Abstract
Superoxide dismutase-rich Tetraselmis chuii (T. chuii) is derived from marine microalgae and has been reported to increase gene expression of nuclear factor erythroid 2-related factor 2 (NRF2) and related antioxidant enzymes in myoblast tissue culture models. Human research has indicated that T. chuii supplementation can improve recovery from exercise-induced muscle damage, but its effects on endurance exercise performance and the molecular bases that may underlie any ergogenic effects are unclear. Healthy participants underwent 14 days of supplementation with 25 mg·day−1 T. chuii and placebo in a randomized, double-blind, crossover experimental design. Prior to and following each supplementation period, participants completed a high-intensity cycling test to assess time to exhaustion and peak oxygen uptake (). A resting skeletal muscle biopsy was collected after both supplementation periods to assess gene expression changes. Compared to pre-supplementation values, was increased following T. chuii (p = 0.013) but not placebo (p = 0.66). Fold-change in glutathione peroxidase 7 [(GPX7) 1.26 ± 1.37], glutathione-disulfide reductase [(GSR) 1.22 ± 1.41], glutathione S-transferase Mu 3 [(GSTM3) 1.34 ± 1.49], peroxiredoxin 6 [(PRDX6) 1.36 ± 1.57], extracellular signal-regulated kinase 3 [(ERK3) 1.92 ± 2.42], NRF2 (1.62 ± 2.16), p38 alpha [(p38a) 1.33 ± 1.58] and sirtuin 1 [(SIRT1) 1.73 ± 2.25] gene expression were higher after T. chuii compared to placebo supplementation (p < 0.05). Short-term T. chuii supplementation increased and skeletal muscle gene expression of key enzymatic antioxidants (GPX7, GSR, GSTM3, and PRDX6), signalling kinases (ERK3 and p38a), post-translational regulators (SIRT1), and transcription factors (NRF2) that may protect against cellular stress insults.
1. Introduction
The ability of the human body to mitigate endogenous and exogenous stressors is required to maintain good health and functional capabilities [1]. When healthy individuals or cells are exposed to such stressors, numerous highly conserved processes can facilitate remodelling towards a phenotype that confers greater resilience to future insults from the same or related stressors through the process of hormesis [2]. One important transcription factor that is activated and orchestrates various adaptive responses upon exposure to cellular stress is nuclear factor erythroid 2-related factor 2 (NRF2) [3]. Indeed, NRF2 controls the expression of more than 200 genes and has been linked to enhancements in a myriad of cellular responses including: detoxication, elimination of damaged macromolecules, and export of xenobiotics; modulating the redox status of the cell by increasing enzymatic antioxidants or synthesizing endogenous reducing agents; regulating apoptosis, cell cycle, and differentiation; elevating chaperones and heat shock proteins; increasing proteins of intercellular adhesion, cytoskeleton, and intracellular transport; regulating ribosomal protein synthesis; bolstering immune and inflammatory responses; and elevating numerous enzymes involved in various aspects of cellular metabolism [4,5]. Accordingly, NRF2 is considered central to cellular homeostasis and the master regulator of the cellular antioxidant response. Data from animal models show that increasing NRF2 activation can enhance exercise capacity [6,7,8], whereas attenuating NRF2 activity compromises exercise capacity [8]. Relatedly, recent data in human skeletal muscle support a role for NRF2 in regulating peak oxygen uptake () and high-intensity exercise performance [9]. Therefore, up-regulating NRF2 activity may confer widespread benefits to local cellular homeostasis, as well as systemic physiological responses and exercise capacity.
In addition to endogenous activation of NRF2, there is great interest in dietary supplements that may activate NRF2, given its potential to improve human health and performance. One supplement reported to transcriptionally activate NRF2 in human myoblasts is superoxide dismutase (SOD)-rich Tetraselmis chuii (T. chuii). SOD-rich T. chuii is a freeze-dried ingredient derived from the green eukaryotic microalgae T. chuii (chlorophyte). In addition to SOD, the ingredient contains a number of potentially bioactive compounds such as poly-unsaturated fatty acids (PUFAs), polyphenols, vitamins, carotenoids, and phytosterols [10,11]. When human myoblasts were acutely treated with T. chuii for 24 h, NRF2 gene expression was increased concomitant with elevated gene expression of the enzymatic antioxidants superoxide dismutase (SOD), catalase (CAT), glutathione peroxidase (GPx), and haem oxygenase 1 (HMOX-1) [12]. However, whilst these preclinical data were encouraging, only a limited number of gene expression targets were assessed. As such, the potential of T. chuii in human skeletal muscle remains unclear, particularly since NRF2 has been implicated in numerous biological processes beyond its role in bolstering the enzymatic antioxidant defense system [4,5]. Using rodent skeletal muscle models, subsequent studies reported that T. chuii administration augmented enzymatic antioxidants (SOD, CAT, and GPx) [13] and myogenic factors regulating satellite cell proliferation [14], and attenuated pro-inflammatory cytokines [15] following endurance training compared to endurance training alone and a control condition without endurance training. Clearly, a limitation of these experiments is that the independent effects of T. chuii supplementation on skeletal muscle redox and inflammatory responses were not evaluated. Moreover, it is currently unclear whether these findings would be reproduced in human skeletal muscle tissue after dietary T. chuii supplementation and, if so, whether this would translate into beneficial effects on systemic physiological responses and exercise capacity. This is a pertinent question, as many purported nutritional NRF2 activators in preclinical models have not been successfully replicated in humans when these nutrients are supplemented [16,17,18].
With regard to the effects of dietary T. chuii supplementation on functional responses in humans, most research to date has assessed its effects on recovery of skeletal muscle function following exercise. Specifically, it was reported that short-term supplementation with 25 mg/day T. chuii expedited the recovery of skeletal muscle function following muscle-damaging exercise or during periods of non-functional overreaching and intensified resistance training [13,14,15]. By contrast, there is limited research examining the effects of T. chuii supplementation on endurance exercise performance and its physiological determinants. In the only study conducted to date, 30 days supplementation with 25 mg/day of T. chuii lowered heart rate and increased [19]. Whilst previous studies have reported beneficial physiological and functional effects after T. chuii supplementation [13,14,15,19,20], the molecular bases for any improvements in endurance exercise performance and [19], as well as post-exercise recovery [13,14,15], have yet to be directly explored in human skeletal muscle tissue. Although reactive oxygen species (ROS) are continually produced by skeletal muscle and other tissues under basal conditions and can serve as beneficial signalling molecules, a series of enzymatic and non-enzymatic antioxidants help to offset the development of dysfunctional redox signalling and oxidative stress [21]. During exercise, ROS production is greatly increased above basal levels, leading to the development of exercise-induced oxidative stress, which has been implicated in fatigue development during exercise [21,22,23]. Therefore, if T. chuii supplementation can augment NRF2 signalling and the up-regulation of endogenous antioxidant enzymes, this could enhance exercise capacity by offsetting the development of exercise-induced oxidative stress.
The purpose of the current study was to assess the effect of 14 days supplementation with 25 mg/day SOD-rich T. chuii on high-intensity endurance exercise capacity, , and numerous genes related to NRF2 signalling and its downstream effects on antioxidant and inflammatory responses (Table 1) in human skeletal muscle tissue using an OpenArray™ system. It was hypothesised that T. chuii supplementation would increase endurance exercise capacity, , and genes related to NRF2 signalling and antioxidant and anti-inflammatory responses in skeletal muscle tissue of healthy adults.

Table 1.
Gene names, gene symbols and ThermoFisher Scientific Assay IDs for human skeletal muscle OpenArray™ gene expression analyses.
