(−)-Methyl-Oleocanthal, a New Oleocanthal Metabolite Reduces LPS-Induced Inflammatory and Oxidative Response: Molecular Signaling Pathways and Histones Epigenetic Modulation
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Instruments
2.3. Isolation of (−)-Oleocanthal (OLE) from Olive Oil
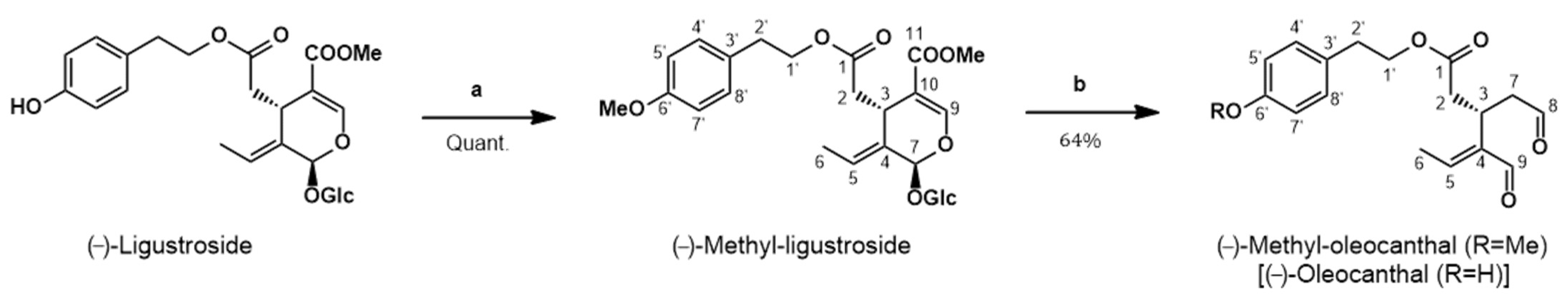
2.4. Synthesis of (−)-Methyl-Oleocanthal (Met-OLE) from (−)-Ligustroside
2.5. Animals
2.6. Murine Macrophages and Spleen Cells Isolation and Culture
2.7. Cell Viability
2.8. Nitric Oxide Production
2.9. Intracellular ROS Production
2.10. Histone Extraction
2.11. Enzyme-Linked Immunosorbent Assay
2.12. Western Blotting
2.13. Statistical Analysis
3. Results
3.1. Chemistry
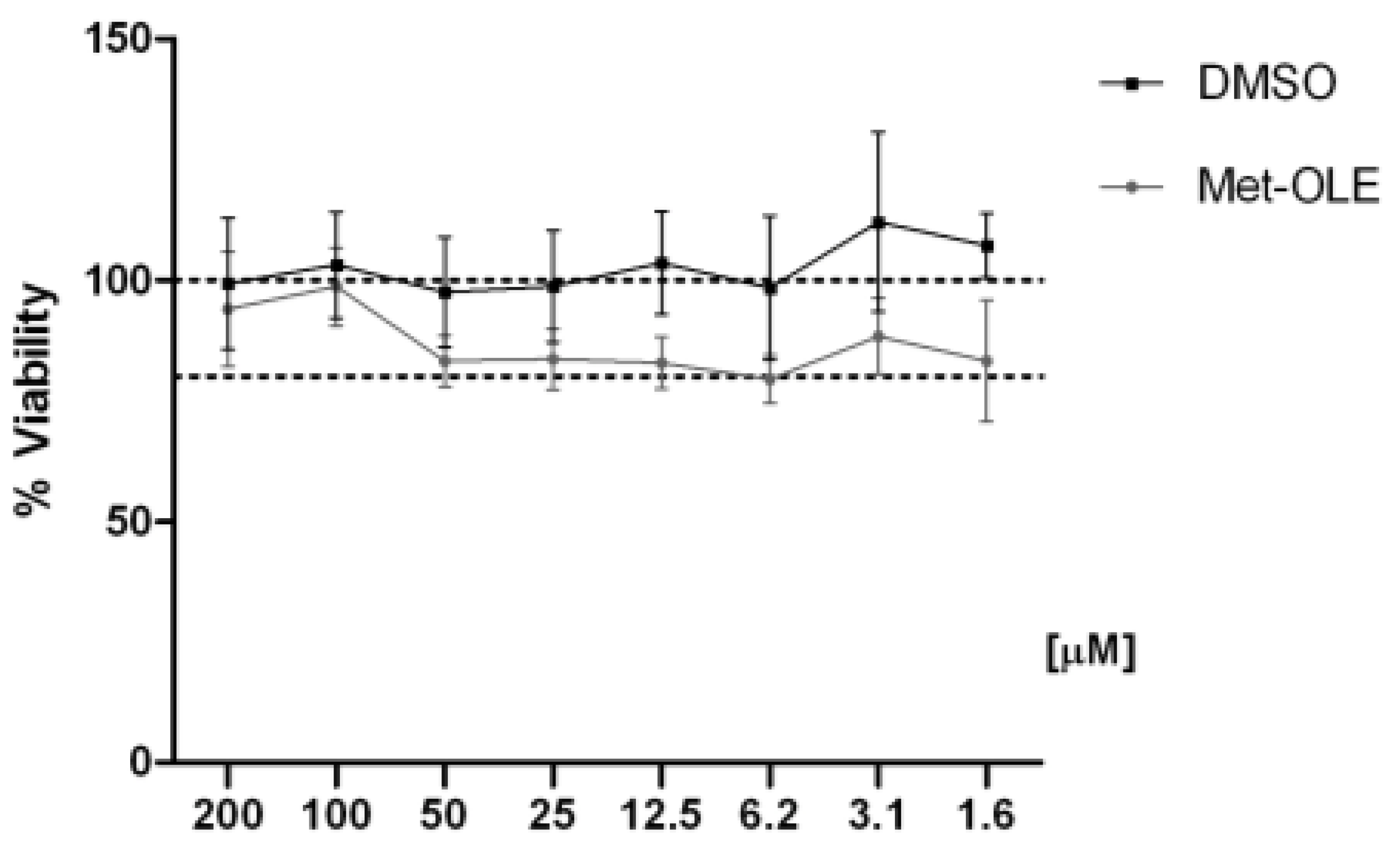
3.2. Effects of Met-OLE on Cell Viability
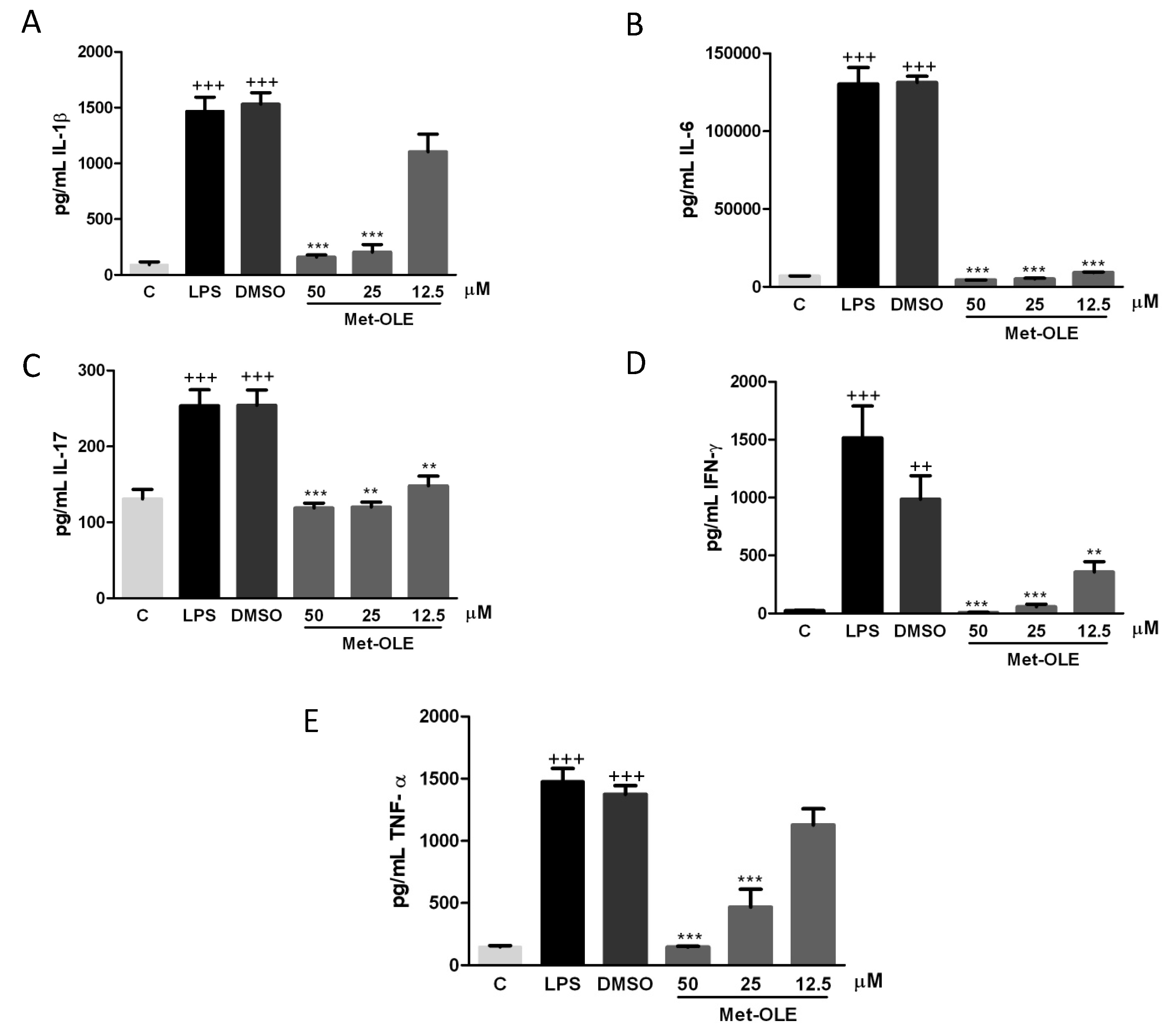
3.3. Effects of Met-OLE on IL-1β, IL-6, IL-17, IFN-γ and TNF-α Production
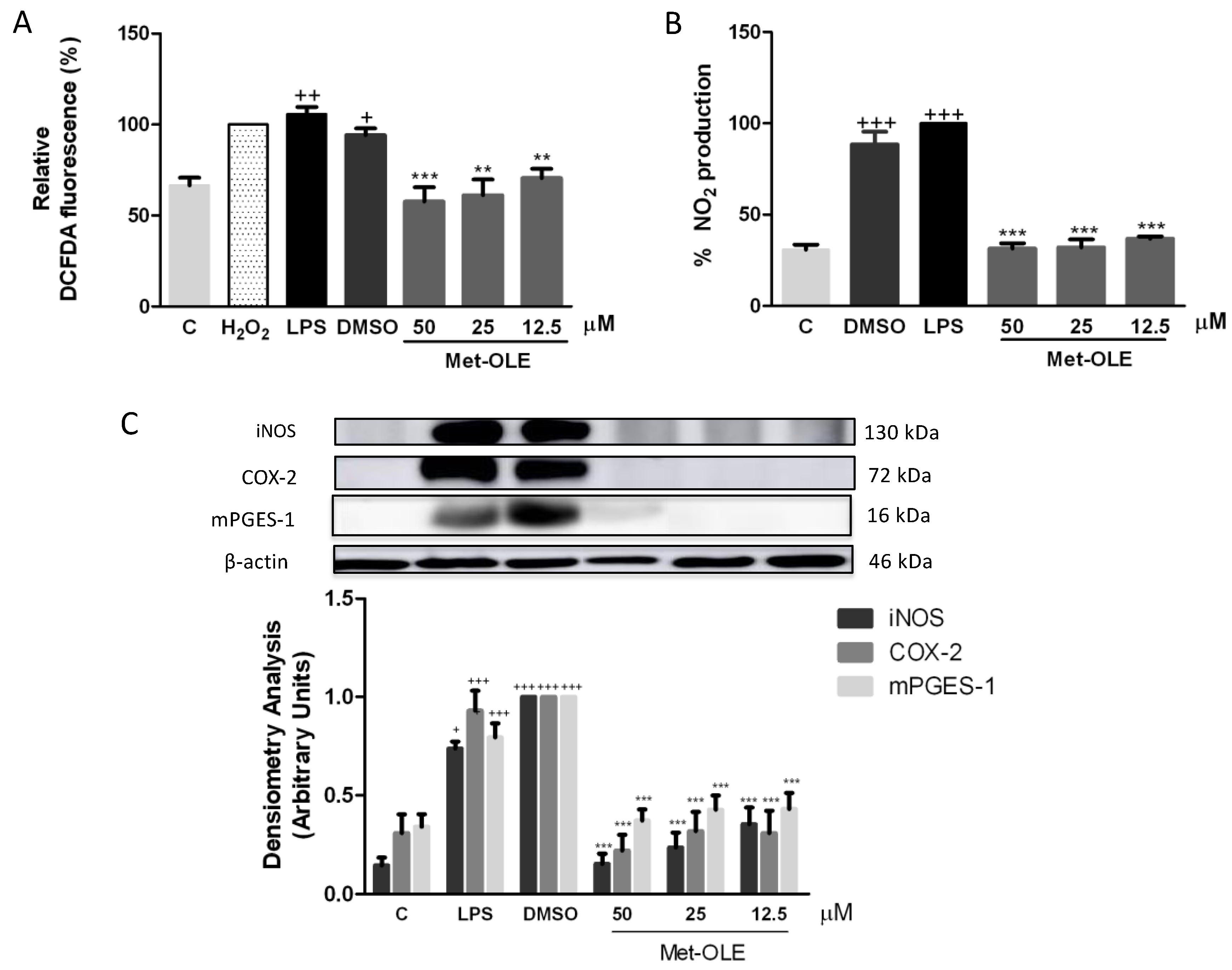
3.4. Effects of Met-OLE on Intracellular ROS and NO Productions
3.5. Met-OLE Down-Regulated iNOS, COX-2 and mPGES-1 Protein Overexpression Induced by LPS in Murine Macrophages
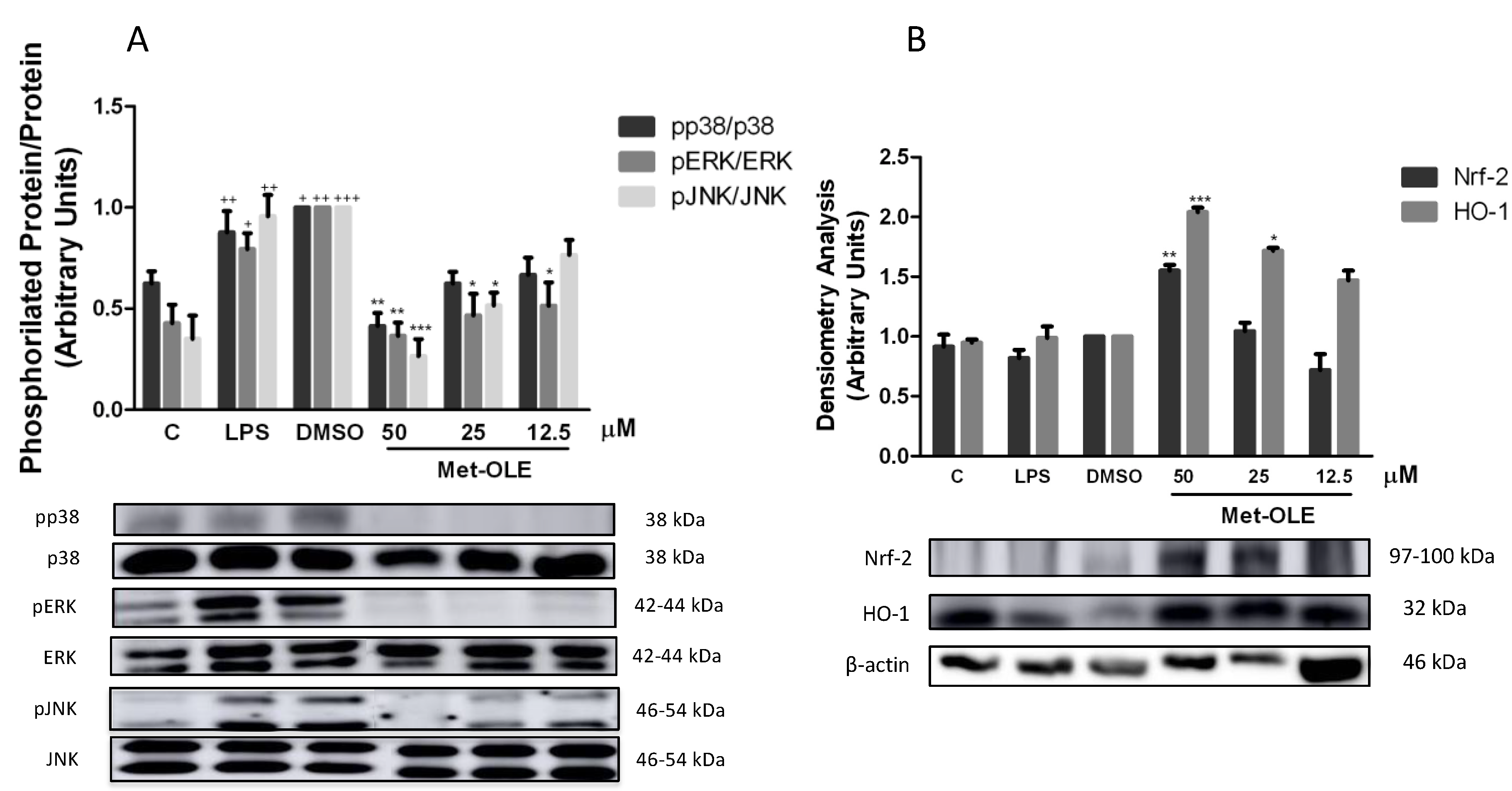
3.6. Effects of Met-OLE on LPS-Induced MAPKs Activation in Murine Peritoneal Macrophages
3.7. Effects of Met-OLE on Nrf2-Mediated Transcriptional Activation and HO-1 Induction in LPS Murine Peritoneal Macrophages
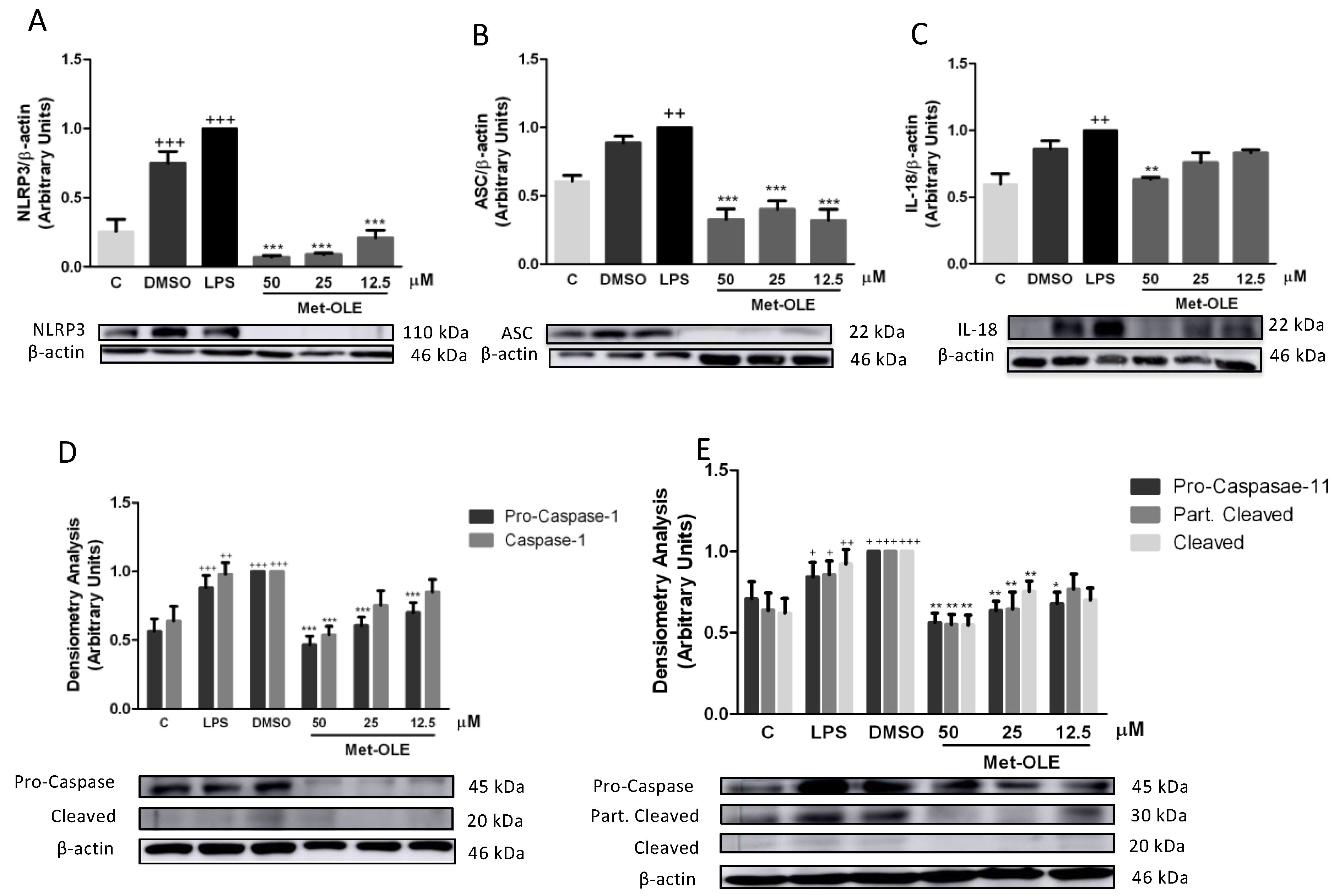
3.8. Effects of Met-OLE on Canonical and Noncanonical Inflammasome Signaling Pathways
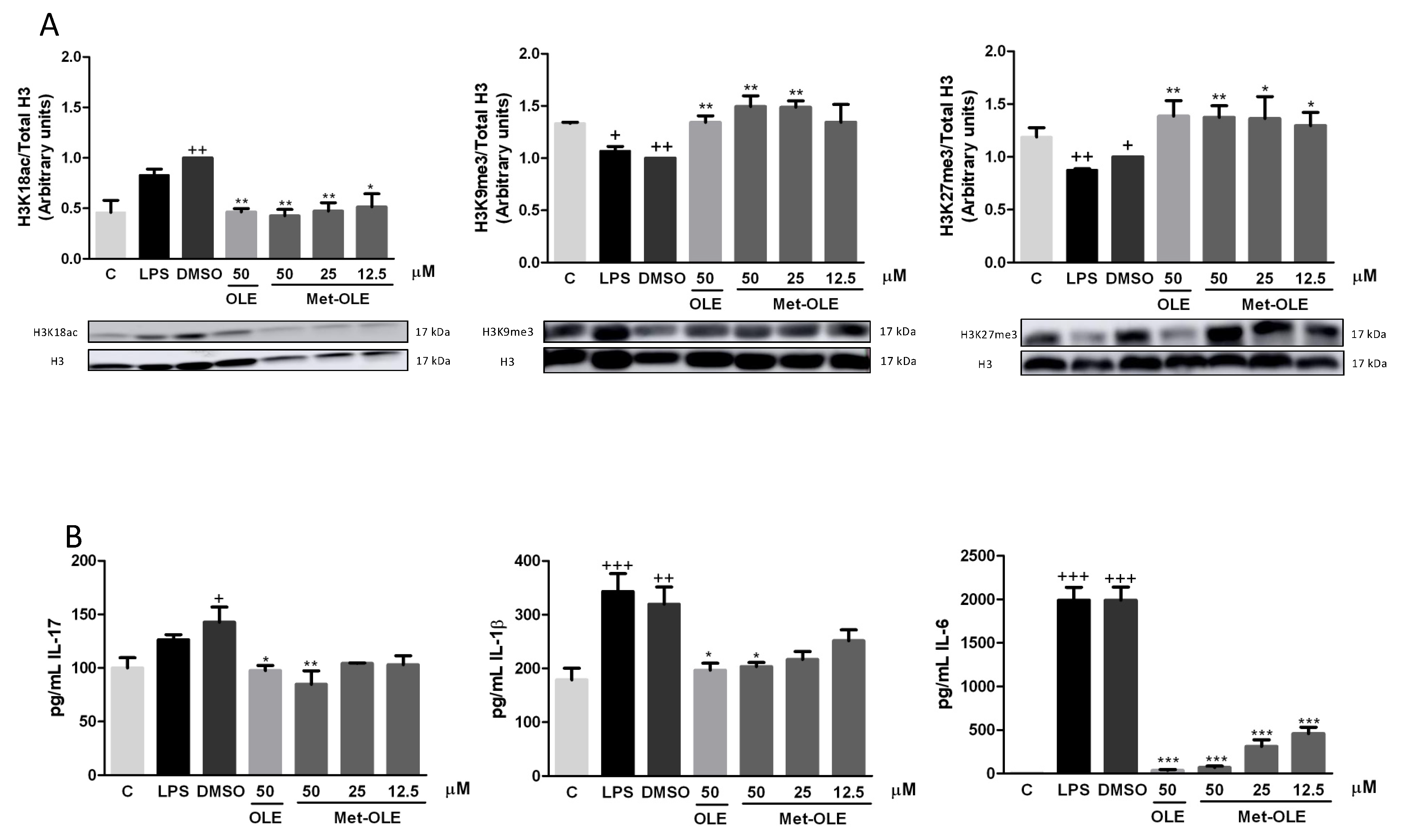
3.9. OLE and Met-OLE Induced Epigenetic Histone Modifications
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wynn, T.A.; Chawla, A.; Pollard, J.W. Macrophage biology in development, homeostasis and disease. Nature 2013, 496, 445–455. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Liu, K.; Zhao, Y.; Hu, X.; Wang, M. Pterostilbene and 4′-Methoxyresveratrol Inhibited Lipopolysaccharide-Induced Inflammatory Response in RAW264.7 Macrophages. Molecules 2018, 23, 1148. [Google Scholar] [CrossRef] [PubMed]
