Quantitative MRI in Neuroimaging: A Review of Techniques, Biomarkers, and Emerging Clinical Applications
Abstract
1. Introduction
2. Scope and Organization of the Review
3. Search Strategy and Selection Criteria
4. Biological and Clinical Ground Truth for qMRI Validation
5. Visualization and Interpretability of qMRI Maps
6. Positioning Relative to Broader qMRI Reviews
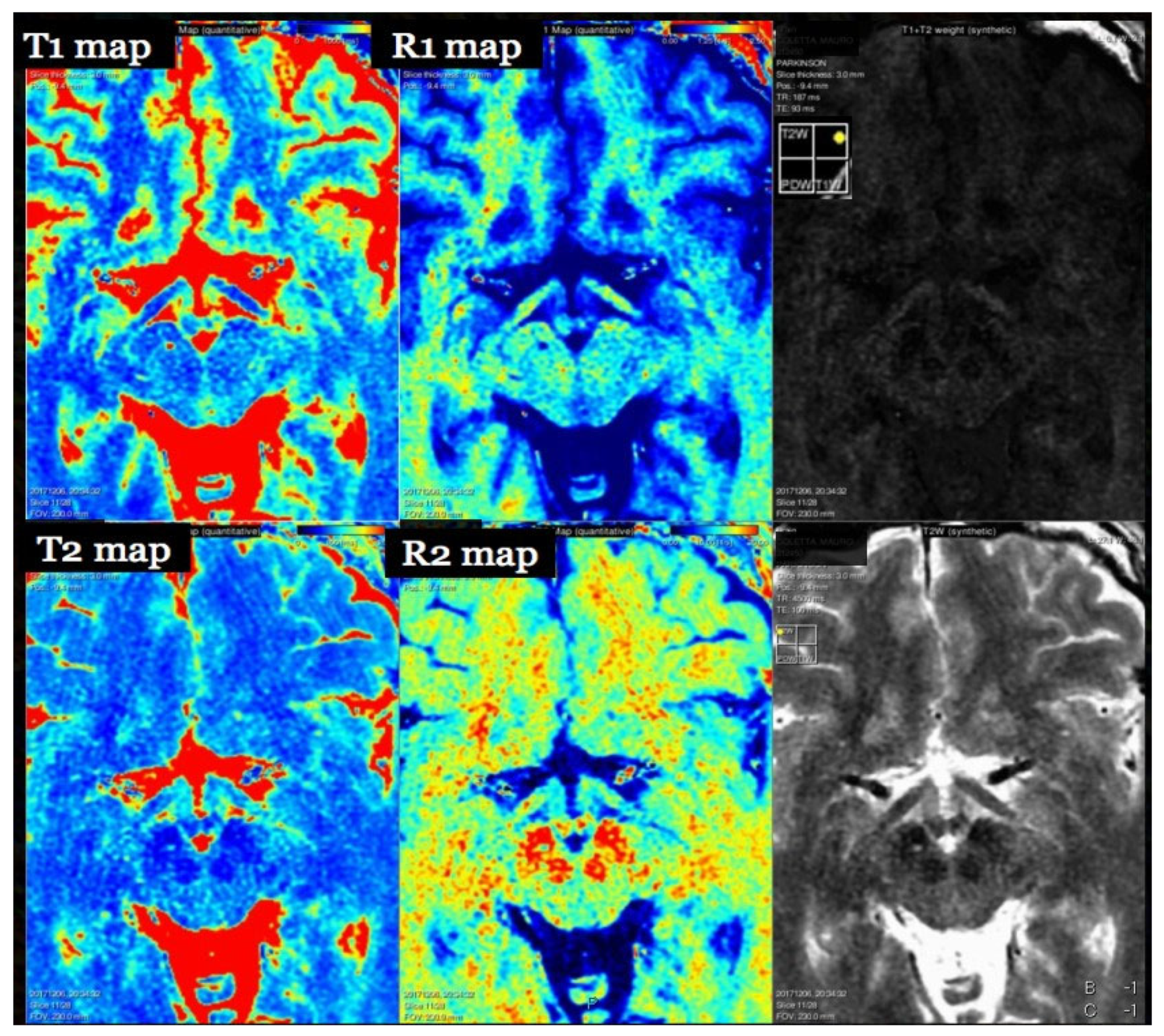
7. T1 Relaxometry
7.1. Physics and Signal Model
7.2. Acquisition and Key Parameters
7.3. Outputs and Units
- R1 = 1/T1, a linear proxy for myelin content,
- T1-normalized intensity, typically normalized to CSF or gray matter for inter-subject comparison,
- Histogram-based features (mean, standard deviation, skewness, kurtosis) of T1 values in NAWM,
- ΔT1 values for longitudinal lesion monitoring,
7.4. Validation and Repeatability
7.5. Clinical Applications
7.6. Multimodal Integration
7.7. Limitations and Pitfalls
8. T2 Relaxometry and Magnetization Transfer
8.1. Physics and Signal Model
8.2. Acquisition and Key Parameters
8.3. Magnetization Transfer (MT) Framework
8.4. Clinical Applications
8.5. Validation and Repeatability
8.6. Multimodal Integration and Future Directions
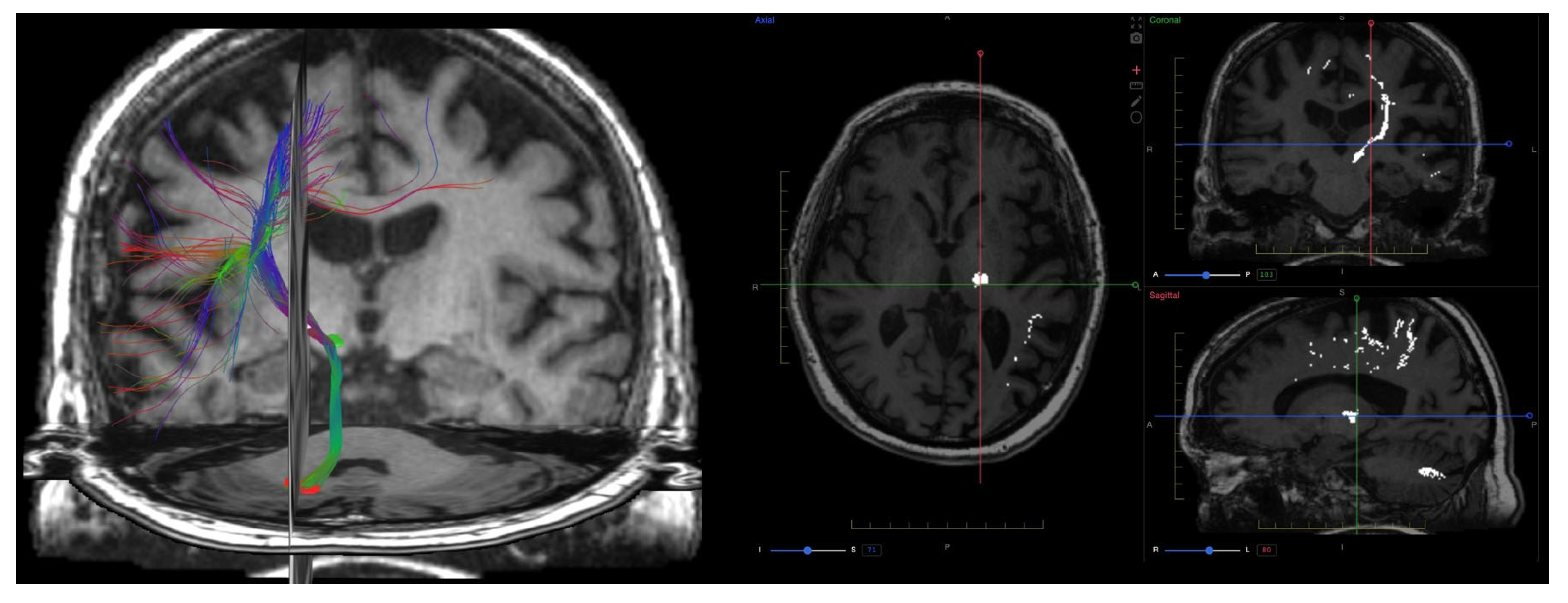
9. Diffusion Imaging (DWI, DTI, DKI)
9.1. Physics and Signal Model
9.2. Acquisition and Key Parameters
9.3. Outputs and Units
- Fractional anisotropy (FA): degree of diffusion directionality,
- Mean diffusivity (MD): average diffusivity, equivalent to ADC but derived from tensor data,
- Axial diffusivity (AD): diffusion parallel to axons,
- From DKI, key parameters include:
- Mean kurtosis (MK): overall measure of tissue complexity,
- Axial kurtosis (AK): non-Gaussianity along the primary fiber axis,
- Radial kurtosis (RK): kurtosis perpendicular to axonal direction, sensitive to myelin integrity.
- In addition, NODDI provides two parameters:
- Neurite density index (NDI): reflects axonal and dendritic density,
- Orientation dispersion index (ODI): measures angular variation in neurites.