2. Materials and Methods
2.1. Participant Characteristics
The data presented in the manuscript are from two separate sets of experiments, which recruited two independent groups of human participants. In the study reporting the effects of SOD-rich T. chuii supplementation on skeletal muscle gene expression, 15 healthy male participants were recruited, with 13 [mean ± SD (range); age, 22 ± 3 (19–28) years; height, 1.78 ± 0.06 (1.67–1.92) m; body mass, 80 ± 9 (68–96) kg; and BMI, 25 ± 2 (22–29) kg·m2] completing the study (Figure 1). This trial was registered with ClinicalTrials.gov ID: NCT06822556. A power calculation was completed using GPower (version 3) using the data of Toro et al. [19]. Based on the increase in reported after T. chuii supplementation compared to baseline and assuming a moderate correlation of 0.5 between groups, an effect size of 0.86 was anticipated. With an α of 0.05 and a power of 0.8, a sample size of 13 participants was required, with 15 participants recruited to account for participant withdrawals. In the study reporting the effects of SOD-rich T. chuii supplementation on exercise tolerance and physiological responses, 15 healthy male participants were also recruited, with 13 [mean ± SD (range); age, 24 ± 4 (19–34) years; height, 1.81 ± 0.08 (1.69–1.91) m; body mass, 82 ± 9 (69–99) kg; BMI, 25 ± 3 (22–32) kg·m2; and , 48 ± 5 (42–59) mL·kg−1·min−1], completing the study (Figure 2). This trial was registered with ClinicalTrials.gov ID: NCT06831656. A power calculation was completed using GPower (Version 3) using the data of Ramírez et al. [12]. Based on the increase in antioxidant gene expression or enzyme activities reported after T. chuii administration compared to baseline and assuming a moderate correlation of 0.5 between groups, an effect size of ≥0.9 was anticipated. With an α of 0.05 and a power of 0.8, a sample size of 12 participants was required, with 15 participants recruited to account for participant withdrawals. All participants were recreationally active, but not highly trained, non-smokers, and were instructed to avoid the consumption of dietary supplements, including antioxidants, and anti-inflammatory medications, including paracetamol and NSAIDs, for the duration of the study. Ethical approval was granted for each set of experiments by Loughborough University Ethics Approvals Committee (Human Participants Sub-Committee) with ethical approval codes of R19-P160 in the study reporting the effects of SOD-rich T. chuii supplementation on skeletal muscle gene expression and 2022-6116-8576 in the study reporting the effects of SOD-rich T. chuii supplementation on exercise tolerance and physiological responses. Before data collection commenced, each participant received a detailed explanation of the experimental procedures, the associated risks, and the potential benefits, and any questions were addressed. Written informed consent was then obtained from all participants prior to commencing data collection. Prior to all laboratory testing sessions, participants were instructed to arrive at the laboratory in a rested and fully hydrated state, having avoided strenuous exercise and alcohol ingestion for 24 h and caffeine ingestion for 12 h. Participants recorded their dietary intake for the 24 h before the first experimental visit and replicated this diet during the 24 h period preceding subsequent visits. All experimental testing was conducted at the same time of day (±2 h) to minimise the influence of diurnal variations on the measurements.
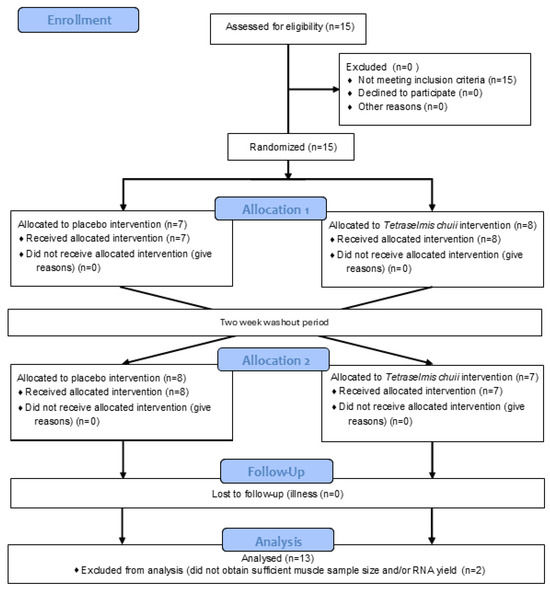
Figure 1.
Flow diagram illustrating the study design for trial NCT06822556.
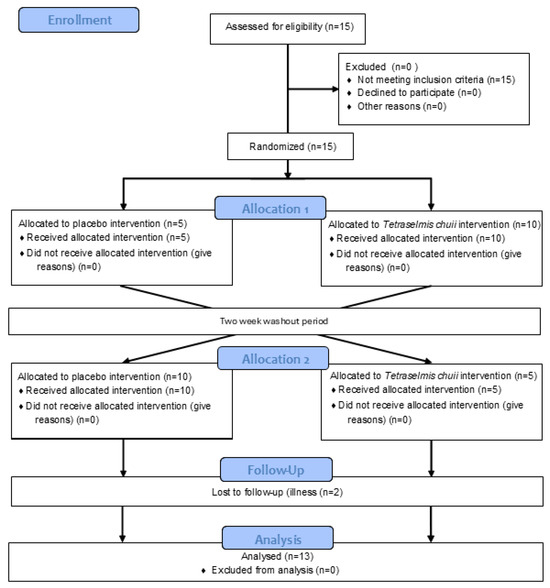
Figure 2.
Flow diagram illustrating the study design for trial NCT06831656.
2.2. Experimental Design
In the study reporting the effects of SOD-rich T. chuii supplementation on skeletal muscle gene expression, a single resting skeletal muscle biopsy was collected on two occasions over a 6–8-week timeframe. In the study reporting the effects of SOD-rich T. chuii supplementation on exercise tolerance and physiological responses, participants reported to the laboratory on 6 occasions over a 10–14-week timeframe. During visit 1, a ramp incremental test was completed for the determination of , gas exchange threshold (GET), and an individualised severe-intensity work rate for use in the subsequent tests. During visit 2, a constant-load step test was completed for familiarisation purposes. During visits 3–6, a constant-load step test was performed for the determination of end-exercise , rating of perceived exertion (RPE), blood lactate concentration (B[La]) and exercise tolerance. Visits 3 and 5 were completed on day 0 whilst visits 4 and 6 were completed on day 14 of each supplementation period. As such, there were 4 experimental conditions: T. chuii-Pre, T. chuii-Post, placebo (PLA)-Pre (PLA-Pre) and PLA-Post. In both studies, participants were assigned in a double-blind, randomised (using a coin toss), crossover experimental design to receive 2 weeks of dietary supplementation with 25 mg/day SOD-rich T. chuii (provided by Fitoplancton Marino, S.L., Spain; a single daily capsule with hemicellulose crystalline as an excipient) or PLA (a single daily capsule containing hemicellulose crystalline only), with supplementation conditions separated by a 2-week washout period. During each supplementation period, participants ingested one capsule on the non-visit days (i.e., days 1–13) in the morning at breakfast, with the visits completed on day 14. The dose and duration of SOD-rich T. chuii supplementation used in this study were based on previous studies reporting ergogenic effects with this supplementation strategy [13,14,15,19,20].
2.3. Data Collection and Analysis Procedures
2.3.1. Skeletal Muscle Sampling
Skeletal muscle biopsies were obtained from the m.vastus lateralis under local anaesthesia (1% lidocaine hydrochloride). After an incision through the skin and underlying fascia site, skeletal muscle tissue was sampled using the Bergstrom-type needle with suction. The samples (~100–250 mg) were minced using scalpels and then submerged in 5× volume RNAlater™ (ThermoFisher Scientific, Paisley, UK). These samples were stored at 4 °C for 24 h and then transferred to −20 °C prior to later RNA isolation.