- Brüne, B.; Dehne, N.; Grossmann, N.; Jung, M.; Namgaladze, D.; Schmid, T.; Von Knethen, A.; Weigert, A. Redox Control of Inflammation in Macrophages. Antioxid. Redox Signal. 2013, 19, 595–637. [Google Scholar] [CrossRef] [PubMed]
- Shanmugam, M.K.; Sethi, G. Role of Epigenetics in Inflammation-Associated Diseases. Epigenetics Dev. Dis. 2013, 61, 627–657. [Google Scholar] [CrossRef]
- Hajji, N.; Joseph, B. Epigenetic regulation of cell life and death decisions and deregulation in cancer. Essays Biochem. 2010, 48, 121–146. [Google Scholar] [CrossRef]
- Zhao, S.; Zhong, Y.; Fu, X.; Wang, Y.; Ye, P.; Cai, J.; Liu, Y.; Sun, J.; Mei, Z.; Jiang, Y.; et al. H3K4 Methylation Regulates LPS-Induced Proinflammatory Cytokine Expression and Release in Macrophages. Shock 2019, 51, 401–406. [Google Scholar] [CrossRef]
- Imuta, H.; Fujita, D.; Oba, S.; Kiyosue, A.; Nishimatsu, H.; Yudo, K.; Suzuki, E. Histone methylation and demethylation are implicated in the transient and sustained activation of the interleukin-1β gene in murine macrophages. Hear. Vessel. 2020, 35, 1746–1754. [Google Scholar] [CrossRef]
- Hachiya, R.; Shiihashi, T.; Shirakawa, I.; Iwasaki, Y. The H3K9 methyltransferase Setdb1 regulates TLR4-mediated inflam-matory responses in macrophages. Sci. Rep. 2016, 28, 28845. [Google Scholar] [CrossRef]
- Bordoni, L.; Fedeli, D.; Fiorini, D.; Gabbianelli, R. Extra Virgin Olive Oil and Nigella sativa Oil Produced in Central Italy: A Comparison of the Nutrigenomic Effects of Two Mediterranean Oils in a Low-Grade Inflammation Model. Antioxidants 2019, 9, 20. [Google Scholar] [CrossRef]
- Castejón, M.L.; Montoya, T.; Alarcón-De-La-Lastra, C.; Sánchez-Hidalgo, M. Potential Protective Role Exerted by Secoiridoids from Olea europaea L. in Cancer, Cardiovascular, Neurodegenerative, Aging-Related, and Immunoinflammatory Diseases. Antioxidants 2020, 9, 149. [Google Scholar] [CrossRef]
