The Interaction of Anthracycline Based Quinone-Chelators with Model Lipid Membranes: 1H NMR and MD Study
Abstract
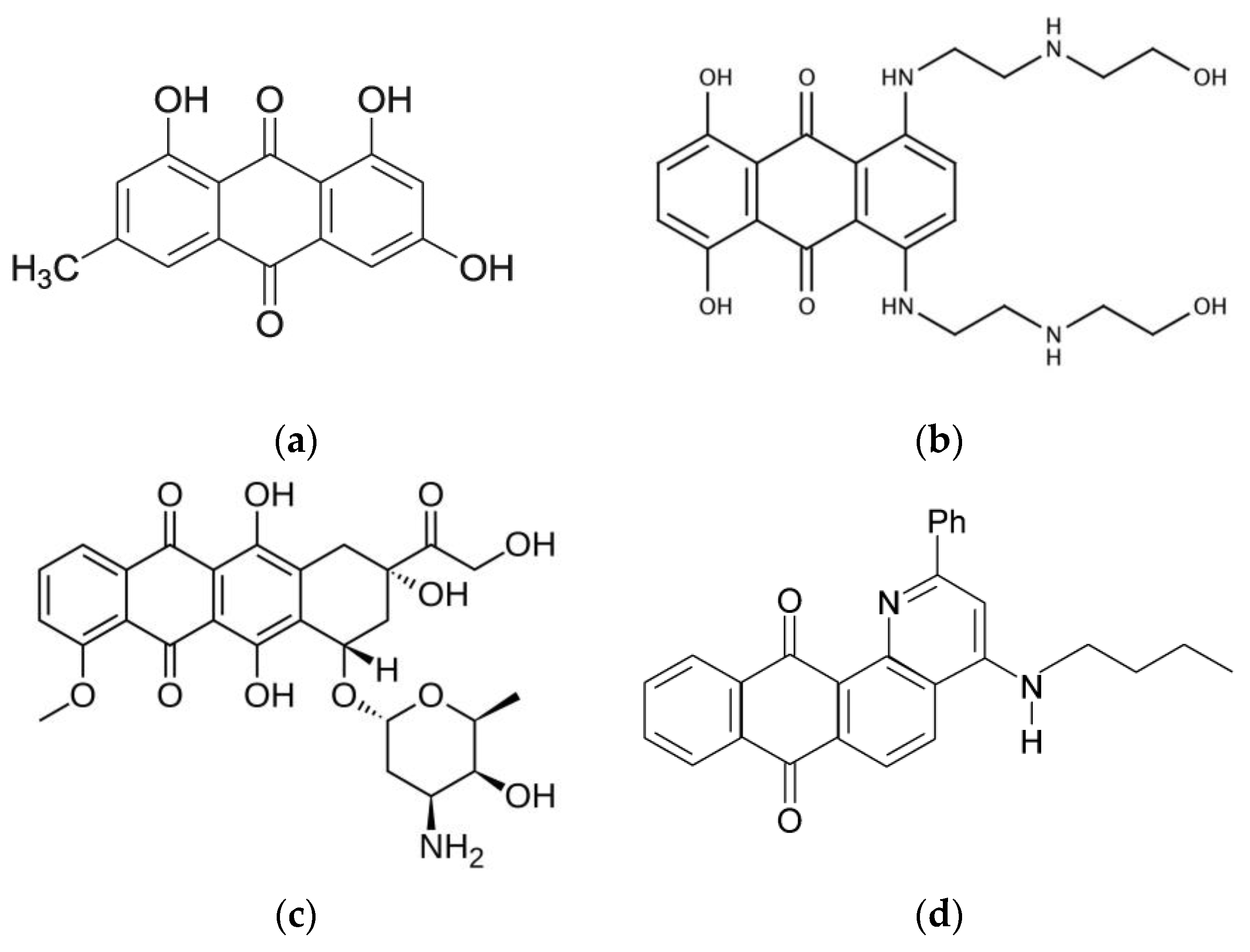
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. NMR Study
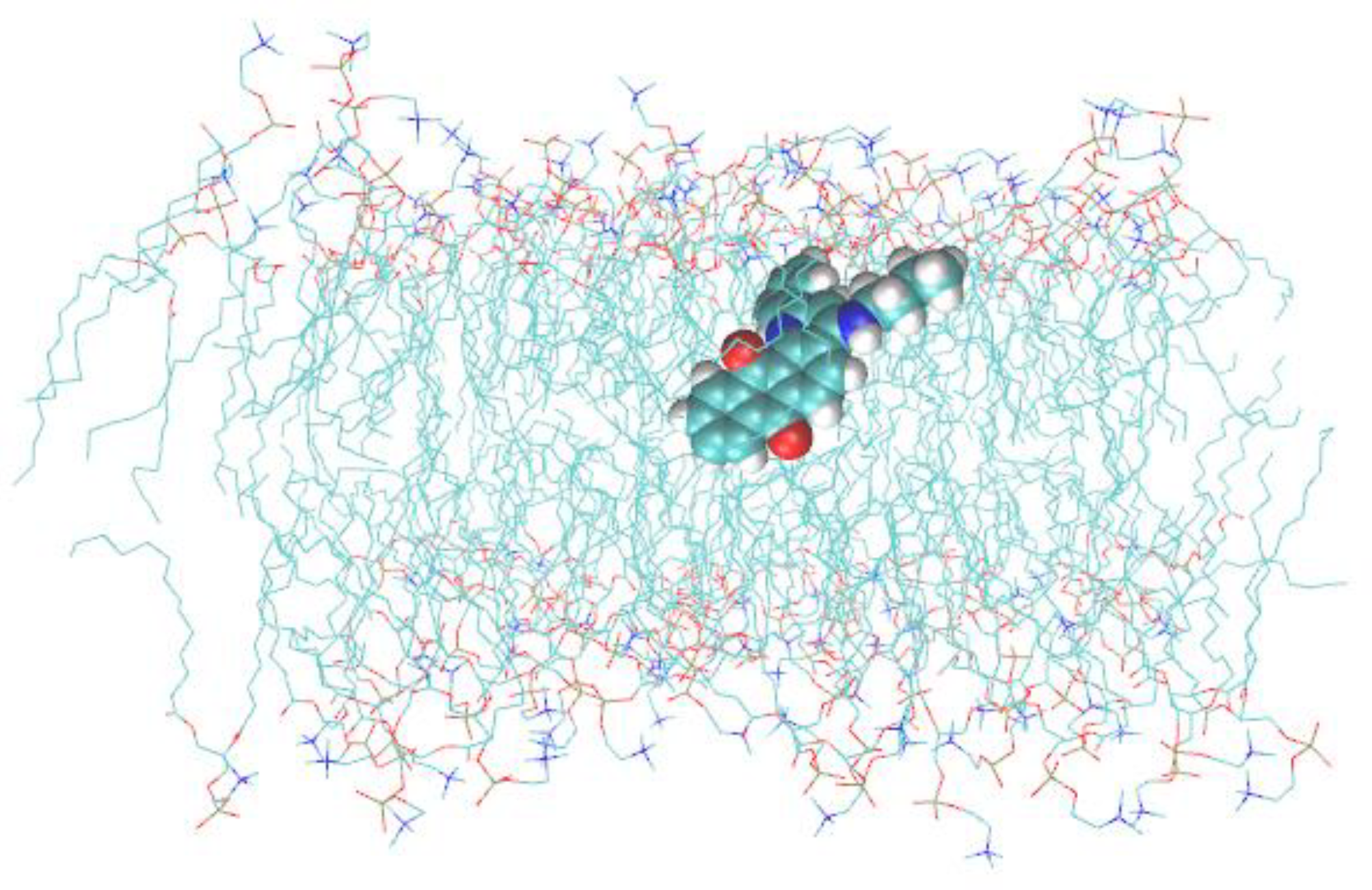
2.3. Molecular Dynamics Simulations
2.4. Relative Lipophilicity (log P) Determination
3. Results and Discussion
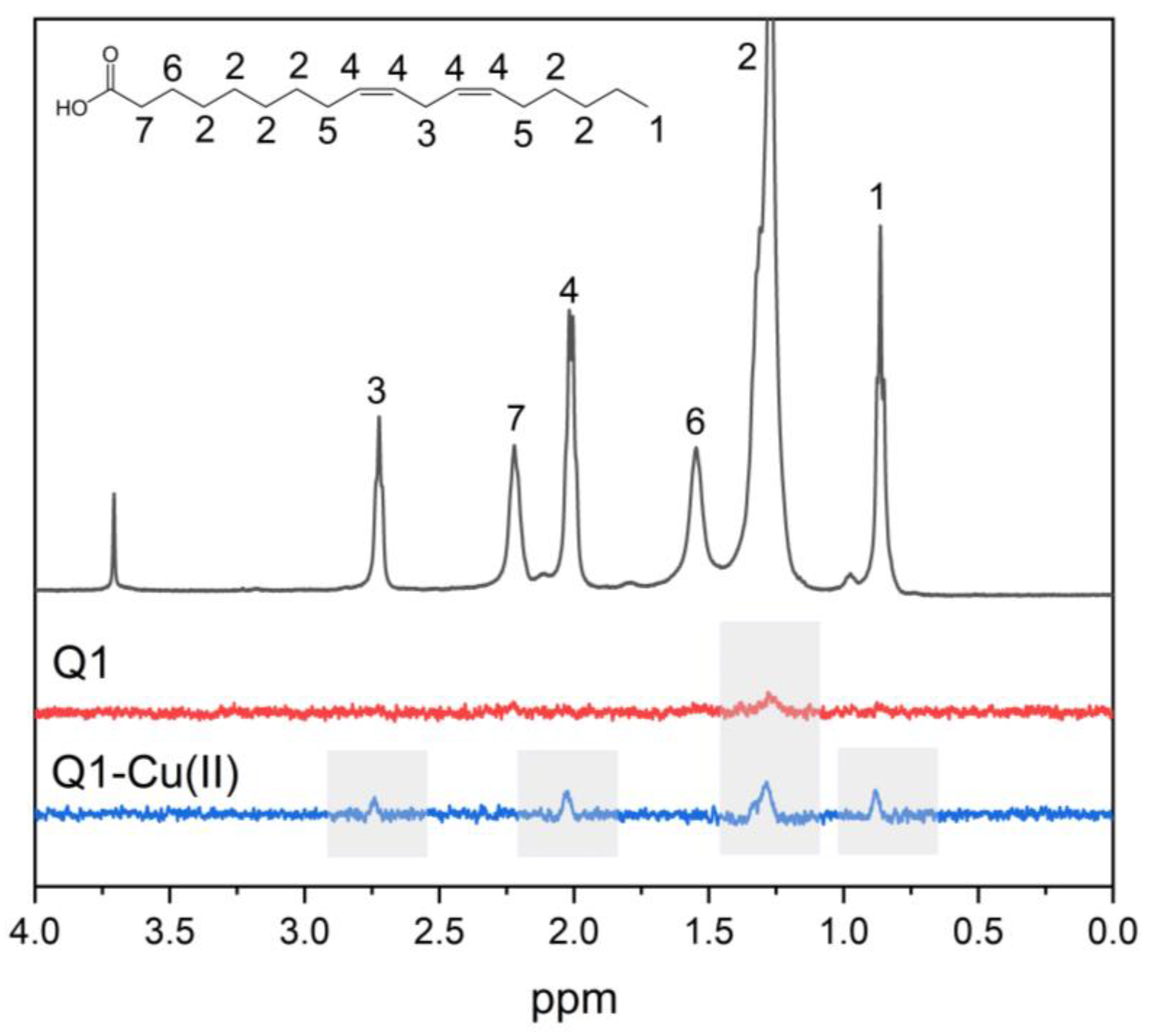
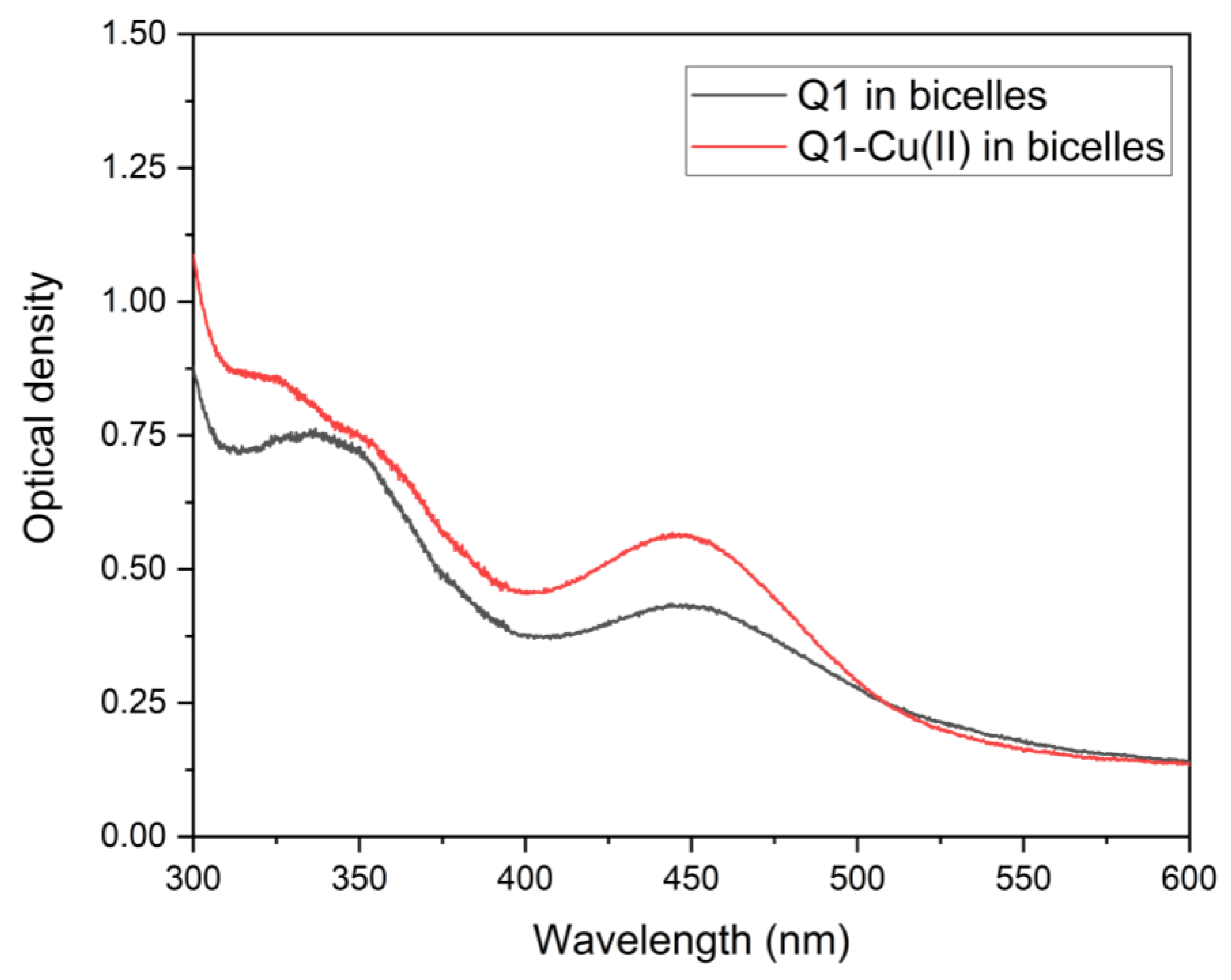
3.1. Q1 and Q1−Cu(II) Interaction with Linoleic Acid Micells
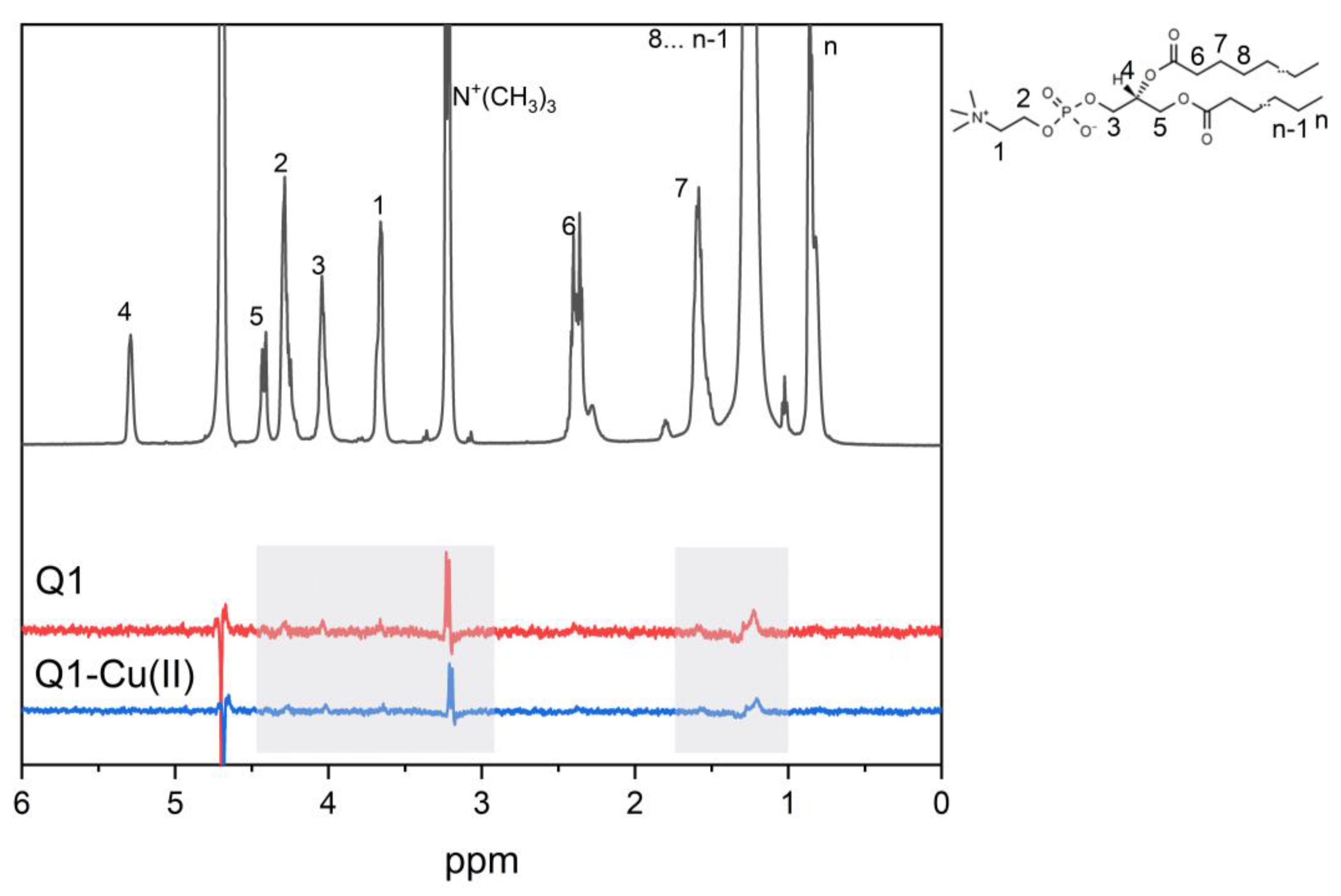
3.2. Q1 and Q1−Cu(II) Interaction with DMPC/DHPC Bicelles
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Frézard, F.; Garnier-Suillerot, A. Permeability of lipid bilayer to anthracycline derivatives. Role of the bilayer composition and of the temperature. Biochim. Biophys. Acta—Lipids Lipid Metab. 1998, 1389, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Toroz, D.; Gould, I.R. A computational study of Anthracyclines interacting with lipid bilayers: Correlation of membrane insertion rates, orientation effects and localisation with cytotoxicity. Sci. Rep. 2019, 9, 2155. [Google Scholar] [CrossRef] [PubMed]
- Powis, G. Free radical formation by antitumor quinones. Free Radic. Biol. Med. 1989, 6, 63–101. [Google Scholar] [CrossRef] [PubMed]
- Kankeu, C.; Clarke, K.; Passante, E.; Huber, H.J. Doxorubicin-induced chronic dilated cardiomyopathy—The apoptosis hypothesis revisited. J. Mol. Med. 2017, 95, 239–248. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, B.; Mukherjee, S.; Bhattacharya, B.; Mukherjee, S. Cancer Therapy Using Antibiotics. J. Cancer Ther. 2015, 6, 849–858. [Google Scholar] [CrossRef]
- Hrelia, S.; Fiorentini, D.; Maraldi, T.; Angeloni, C.; Bordoni, A.; Biagi, P.L.; Hakim, G. Doxorubicin induces early lipid peroxidation associated with changes in glucose transport in cultured cardiomyocytes. Biochim. Biophys. Acta—Biomembr. 2002, 1567, 150–156. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Guo, F.; Guan, Y.; Chen, T.; Ma, K.; Zhang, L.; Wang, Z.; Su, Q.; Feng, L.; Liu, Y.; et al. Novel Anthraquinone Compounds Inhibit Colon Cancer Cell Proliferation via the Reactive Oxygen Species/JNK Pathway. Molecules 2020, 25, 1672. [Google Scholar] [CrossRef]