2.3.2. RNA Isolation
Muscle lysis was first performed using TRIsure™ (Bioline, London, UK), Lysing Matrix D columns, and a FastPrep™ Lysis System (MP Biomedicals, Santa Ana, CA, USA). Phase separation, using chloroform, was then performed, with the upper aqueous phase recovered for RNA isolation. RNA was then isolated using a spin column procedure. Here, an RNeasy® Mini Kit was used, with on-column DNA digestion using an RNase-free DNase Set and RNA re-purification and concentration using an RNeasy® MinElute® Cleanup Set (Qiagen, Hilden, Germany). The procedure was performed in duplicate per sample and following the manufacturer’s recommended protocol. Sterile conditions were achieved by cleaning the work surfaces and utensils with RNaseZap™ RNase Decontamination Solution (Invitrogen, Carlsbad, CA, USA). Furthermore, RNase-free collection tubes and pipette tips were used throughout. The content, concentration, and quality of RNA isolates were determined using a NanoDrop 2000 spectrophotometer (ThermoFisher Scientific, Paisley, UK), with samples attaining an A260/A230 ratio < 1.8 undergoing a further clean-up kit procedure. RNA isolates were then stored at −80 °C, where they remained until the measurement of gene expression.
2.3.3. OpenArray™ Gene Expression Measurement
Initially, RNA isolates were reverse transcribed using a SuperScript™ VILO™ cDNA Synthesis Kit (ThermoFisher Scientific, Paisley, UK). Here, a master mix was first prepared for each sample, which consisted of 4 μL 5× VILO™ Reaction Mix, 2 μL 10× SuperScript™ Enzyme Mix, 2.5 μg RNA, and diethylpyrocarbonate (DEPC)-treated H2O up to a total volume of 20 μL. Following gentle mixing, the cDNA synthesis reaction proceeded by incubation steps, which included 10 min at 25 °C, 60 min at 42 °C, and 5 min at 85 °C. Samples were then stored at −20 °C until the measurement of gene expression.
Gene expression was determined using the QuantStudio™ 12K Flex OpenArray™ System (ThermoFisher Scientific, Paisley, UK), with the 112 × 24 format selected. The genes and their abbreviations and ThermoFisher Scientific Assay ID numbers can be found in Table 1. Targets were selected based on their functions and expression in human skeletal muscle using the Human Protein Atlas database (proteinatlas.org) [24]. ACTB, B2M, GAPDH, PPP1CA, and TBP were selected as candidate reference genes. 5 μL samples, which consisted of 3.8 μL OpenArray™ PCR MasterMix and 1.2 μL cDNA (20 ng/μL), were first generated. Reactions were then performed in triplicate using 33 nL volumes (~1 ng cDNA) in 384-well Applied Biosystems™ OpenArray™ plates (ThermoFisher Scientific, Paisley, UK). The quantification cycle (Cq; the cycle number at which the fluorescence level from accumulating amplicons crosses a defined threshold; also known as the threshold cycle, Ct) was quantified using the relative quantification (Crq; also known as the relative threshold, Crt) method. Here, the amplification curve is first set to a relative scale that ranges from 0 (minimum fluorescence) to 1 (maximum fluorescence). Using proprietary techniques, a curve that models the reaction efficiency is calculated, and Cq is then determined in four steps: (1) an internal reference efficiency level is used to determine the fractional cycle (Ce) whereby the reaction efficiency curve reaches a predetermined value; (2) the fluorescence level (Fe) corresponding to the Ce is then calculated; (3) the relative fluorescence threshold is computed as a specific percentage of Fe; and (4) Cq is then determined as the cycle number at which the amplification curve crosses the relative fluorescence threshold.
2.3.4. Gene Expression Data Analysis
Gene expression Cq values were obtained using the Thermo Fisher Connect™ online platform and the RQ module (ThermoFisher Scientific, Paisley, UK). Here, the data were first reviewed for quality assurance, with all data represented by a Cq confidence score < 0.8, AMP Score < 1.24, and AMP Status not equal to “AMP” checked and retained only if a valid PCR profile was evident. Also, all data with a replicate SD > 0.5 were checked, and outliers were omitted. The remaining raw data were then exported for further analysis. Here, the data were initially filtered to exclude Cq values < 6 and >28 (the RQ module’s default analysis setting). Also, to build upon the replicate SD check within the RQ module, which can filter errant replicates but not errant samples, the data were filtered based on a range of expected Cq values relative to the condition and participant mean. Here, all data with both a condition and participant SD > 2 were checked (determined by trial and error and intended to omit large errors only), and replicates > ±1 SD from both the condition and participant mean were omitted. Where the review and filtering procedures had removed more than a third of the samples, the gene target was omitted. The replicate Cq values were then averaged, with gene expression (fold-change) determined using the 2−ΔΔCq method [25], with the exception that ΔΔCq was calculated relative to the participant’s control condition ΔCq and not to a group average control condition ΔCq as used in the original method. ΔCq was calculated relative to the geometric mean (GM) of the three most stable reference genes (as determined using the ΔCq method) [26]. GM and geometric standard deviations (GSD) are reported to ensure symmetry for up- and down-regulated genes. Similarly, whilst the data are expressed as 2−ΔΔCq, statistical analyses were performed using Log2(2−ΔCq) [27]. Finally, the gene expression in the control condition (i.e., PLA) was compared to the difference in gene expression from the control condition to the experimental condition (i.e., ΔPLA-T. chuii) to determine if the effect of T. chuii was related to basal gene expression.
2.3.5. Exercise Testing Procedures
All exercise tests were performed on an electronically braked (Excalibur Sport) cycle ergometer (Lode, Groningen, The Netherlands), and the saddle and handlebar spatial configuration was recorded during visit 1 and reproduced during the subsequent visits. Participants were first fitted with a facemask to enable the collection of pulmonary gas exchange and ventilation data. All tests began with 5 min of baseline cycling at 50 W. Following this, the work rate increased linearly by 30 W/min during the ramp incremental test (visit 1) or transitioned abruptly to the individualised work rate during the constant-load step test completed in all subsequent visits. Participants cycled at a self-selected cadence (70–90 rpm). All tests continued until the limit of tolerance (Tlim), which was determined as either volitional exhaustion or when the pedal rate dropped by >10 rpm for 5 consecutive s, despite strong verbal encouragement.
For the constant-load step tests, venous blood was collected within 5 min post-exercise from an antecubital forearm vein by the venepuncture technique. Samples were drawn into 6 mL Vacutainer® tubes (Becton-Dickinson Vacutainer, Franklin Lakes, NJ, USA) containing EDTA as the anticoagulant and mixed gently by inversion. Twenty microliters of whole blood was then mixed in 1 mL of Haemolyising Solution (HaB Direct, Southam, UK) in duplicate and stored on ice for same-day B[La] analysis. End-exercise B[La] was measured in duplicate using an automated Biosen C-Line Clinic analyser (EKF Diagnostics, Cardiff, Wales). The analyser was first calibrated using a multi-standard solution tube (HaB Direct, Southam, UK) that contained 12 mmol/L [La].
Breath-by-breath pulmonary gas exchange and ventilation were measured continuously during all exercise tests using the Vyntus™ CPX gas analysis system (CareFusion, Höchberg, Germany). Participants wore a Hans Rudolph 7450 Series V2 Mask and breathed through a flat-fan system that included a digital volume transducer (CareFusion, Höchberg, Germany). The inspired and expired gas volume signal was sampled continuously via a capillary line connected to the flat-fan system. The gas analyser was calibrated immediately before each use with both ambient air and gas of known concentration (16% O2 and 5% CO2), and the volume transducer was calibrated with a 3 L syringe (Hans Rudolph, Kansas City, MO, USA).