- Fabiani, R.; Vella, N.; Rosignoli, P. Epigenetic Modifications Induced by Olive Oil and Its Phenolic Compounds: A Systematic Review. Molecules 2021, 26, 273. [Google Scholar] [CrossRef] [PubMed]
- Cicerale, S.; Conlan, X.; Barnett, N.W.; Keast, R. Storage of extra virgin olive oil and its effect on the biological activity and concentration of oleocanthal. Food Res. Int. 2013, 50, 597–602. [Google Scholar] [CrossRef]
- Segura-Carretero, A.; Curiel, J.A. Current Disease-Targets for Oleocanthal as Promising Natural Therapeutic Agent. Int. J. Mol. Sci. 2018, 19, 2899. [Google Scholar] [CrossRef]
- Montoya, T.; Castejón, M.L.; Sánchez-Hidalgo, M.; González-Benjumea, A.; Fernández-Bolaños, J.G.; De-La-Lastra, C.A. Oleocanthal Modulates LPS-Induced Murine Peritoneal Macrophages Activation via Regulation of Inflammasome, Nrf-2/HO-1, and MAPKs Signaling Pathways. J. Agric. Food Chem. 2019, 67, 5552–5559. [Google Scholar] [CrossRef]
- Montoya, T.; Sánchez-Hidalgo, M.; Castejón, M.; Rosillo, M.; González-Benjumea, A.; Alarcón-De-La-Lastra, C. Dietary Oleocanthal Supplementation Prevents Inflammation and Oxidative Stress in Collagen-Induced Arthritis in Mice. Antioxidants 2021, 10, 650. [Google Scholar] [CrossRef] [PubMed]
- López-Yerena, A.; Vallverdú-Queralt, A.; Jáuregui, O.; Garcia-Sala, X.; Lamuela-Raventós, R.; Escribano-Ferrer, E. Tissue Distribution of Oleocanthal and Its Metabolites after Oral Ingestion in Rats. Antioxidants 2021, 10, 688. [Google Scholar] [CrossRef] [PubMed]
- Wen, L.; Jiang, Y.; Yang, J.; Zhao, Y.; Tian, M.; Yang, B. Structure, bioactivity, and synthesis of methylated flavonoids. Ann. N. Y. Acad. Sci. 2017, 1398, 120–129. [Google Scholar] [CrossRef] [PubMed]