- Malik, M.S.; Alsantali, R.I.; Jassas, R.S.; Alsimaree, A.A.; Syed, R.; Alsharif, M.A.; Kalpana, K.; Morad, M.; Althagafi, I.I.; Ahmed, S.A. Journey of anthraquinones as anticancer agents—A systematic review of recent literature. RSC Adv. 2021, 11, 35806–35827. [Google Scholar] [CrossRef]
- Pang, M.J.; Yang, Z.; Zhang, X.L.; Liu, Z.F.; Fan, J.; Zhang, H.Y. Physcion, a naturally occurring anthraquinone derivative, induces apoptosis and autophagy in human nasopharyngeal carcinoma. Acta Pharmacol. Sin. 2016, 37, 1623–1640. [Google Scholar] [CrossRef]
- Abdella, B.R.; Fisher, J. A chemical perspective on the anthracycline antitumor antibiotics. Environ. Health Perspect. 1985, 64, 4. [Google Scholar] [CrossRef]
- Wheeler, C.; Rader, R.; Kessel, D. Membrane alterations associated with progressive adriamycin resistance. Biochem. Pharmacol. 1982, 31, 2691–2693. [Google Scholar] [CrossRef] [PubMed]
- Alves, D.S.; Pérez-Fons, L.; Estepa, A.; Micol, V. Membrane-related effects underlying the biological activity of the anthraquinones emodin and barbaloin. Biochem. Pharmacol. 2004, 68, 549–561. [Google Scholar] [CrossRef] [PubMed]
- Siegfried, J.M.; Burke, T.G.; Tritton, T.R. Cellular transport of anthracyclines by passive diffusion: Implications for drug resistance. Biochem. Pharmacol. 1985, 34, 593–598. [Google Scholar] [CrossRef] [PubMed]
- Siegfried, J.A.; Kennedy, K.A.; Sartorelli, A.C.; Tritton, T.R. The role of membranes in the mechanism of action of the antineoplastic agent adriamycin. Spin-labeling studies with chronically hypoxic and drug-resistant tumor cells. J. Biol. Chem. 1983, 258, 339–343. [Google Scholar] [CrossRef] [PubMed]
- Escriba, P.V.; Ferrer-Montiel, A.V.; Ferragut, J.A.; Gonzalez-Ros, J.M. Role of membrane lipids in the interaction of daunomycin with plasma membranes from tumor cells: Implications in drug-resistance phenomena. Biochemistry 1990, 29, 7275–7282. [Google Scholar] [CrossRef] [PubMed]