2.3.6. Exercise Physiology and Tolerance Data Analysis Procedures
For the ramp incremental test, the pulmonary gas exchange and ventilation data were exported as consecutive 10 s averages. The was then denoted as the greatest 30 s rolling average attained before Tlim, whilst GET was determined from a cluster of measurements used previously [28]. These were: (1) the first disproportionate increase in CO2 production () from visual inspection of individual plots of vs. ; (2) an increase in expired ventilation ()/ with no increase in /; and (3) an increase in end-tidal O2 tension with no fall in end-tidal CO2 tension.
Once the and GET had been determined, an individualised severe-intensity work rate was calculated. Here, the power outputs associated with the and GET were first adjusted to account for the mean response time to ramp incremental exercise (i.e., two-thirds of the ramp rate was subtracted from each power output) [29]. The work rate was then set to 70% of the difference (70%Δ) between these two adjusted power outputs.
For the constant-load step tests, the pulmonary gas exchange and ventilation data were exported as breath-by-breath data. These were first examined to exclude errant breaths caused by coughing, swallowing, sighing, etc., and those values lying more than four standard deviations from the local mean were removed. The data were then linearly interpolated to provide second-by-second values and time-aligned to the start of the step transition. End-exercise was then denoted as the greatest 30 s rolling average attained before Tlim. All constant-load step tests continued until Tlim, which was determined as either volitional exhaustion or when the pedal rate dropped by >10 rpm for 5 consecutive s. When this occurred, Tlim (s) was noted. For the constant-load step tests, the end-exercise RPE was measured at the Tlim using the Borg 6–20 scale.
2.4. Statistical Analysis Procedures
Statistical analyses were performed using SPSS® Statistics version 28 (IBM, New York, NY, USA). All data were first explored for normal distribution using Shapiro-Wilk tests (p > 0.05) supported by skewness and kurtosis and visual inspection of histograms. Differences in gene expression between the experimental conditions (PLA and T. chuii) were assessed using paired samples t-tests (parametric data) or Wilcoxon signed-rank tests (non-parametric data). Differences in end-exercise , RPE and B[La], and Tlim between the experimental conditions (i.e., T. chuii-Pre, T. chuii-Post, PLA-Pre and PLA-Post) were assessed using two-way repeated-measures ANOVAs [supplement (PLA and T. chuii) × time (Pre and Post)] (parametric data) or Friedman’s test (non-parametric data). Additionally, for the two-way repeated-measures ANOVAs only, Greenhouse–Geisser corrections were applied when sphericity was violated, and significant interaction effects were explored further using post hoc paired samples t-tests, with family-wise error rate controlled using Holm–Bonferroni corrections. Post hoc effect sizes are presented using Cohen’s dz (parametric data) or Rosenthal’s r (non-parametric data) and interpreted as “small” (0.20–0.49), “moderate” (0.50–0.79), and “large” (≥0.80). Omnibus test effect sizes are presented using Partial Eta Squared (np2; parametric data) or Kendall’s w (non-parametric data). Relationships between variables were explored using Pearson’s r (parametric data) or Spearman’s ρ (non-parametric data). Data are presented as the mean ± SD unless stated otherwise. Statistical significance was accepted when p < 0.05.
3. Results
All participants self-reported 100% compliance in avoiding potentially confounding factors (i.e., not using medication or dietary supplements throughout the study and avoiding strenuous exercise and alcohol ingestion for 24 h, and caffeine ingestion for 12 h before the experimental visits). Furthermore, all participants self-reported consistent dietary habits throughout. The T. chuii and PLA supplements were well tolerated, with no side effects reported.
3.1. Human Skeletal Muscle Gene Expression
The ΔCq was calculated relative to the geometric mean (GM) of ACTB, B2M and PPP1CA as the three most stable reference genes and subsequently used to calculate Log2(2−ΔCq) for statistical analyses of gene expression data.
Expression of 15 genes was up-regulated for PLA-T. chuii (ΔPLA-T. chuii) with medium to large effect sizes (p < 0.05; Table 2). A further 12 genes were up-regulated and 2 genes were down-regulated without statistical significance but with medium effect sizes (p = 0.05–0.10, in all instances; Table 2). There were no significant differences in the remaining 66 genes, and effect sizes were small (p > 0.05).

Table 2.
Gene expression in RNA isolated from resting human skeletal muscle following dietary supplementation for 2 weeks with 25 mg/day T. chuii with hemicellulose crystalline as an excipient compared to a placebo containing hemicellulose crystalline only.
Correlation analyses revealed strong negative relationships for PLA-(ΔPLA-T. chuii) in 25 genes (p < 0.05; Table 3). Moderate negative relationships were revealed in a further 28 genes (p < 0.05; Table 3). An additional 23 genes showed moderate negative relationships but without statistical significance (p > 0.05). The remaining 18 genes showed weak relationships without statistical significance (p > 0.05).

Table 3.
Correlations between gene expression in RNA isolated from resting human skeletal muscle following dietary supplementation for 2 weeks with a placebo containing hemicellulose crystalline only compared to the difference in gene expression between placebo and dietary supplementation for 2 weeks with 25 mg/day T. chuii with hemicellulose crystalline as an excipient.
3.2. Exercise Physiology and Tolerance
There was a supplement × time interaction effect for end-exercise (F1,12 = 6.395, p = 0.026; np2 = 0.348), but no main effect for time (F1,12 = 1.927, p = 0.190; np2 = 0.138) or supplement (F1,12 = 0.270, p = 0.613; np2 = 0.022). Post hoc paired samples t-tests revealed that end-exercise was greater for T. chuii-Post compared to T. chuii-Pre (p = 0.013; dz = 0.8; Table 4; Figure 3). End-exercise was not different between PLA-Pre and PLA-Post (p = 0.66; dz = 0.1) and was not different between PLA-Pre and T. chuii-Pre (p = 0.06; dz = 0.6) or PLA-Post and T. chuii-Post (p = 0.11; dz = 0.5). Friedman’s test revealed that end-exercise RPE was unchanged (p = 0.34; w = 0.1; Table 4). There was a main effect for time (F1,12 = 9.238, p = 0.013; np2 = 0.412), but no main effect for supplement (F1,12 = 0.877, p = 0.367; np =0.068) and no supplement × time interaction effect (F1,12 = 0.026, p = 0.847; np2 = 0.002; Table 4) for end-exercise B[La]. Similarly, there was a main effect for time (F1,12 = 9.238, p = 0.010; np2 = 0.435), but no main effect for supplement (F1,12 = 0.270, p = 0.613; np2 = 0.022) and no supplement × time interaction effect (F1,12 = 0.692, p = 0.422; np2 = 0.055) (Table 4) for Tlim.

Table 4.
End-exercise pulmonary oxygen uptake, rating of perceived exertion, blood lactate concentration, and exercise tolerance following a step increment in work rate from baseline to severe-intensity cycling prior to and following dietary supplementation for 2 weeks with T. chuii with hemicellulose crystalline as an excipient and a placebo containing hemicellulose crystalline only.
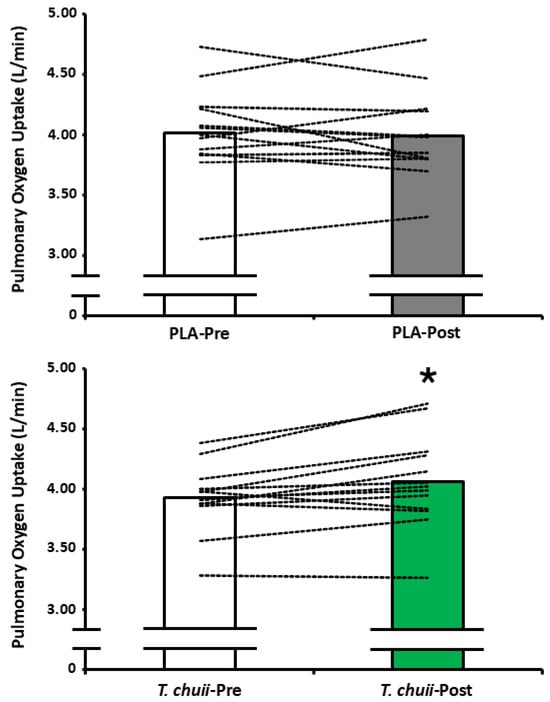
Figure 3.