- In neuro-oncology, ADC has proven valuable in:
- Grading gliomas (high vs. low grade),
- Distinguishing gliomas from metastases,
- Differentiating tumor progression from pseudo-progression, and
9.4. Clinical Applications
9.5. Validation and Repeatability
9.6. Emerging Techniques and Integration
9.7. Summary and Outlook
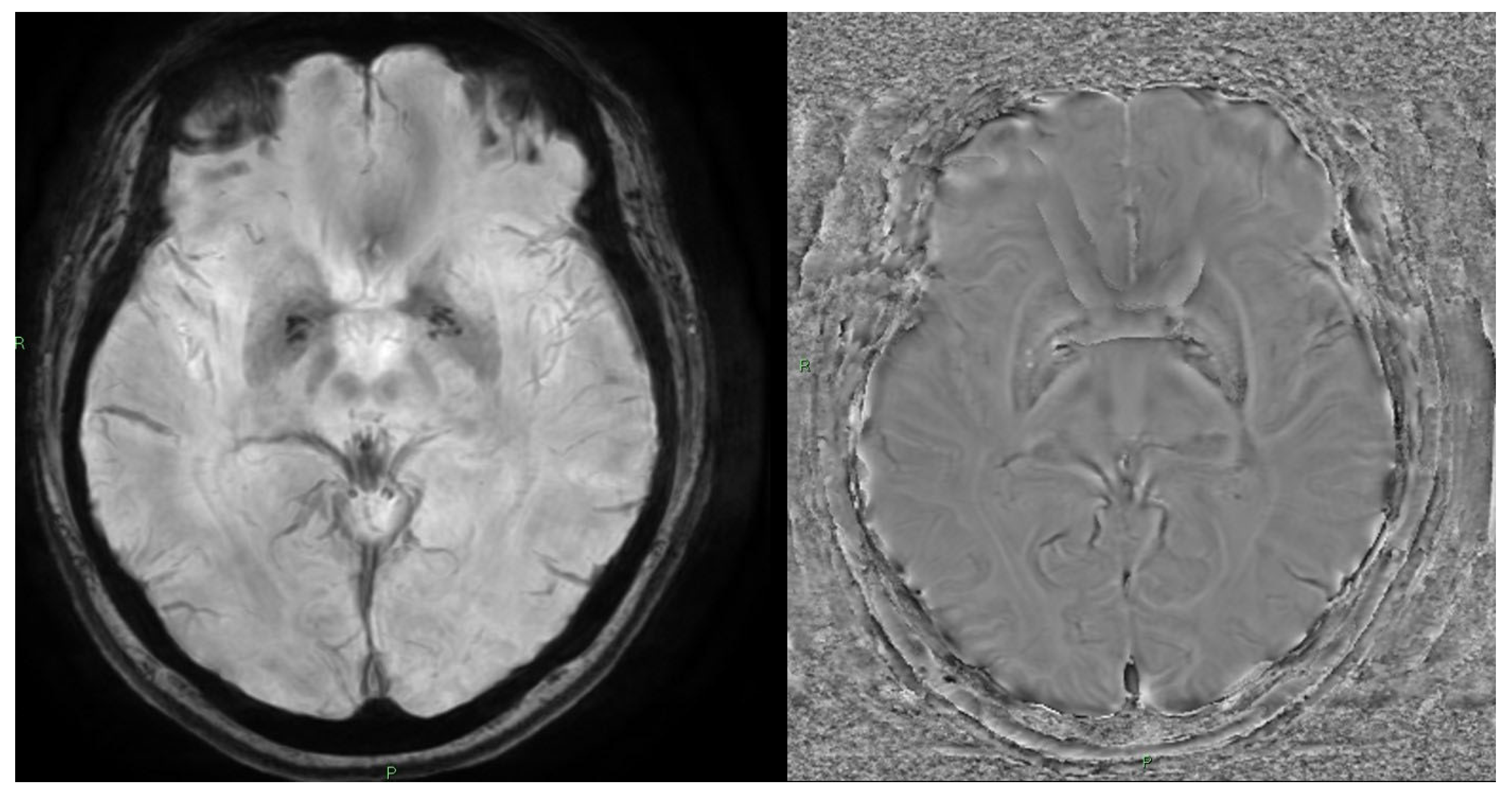
10. Quantitative Susceptibility Mapping (QSM) and Susceptibility-Weighted Imaging (SWI)
10.1. Physics and Signal Model
10.2. Acquisition and Key Parameters
10.3. Outputs and Units
10.4. Clinical Applications
10.5. Limitations and Pitfalls
- Lack of standardization among reconstruction algorithms and no universally accepted processing pipeline;
- Offline post-processing requirements that are complex and time-consuming;
- Limited vendor integration, although standard GRE sequences used for SWI or T2* can often be repurposed for QSM if phase images are preserved.
10.6. Future Outlook
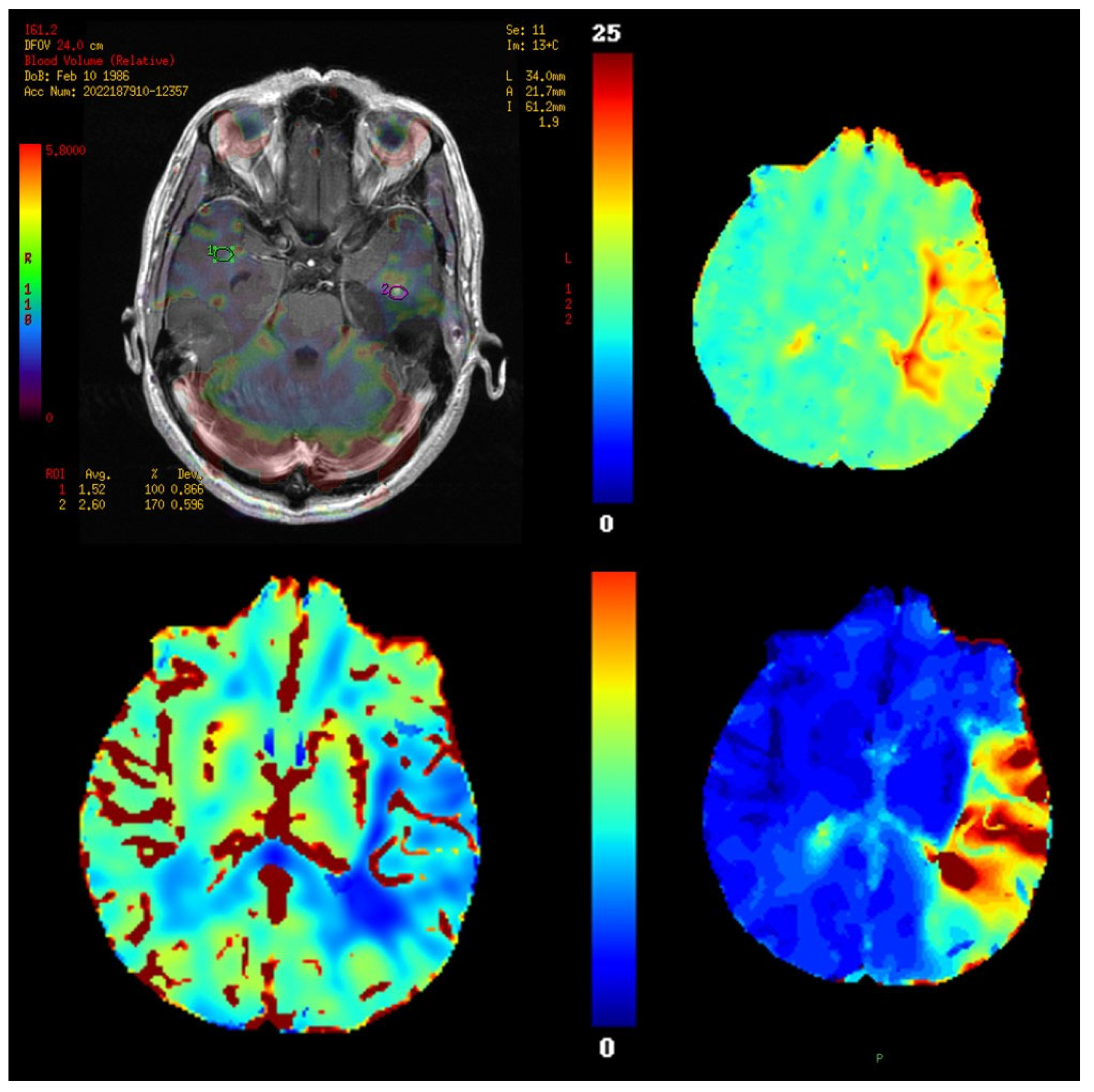
11. Perfusion Imaging
11.1. Physics and Signal Model
11.2. Acquisition and Key Parameters
11.3. Outputs and Units
11.4. Validation and Quantification Considerations
11.5. Clinical Applications
- Tumor grading and characterization,
11.6. Limitations and Pitfalls
- Model- and software-dependent variability in DCE parameter estimation,
- AIF inaccuracies and technical demands of T1 mapping,
- Lack of universal rCBV thresholds and consistent leakage correction methods,
- Parameter fitting instability in multi-compartment models,
- Absence of digital phantoms for cross-platform validation.