- Mandal, B.; Singha, S.; Dey, S.K.; Mazumdar, S.; Kumar, S.; Karmakar, P.; Das, S. CuII complex of emodin with improved anticancer activity as demonstrated by its performance on HeLa and Hep G2 cells. RSC Adv. 2017, 7, 41403–41418. [Google Scholar] [CrossRef]
- Jabłońska-Trypuć, A.; Świderski, G.; Krętowski, R.; Lewandowski, W. Newly Synthesized Doxorubicin Complexes with Selected Metals-Synthesis, Structure and Anti-Breast Cancer Activity. Molecules 2017, 22, 1106. [Google Scholar] [CrossRef]
- Christidi, E.; Brunham, L.R. Regulated cell death pathways in doxorubicin-induced cardiotoxicity. Cell Death Dis. 2021, 12, 339. [Google Scholar] [CrossRef]
- Rahimipour, S.; Gescheidt, G.; Bilkis, I.; Fridkin, M.; Weiner, L. Towards the Efficiency of Pharmacologically Active Quinoid Compounds: Electron Transfer and Formation of Reactive Oxygen Species. Appl. Magn. Reson. 2009, 37, 629–648. [Google Scholar] [CrossRef]
- Polyakov, N.; Leshina, T.; Fedenok, L.; Slepneva, I.; Kirilyuk, I.; Furso, J.; Olchawa, M.; Sarna, T.; Elas, M.; Bilkis, I.; et al. Redox-Active Quinone Chelators: Properties, Mechanisms of Action, Cell Delivery, and Cell Toxicity. Antioxid. Redox Signal. 2018, 28, 1394–1403. [Google Scholar] [CrossRef]
- Denoyer, D.; Masaldan, S.; La Fontaine, S.; Cater, M.A. Targeting copper in cancer therapy: “Copper That Cancer”. Metallomics 2015, 7, 1459–1476. [Google Scholar] [CrossRef] [PubMed]
- Fukai, T.; Ushio-Fukai, M. Superoxide dismutases: Role in redox signaling, vascular function, and diseases. Antioxid. Redox Signal. 2011, 15, 1583–1606. [Google Scholar] [CrossRef] [PubMed]
- Gupte, A.; Mumper, R.J. Elevated copper and oxidative stress in cancer cells as a target for cancer treatment. Cancer Treat. Rev. 2009, 35, 32–46. [Google Scholar] [CrossRef] [PubMed]
- Lovejoy, D.B.; Jansson, P.J.; Brunk, U.T.; Wong, J.; Ponka, P.; Richardson, D.R. Antitumor activity of metal-chelating compound Dp44mT is mediated by formation of a redox-active copper complex that accumulates in lysosomes. Cancer Res. 2011, 71, 5871–5880. [Google Scholar] [CrossRef] [PubMed]