Pulmonary oxygen update during the final 30 s of a high-intensity cycling test continued to exhaustion. Bars represent group mean data whilst dashed lines represent individual participant data. T. chuii, 25 mg/day T. chuii with hemicellulose crystalline as an excipient; placebo (PLA), PLA containing hemicellulose crystalline only; pre, pre-supplementation baseline; post, post-supplementation for 2 weeks. * denotes significant different (p < 0.05) from T. chuii-Pre.
4. Discussion
The principal original findings of the current study were that 2 weeks of T. chuii supplementation increased gene expression of NRF2, antioxidant enzymes (GPX7, GSR, GSTM3 and PRDX6), signalling kinases (ERK3 and p38a) and post-translational regulators (SIRT1) in human skeletal muscle tissue. Moreover, the magnitude to which T. chuii supplementation increased gene expression related to NRF2 signalling, transcription factors, redox balance and inflammation was inversely related to baseline gene expression; that is, those individuals that typically presented with the smallest basal gene expression exhibited a greater increase in gene expression following T. chuii supplementation. Supplementation with T. chuii also increased end-exercise during an exhaustive severe-intensity exercise test. These observations are broadly consistent with the experimental hypotheses and suggest that T. chuii supplementation can positively modulate NRF2 and antioxidant enzyme gene expression in skeletal muscle and increase in healthy adults. As such, T. chuii supplementation has the potential to confer beneficial effects on local skeletal muscle signalling and functional systemic physiological processes.
Whilst increased NRF2 gene expression has previously been reported in human myoblasts after T. chuii treatment [12], an important novel contribution of the current study is the observation that short-term dietary T. chuii supplementation increased NRF2 gene expression in human muscle tissue. A further important expansion of the data presented in the current manuscript compared to the previous paper by Ramírez and colleagues [12] is the use of OpenArray™ to assess expression of a far greater number of genes relating to redox and inflammatory signalling as well as transcripts with the potential to modulate NRF2 signalling. Principally, NRF2 is recognised as the master regulator of the cellular antioxidant response [30]. Accordingly, T. chuii supplementation increased skeletal muscle gene expression of the enzymatic antioxidants GPX7 and PRDX6 [31,32], which are integral to the elimination of the ROS, hydrogen peroxide (H2O2) [33]. GPX7 is an important sensor of oxidative stress and confers antioxidant effects indirectly through increasing protein activity of downstream targets [31]. On the other hand, PRDX6 can directly reduce H2O2 to water using reduced glutathione (GSH) as the reducing substrate, forming oxidised glutathione (GSSG) [32]. In addition, T. chuii supplementation increased GSR gene expression, which is important to sustain the flux of H2O2 reduction through PRDX6, as well as some other enzymatic antioxidants that use GSH as a reduction substrate (e.g., GPX and TXN), as GSR catalyses the reduction of GSSG of GSH [34]. There was also an increase in GSTM3 gene expression after T. chuii supplementation in the current study. Hydroperoxides are ROS that can be detoxified by PRDX6 [32] and via biotransformation (i.e., conjugation with GSH) by GSTM3 in the cytosol [35,36]. Therefore, the increases in skeletal muscle GPX7, PDRX6, GSR and GSTM3 gene expression after T. chuii supplementation indicates that muscle may be better equipped with enzymatic antioxidants to preserve a more optimal redox balance when challenged by pro-oxidative insults.
An interesting observation of the current study is an increase in skeletal muscle gene expression of PLA2G12A and PLA2G16 after T. chuii supplementation. Since PLA2 is recognised as a direct source of ROS in skeletal muscle and can indirectly increase ROS production through activation of nicotinamide adenine dinucleotide phosphate oxidase (NADPH) oxidases (NOX) [37], an increase in pro-oxidant enzymes (PLA2G12A and PLA2G16) alongside increases in antioxidant enzymes (GPX7, PDRX6, GSR and GSTM3) gene expression after T. chuii supplementation appears somewhat paradoxical. However, a physiological and low-dose increase in ROS production can increase NRF2 signalling to promote an up-regulation in endogenous antioxidant enzymes, consistent with the principle of hormetic nutrition, as reported with other purported dietary antioxidants such as (poly)phenols [38,39]. Increased skeletal muscle PLA2G12A and PLA2G16 may also have contributed to the activation of the redox-sensitive transcripts, ERK3 (MAPK6) and p38a (MAPK14), which contribute to the regulation of enzymatic antioxidants [40,41]. There were also increases in JUN and SIRT1 gene expression after T. chuii supplementation, which can aid NRF2 signalling. Indeed, JUN can dimerise with NRF2 to augment antioxidant response element (ARE)-mediated increases in expression of enzymatic antioxidants [42,43,44]. In addition, SIRT1 is positively associated with NRF2 content and deacetylates NRF2, which increases the stability and transcriptional activity of NRF2 and stimulates the transport of NRF2 to the nucleus to promote ARE effects [45]. Whilst these gene expression changes may collectively indicate that skeletal muscle NRF2 signalling was increased, CUL3 expression was also increased following T. chuii supplementation, with CUL3 being part of the KEAP1-CUL3-RBX1 E3 ubiquitin ligase complex which targets NRF2 for ubiquitination and proteasomal degradation [46,47]. Increased CUL3 expression, therefore, indicates a potential negative feedback response following an increase in NRF2 gene expression after T. chuii supplementation to modulate production of enzymatic antioxidants, which could promote an overly reduced cellular redox balance and compromise aspects of skeletal muscle function [48]. However, there is also evidence that CUL3 can ubiquitinate and subsequently degrade KEAP1 [49], which could aid NRF2 migration to the nucleus and up-regulate enzymatic antioxidants. Therefore, in aggregate, the gene expression data imply that muscle NRF2 signalling and antioxidant enzymes may have been increased after short-term T. chuii supplementation. These observations are important since previous purported nutritional NRF2 activators have largely shown no effect in human nutritional trials [16,17,18].