11.7. Future Directions
12. Arterial Spin Labeling (ASL)
12.1. Physics and Signal Model
12.2. Acquisition and Key Parameters
- Pulsed ASL (PASL): uses a short, high-powered pulse to invert a thick slab of arterial blood proximal to the imaging volume;
- Continuous ASL (CASL): applies a long, uninterrupted RF pulse and gradient field to continuously invert blood across a fixed labeling plane;
- Labeling duration: ~1.5–2.0 s
- Post-labeling delay (PLD): ~1.5–2.0 s
- 3D acquisition (e.g., GRASE or spiral)
- Background suppression pulses for artifact reduction
- Optional multi-delay protocols for estimation of ATT, particularly in cerebrovascular disorders
12.3. Outputs and Quantification
12.4. Clinical Applications and Biomarkers
- Reduced CBF in NAWM and cortical gray matter;
- Associations between low CBF and increased disability scores (EDSS), cognitive impairment, and atrophy;
12.5. Advantages, Limitations, and Future Directions
13. Brain Volume Quantification
13.1. Scope and Overview
13.2. Acquisition Physics and Pre-Processing
13.3. Acquisition and Processing Pipeline
- Voxel-based morphometry (VBM): detects regional differences in GM/WM density,
- Surface-based morphometry (SBM): estimates cortical thickness and curvature using 3D cortical meshes,
- Deep learning algorithms: allow for rapid and accurate segmentation, even in the presence of artifacts or lesions.
13.4. Outputs and Units
13.5. Clinical Applications—Dementia
13.6. Clinical Applications—Multiple Sclerosis
13.7. Validation and Repeatability
13.8. Limitations and Pitfalls
13.9. Future Directions
14. Conclusions
- The lack of standardized acquisition protocols across vendors and platforms,
- Limited availability of robust, validated software for map reconstruction and biomarker extraction,
- Absence of large normative datasets and clinically validated pathological cut-off values,
- The need for multicenter clinical validation studies directly comparing qMRI metrics to established clinical, histological, or molecular outcomes.
- Motion-compensated acquisition strategies and physiological gating,
- Dedicated hardware (e.g., optimized phased-array coils),
- Advanced software for region-of-interest (ROI) localization and signal modeling.
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Smits, M. MRI biomarkers in neuro-oncology. Nat. Rev. Neurol. 2021, 17, 486–500. [Google Scholar] [CrossRef]
- Yousaf, T.; Dervenoulas, G.; Politis, M. Advances in MRI Methodology. Int. Rev. Neurobiol. 2018, 141, 31–76. [Google Scholar]
- Aida, N. 1H-MR Spectroscopy of the Early Developmental Brain, Neonatal Encephalopathies, and Neurometabolic Disorders. Magn. Reson. Med Sci. 2022, 21, 9–28. [Google Scholar] [CrossRef]
- Matsoukas, S.; Scaggiante, J.; Schuldt, B.R.; Smith, C.J.; Chennareddy, S.; Kalagara, R.; Majidi, S.; Bederson, J.B.; Fifi, J.T.; Mocco, J.; et al. Accuracy of artificial intelligence for the detection of intracranial hemorrhage and chronic cerebral microbleeds: A systematic review and pooled analysis. La Radiol. medica 2022, 127, 1106–1123. [Google Scholar] [CrossRef]
- Shukla-Dave, A.; Obuchowski, N.A.; Chenevert, T.L.; Jambawalikar, S.; Schwartz, L.H.; Malyarenko, D.; Huang, W.; Noworolski, S.M.; Young, R.J.; Shiroishi, M.S.; et al. Quantitative imaging biomarkers alliance (QIBA) recommendations for improved precision of DWI and DCE-MRI derived biomarkers in multicenter oncology trials. J. Magn. Reson. Imaging 2018, 49, e101–e121. [Google Scholar] [CrossRef]
- Albano, D.; Stecco, A.; Micci, G.; Sconfienza, L.M.; Colagrande, S.; Reginelli, A.; Grassi, R.; Carriero, A.; Midiri, M.; Lagalla, R.; et al. Whole-body magnetic resonance imaging (WB-MRI) in oncology: An Italian survey. La Radiol. medica 2020, 126, 299–305. [Google Scholar] [CrossRef]
- Bonatti, M.; Valletta, R.; Lombardo, F.; Zamboni, G.A.; Turri, E.; Avesani, G.; Mansueto, G.; Manfredi, R.; Schifferle, G. Accuracy of unenhanced CT in the diagnosis of cerebral venous sinus thrombosis. La Radiol. medica 2020, 126, 399–404. [Google Scholar] [CrossRef]
- Combes, A.J.; Clarke, M.A.; O’Grady, K.P.; Schilling, K.G.; Smith, S.A. Advanced spinal cord MRI in multiple sclerosis: Current techniques and future directions. NeuroImage: Clin. 2022, 36, 103244. [Google Scholar] [CrossRef]
- Bellisari, F.C.; De Marino, L.; Arrigoni, F.; Mariani, S.; Bruno, F.; Palumbo, P.; De Cataldo, C.; Sgalambro, F.; Catallo, N.; Zugaro, L.; et al. T2-mapping MRI evaluation of patellofemoral cartilage in patients submitted to intra-articular platelet-rich plasma (PRP) injections. La Radiol. medica 2021, 126, 1085–1094. [Google Scholar] [CrossRef]
- Troudi, A.; Tensaouti, F.; Baudou, E.; Péran, P.; Laprie, A. Arterial Spin Labeling Perfusion in Pediatric Brain Tumors: A Review of Techniques, Quality Control, and Quantification. Cancers 2022, 14, 4734. [Google Scholar] [CrossRef]
- Fasen, B.A.C.M.; Berendsen, R.C.M.; Kwee, R.M. Artificial intelligence software for diagnosing intracranial arterial occlusion in patients with acute ischemic stroke. Neuroradiology 2022, 64, 1579–1583. [Google Scholar] [CrossRef]
- Berardo, S.; Sukhovei, L.; Andorno, S.; Carriero, A.; Stecco, A. Quantitative bone marrow magnetic resonance imaging through apparent diffusion coefficient and fat fraction in multiple myeloma patients. La Radiol. medica 2020, 126, 445–452. [Google Scholar] [CrossRef]
- Alger, J.R. Quantitative Proton Magnetic Resonance Spectroscopy and Spectroscopic Imaging of the Brain. Top. Magn. Reson. Imaging 2010, 21, 115–128. [Google Scholar] [CrossRef]
- Zhu, H.; Barker, P.B. MR spectroscopy and spectroscopic imaging of the brain. Methods Mol. Biol. 2011, 711, 203–226. [Google Scholar]
- Bracco, S.; Zanoni, M.; Casseri, T.; Castellano, D.; Cioni, S.; Vallone, I.M.; Gennari, P.; Mazzei, M.A.; Romano, D.G.; Piano, M.; et al. Endovascular treatment of acute ischemic stroke due to tandem lesions of the anterior cerebral circulation: A multicentric Italian observational study. La Radiol. medica 2021, 126, 804–817. [Google Scholar] [CrossRef]
- Cellina, M.; Pirovano, M.; Ciocca, M.; Gibelli, D.; Floridi, C.; Oliva, G. Radiomic analysis of the optic nerve at the first episode of acute optic neuritis: An indicator of optic nerve pathology and a predictor of visual recovery? La Radiol. medica 2021, 126, 698–706. [Google Scholar] [CrossRef]
- Handa, P.; Samkaria, A.; Sharma, S.; Arora, Y.; Mandal, P.K. Comprehensive Account of Sodium Imaging and Spectroscopy for Brain Research. ACS Chem. Neurosci. 2022, 13, 859–875. [Google Scholar] [CrossRef]
- Ceravolo, I.; Barchetti, G.; Biraschi, F.; Gerace, C.; Pampana, E.; Pingi, A.; Stasolla, A. Early stage glioblastoma: Retrospective multicentric analysis of clinical and radiological features. La Radiol. medica 2021, 126, 1468–1476. [Google Scholar] [CrossRef]
- Coppola, F.; Faggioni, L.; Regge, D.; Giovagnoni, A.; Golfieri, R.; Bibbolino, C.; Miele, V.; Neri, E.; Grassi, R. Artificial intelligence: Radiologists’ expectations and opinions gleaned from a nationwide online survey. La Radiol. medica 2020, 126, 63–71. [Google Scholar] [CrossRef]
- Ahlawat, S.; Blakeley, J.O.; Langmead, S.; Belzberg, A.J.; Fayad, L.M. Current status and recommendations for imaging in neurofibromatosis type 1, neurofibromatosis type 2, and schwannomatosis. Skelet. Radiol. 2019, 49, 199–219. [Google Scholar] [CrossRef]