- Fiallo, M.M.L.; Garnier-Suillerot, A. Metal Anthracycline Complexes as a New Class of Anthracycline Derivatives. Pd(II)-Adriamycin and Pd(II)-Daunorubicin Complexes: Physicochemical Characteristics and Antitumor Activity. Biochemistry 1986, 25, 924–930. [Google Scholar] [CrossRef]
- Eliot, H.; Gianni, L.; Myers, C. Oxidative Destruction of DNA by the Adriamycin-iron Complex. Biochemistry 1984, 23, 928–936. [Google Scholar] [CrossRef]
- Myers, C.E.; Gianni, L.; Simone, C.B.; Klecker, R.; Greene, R. Oxidative destruction of erythrocyte ghost membranes catalyzed by the doxorubicin-iron complex. Biochemistry 1982, 21, 1707–1713. [Google Scholar] [CrossRef]
- Mizutani, H.; Oikawa, S.; Hiraku, Y.; Murata, M.; Kojima, M.; Kawanishi, S. Distinct mechanisms of site-specific oxidative DNA damage by doxorubicin in the presence of copper(II) and NADPH-cytochrome P450 reductase. Cancer Sci. 2003, 94, 686–691. [Google Scholar] [CrossRef]
- Mizutani, H.; Nishimoto, A.; Hotta, S.; Ikemura, K.; Imai, M.; Miyazawa, D.; Ohta, K.; Ikeda, Y.; Maeda, T.; Yoshikawa, M.; et al. Oxidative DNA Damage Induced by Pirarubicin, an Anthracycline Anticancer Agent, in the Presence of Copper(II). Anticancer Res. 2018, 38, 2643–2648. [Google Scholar] [CrossRef]
- Yang, P.; Wang, H.; Gao, F.; Yang, B. Antitumor activity of the Cu(II)-mitoxantrone complex and its interaction with deoxyribonucleic acid. J. Inorg. Biochem. 1996, 62, 137–145. [Google Scholar] [CrossRef]
- Rao, V.A. Iron Chelators with Topoisomerase-Inhibitory Activity and Their Anticancer Applications. Antioxid. Redox Signal. 2013, 18, 930. [Google Scholar] [CrossRef] [PubMed]
- Monti, E.; Paracchini, L.; Piccinini, F.; Malatesta, V.; Morazzoni, F.; Supino, R. Cardiotoxicity and antitumor activity of a copper(II)-doxorubicin chelate. Cancer Chemother. Pharmacol. 1990, 25, 333–336. [Google Scholar] [CrossRef] [PubMed]
- Dikalov, S.; Alov, P.; Rangelova, D. Role of Iron Ion Chelation by Quinones in Their Reduction, OH-Radical Generation, and Lipid Peroxidation. Biochem. Biophys. Res. Commun. 1993, 195, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Selyutina, O.Y.; Kononova, P.A.; Koshman, V.E.; Fedenok, L.G.; Polyakov, N.E. The Interplay of Ascorbic Acid with Quinones-Chelators—Influence on Lipid Peroxidation: Insight into Anticancer Activity. Antioxidants 2022, 11, 376. [Google Scholar] [CrossRef] [PubMed]