- Diez-Bello, R.; Jardin, I.; Lopez, J.; El Haouari, M.; Ortega-Vidal, J.; Altarejos, J.; Salido, G.; Salido, S.; Rosado, J. (−)-Oleocanthal inhibits proliferation and migration by modulating Ca2+ entry through TRPC6 in breast cancer cells. Biochim. et Biophys. Acta (BBA)-Bioenerg. 2019, 1866, 474–485. [Google Scholar] [CrossRef]
- Pérez-Bonilla, M.; Salido, S.; van Beek, T.A.; Linares-Palomino, P.J.; Altarejos, J.; Nogueras, M.; Sánchez, A. Isolation and identification of radical scavengers in olive tree (Olea europaea) wood. J. Chromatogr. A 2006, 1112, 311–318. [Google Scholar] [CrossRef]
- Chang, H.C.; Wang, S.W.; Chen, C.Y.; Hwang, T.L.; Cheng, M.J.; Sung, P.J.; Liao, K.W.; Chen, J.J. Secoiridoid glucosides and an-ti-inflammatory constituents from the stem bark of Fraxinus chinensis. Molecules 2020, 25, 5911. [Google Scholar] [CrossRef]
- Skehan, P.; Storeng, R.; Scudiero, D.; Monks, A.; McMahon, J.M.; Vistica, D.; Warren, J.T.; Bokesch, H.; Kenney, S.; Boyd, M.R. New Colorimetric Cytotoxicity Assay for Anticancer-Drug Screening. JNCI J. Natl. Cancer Inst. 1990, 82, 1107–1112. [Google Scholar] [CrossRef] [PubMed]
- Montoya, T.; Aparicio-Soto, M.; Castejón, M.L.; Rosillo, M.A.; Sánchez-Hidalgo, M.; Begines, P.; Férnandez-Bolaños, J.G.; Alarcón-de-la-Lastra, C. Peracetylated-Hydroxytyrosol, a new hydroxytyrosol derivate, attenuates LPS-induced inflammato-ry response in murine peritoneal macrophages via regulation of non-canonical inflammasome, Nrf2/HO1 and JAK/STAT signaling pathways. J. Nutr. Biochem. 2018, 57, 110–120. [Google Scholar] [CrossRef] [PubMed]
- Vougogiannopoulou, K.; Lemus, C.; Halabalaki, M.; Pergola, C.; Werz, O.; Smith, A.B., III; Michel, S.; Skaltsounis, L.; Deguin, B. One-step semisynthesis of olea-cein and the determination as a 5-lipoxygenase inhibitor. J. Nat. Prod. 2014, 77, 441–445. [Google Scholar] [CrossRef] [PubMed]
- Costanzo, P.; Bonacci, S.; Cariati, L.; Nardi, M.; Oliverio, M.; Procopio, A. Simple and efficient sustainable semi-synthesis of oleacein [2-(3,4-hydroxyphenyl) ethyl (3S,4E)-4-formyl-3-(2-oxoethyl)hex-4-enoate] as potential additive for edible oils. Food Chem. 2018, 245, 410–414. [Google Scholar] [CrossRef] [PubMed]