As well as increasing skeletal muscle gene expression of enzymatic antioxidants and associated signalling cascades, short-term T. chuii supplementation increased . This observation is consistent with a previous study which reported increased after 30 days of supplementation with 25 mg/day T. chuii [19]. Our findings expand this previous observation by indicating that a shorter duration of T. chuii supplementation can also increase . The concomitant increases in and gene expression of NRF2 and enzymatic antioxidants are also consistent with previous reports suggesting that these variables are positively associated [9,50]. In addition to its role as a regulator of antioxidant responses, NRF2 has been implicated in mitochondrial biogenesis [51]. Moreover, SIRT1 and MAPKs are also recognised transcripts to orchestrate mitochondrial biogenesis [52]. It is possible, therefore, that the increased after T. chuii supplementation could have been mediated by increased mitochondrial biogenesis. Whilst PGC1α is considered to be the master regulator of mitochondrial biogenesis and was not increased after T. chuii supplementation in the current study, alternative signalling mechanisms can contribute to mitochondrial biogenesis, including NRF2, SIRT1 and MAPKs, which were elevated after T. chuii supplementation in this study [52,53]. Whether NRF2, SIRT1 and MAPKs can increase mitochondrial biogenesis independent of PGC1α is unclear. It is also possible that gene expression of alternative transcription factors could have been increased after T. chuii supplementation in the current study and could have potentially elicited mitochondrial biogenesis via PGC1α-dependent or PGC1α-independent mechanisms. Further research is required to explore these possibilities. It is recognised that the lack of assessment of mitochondrial respiration and content markers is a limitation of this study. We also cannot exclude the possibility that increased after T. chuii supplementation could have been linked to improved skeletal muscle perfusion or that the supplement elicited effects in different organs to skeletal muscle, but further research is required to assess these possibilities. It is also acknowledged that since skeletal muscle gene expression and were assessed in two separate studies after T. chuii supplementation, this compromises direct comparison of the link between these responses in the current manuscript. Further research is also required to assess whether T. chuii supplementation can improve other cardiorespiratory variables during exercise, such as ventilatory efficiency (/) [54]. The increase in is of functional significance since it is positively associated with exercise capacity and performance [55] and inversely associated with ill health and mortality [56,57]. Given the increase in , it is surprising, therefore, that T. chuii supplementation did not significantly increase Tlim in the current study. It is possible that we were statistically underpowered to detect this 17% increase in Tlim compared to baseline and, as such, further research is required to assess the ergogenic potential of T. chuii supplementation in different exercise settings. It is also possible that, since PLA elicited a non-significant increase in Tlim of 11% compared to baseline, there may have been an insufficient washout between conditions for participants who underwent the T. chuii supplementation arm of the study first.
Whilst the current study observed increases in skeletal muscle gene expression of key enzymatic antioxidants (GPX7, GSR, GSTM3, and PRDX6), signalling kinases (ERK3 and p38a), post-translational regulators (SIRT1), and transcription factors (NRF2), a limitation of the current study is that proteomic analyses were not completed to determine whether these transcriptional responses translated into increased protein expression. Further research is therefore required to probe the molecular bases by which T. chuii supplementation can improve cell signalling in skeletal muscle, and other tissues, alongside the potential for wider health benefits. It is also recognised that, since the participants in the current study were all healthy and active, the findings of the current study may not translate to different populations. It is of interest to determine whether individuals presenting with a more pro-oxidative or proinflammatory state, such as older adults or clinical populations, would benefit more from NRF2 activation and associated antioxidant responses that may ensue following T. chuii supplementation. Indeed, participants presenting with depleted antioxidant status appear more likely to improve exercise capacity and skeletal muscle function after dietary antioxidant supplementation [58]. Consistent with this, there was an inverse correlation between basal expression of 53 genes and the magnitude of increase in these genes after T. chuii supplementation. Moreover, some of the skeletal muscle gene expression changes reported herein may improve understanding of the molecular bases for improved post-exercise recovery reported in previous studies after the same T. chuii supplementation protocol administered herein [13,14,15].
5. Conclusions
Short-term supplementation with T. chuii increased skeletal muscle gene expression of NRF2, positive NRF2 signalling mediators (JUN and SIRT1) and antioxidant enzymes (GPX7, PDRX6, GSR and GSTM3). T. chuii therefore appears to hold promise as a nutritional NRF2 signalling and antioxidant precursor, at least in healthy human skeletal muscle. In addition, T. chuii supplementation increased and since the supplement also increased skeletal muscle NRF2, MAPK, and SIRT1 gene expression, which are key mediators of mitochondrial biogenesis, the increased could be a function of enhanced mitochondrial capacity. Therefore, these data suggest T. chuii may represent a natural and blue-food supplement with the potential to improve skeletal muscle antioxidant responses and systemic oxidative metabolism in healthy adult humans.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/antiox14040435/s1, File S1. CONSORT 2010 checklist of information to include when reporting a randomised trial.
Author Contributions
Conceptualization, S.P.C., N.C.B., N.R.W.M., A.J.W. and S.J.B.; methodology, S.P.C., J.P.A., N.S.A., R.A.F., N.C.B., N.R.W.M., C.I., S.T., C.L., L.M., M.M., A.J.W. and S.J.B.; formal analysis, S.P.C. and S.J.B.; investigation, S.P.C., J.P.A., N.S.A., T.C., R.A.F., N.C.B., N.R.W.M., A.J.W. and S.J.B.; resources, M.M.; data curation, S.P.C. and S.J.B.; writing—original draft preparation, S.P.C. and S.J.B.; writing—review and editing, S.P.C., J.P.A., N.S.A., T.C., R.A.F., N.C.B., N.R.W.M., C.I., L.M., M.M., A.J.W. and S.J.B.; methodology, S.P.C., J.P.A., N.S.A., R.A.F., N.C.B., N.R.W.M., C.I., S.T., C.L., L.M., M.M., A.J.W. and S.J.B.; visualization, S.P.C. and S.J.B.; supervision, R.A.F., N.C.B., N.R.W.M. and S.J.B.; funding acquisition, S.J.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by a research grant to S.J.B. from Fitoplancton Marino, S.L.
Institutional Review Board Statement
All experimental procedures were approved by Loughborough University Research Ethics Approvals Human Participants Sub Committee.
Informed Consent Statement
Participants were fully informed of the risks and discomforts associated with all trials and provided written, informed consent.
Data Availability Statement
The original contributions presented in this study are included in the article/Supplementary Materials. Further inquiries can be directed to the corresponding author(s).
Acknowledgments
This research was supported by the NIHR Leicester Biomedical Research Centre.
Conflicts of Interest
C.I., S.T., C.L. and L.M. are employed by Fitoplancton Marino who funded the current work. The funders had no role in the design of the study; in the collection or interpretation of data; or in the decision to publish the results. Whilst the funders did assist in the OpenArray™ gene expression measurements, all measures were conducted with the funders blinded to the supplement order. The funders provided some comments on the manuscript draft but the final decision on the manuscript content was made by S.J.B.