- D’aRgento, F.; Pedicelli, A.; Ciardi, C.; Leone, E.; Scarabello, M.; Infante, A.; Alexandre, A.; Lozupone, E.; Valente, I.; Colosimo, C. Intra- and inter-observer variability in intracranial aneurysm segmentation: Comparison between CT angiography (semi-automated segmentation software stroke VCAR) and digital subtraction angiography (3D rotational angiography). La Radiol. medica 2020, 126, 484–493. [Google Scholar] [CrossRef]
- Counsell, S.J.; Arichi, T.; Arulkumaran, S.; Rutherford, M.A. Fetal and neonatal neuroimaging. Handb. Clin. Neurol. 2019, 162, 67–103. [Google Scholar]
- Detti, B.; Scoccianti, S.; Teriaca, M.A.; Maragna, V.; Lorenzetti, V.; Lucidi, S.; Bellini, C.; Greto, D.; Desideri, I.; Livi, L. Bevacizumab in recurrent high-grade glioma: A single institution retrospective analysis on 92 patients. La Radiol. medica 2021, 126, 1249–1254. [Google Scholar] [CrossRef]
- Vargas, M.; Boto, J.; Meling, T. Imaging of the spine and spinal cord: An overview of magnetic resonance imaging (MRI) techniques. Rev. Neurol. 2021, 177, 451–458. [Google Scholar] [CrossRef]
- Feraco, P.; Piccinini, S.; Gagliardo, C. Imaging of inner ear malformations: A primer for radiologists. La Radiol. medica 2021, 126, 1282–1295. [Google Scholar] [CrossRef]
- Di Giuliano, F.; Minosse, S.; Picchi, E.; Ferrazzoli, V.; Da Ros, V.; Muto, M.; Pistolese, C.A.; Garaci, F.; Floris, R. Qualitative and quantitative analysis of 3D T1 Silent imaging. La Radiol. medica 2021, 126, 1207–1215. [Google Scholar] [CrossRef]
- MacDonald, M.E.; Pike, G.B. MRI of healthy brain aging: A review. NMR Biomed. 2021, 34, e4564. [Google Scholar] [CrossRef]
- Pope, W.B.; Djoukhadar, I.; Jackson, A. Neuroimaging. Handb. Clin. Neurol. 2016, 134, 27–50. [Google Scholar]
- Jiang, W.; Zhao, Z.; Wu, Q.; Wang, L.; Zhou, L.; Li, D.; He, L.; Tan, Y. Study on brain structure network of patients with delayed encephalopathy after carbon monoxide poisoning: Based on diffusion tensor imaging. La Radiol. medica 2020, 126, 133–141. [Google Scholar] [CrossRef]
- Liheng, M.; Guofan, X.; Balzano, R.F.; Yuying, L.; Weifeng, H.; Ning, Y.; Yayun, J.; Mouyuan, L.; Guglielmi, G. The value of DTI: Achieving high diagnostic performance for brain metastasis. La Radiol. medica 2020, 126, 291–298. [Google Scholar] [CrossRef]
- Paoletti, M.; Muzic, S.I.; Marchetti, F.; Farina, L.M.; Bastianello, S.; Pichiecchio, A. Differential imaging of atypical demyelinating lesions of the central nervous system. La Radiol. medica 2021, 126, 827–842. [Google Scholar] [CrossRef]
- Romano, N.; Castaldi, A. Imaging of intracranial fat: From normal findings to pathology. La Radiol. medica 2021, 126, 971–978. [Google Scholar] [CrossRef]
- Salaffi, F.; Ceccarelli, L.; Carotti, M.; Di Carlo, M.; Polonara, G.; Facchini, G.; Golfieri, R.; Giovagnoni, A. Differentiation between infectious spondylodiscitis versus inflammatory or degenerative spinal changes: How can magnetic resonance imaging help the clinician? La Radiol. medica 2021, 126, 843–859. [Google Scholar] [CrossRef]
- Scapicchio, C.; Gabelloni, M.; Barucci, A.; Cioni, D.; Saba, L.; Neri, E. A deep look into radiomics. La Radiol. medica 2021, 126, 1296–1311. [Google Scholar] [CrossRef]
- Turna, O.; Turna, I.F. Quantitative assessment of cervical spinal cord by diffusion tensor tractography in 3.0 T. La Radiol. medica 2020, 126, 83–88. [Google Scholar] [CrossRef]
- Romano, A.; Moltoni, G.; Guarnera, A.; Pasquini, L.; Di Napoli, A.; Napolitano, A.; Espagnet, M.C.R.; Bozzao, A. Single brain metastasis versus glioblastoma multiforme: A VOI-based multiparametric analysis for differential diagnosis. La Radiol. medica 2022, 127, 490–497. [Google Scholar] [CrossRef]
- Talwar, P.; Kushwaha, S.; Chaturvedi, M.; Mahajan, V. Systematic Review of Different Neuroimaging Correlates in Mild Cognitive Impairment and Alzheimer’s Disease. Clin. Neuroradiol. 2021, 31, 953–967. [Google Scholar] [CrossRef]
- Varrassi, M.; Corridore, A.; Tommasino, E.; Saltelli, G.; Bruno, F.; Di Sibio, A.; Splendiani, A.; Di Cesare, E.; Masciocchi, C. MR imaging of cerebral involvement of Rosai–Dorfman disease: A single-centre experience with review of the literature. La Radiol. medica 2020, 126, 89–98. [Google Scholar] [CrossRef]
- Yang, H.; Yang, X.; Liu, H.; Long, H.; Hu, H.; Wang, Q.; Huang, R.; Shan, D.; Li, K.; Lai, W. Placebo modulation in orthodontic pain: A single-blind functional magnetic resonance study. La Radiol. medica 2021, 126, 1356–1365. [Google Scholar] [CrossRef]
- Granziera, C.; Wuerfel, J.; Barkhof, F.; Calabrese, M.; De Stefano, N.; Enzinger, C.; Evangelou, N.; Filippi, M.; Geurts, J.J.G.; Reich, D.S.; et al. Quantitative magnetic resonance imaging towards clinical application in multiple sclerosis. Brain 2021, 144, 1296–1311. [Google Scholar] [CrossRef]
- Goncalves, F.G.; Serai, S.D.; Zuccoli, G. Synthetic Brain MRI: Review of Current Concepts and Future Directions. Top. Magn. Reson. Imaging 2018, 27, 387–393. [Google Scholar] [CrossRef]
- André, J.; Barrit, S.; Jissendi, P. Synthetic MRI for stroke: A qualitative and quantitative pilot study. Sci. Rep. 2022, 12, 1–8. [Google Scholar] [CrossRef]
- Avesani, G.; Perazzolo, A.; Elia, L.; Anghelone, A.G.; Gaudino, S.; Russo, L.; Genco, E.; Di Paola, V.; Massimi, L.; De Santis, M.; et al. Fetal MRI prior to intrauterine surgery of open neural tube defects: What does the radiologist need to know. La Radiol. medica 2022, 128, 1–12. [Google Scholar] [CrossRef]
- Barbiera, F.; Cosentino, G.; La Seta, F.; Vetrano, E.; Murmura, B.; Avenali, M.; Alfonsi, E.; Tassorelli, C. A narrative review on the role and main findings of the Videofluoroscopic Study of Swallowing in Parkison’s disease. La Radiol. medica 2022, 128, 1–8. [Google Scholar] [CrossRef]
- Calloni, S.F.; Panni, P.; Calabrese, F.; del Poggio, A.; Roveri, L.; Squarza, S.; Pero, G.C.; Paolucci, A.; Filippi, M.; Falini, A.; et al. Cerebral hyperdensity on CT imaging (CTHD) post-reperfusion treatment in patients with acute cerebral stroke: Understanding its clinical meaning. La Radiol. medica 2022, 127, 973–980. [Google Scholar] [CrossRef]
- Cannavale, A.; Santoni, M.; Nardis, P.; Lucatelli, P.; Corona, M.; Cannavale, G.; Catalano, C.; Ricci, P. Role of CT and MR imaging in the assessment of suspected spondylodiscitis and planning of needle biopsy. La Radiol. medica 2022, 127, 1023–1031. [Google Scholar] [CrossRef]
- Cirillo, L.; Rustici, A.; Toni, F.; Zoli, M.; Bartiromo, F.; Gramegna, L.L.; Cicala, D.; Tonon, C.; Caranci, F.; Lodi, R. Vessel Wall MRI: Clinical implementation in cerebrovascular disorders—technical aspects. La Radiol. medica 2022, 127, 645–651. [Google Scholar] [CrossRef]
- Rossi, M.M.; Ruottinen, H.; Elovaara, I.; Ryymin, P.M.; Soimakallio, S.; Eskola, H.; Dastidar, P. Brain Iron Deposition and Sequence Characteristics in Parkinsonism. Investig. Radiol. 2010, 45, 795–802. [Google Scholar] [CrossRef]
- Gitto, S.; Bologna, M.; Corino, V.D.A.; Emili, I.; Albano, D.; Messina, C.; Armiraglio, E.; Parafioriti, A.; Luzzati, A.; Mainardi, L.; et al. Diffusion-weighted MRI radiomics of spine bone tumors: Feature stability and machine learning-based classification performance. La Radiol. medica 2022, 127, 518–525. [Google Scholar] [CrossRef]