- Mason, J.D.; Murphree, S.S. Microwave-Assisted Aqueous Krapcho Decarboxylation. Synlett 2013, 24, 1391–1394. [Google Scholar] [CrossRef][Green Version]
- Fernández-Bolaños, J.M.; Maya-Castilla, I.; González-Benjumea, A. Use of DMSO for the Synthesis of Oleacein and Oleocanthal. WO2018162769A2, 13 September 2018. [Google Scholar]
- Karkoula, E.; Skantzari, A.; Melliou, E.; Magiatis, P. Quantitative Measurement of Major Secoiridoid Derivatives in Olive Oil Using qNMR. Proof of the Artificial Formation of Aldehydic Oleuropein and Ligstroside Aglycon Isomers. J. Agric. Food Chem. 2014, 62, 600–607. [Google Scholar] [CrossRef] [PubMed]
- Angelis, A.; Michailidis, D.; Antoniadi, L.; Stathopoulos, P.; Tsantila, V.; Nuzillard, J.-M.; Renault, J.-H.; Skaltsounis, L.A. Pilot continuous centrifugal liquid-liquid extraction of extra virgin olive oil biophenols and gram-scale recovery of pure oleocanthal, oleacein, MFOA, MFLA and hydroxytyrosol. Sep. Purif. Technol. 2021, 255, 117692. [Google Scholar] [CrossRef]
- During, A.; Larondelle, Y. The O-methylation of chrysin markedly improves its intestinal anti-inflammatory properties: Structure–activity relationships of flavones. Biochem. Pharmacol. 2013, 86, 1739–1746. [Google Scholar] [CrossRef]
- Koirala, N.; Thuan, N.H.; Ghimire, G.P.; Van Thang, D.; Sohng, J.K. Methylation of flavonoids: Chemical structures, bioactivities, progress and perspectives for biotechnological production. Enzym. Microb. Technol. 2016, 86, 103–116. [Google Scholar] [CrossRef]
- Castejón, M.L.; Montoya, T.; Alarcón-De-La-Lastra, C.; González-Benjumea, A.; Vázquez-Román, M.V.; Sánchez-Hidalgo, M. Dietary oleuropein and its acyl derivative ameliorate inflammatory response in peritoneal macrophages from pristane-induced SLE mice via canonical and noncanonical NLRP3 inflammasomes pathway. Food Funct. 2020, 11, 6622–6631. [Google Scholar] [CrossRef]