- Sanders, C.R.; Prosser, R.S. Bicelles: A model membrane system for all seasons? Structure 1998, 6, 1227–1234. [Google Scholar] [CrossRef] [PubMed]
- Hutchison, J.M.; Shih, K.C.; Scheidt, H.A.; Fantin, S.M.; Parson, K.F.; Pantelopulos, G.A.; Harrington, H.R.; Mittendorf, K.F.; Qian, S.; Stein, R.A.; et al. Bicelles Rich in both Sphingolipids and Cholesterol and Their Use in Studies of Membrane Proteins. J. Am. Chem. Soc. 2020, 142, 12715–12729. [Google Scholar] [CrossRef] [PubMed]
- Dikalov, S.I.; Rumyantseva, G.V.; Piskunov, A.V.; Weiner, L.M. Role of Quinone-Iron(III) Interaction in NADPH-Dependent Enzymatic Generation of Hydroxyl Radicals. Biochemistry 1992, 31, 8947–8953. [Google Scholar] [CrossRef]
- Stroet, M.; Caron, B.; Visscher, K.M.; Geerke, D.P.; Malde, A.K.; Mark, A.E. Automated Topology Builder Version 3.0: Prediction of Solvation Free Enthalpies in Water and Hexane. J. Chem. Theory Comput. 2018, 14, 5834–5845. [Google Scholar] [CrossRef]
- Poger, D.; Mark, A.E. On the validation of molecular dynamics simulations of saturated and cis-monounsaturated phosphatidylcholine lipid bilayers: A comparison with experiment. J. Chem. Theory Comput. 2010, 6, 325–336. [Google Scholar] [CrossRef]
- Parrinello, M.; Rahman, A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Hoover, W.G. Canonical dynamics: Equilibrium phase-space distributions. Phys. Rev. A 1985, 31, 1695–1697. [Google Scholar] [CrossRef] [PubMed]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef]
- da Cunha, A.R.; Duarte, E.L.; Stassen, H.; Lamy, M.T.; Coutinho, K. Experimental and theoretical studies of emodin interacting with a lipid bilayer of DMPC. Biophys. Rev. 2017, 9, 729–745. [Google Scholar] [CrossRef] [PubMed]
- Richardson, D.R.; Wis Vitolo, L.M.; Hefter, G.T.; May, P.M.; Clare, B.W.; Webb, J.; Wilairat, P. Iron chelators of the pyridoxal isonicotinoyl hydrazone class Part I. Ionisation characteristics of the ligands and their relevance to biological properties. Inorg. Chim. Acta 1990, 170, 165–170. [Google Scholar] [CrossRef]