- Hanna, J.M.; Temares, D.; Hyder, F.; Rothman, D.L.; Fulbright, R.K.; Chiang, V.L.; Coman, D. Prognosticating brain tumor patient survival after laser thermotherapy: Comparison between neuroradiological reading and semi-quantitative analysis of MRI data. Magn. Reson. Imaging 2020, 65, 45–54. [Google Scholar] [CrossRef]
- Meloni, A.; Pistoia, L.; Restaino, G.; Missere, M.; Positano, V.; Spasiano, A.; Casini, T.; Cossu, A.; Cuccia, L.; Massa, A.; et al. Quantitative T2* MRI for bone marrow iron overload: Normal reference values and assessment in thalassemia major patients. La Radiol. medica 2022, 127, 1199–1208. [Google Scholar] [CrossRef]
- Autorino, R.; Gui, B.; Panza, G.; Boldrini, L.; Cusumano, D.; Russo, L.; Nardangeli, A.; Persiani, S.; Campitelli, M.; Ferrandina, G.; et al. Radiomics-based prediction of two-year clinical outcome in locally advanced cervical cancer patients undergoing neoadjuvant chemoradiotherapy. La Radiol. medica 2022, 127, 498–506. [Google Scholar] [CrossRef]
- Zhu, Y.; Young, G.S.; Xue, Z.; Huang, R.Y.; You, H.; Setayesh, K.; Hatabu, H.; Cao, F.; Wong, S.T. Semi-Automatic Segmentation Software for Quantitative Clinical Brain Glioblastoma Evaluation. Acad. Radiol. 2012, 19, 977–985. [Google Scholar] [CrossRef]
- Viallon, M.; Cuvinciuc, V.; Delattre, B.; Merlini, L.; Barnaure-Nachbar, I.; Toso-Patel, S.; Becker, M.; Lovblad, K.-O.; Haller, S. State-of-the-art MRI techniques in neuroradiology: Principles, pitfalls, and clinical applications. Neuroradiology 2015, 57, 441–467. [Google Scholar] [CrossRef]
- Colombo, E.; Fick, T.; Esposito, G.; Germans, M.; Regli, L.; van Doormaal, T. Segmentation techniques of brain arteriovenous malformations for 3D visualization: A systematic review. La Radiol. medica 2022, 127, 1333–1341. [Google Scholar] [CrossRef] [PubMed]
- De Vito, A.; Mankad, K.; Pujar, S.; Chari, A.; Ippolito, D.; D’aRco, F. Narrative review of epilepsy: Getting the most out of your neuroimaging. Transl. Pediatr. 2021, 10, 1078–1099. [Google Scholar] [CrossRef] [PubMed]
- Shah, A.D.; Konar, A.S.; Paudyal, R.; Oh, J.H.; LoCastro, E.; Nuñez, D.A.; Swinburne, N.; Vachha, B.; Ulaner, G.A.; Young, R.J.; et al. Diffusion and Perfusion MRI Predicts Response Preceding and Shortly After Radiosurgery to Brain Metastases: A Pilot Study. J. Neuroimaging 2020, 31, 317–323. [Google Scholar] [CrossRef] [PubMed]
- Meoded, A.; Orman, G.; Huisman, T.A. Diffusion Weighted and Diffusion Tensor MRI in Pediatric Neuroimaging Including Connectomics: Principles and Applications. Semin. Pediatr. Neurol. 2020, 33, 100797. [Google Scholar] [CrossRef]
- Jeong, K.; Shah, L.M.; Lee, Y.-J.; Thapa, B.; Sapkota, N.; Bisson, E.; Carlson, N.G.; Jeong, E.; Rose, J.W. High-b diffusivity of MS lesions in cervical spinal cord using ultrahigh-b DWI (UHb-DWI). NeuroImage: Clin. 2021, 30, 102610. [Google Scholar] [CrossRef]
- Freund, P.; Seif, M.; Weiskopf, N.; Friston, K.; Fehlings, M.G.; Thompson, A.J.; Curt, A. MRI in traumatic spinal cord injury: From clinical assessment to neuroimaging biomarkers. Lancet Neurol. 2019, 18, 1123–1135. [Google Scholar] [CrossRef]
- Seif, M.; David, G.; Huber, E.; Vallotton, K.; Curt, A.; Freund, P. Cervical Cord Neurodegeneration in Traumatic and Non-Traumatic Spinal Cord Injury. J. Neurotrauma 2020, 37, 860–867. [Google Scholar] [CrossRef]
- Fan, Y.; Zhao, Z.; Wang, X.; Ai, H.; Yang, C.; Luo, Y.; Jiang, X. Radiomics for prediction of response to EGFR-TKI based on metastasis/brain parenchyma (M/BP)-interface. La Radiol. medica 2022, 127, 1342–1354. [Google Scholar] [CrossRef]
- Fushimi, Y.; Yoshida, K.; Okawa, M.; Maki, T.; Nakajima, S.; Sakata, A.; Okuchi, S.; Hinoda, T.; Kanagaki, M.; Nakamoto, Y. Vessel wall MR imaging in neuroradiology. La Radiol. medica 2022, 127, 1032–1045. [Google Scholar] [CrossRef]
- Smith, E.E.; Beaudin, A.E. New insights into cerebral small vessel disease and vascular cognitive impairment from MRI. Curr. Opin. Neurol. 2018, 31, 36–43. [Google Scholar] [CrossRef] [PubMed]
- Christidi, F.; Karavasilis, E.; Samiotis, K.; Bisdas, S.; Papanikolaou, N. Fiber tracking: A qualitative and quantitative comparison between four different software tools on the reconstruction of major white matter tracts. Eur. J. Radiol. Open 2016, 3, 153–161. [Google Scholar] [CrossRef]
- Giammello, F.; De Martino, S.R.M.; Simonetti, L.; Agati, R.; Battaglia, S.; Cirillo, L.; Gentile, M.; Migliaccio, L.; Forlivesi, S.; Romoli, M.; et al. Predictive value of Tmax perfusion maps on final core in acute ischemic stroke: An observational single-center study. La Radiol. medica 2022, 127, 414–425. [Google Scholar] [CrossRef] [PubMed]
- David, G.; Mohammadi, S.; Martin, A.R.; Cohen-Adad, J.; Weiskopf, N.; Thompson, A.; Freund, P. Traumatic and nontraumatic spinal cord injury: Pathological insights from neuroimaging. Nat. Rev. Neurol. 2019, 15, 718–731. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Li, W.; Tong, K.A.; Yeom, K.W.; Kuzminski, S. Susceptibility-weighted imaging and quantitative susceptibility mapping in the brain. J. Magn. Reson. Imaging 2014, 42, 23–41. [Google Scholar] [CrossRef]
- Vallotton, K.; David, G.; Hupp, M.; Pfender, N.; Cohen-Adad, J.; Fehlings, M.G.; Samson, R.S.; Wheeler-Kingshott, C.A.G.; Curt, A.; Freund, P.; et al. Tracking White and Gray Matter Degeneration along the Spinal Cord Axis in Degenerative Cervical Myelopathy. J. Neurotrauma 2021, 38, 2978–2987. [Google Scholar] [CrossRef]
- Hasan, K.M.; Keser, Z.; Schulz, P.E.; Wilde, E.A. Multimodal Advanced Imaging for Concussion. Neuroimaging Clin. North Am. 2018, 28, 31–42. [Google Scholar] [CrossRef]
- Ahmad, F.U.; Li, D.C.; Malcolm, J.G.; Rindler, R.S.; Baum, G.R.; Rao, A.; Khurpad, S.N. The role of diffusion tensor imaging in spinal pathology: A review. Neurol. India 2017, 65, 982–992. [Google Scholar] [CrossRef]
- Xu, S.; Yao, X.; Han, L.; Lv, Y.; Bu, X.; Fan, Y.; Yu, T.; Huang, G. Brain network analyses of diffusion tensor imaging for brain aging. Math. Biosci. Eng. 2021, 18, 6066–6078. [Google Scholar] [CrossRef]
- Leone, E.; Ferrari, R.; Trinci, M.; Cingolani, E.; Galluzzo, M. Imaging features of electric scooter trauma: What an emergency radiologist needs to know. La Radiol. medica 2022, 127, 872–880. [Google Scholar] [CrossRef]
- Berghe, S.V.; Cappelle, S.; De Keyzer, F.; Peeters, R.; Coursier, K.; Depotter, A.; Van Cauter, S.; Ameloot, K.; Dens, J.; Lemmens, R.; et al. Qualitative and quantitative analysis of diffusion-weighted brain MR imaging in comatose survivors after cardiac arrest. Neuroradiology 2020, 62, 1361–1369. [Google Scholar] [CrossRef]
- Shannon, B.A.; Ahlawat, S.; Morris, C.D.; Levin, A.S.; Fayad, L.M. Do contrast-enhanced and advanced MRI sequences improve diagnostic accuracy for indeterminate lipomatous tumors? La Radiol. medica 2021, 127, 90–99. [Google Scholar] [CrossRef] [PubMed]
- Voicu, I.P.; Pravatà, E.; Panara, V.; Navarra, R.; Mattei, P.A.; Caulo, M. Differentiating solitary brain metastases from high-grade gliomas with MR: Comparing qualitative versus quantitative diagnostic strategies. La Radiol. medica 2022, 127, 891–898. [Google Scholar] [CrossRef] [PubMed]
- Perillo, T.; Paolella, C.; Perrotta, G.; Serino, A.; Caranci, F.; Manto, A. Reversible cerebral vasoconstriction syndrome: Review of neuroimaging findings. La Radiol. medica 2022, 127, 981–990. [Google Scholar] [CrossRef] [PubMed]