- Scotece, M.; Gomez, R.; Conde, J.; Lopez, V.; Gomez-Reino, J.; Lago, F.; Iii, A.S.; Gualillo, O. Oleocanthal Inhibits Proliferation and MIP-1α Expression in Human Multiple Myeloma Cells. Curr. Med. Chem. 2013, 20, 2467–2475. [Google Scholar] [CrossRef]
- Iacono, A.; Gómez, R.; Sperry, J.; Conde, J.; Bianco, G.; Meli, R.; Gómez-Reino, J.J.; Smith, A.B.; Gualillo, O. Effect of oleocanthal and its derivatives on inflammatory response induced by lipopolysaccharide in a murine chondrocyte cell line. Arthritis Rheum. 2010, 62, 1675–1682. [Google Scholar] [CrossRef] [PubMed]
- Gallo, K.A.; Ellsworth, E.; Stoub, H.; Conrad, S.E. Therapeutic potential of targeting mixed lineage kinases in cancer and inflammation. Pharmacol. Ther. 2020, 207, 107457. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; He, J.; Li, S.; Song, L.; Guo, X.; Yao, W.; Zou, D.; Gao, X.; Liu, Y.; Bai, F.; et al. Fibroblast growth factor 21 (FGF21) inhibits macrophage-mediated inflammation by activating Nrf2 and suppressing the NF-κB signaling pathway. Int. Immunopharmacol. 2016, 38, 144–152. [Google Scholar] [CrossRef]
- Sharma, D.; Kanneganti, T.-D. The cell biology of inflammasomes: Mechanisms of inflammasome activation and regulation. J. Cell Biol. 2016, 213, 617–629. [Google Scholar] [CrossRef]
- Tschopp, J.; Schroder, K. NLRP3 inflammasome activation: The convergence of multiple signaling pathways on ROS produc-tion. Nat. Rev. Immunol. 2010, 10, 210–215. [Google Scholar] [CrossRef]
- Kumari, A.; Bhawal, S.; Kapila, S.; Yadav, H.; Kapila, R. Health-promoting role of dietary bioactive compounds through epigenetic modulations: A novel prophylactic and therapeutic approach. Crit. Rev. Food Sci. Nutr. 2020, 21, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Ideraabdullah, F.; Zeisel, S.H. Dietary Modulation of the Epigenome. Physiol. Rev. 2018, 98, 667–695. [Google Scholar] [CrossRef]