- Richardson, D.R.; Tran, E.H.; Ponka, P. The potential of iron chelators of the pyridoxal isonicotinoyl hydrazone class as effective antiproliferative agents. Blood 1995, 86, 4295–4306. [Google Scholar] [CrossRef]
- Alrushaid, S.; Sayre, C.L.; Yáñez, J.A.; Forrest, M.L.; Senadheera, S.N.; Burczynski, F.J.; Löbenberg, R.; Davies, N.M. Pharmacokinetic and Toxicodynamic Characterization of a Novel Doxorubicin Derivative. Pharmaceutics 2017, 9, 35. [Google Scholar] [CrossRef]
- Selyutina, O.Y.; Kononova, P.A.; Koshman, V.E.; Shelepova, E.A.; Azad, M.G.; Afroz, R.; Dharmasivam, M.; Bernhardt, P.V.; Polyakov, N.E.; Richardson, D.R. Ascorbate-and iron-driven redox activity of Dp44mT and Emodin facilitates peroxidation of micelles and bicelles. Biochim. Biophys. Acta—Gen. Subj. 2022, 1866, 130078. [Google Scholar] [CrossRef]
- Burns, C.P.; Haugstad, B.N.; Mossman, C.J.; North, J.A.; Ingraham, L.M. Membrane lipid alteration: Effect on cellular uptake of mitoxantrone. Lipids 1988, 23, 393–397. [Google Scholar] [CrossRef]
- Stefani, C.; Jansson, P.J.; Gutierrez, E.; Bernhardt, P.V.; Richardson, D.R.; Kalinowski, D.S. Alkyl substituted 2′-benzoylpyridine thiosemicarbazone chelators with potent and selective anti-neoplastic activity: Novel ligands that limit methemoglobin formation. J. Med. Chem. 2013, 56, 357–370. [Google Scholar] [CrossRef]
- Stefani, C.; Punnia-Moorthy, G.; Lovejoy, D.B.; Jansson, P.J.; Kalinowski, D.S.; Sharpe, P.C.; Bernhardt, P.V.; Richardson, D.R. Halogenated 2′-benzoylpyridine thiosemicarbazone (XBpT) chelators with potent and selective anti-neoplastic activity: Relationship to intracellular redox activity. J. Med. Chem. 2011, 54, 6936–6948. [Google Scholar] [CrossRef]
- Selyutina, O.Y.; Kononova, P.A.; Polyakov, N.E. Experimental and Theoretical Study of Emodin Interaction with Phospholipid Bilayer and Linoleic Acid. Appl. Magn. Reson. 2020, 51, 951–960. [Google Scholar] [CrossRef]