- Pizzini, F.B.; Conti, E.; Bianchetti, A.; Splendiani, A.; Fusco, D.; Caranci, F.; Bozzao, A.; Landi, F.; Gandolfo, N.; Farina, L.; et al. Radiological assessment of dementia: The Italian inter-society consensus for a practical and clinically oriented guide to image acquisition, evaluation, and reporting. La Radiol. medica 2022, 127, 998–1022. [Google Scholar] [CrossRef]
- Liu, S.; Buch, S.; Chen, Y.; Choi, H.; Dai, Y.; Habib, C.; Hu, J.; Jung, J.; Luo, Y.; Utriainen, D.; et al. Susceptibility-weighted imaging: Current status and future directions. NMR Biomed. 2016, 30. [Google Scholar] [CrossRef]
- Reichenbach, J.R.; Schweser, F.; Serres, B.; Deistung, A. Quantitative Susceptibility Mapping: Concepts and Applications. Clin. Neuroradiol. 2015, 25, 225–230. [Google Scholar] [CrossRef]
- Scola, E.; Desideri, I.; Bianchi, A.; Gadda, D.; Busto, G.; Fiorenza, A.; Amadori, T.; Mancini, S.; Miele, V.; Fainardi, E. Assessment of brain tumors by magnetic resonance dynamic susceptibility contrast perfusion-weighted imaging and computed tomography perfusion: A comparison study. La Radiol. medica 2022, 127, 664–672. [Google Scholar] [CrossRef]
- Zhang, S.; Chiang, G.C.-Y.; Knapp, J.M.; Zecca, C.M.; He, D.; Ramakrishna, R.; Magge, R.S.; Pisapia, D.J.; Fine, H.A.; Tsiouris, A.J.; et al. Grading meningiomas utilizing multiparametric MRI with inclusion of susceptibility weighted imaging and quantitative susceptibility mapping. J. Neuroradiol. 2020, 47, 272–277. [Google Scholar] [CrossRef]
- Saini, J.; Gupta, P.K.; Sahoo, P.; Singh, A.; Patir, R.; Ahlawat, S.; Beniwal, M.; Thennarasu, K.; Santosh, V.; Gupta, R.K. Differentiation of grade II/III and grade IV glioma by combining “T1 contrast-enhanced brain perfusion imaging” and susceptibility-weighted quantitative imaging. Neuroradiology 2017, 60, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Vinayagamani, S.; Sabarish, S.; Nair, S.S.; Tandon, V.; Kesavadas, C.; Thomas, B. Quantitative susceptibility-weighted imaging in predicting disease activity in multiple sclerosis. Neuroradiology 2021, 63, 1061–1069. [Google Scholar] [CrossRef] [PubMed]
- Eilaghi, A.; Yeung, T.; D’Esterre, C.; Bauman, G.; Yartsev, S.; Easaw, J.; Fainardi, E.; Lee, T.-Y.; Frayne, R. Quantitative Perfusion and Permeability Biomarkers in Brain Cancer from Tomographic CT and MR Images. Biomarkers Cancer 2016, 8s2, BIC.S31801–BIC.S31859. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Park, J.E.; Chung, M.S.; Oh, S.W.; Moon, W.-J. Expert Opinions and Recommendations for the Clinical Use of Quantitative Analysis Software for MRI-Based Brain Volumetry. J. Korean Soc. Radiol. 2021, 82, 1124–1139. [Google Scholar] [CrossRef] [PubMed]
- Sozzi, C.; Trentadue, M.; Nicolì, L.; Tavani, F.; Piovan, E. Utility of vertebral biopsy before vertebroplasty in patients with diagnosis of vertebral compression fracture. La Radiol. medica 2021, 126, 956–962. [Google Scholar] [CrossRef]
- Zaki, L.A.M.; Vernooij, M.W.; Smits, M.; Tolman, C.; Papma, J.M.; Visser, J.J.; Steketee, R.M.E. Comparing two artificial intelligence software packages for normative brain volumetry in memory clinic imaging. Neuroradiology 2022, 64, 1359–1366. [Google Scholar] [CrossRef]
- Sullivan, D.C.; Obuchowski, N.A.; Kessler, L.G.; Raunig, D.L.; Gatsonis, C.; Huang, E.P.; Kondratovich, M.; McShane, L.M.; Reeves, A.P.; Barboriak, D.P.; et al. Metrology Standards for Quantitative Imaging Biomarkers. Radiology 2015, 277, 813–825. [Google Scholar] [CrossRef]
- Raunig, D.L.; McShane, L.M.; Pennello, G.; Gatsonis, C.; Carson, P.L.; Voyvodic, J.T.; Wahl, R.L.; Kurland, B.F.; Schwarz, A.J.; Gönen, M.; et al. Quantitative imaging biomarkers: A review of statistical methods for technical performance assessment. Stat. Methods Med Res. 2014, 24, 27–67. [Google Scholar] [CrossRef]
- Cashmore, M.T.; McCann, A.J.; Wastling, S.J.; McGrath, C.; Thornton, J.; Hall, M.G. Clinical quantitative MRI and the need for metrology. Br. J. Radiol. 2021, 94, 20201215. [Google Scholar] [CrossRef]
- Minosse, S.; Picchi, E.; Ferrazzoli, V.; Pucci, N.; Da Ros, V.; Giocondo, R.; Floris, R.; Garaci, F.; Di Giuliano, F. Influence of scan duration on dynamic contrast -enhanced magnetic resonance imaging pharmacokinetic parameters for brain lesions. Magn. Reson. Imaging 2023, 105, 46–56. [Google Scholar] [CrossRef]
- LoCastro, E.; Paudyal, R.; Konar, A.S.; LaViolette, P.S.; Akin, O.; Hatzoglou, V.; Goh, A.C.; Bochner, B.H.; Rosenberg, J.; Wong, R.J.; et al. A Quantitative Multiparametric MRI Analysis Platform for Estimation of Robust Imaging Biomarkers in Clinical Oncology. Tomography 2023, 9, 2052–2066. [Google Scholar] [CrossRef] [PubMed]
- Chauvie, S.; Mazzoni, L.N.; O’doherty, J. A Review on the Use of Imaging Biomarkers in Oncology Clinical Trials: Quality Assurance Strategies for Technical Validation. Tomography 2023, 9, 1876–1902. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, T. Quantitative susceptibility mapping (QSM): Decoding MRI data for a tissue magnetic biomarker. Magn. Reson. Med. 2014, 73, 82–101. [Google Scholar] [CrossRef] [PubMed]
- Boss, M.A.; Malyarenko, D.; Partridge, S.; Obuchowski, N.; Shukla-Dave, A.; Winfield, J.M.; Fuller, C.D.; Miller, K.; Mishra, V.; Ohliger, M.; et al. The QIBA Profile for Diffusion-Weighted MRI: Apparent Diffusion Coefficient as a Quantitative Imaging Biomarker. Radiology 2024, 313, e233055. [Google Scholar] [CrossRef]
- Keenan, K.E.; Ainslie, M.; Barker, A.J.; Boss, M.A.; Cecil, K.M.; Charles, C.; Chenevert, T.L.; Clarke, L.; Evelhoch, J.L.; Finn, P.; et al. Quantitative magnetic resonance imaging phantoms: A review and the need for a system phantom. Magn. Reson. Med. 2017, 79, 48–61. [Google Scholar] [CrossRef]
- Kessler, L.G.; Barnhart, H.X.; Buckler, A.J.; Choudhury, K.R.; Kondratovich, M.V.; Toledano, A.; Guimaraes, A.R.; Filice, R.; Zhang, Z.; Sullivan, D.C.; et al. The emerging science of quantitative imaging biomarkers terminology and definitions for scientific studies and regulatory submissions. Stat. Methods Med Res. 2014, 24, 9–26. [Google Scholar] [CrossRef]
- Cristinacce, P.L.H.; Keaveney, S.; Aboagye, E.O.; Hall, M.G.; Little, R.A.; O’COnnor, J.P.; Parker, G.J.; Waterton, J.C.; Winfield, J.M.; Jauregui-Osoro, M. Clinical translation of quantitative magnetic resonance imaging biomarkers – An overview and gap analysis of current practice. Phys. Medica 2022, 101, 165–182. [Google Scholar] [CrossRef]
- Zwanenburg, A.; Vallières, M.; Abdalah, M.A.; Aerts, H.J.W.L.; Andrearczyk, V.; Apte, A.; Ashrafinia, S.; Bakas, S.; Beukinga, R.J.; Boellaard, R.; et al. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology 2020, 295, 328–338. [Google Scholar] [CrossRef]
- Marques, J.P.; Kober, T.; Krueger, G.; van der Zwaag, W.; Van de Moortele, P.-F.; Gruetter, R. MP2RAGE, a self bias-field corrected sequence for improved segmentation and T1-mapping at high field. NeuroImage 2010, 49, 1271–1281. [Google Scholar] [CrossRef]
- Tambalo, S.; Finora, A.; Cavalli, D.; Jovicich, J. Fast T1 Mapping: Accuracy and Reproducibility of Volumetric Sequences (IR, MP2RAGE, csMP2RAGE). Available online: https://cds.ismrm.org/protected/21MPresentations/abstracts/1056.html (accessed on 3 October 2025).