References
- Rattan, S.I. Hormesis in aging. Ageing Res. Rev. 2008, 7, 63–78. [Google Scholar] [CrossRef] [PubMed]
- Calabrese, E.J.; Dhawan, G.; Kapoor, R.; Iavicoli, I.; Calabrese, V. HORMESIS: A Fundamental Concept with Widespread Biological and Biomedical Applications. Gerontology 2016, 62, 530–535. [Google Scholar] [PubMed]
- McCord, J.M.; Gao, B.; Hybertson, B.M. The Complex Genetic and Epigenetic Regulation of the Nrf2 Pathways: A Review. Antioxidants 2023, 12, 366. [Google Scholar] [CrossRef]
- Hayes, J.D.; Dinkova-Kostova, A.T. The Nrf2 Regulatory Network Provides an Interface between Redox and Intermediary Metabolism. Trends Biochem. Sci. 2014, 39, 199–218. [Google Scholar] [CrossRef]
- He, F.; Ru, X.; Wen, T. NRF2, a Transcription Factor for Stress Response and Beyond. Int. J. Mol. Sci. 2020, 21, 4777. [Google Scholar] [CrossRef]
- Onoki, T.; Izumi, Y.; Takahashi, M.; Murakami, S.; Matsumaru, D.; Ohta, N.; Wati, S.M.; Hatanaka, N.; Katsuoka, F.; Okutsu, M.; et al. Skeletal muscle-specific Keap1 disruption modulates fatty acid utilization and enhances exercise capacity in female mice. Redox Biol. 2021, 43, 101966. [Google Scholar] [CrossRef]
- Wang, L.; Yang, S.; Yan, L.; Wei, H.; Wang, J.; Yu, S.; Kong, A.T.; Zhang, Y. Hypoxia preconditioning promotes endurance exercise capacity of mice by activating skeletal muscle Nrf2. J. Appl. Physiol. 2019, 127, 1267–1277. [Google Scholar] [CrossRef]
- Gao, L.; Kumar, V.; Vellichirammal, N.N.; Park, S.Y.; Rudebush, T.L.; Yu, L.; Son, W.M.; Pekas, E.J.; Wafi, A.M.; Hong, J.; et al. Functional, proteomic and bioinformatic analyses of Nrf2- and Keap1- null skeletal muscle. J. Physiol. 2020, 598, 5427–5451. [Google Scholar] [CrossRef]
- Martinez-Canton, M.; Galvan-Alvarez, V.; Martin-Rincon, M.; Calbet, J.A.L.; Gallego-Selles, A. Unlocking peak performance: The role of Nrf2 in enhancing exercise outcomes and training adaptation in humans. Free Radic. Biol. Med. 2024, 224, 168–181. [Google Scholar] [CrossRef]
- Cokdinleyen, M.; Alvarez-Rivera, G.; Tejera, J.L.G.; Mendiola, J.A.; Valdés, A.; Kara, H.; Ibáñez, E.; Cifuentes, A. Tetraselmis chuii Edible Microalga as a New Source of Neuroprotective Compounds Obtained Using Fast Biosolvent Extraction. Int. J. Mol. Sci. 2024, 25, 3897. [Google Scholar] [CrossRef]
- Mantecón, L.; Moyano, R.; Cameán, A.M.; Jos, A. Safety assessment of a lyophilized biomass of Tetraselmis chuii (TetraSOD®) in a 90 day feeding study. Food Chem. Toxicol. 2019, 133, 110810. [Google Scholar] [CrossRef]
- Ramírez, P.; Torres, S.; Lama, C.; Mantecón, L.; Unamunzaga, C.; Infante, C. TetraSOD® activates the antioxidant response pathway in human cells: An in vitro approach. Afr. J. Biotechnol. 2020, 19, 367–373. [Google Scholar]
- Sharp, M.; Sahin, K.; Stefan, M.; Orhan, C.; Gheith, R.; Reber, D.; Sahin, N.; Tuzcu, M.; Lowery, R.; Durkee, S.; et al. Phytoplankton Supplementation Lowers Muscle Damage and Sustains Performance across Repeated Exercise Bouts in Humans and Improves Antioxidant Capacity in a Mechanistic Animal. Nutrients 2020, 12, 1990. [Google Scholar] [CrossRef]
- Sharp, M.H.; Sahin, K.; Stefan, M.W.; Gheith, R.H.; Reber, D.D.; Ottinger, C.R.; Orhan, C.; Tuzcu, M.; Sahin, N.; Lowery, R.P.; et al. Marine Phytoplankton Improves Exercise Recovery in Humans and Activates Repair Mechanisms in Rats. Int. J. Sports Med. 2021, 42, 1070–1082. [Google Scholar] [CrossRef]
- Sharp, M.; Wilson, J.; Stefan, M.; Gheith, R.; Lowery, R.; Ottinger, C.; Reber, D.; Orhan, C.; Sahin, N.; Tuzcu, M.; et al. Marine phytoplankton improves recovery and sustains immune function in humans and lowers proinflammatory immunoregulatory cytokines in a rat model. Phys. Act. Nutr. 2021, 25, 42–55. [Google Scholar] [CrossRef]
- Clifford, T.; Acton, J.P.; Cocksedge, S.P.; Davies, K.A.B.; Bailey, S.J. The effect of dietary phytochemicals on nuclear factor erythroid 2-related factor 2 (Nrf2) activation: A systematic review of human intervention trials. Mol. Biol. Rep. 2021, 48, 1745–1761. [Google Scholar] [CrossRef]
- Thorley, J.; Thomas, C.; Thon, N.; Nuttall, H.; Martin, N.R.W.; Bishop, N.; Bailey, S.J.; Clifford, T. Combined effects of green tea supplementation and eccentric exercise on nuclear factor erythroid 2-related factor 2 activity. Eur. J. Appl. Physiol. 2024, 124, 245–256. [Google Scholar] [CrossRef]
- Thorley, J.; Alhebshi, A.; Rodriguez-Mateos, A.; Zhang, Z.; Bailey, S.J.; Martin, N.R.W.; Bishop, N.C.; Clifford, T. Acute supplementation with a curcuminoid-based formulation fails to enhance resting or exercise-induced NRF2 activity in males and females. Food Funct. 2024, 15, 10782–10794. [Google Scholar] [CrossRef]
- Toro, V.; Siquier-Coll, J.; Bartolomé, I.; Robles-Gil, M.C.; Rodrigo, J.; Maynar-Mariño, M. Effects of Tetraselmis chuii Microalgae Supplementation on Ergospirometric, Haematological and Biochemical Parameters in Amateur Soccer Players. Int. J. Environ. Res. Public Health 2020, 17, 6885. [Google Scholar] [CrossRef]
- García, Á.; Toro-Román, V.; Siquier-Coll, J.; Bartolomé, I.; Muñoz, D.; Maynar-Mariño, M. Effects of Tetraselmis chuii Microalgae Supplementation on Anthropometric, Hormonal and Hematological Parameters in Healthy Young Men: A Double-Blind Study. Int. J. Environ. Res. Public Health 2022, 19, 6060. [Google Scholar] [CrossRef]
- Sies, H.; Jones, D.P. Reactive oxygen species (ROS) as pleiotropic physiological signalling agents. Nat. Rev. Mol. Cell Biol. 2020, 21, 363–383. [Google Scholar] [CrossRef]
- Nikolaidis, M.G.; Margaritelis, N.V.; Matsakas, A. Quantitative Redox Biology of Exercise. Int. J. Sports Med. 2020, 41, 633–645. [Google Scholar] [CrossRef]
- Westerblad, H.; Allen, D.G. Emerging roles of ROS/RNS in muscle function and fatigue. Antioxid. Redox Signal. 2011, 15, 2487–2499. [Google Scholar] [CrossRef]
- Uhlen, M.; Oksvold, P.; Fagerberg, L.; Lundberg, E.; Jonasson, K.; Forsberg, M.; Zwahlen, M.; Kampf, C.; Wester, K.; Hober, S.; et al. Towards a knowledge-based Human Protein Atlas. Nat. Biotechnol. 2010, 28, 1248–1250. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔC(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Silver, N.; Best, S.; Jiang, J.; Thein, S.L. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 2006, 7, 33. [Google Scholar] [CrossRef]
- Kuang, J.; McGinley, C.; Lee, M.J.-C.; Saner, N.J.; Garnham, A.; Bishop, D.J. Interpretation of exericse-induced changes in human skeletal muscle mRNA expression depends on the timing of the post-exercise biopsies. Peer J. 2022, 10, e12856. [Google Scholar] [CrossRef]
- Bailey, S.J.; Wilkerson, D.P.; Dimenna, F.J.; Jones, A.M. Influence of repeated sprint training on pulmonary O2 uptake and muscle deoxygenation kinetics in humans. J. Appl. Physiol. 2009, 106, 1875–1887. [Google Scholar] [CrossRef]