- Spiteller, G. Linoleic acid peroxidation—The dominant lipid peroxidation process in low density lipoprotein—And its relationship to chronic diseases. Chem. Phys. Lipids 1998, 95, 105–162. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C. An update on products and mechanisms of lipid peroxidation. Mol. Nutr. Food Res. 2009, 53, 315–321. [Google Scholar] [CrossRef] [PubMed]
- Cotticelli, M.G.; Crabbe, A.M.; Wilson, R.B.; Shchepinov, M.S. Insights into the role of oxidative stress in the pathology of Friedreich ataxia using peroxidation resistant polyunsaturated fatty acids. Redox Biol. 2013, 1, 398–404. [Google Scholar] [CrossRef]
- Struppe, J.; Whiles, J.A.; Void, R.R. Acidic Phospholipid Bicelles: A Versatile Model Membrane System. Biophys. J. 2000, 78, 281–289. [Google Scholar] [CrossRef]
- Mastova, A.V.; Selyutina, O.Y.; Evseenko, V.I.; Polyakov, N.E. Photoinduced Oxidation of Lipid Membranes in the Presence of the Nonsteroidal Anti-Inflammatory Drug Ketoprofen. Membranes 2022, 12, 251. [Google Scholar] [CrossRef]
- Selyutina, O.Y.; Kononova, P.A.; Polyakov, N.E. Effect of glycyrrhizic acid on phospholipid membranes in media with different pH. Russ. Chem. Bull. 2022, 70, 2434–2439. [Google Scholar] [CrossRef]
- Ellena, J.F.; Lepore, L.S.; Cafiso, D.S. Estimating lipid lateral diffusion in phospholipid vesicles from carbon-13 spin-spin relaxation. J. Phys. Chem. 1993, 97, 2952–2957. [Google Scholar] [CrossRef]
- Lepore, L.S.; Ellena, J.F.; Cafiso, D.S. Comparison of the lipid acyl chain dynamics between small and large unilamellar vesicles. Biophys. J. 1992, 61, 767–775. [Google Scholar] [CrossRef]
- Richards, M.J.; Hsia, C.Y.; Singh, R.R.; Haider, H.; Kumpf, J.; Kawate, T.; Daniel, S. Membrane Protein Mobility and Orientation Preserved in Supported Bilayers Created Directly from Cell Plasma Membrane Blebs. Langmuir 2016, 32, 2963–2974. [Google Scholar] [CrossRef]
- Tian, J.; Sethi, A.; Swanson, B.I.; Goldstein, B.; Gnanakaran, S. Taste of Sugar at the Membrane: Thermodynamics and Kinetics of the Interaction of a Disaccharide with Lipid Bilayers. Biophys. J. 2013, 104, 622. [Google Scholar] [CrossRef] [PubMed]