- Saunders, A.M.; Kim, M.E.; Gao, C.; Remedios, L.W.; Krishnan, A.R.; Schilling, K.G.; O’GRady, K.P.; Smith, S.A.; Landman, B.A. Comparison and calibration of MP2RAGE quantitative T1 values to multi-TI inversion recovery T1 values. Magn. Reson. Imaging 2025, 117, 110322. [Google Scholar] [CrossRef]
- Rowley, C.D.; Nelson, M.C.; Campbell, J.S.W.; Leppert, I.R.; Pike, G.B.; Tardif, C.L. Correcting for T1 bias in Mag-netization Transfer Saturation (MTsat) Maps Using Sparse-MP2RAGE (Version 1). arXiv 2023, arXiv:2310.09102. [Google Scholar] [CrossRef]
- Kraff, O.; May, M.W. Multi-center QA of ultrahigh-field systems. Magn. Reson. Mater. Physics, Biol. Med. 2025, 38, 519–532. [Google Scholar] [CrossRef] [PubMed]
- Warntjes, J.; Leinhard, O.D.; West, J.; Lundberg, P. Rapid magnetic resonance quantification on the brain: Optimization for clinical usage. Magn. Reson. Med. 2008, 60, 320–329. [Google Scholar] [CrossRef]
- Schmidbauer, V.; Geisl, G.; Diogo, M.; Weber, M.; Goeral, K.; Klebermass-Schrehof, K.; Berger, A.; Prayer, D.; Kasprian, G. SyMRI detects delayed myelination in preterm neonates. Eur. Radiol. 2019, 29, 7063–7072. [Google Scholar] [CrossRef]
- Lee, S.M.; Choi, Y.H.; You, S.-K.; Lee, W.K.; Kim, W.H.; Kim, H.J.; Lee, S.Y.; Cheon, H. Age-Related Changes in Tissue Value Properties in Children. Investig. Radiol. 2018, 53, 236–245. [Google Scholar] [CrossRef]
- Hagiwara, A.; Warntjes, M.; Hori, M.; Andica, C.; Nakazawa, M.; Kumamaru, K.K.; Abe, O.; Aoki, S. SyMRI of the Brain. Investig. Radiol. 2017, 52, 647–657. [Google Scholar] [CrossRef]
- Tanenbaum, L.; Tsiouris, A.; Johnson, A.; Naidich, T.; DeLano, M.; Melhem, E.; Quarterman, P.; Parameswaran, S.; Shankaranarayanan, A.; Goyen, M.; et al. Synthetic MRI for Clinical Neuroimaging: Results of the Magnetic Resonance Image Compilation (MAGiC) Prospective, Multicenter, Multireader Trial. Am. J. Neuroradiol. 2017, 38, 1103–1110. [Google Scholar] [CrossRef]
- Deoni, S.C.L.; Peters, T.M.; Rutt, B.K. High-resolution T1 and T2 mapping of the brain in a clinically acceptable time with DESPOT1 and DESPOT2. Magn. Reson. Med. 2004, 53, 237–241. [Google Scholar] [CrossRef]
- Liao, C.; Cao, X.; Iyer, S.S.; Schauman, S.; Zhou, Z.; Yan, X.; Chen, Q.; Li, Z.; Wang, N.; Gong, T.; et al. High-resolution myelin-water fraction and quantitative relaxation mapping using 3D ViSTa-MR fingerprinting. Magn. Reson. Med. 2023, 91, 2278–2293. [Google Scholar] [CrossRef]
- Arshad, N.H.; Abu Hassan, H. Quantifying myelin in neonates using magnetic resonance imaging: A systematic literature review. Clin. Exp. Pediatr. 2023, 67, 371–385. [Google Scholar] [CrossRef] [PubMed]
- Zöllei, L.; Iglesias, J.E.; Ou, Y.; Grant, P.E.; Fischl, B. Infant FreeSurfer: An automated segmentation and surface extraction pipeline for T1-weighted neuroimaging data of infants 0–2 years. NeuroImage 2020, 218, 116946. [Google Scholar] [CrossRef] [PubMed]
- Wada, A.; Saito, Y.; Irie, R.; Kamagata, K.; Maekawa, T.; Fujita, S.; Hagiwara, A.; Kumamaru, K.K.; Suzuki, M.; Nakanishi, A.; et al. Convolutional Neural Networks for Estimation of Myelin Maturation in Infant Brain. arXiv 2019, arXiv:1909.03986. [Google Scholar]
- Deoni, S.C.; Kolind, S.H. Investigating the stability of mcDESPOT myelin water fraction values derived using a stochastic region contraction approach. Magn. Reson. Med. 2014, 73, 161–169. [Google Scholar] [CrossRef]
- Bouhrara, M.; Rejimon, A.C.; Cortina, L.E.; Khattar, N.; Bergeron, C.M.; Ferrucci, L.; Resnick, S.M.; Spencer, R.G. Adult brain aging investigated using BMC-mcDESPOT–based myelin water fraction imaging. Neurobiol. Aging 2020, 85, 131–139. [Google Scholar] [CrossRef]
- Gong, Z.; Khattar, N.; Kiely, M.; Triebswetter, C.; Bouhrara, M. REUSED: A deep neural network method for rapid whole-brain high-resolution myelin water fraction mapping from extremely under-sampled MRI. Comput. Med Imaging Graph. 2023, 108, 102282. [Google Scholar] [CrossRef]
- Faulkner, M.E.; Gong, Z.; Guo, A.; Laporte, J.P.; Bae, J.; Bouhrara, M. Harnessing myelin water fraction as an imaging biomarker of human cerebral aging, neurodegenerative diseases, and risk factors influencing myelination: A review. J. Neurochem. 2024, 168, 2243–2263. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.J.; Scheuren, P.S.; Liu, H.; Loke, R.W.J.; Laule, C.; Loucks, C.M.; Kramer, J.L. The myelin water imaging transcriptome: Myelin water fraction regionally varies with oligodendrocyte-specific gene expression. Mol. Brain 2024, 17, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Yarnykh, V.L. Fast macromolecular proton fraction mapping from a single off-resonance magnetization transfer measurement. Magn. Reson. Med. 2011, 68, 166–178. [Google Scholar] [CrossRef] [PubMed]
- York, E.N.; Thrippleton, M.J.; Meijboom, R.; Hunt, D.P.J.; Waldman, A.D. Quantitative magnetization transfer imaging in relapsing-remitting multiple sclerosis: A systematic review and meta-analysis. Brain Commun. 2022, 4, fcac088. [Google Scholar] [CrossRef]
- Mehrabian, H.; Myrehaug, S.; Soliman, H.; Sahgal, A.; Stanisz, G.J. Quantitative Magnetization Transfer in Monitoring Glioblastoma (GBM) Response to Therapy. Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef]
- Assländer, J.; Gultekin, C.; Mao, A.; Zhang, X.; Duchemin, Q.; Liu, K.; Charlson, R.W.; Shepherd, T.M.; Fernandez-Granda, C.; Flassbeck, S. Rapid quantitative magnetization transfer imaging: Utilizing the hybrid state and the generalized Bloch model. Magn. Reson. Med. 2023, 91, 1478–1497. [Google Scholar] [CrossRef]
- Rowley, C.D.; Campbell, J.S.W.; Leppert, I.R.; Nelson, M.C.; Pike, G.B.; Tardif, C.L. Optimization of acquisition parameters for cortical inhomogeneous magnetization transfer (ihMT) imaging using a rapid gradient echo readout. Magn. Reson. Med. 2023, 90, 1762–1775. [Google Scholar] [CrossRef]
- Soustelle, L.; Troalen, T.; Hertanu, A.; Ranjeva, J.; Guye, M.; Varma, G.; Alsop, D.C.; Duhamel, G.; Girard, O.M. Quantitative magnetization transfer MRI unbiased by on-resonance saturation and dipolar order contributions. Magn. Reson. Med. 2023, 90, 875–893. [Google Scholar] [CrossRef]
- Chen, X.; Roberts, N.; Zheng, Q.; Peng, Y.; Han, Y.; Luo, Q.; Feng, J.; Luo, T.; Li, Y. Comparison of diffusion tensor imaging (DTI) tissue characterization parameters in white matter tracts of patients with multiple sclerosis (MS) and neuromyelitis optica spectrum disorder (NMOSD). Eur. Radiol. 2024, 34, 5263–5275. [Google Scholar] [CrossRef] [PubMed]
- Deoni, S.C.L.; Mercure, E.; Blasi, A.; Gasston, D.; Thomson, A.; Johnson, M.; Williams, S.C.R.; Murphy, D.G.M. Mapping Infant Brain Myelination with Magnetic Resonance Imaging. J. Neurosci. 2011, 31, 784–791. [Google Scholar] [CrossRef] [PubMed]