- Whipp, B.J.; Davis, J.A.; Torres, F.; Wasserman, K. A test to determine parameters of aerobic function during exercise. J. Appl. Physiol. Respir. Environ. Exerc. Physiol. 1981, 50, 217–221. [Google Scholar] [CrossRef]
- Vomund, S.; Schäfer, A.; Parnham, M.J.; Brüne, B.; von Knethen, A. Nrf2, the Master Regulator of Anti-Oxidative Responses. Int. J. Mol. Sci. 2017, 18, 2772. [Google Scholar] [CrossRef]
- Pei, J.; Pan, X.; Wei, G.; Hua, Y. Research Progress of Glutathione Peroxidase Family (GPX) in Redoxidation. Front. Pharmacol. 2023, 14, 1147414. [Google Scholar] [CrossRef]
- Manevich, Y.; Fisher, A.B. Peroxiredoxin 6, a 1-Cys peroxiredoxin, functions in antioxidant defense and lung phospholipid metabolism. Free Radic. Biol. Med. 2005, 38, 1422–1432. [Google Scholar] [CrossRef]
- Wadley, A.J.; Aldred, S.; Coles, S.J. An unexplored role for Peroxiredoxin in exercise-induced redox signalling? Redox Biol. 2016, 8, 51–58. [Google Scholar] [CrossRef]
- Averill-Bates, D.A. The antioxidant glutathione. Vitam. Horm. 2023, 121, 109–141. [Google Scholar] [CrossRef]
- Grillo, M.P. Bioactivation by Phase-II-Enzyme-Catalyzed Conjugation of Xenobiotics. In Encyclopedia of Drug Metabolism and Interactions; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2012. [Google Scholar] [CrossRef]
- Hayes, J.D.; McLellan, L.I. Glutathione and glutathione-dependent enzymes represent a co-ordinately regulated defence against oxidative stress. Free Radic. Res. 1999, 31, 273–300. [Google Scholar] [CrossRef]
- Barbieri, E.; Sestili, P. Reactive oxygen species in skeletal muscle signaling. J. Signal Transduct. 2012, 2012, 982794. [Google Scholar] [CrossRef]
- Murakami, A. Non-specific protein modifications may be novel mechanism underlying bioactive phytochemicals. J. Clin. Biochem. Nutr. 2018, 62, 115–123. [Google Scholar] [CrossRef]
- Scuto, M.; Rampulla, F.; Reali, G.M.; Spanò, S.M.; Trovato Salinaro, A.; Calabrese, V. Hormetic Nutrition and Redox Regulation in Gut-Brain Axis Disorders. Antioxidants 2024, 13, 484. [Google Scholar] [CrossRef]
- L’honoré, A.; Commère, P.H.; Negroni, E.; Pallafacchina, G.; Friguet, B.; Drouin, J.; Buckingham, M.; Montarras, D. The role of Pitx2 and Pitx3 in muscle stem cells gives new insights into P38α MAP kinase and redox regulation of muscle regeneration. eLife 2018, 7, e32991. [Google Scholar] [CrossRef]
- Ji, L.L.; Kang, C.; Zhang, Y. Exercise-induced hormesis and skeletal muscle health. Free Radic. Biol. Med. 2016, 98, 113–122. [Google Scholar] [CrossRef]
- Fittipaldi, S.; Mercatelli, N.; Dimauro, I.; Jackson, M.J.; Paronetto, M.P.; Caporossi, D. Alpha B-crystallin induction in skeletal muscle cells under redox imbalance is mediated by a JNK-dependent regulatory mechanism. Free Radic. Biol. Med. 2015, 86, 331–342. [Google Scholar] [CrossRef] [PubMed]
- Jeyapaul, J.; Jaiswal, A.K. Nrf2 and c-Jun regulation of antioxidant response element (ARE)-mediated expression and induction of gamma-glutamylcysteine synthetase heavy subunit gene. Biochem. Pharmacol. 2000, 59, 1433–1439. [Google Scholar] [CrossRef]
- Venugopal, R.; Jaiswal, A.K. Nrf2 and Nrf1 in association with Jun proteins regulate antioxidant response element-mediated expression and coordinated induction of genes encoding detoxifying enzymes. Oncogene 1998, 17, 3145–3156. [Google Scholar] [CrossRef]
- Singh, V.; Ubaid, S. Role of Silent Information Regulator 1 (SIRT1) in Regulating Oxidative Stress and Inflammation. Inflammation 2020, 43, 1589–1598. [Google Scholar] [CrossRef]
- Baird, L.; Yamamoto, M. The Molecular Mechanisms Regulating the NRF2 Pathway. Mol. Cell Biol. 2020, 40, e00099-20. [Google Scholar] [CrossRef]
- Dinkova-Kostova, A.T.; Holtzclaw, W.D.; Kensler, T.W. The role of Keap1 in cellular protective responses. Chem. Res. Toxicol. 2005, 18, 1779–1791. [Google Scholar] [CrossRef]
- Reid, M.B. Invited Review: Redox modulation of skeletal muscle contraction: What we know and what we don’t. J. Appl. Physiol. 2001, 90, 724–731. [Google Scholar] [CrossRef]
- Zhang, D.D.; Lo, S.C.; Sun, Z.; Habib, G.M.; Lieberman, M.W.; Hannink, M. Ubiquitination of Keap1, a BTB-Kelch Substrate Adaptor Protein for Cul3, Targets Keap1 for Degradation by a Proteasome-Independent Pathway. J. Biol. Chem. 2005, 280, 30091–30099. [Google Scholar] [CrossRef]
- Tranah, G.J.; Barnes, H.N.; Cawthon, P.M.; Coen, P.M.; Esser, K.A.; Hepple, R.T.; Huo, Z.; Kramer, P.A.; Toledo, F.G.S.; Zhang, X.; et al. Expression of mitochondrial oxidative stress response genes in muscle is associated with mitochondrial respiration, physical performance, and muscle mass in the Study of Muscle, Mobility, and Aging. Aging Cell 2024, 23, e14114. [Google Scholar] [CrossRef]
- Islam, H.; Bonafiglia, J.T.; Turnbull, P.C.; Simpson, C.A.; Perry, C.G.R.; Gurd, B.J. The impact of acute and chronic exercise on Nrf2 expression in relation to markers of mitochondrial biogenesis in human skeletal muscle. Eur. J. Appl. Physiol. 2020, 120, 149–160. [Google Scholar] [CrossRef]
- Russell, A.P.; Foletta, V.C.; Snow, R.J.; Wadley, G.D. Skeletal muscle mitochondria: A major player in exercise, health and disease. Biochim. Biophys. Acta 2014, 1840, 1276–1284. [Google Scholar] [CrossRef]
- Islam, H.; Hood, D.A.; Gurd, B.J. Looking beyond PGC-1α: Emerging regulators of exercise-induced skeletal muscle mitochondrial biogenesis and their activation by dietary compounds. Appl. Physiol. Nutr. Metab. 2020, 45, 11–23. [Google Scholar] [CrossRef]
- Kasiak, P.; Kowalski, T.; Rębiś, K.; Klusiewicz, A.; Ładyga, M.; Sadowska, D.; Wilk, A.; Wiecha, S.; Barylski, M.; Poliwczak, A.R.; et al. Is the Ventilatory Efficiency in Endurance Athletes Different?-Findings from the NOODLE Study. J. Clin. Med. 2024, 13, 490. [Google Scholar] [CrossRef]
- Joyner, M.J.; Coyle, E.F. Endurance exercise performance: The physiology of champions. J. Physiol. 2008, 586, 35–44. [Google Scholar]
- Kurl, S.; Hakkarainen, P.; Voutilainen, A.; Lönnroos, E. Combined effects of maximal oxygen uptake and glucose status on mortality: The Prospective KIHD cohort study. Scand. J. Med. Sci. Sports 2022, 32, 913–923. [Google Scholar]
- Ung, G.A.; Nguyen, K.H.; Hui, A.; Wong, N.D.; Dineen, E.H. Impact of cardiorespiratory fitness and diabetes status on cardiovascular disease and all-cause mortality: An NHANES retrospective cohort study. Am. Heart J. Plus 2024, 42, 100395. [Google Scholar] [CrossRef]
- Margaritelis, N.V.; Paschalis, V.; Theodorou, A.A.; Kyparos, A.; Nikolaidis, M.G. Antioxidant supplementation, redox deficiencies and exercise performance: A falsification design. Free Radic. Biol. Med. 2020, 158, 44–52. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).