- Bogdanov, M.; Mileykovskaya, E.; Dowhan, W. Lipids in the Assembly of Membrane Proteins and Organization of Protein Supercomplexes: Implications for Lipid-Linked Disorders. Subcell. Biochem. 2008, 49, 197. [Google Scholar] [CrossRef] [PubMed]
- Alves, A.C.; Magarkar, A.; Horta, M.; Lima, J.L.F.C.; Bunker, A.; Nunes, C.; Reis, S. Influence of doxorubicin on model cell membrane properties: Insights from in vitro and in silico studies. Sci. Rep. 2017, 7, 6343. [Google Scholar] [CrossRef] [PubMed]
- Van Hell, A.J.; Melo, M.N.; Van Blitterswijk, W.J.; Gueth, D.M.; Braumuller, T.M.; Pedrosa, L.R.C.; Song, J.Y.; Marrink, S.J.; Koning, G.A.; Jonkers, J.; et al. Defined lipid analogues induce transient channels to facilitate drug-membrane traversal and circumvent cancer therapy resistance. Sci. Rep. 2013, 3, 1949. [Google Scholar] [CrossRef]
- Tritton, T.R.; Yee, G. The Anticancer Agent Adriamycin Can Be Actively Cytotoxic Without Entering Cells. Science 1982, 217, 248–250. [Google Scholar] [CrossRef]



| N+(CH3)3 | CH2 | CH3 | |
|---|---|---|---|
| w/o Q1 | 0.81 ± 0.06 s | 1.120 ± 0.04 s | 1.370 ± 0.07 s |
| with Q1 | 0.7 ± 0.07 s | 0.8 ± 0.08 s | 1.170 ± 0.1 s |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Selyutina, O.Y.; Mastova, A.V.; Polyakov, N.E. The Interaction of Anthracycline Based Quinone-Chelators with Model Lipid Membranes: 1H NMR and MD Study. Membranes 2023, 13, 61. https://doi.org/10.3390/membranes13010061
Selyutina OY, Mastova AV, Polyakov NE. The Interaction of Anthracycline Based Quinone-Chelators with Model Lipid Membranes: 1H NMR and MD Study. Membranes. 2023; 13(1):61. https://doi.org/10.3390/membranes13010061
Chicago/Turabian StyleSelyutina, Olga Yu., Anna V. Mastova, and Nikolay E. Polyakov. 2023. "The Interaction of Anthracycline Based Quinone-Chelators with Model Lipid Membranes: 1H NMR and MD Study" Membranes 13, no. 1: 61. https://doi.org/10.3390/membranes13010061
APA StyleSelyutina, O. Y., Mastova, A. V., & Polyakov, N. E. (2023). The Interaction of Anthracycline Based Quinone-Chelators with Model Lipid Membranes: 1H NMR and MD Study. Membranes, 13(1), 61. https://doi.org/10.3390/membranes13010061









