host Nuclear Lamina 1 | SL-0416 | Equivalent of SL-0180 for viral proteins that target the nuclear lamina of host cells. | 7 |
| Host Perinuclear Region 1 | SL-0382 | Equivalent of SL-0198 for viral proteins that target the perinuclear region of host cells. | 231 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baltoumas, F.A.; Sofras, D.; Apostolakou, A.E.; Litou, Z.I.; Iconomidou, V.A. NucEnvDB: A Database of Nuclear Envelope Proteins and Their Interactions. Membranes 2023, 13, 62. https://doi.org/10.3390/membranes13010062
Baltoumas FA, Sofras D, Apostolakou AE, Litou ZI, Iconomidou VA. NucEnvDB: A Database of Nuclear Envelope Proteins and Their Interactions. Membranes. 2023; 13(1):62. https://doi.org/10.3390/membranes13010062
Chicago/Turabian StyleBaltoumas, Fotis A., Dimitrios Sofras, Avgi E. Apostolakou, Zoi I. Litou, and Vassiliki A. Iconomidou. 2023. "NucEnvDB: A Database of Nuclear Envelope Proteins and Their Interactions" Membranes 13, no. 1: 62. https://doi.org/10.3390/membranes13010062
APA StyleBaltoumas, F. A., Sofras, D., Apostolakou, A. E., Litou, Z. I., & Iconomidou, V. A. (2023). NucEnvDB: A Database of Nuclear Envelope Proteins and Their Interactions. Membranes, 13(1), 62. https://doi.org/10.3390/membranes13010062