- Caradonna, F.; Consiglio, O.; Luparello, C.; Gentile, C. Science and Healthy Meals in the World: Nutritional Epigenomics and Nutrigenetics of the Mediterranean Diet. Nutrients 2020, 12, 1748. [Google Scholar] [CrossRef]
- Álvarez-Errico, D.; Vento-Tormo, R.; Sieweke, M.; Ballestar, E. Epigenetic control of myeloid cell differentiation, identity and function. Nat. Rev. Immunol. 2015, 15, 7–17. [Google Scholar] [CrossRef]
- Rohraff, D.M.; He, Y.; Farkash, E.A.; Schonfeld, M.; Tsou, P.; Sawalha, A.H. Inhibition of EZH2 Ameliorates Lupus—Like Disease in MRL/lpr Mice. Arthritis Rheumatol. 2019, 71, 1681–1690. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Wan, D.; Li, J.; Chen, H.; Huang, K.; Zheng, L. Histone acetyltransferase PCAF regulates inflammatory molecules in the development of renal injury. Epigenetics 2015, 101, 62–72. [Google Scholar] [CrossRef]
- Yi, Y.-S. Functional Interplay between Methyltransferases and Inflammasomes in Inflammatory Responses and Diseases. Int. J. Mol. Sci. 2021, 22, 7580. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Wang, J.; Liu, Z.; Jiang, A.; Li, S.; Wu, D. Sodium butyrate alleviates lipopolysaccharide-induced inflammatory responses by down-Regulation of NF-κB, NLRP3 Signaling Pathway, and activating histone acetylation in bovine macro-phages. Front. Vet. Sci. 2020, 7, 579674. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.-J.; Ren, X.-S.; Xiong, X.-Q.; Chen, Y.-Z.; Zhao, M.-X.; Wang, J.-J.; Zhou, Y.-B.; Han, Y.; Chen, Q.; Li, Y.-H.; et al. NLRP3 inflammasome activation contributes to VSMC phenotypic transformation and proliferation in hypertension. Cell Death Dis. 2017, 8, e3074. [Google Scholar] [CrossRef]
- Fang, P.; Chen, C.; Zheng, F.; Jia, J.; Chen, T.; Zhu, J.; Chang, J.; Zhang, Z. NLRP3 inflammasome inhibition by histone acetylation amelio-rates sevoflurane-induced cognitive impairment in aged mice by activating the autophagy pathway. Brain Res. Bull. 2021, 172, 79–88. [Google Scholar] [CrossRef]
- Liu, C.-C.; Huang, Z.-X.; Li, X.; Shen, K.-F.; Liu, M.; Ouyang, H.-D.; Zhang, S.-B.; Ruan, Y.-T.; Zhang, X.-L.; Wu, S.-L.; et al. Upregulation of NLRP3 via STAT3-dependent histone acetylation contributes to painful neuropathy induced by bortezomib. Exp. Neurol. 2018, 302, 104–111. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Montoya, T.; Alarcón-de-la-Lastra, C.; Castejón, M.L.; Ortega-Vidal, J.; Altarejos, J.; Sánchez-Hidalgo, M. (−)-Methyl-Oleocanthal, a New Oleocanthal Metabolite Reduces LPS-Induced Inflammatory and Oxidative Response: Molecular Signaling Pathways and Histones Epigenetic Modulation. Antioxidants 2022, 11, 56. https://doi.org/10.3390/antiox11010056
Montoya T, Alarcón-de-la-Lastra C, Castejón ML, Ortega-Vidal J, Altarejos J, Sánchez-Hidalgo M. (−)-Methyl-Oleocanthal, a New Oleocanthal Metabolite Reduces LPS-Induced Inflammatory and Oxidative Response: Molecular Signaling Pathways and Histones Epigenetic Modulation. Antioxidants. 2022; 11(1):56. https://doi.org/10.3390/antiox11010056
Chicago/Turabian StyleMontoya, Tatiana, Catalina Alarcón-de-la-Lastra, María Luisa Castejón, Juan Ortega-Vidal, Joaquín Altarejos, and Marina Sánchez-Hidalgo. 2022. "(−)-Methyl-Oleocanthal, a New Oleocanthal Metabolite Reduces LPS-Induced Inflammatory and Oxidative Response: Molecular Signaling Pathways and Histones Epigenetic Modulation" Antioxidants 11, no. 1: 56. https://doi.org/10.3390/antiox11010056
APA StyleMontoya, T., Alarcón-de-la-Lastra, C., Castejón, M. L., Ortega-Vidal, J., Altarejos, J., & Sánchez-Hidalgo, M. (2022). (−)-Methyl-Oleocanthal, a New Oleocanthal Metabolite Reduces LPS-Induced Inflammatory and Oxidative Response: Molecular Signaling Pathways and Histones Epigenetic Modulation. Antioxidants, 11(1), 56. https://doi.org/10.3390/antiox11010056