- Laule, C.; Kozlowski, P.; Leung, E.; Li, D.K.; MacKay, A.L.; Moore, G.W. Myelin water imaging of multiple sclerosis at 7 T: Correlations with histopathology. NeuroImage 2008, 40, 1575–1580. [Google Scholar] [CrossRef] [PubMed]
- Chuhutin, A.; Hansen, B.; Wlodarczyk, A.; Owens, T.; Shemesh, N.; Jespersen, S.N. Diffusion Kurtosis Imaging maps neural damage in the EAE model of multiple sclerosis. NeuroImage 2019, 208, 116406. [Google Scholar] [CrossRef]
- Lu, D.; Jiang, Y.; Ji, Y.; Zhou, I.Y.; Mandeville, E.; Lo, E.H.; Wang, X.; Sun, P.Z. JOURNAL CLUB: Evaluation of Diffusion Kurtosis Imaging of Stroke Lesion With Hemodynamic and Metabolic MRI in a Rodent Model of Acute Stroke. Am. J. Roentgenol. 2018, 210, 720–727. [Google Scholar] [CrossRef]
- Jensen, J.H.; Helpern, J.A. MRI quantification of non-Gaussian water diffusion by kurtosis analysis. NMR Biomed. 2010, 23, 698–710. [Google Scholar] [CrossRef]
- Fieremans, E.; Jensen, J.H.; Helpern, J.A. Diffusion kurtosis imaging in brain tumors: Initial evidence for glioma grading. AJNR Am. J. Neuroradiol. 2019, 40, 1531–1538. [Google Scholar]
- Hui, E.S.; Fieremans, E.; Jensen, J.H.; Tabesh, A.; Feng, W.; Bonilha, L.; Spampinato, M.V.; Adams, R.; Helpern, J.A. Stroke Assessment With Diffusional Kurtosis Imaging. Stroke 2012, 43, 2968–2973. [Google Scholar] [CrossRef]
- Nilsson, M.; Englund, E.; Szczepankiewicz, F.; van Westen, D.; Sundgren, P.C. Imaging brain tumour microstructure. NeuroImage 2018, 182, 232–250. [Google Scholar] [CrossRef]
- Zerweck, L.; Würtemberger, U.; Klose, U.; Reisert, M.; Richter, V.; Nägele, T.; Staber, D.; Han, T.; Shen, M.; Xie, C.; et al. Performance Comparison of Diffusion Kurtosis Imaging (DKI), Neurite Orientation Dispersion and Density Imaging (NODDI), and Diffusion Microstructure Imaging (DMI) in Predicting Adult-Type Glioma Subtype—A Pilot Study. Cancers 2025, 17, 876. [Google Scholar] [CrossRef] [PubMed]
- Panagiotaki, E.; Walker-Samuel, S.; Siow, B.; Johnson, S.P.; Rajkumar, V.; Pedley, R.B.; Lythgoe, M.F.; Alexander, D.C. Noninvasive Quantification of Solid Tumor Microstructure Using VERDICT MRI. Cancer Res. 2014, 74, 1902–1912. [Google Scholar] [CrossRef] [PubMed]
- Palombo, M.; Valindria, V.; Singh, S.; Chiou, E.; Giganti, F.; Pye, H.; Whitaker, H.C.; Atkinson, D.; Punwani, S.; Alexander, D.C.; et al. Joint estimation of relaxation and diffusion tissue parameters for prostate cancer with relaxation-VERDICT MRI. Sci. Rep. 2023, 13, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Zaccagna, F.; Riemer, F.; Priest, A.N.; McLean, M.A.; Allinson, K.; Grist, J.T.; Dragos, C.; Matys, T.; Gillard, J.H.; Watts, C.; et al. Non-invasive assessment of glioma microstructure using VERDICT MRI: Correlation with histology. Eur. Radiol. 2019, 29, 5559–5566. [Google Scholar] [CrossRef]
- Snoussi, H.; Cohen-Adad, J.; Combès, B.; Bannier, É.; Tounekti, S.; Kerbrat, A.; Barillot, C.; Caruyer, E. Effectiveness of regional diffusion MRI measures in distinguishing multiple sclerosis abnormalities within the cervical spinal cord. Brain Behav. 2023, 13, e3159. [Google Scholar] [CrossRef]
- Trò, R.; Roascio, M.; Tortora, D.; Severino, M.; Rossi, A.; Cohen-Adad, J.; Fato, M.M.; Arnulfo, G. Diffusion Kurtosis Imaging of Neonatal Spinal Cord in Clinical Routine. Front. Radiol. 2022, 2. [Google Scholar] [CrossRef]
- Jespersen, S.N.; Olesen, J.L.; Hansen, B.; Shemesh, N. Diffusion time dependence of microstructural parameters in fixed spinal cord. NeuroImage 2018, 182, 329–342. [Google Scholar] [CrossRef]
- Henriques, R.N.; Jespersen, S.N.; Shemesh, N. Evidence for microscopic kurtosis in neural tissue revealed by correlation tensor MRI. Magn. Reson. Med. 2021, 86, 3111–3130. [Google Scholar] [CrossRef]
- Martinez-Heras, E.; Grussu, F.; Prados, F.; Solana, E.; Llufriu, S. Diffusion-Weighted Imaging: Recent Advances and Applications. Semin. Ultrasound, CT MRI 2021, 42, 490–506. [Google Scholar] [CrossRef]
- Barnes, N.; Dkhar, W.; Kadavigere, R.; Sukumar, S.; K, V.; Pradhan, A. A narrative review of diffusion kurtosis MRI parameters in diagnosing degenerative spine diseases in animal models. Open Veter- J. 2024, 14, 3181–3188. [Google Scholar] [CrossRef]
- Tang, L.; Zhou, X.J. Diffusion MRI of cancer: From low to high b-values. J. Magn. Reson. Imaging 2018, 49, 23–40. [Google Scholar] [CrossRef]
- O’COnnor, J.P.B.; Aboagye, E.O.; Adams, J.E.; Aerts, H.J.W.L.; Barrington, S.F.; Beer, A.J.; Boellaard, R.; Bohndiek, S.E.; Brady, M.; Brown, G.; et al. Imaging biomarker roadmap for cancer studies. Nat. Rev. Clin. Oncol. 2016, 14, 169–186. [Google Scholar] [CrossRef] [PubMed]
- Mito, R.; Cole, J.H.; Genc, S.; Jackson, G.D.; Zalesky, A. Towards precision MRI biomarkers in epilepsy with normative modelling. Brain 2025, 148, 2247–2261. [Google Scholar] [CrossRef] [PubMed]
- Schweser, F.; Deistung, A.; Lehr, B.W.; Reichenbach, J.R. Quantitative imaging of intrinsic magnetic tissue properties using MRI signal phase: An approach to in vivo brain iron metabolism? NeuroImage 2011, 54, 2789–2807. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Avram, A.V.; Wu, B.; Xiao, X.; Liu, C. Integrated Laplacian-based phase unwrapping and background phase removal for quantitative susceptibility mapping. NMR Biomed. 2013, 27, 219–227. [Google Scholar] [CrossRef]
- Sun, H.; Wilman, A.H. Background field removal using spherical mean value filtering and Tikhonov regularization. Magn. Reson. Med. 2013, 71, 1151–1157. [Google Scholar] [CrossRef]
- Li, W.; Wu, B.; Liu, C. Quantitative susceptibility mapping of human brain reflects spatial variation in tissue composition. NeuroImage 2011, 55, 1645–1656. [Google Scholar] [CrossRef]
- QSM Consensus Organization Committee; Bilgic, B.; Costagli, M.; Chan, K.; Duyn, J.; Langkammer, C.; Lee, J.; Li, X.; Liu, C.; Marques, J.P.; et al. Recommended implementation of quantitative susceptibility mapping for clinical research in the brain: A consensus of the ISMRM electro-magnetic tissue properties study group. Magn. Reson. Med. 2024, 91, 1834–1862. [Google Scholar] [CrossRef]
- Zhang, Y.; Gauthier, S.; Gupta, A.; Tu, L.; Comunale, J.; Chiang, G.-Y.; Chen, W.; Salustri, C.; Zhu, W.; Wang, Y. Magnetic Susceptibility from Quantitative Susceptibility Mapping Can Differentiate New Enhancing from Nonenhancing Multiple Sclerosis Lesions without Gadolinium Injection. Am. J. Neuroradiol. 2016, 37, 1794–1799. [Google Scholar] [CrossRef]
- Absinta, M.; Sati, P.; Masuzzo, F.; Nair, G.; Sethi, V.; Kolb, H.; Ohayon, J.; Wu, T.; Cortese, I.C.M.; Reich, D.S. Association of Chronic Active Multiple Sclerosis Lesions With Disability In Vivo. JAMA Neurol. 2019, 76, 1474–1483. [Google Scholar] [CrossRef]
- Karsa, A.; Punwani, S.; Shmueli, K. The effect of low resolution and coverage on the accuracy of susceptibility mapping. Magn. Reson. Med. 2018, 81, 1833–1848. [Google Scholar] [CrossRef]
- Ravanfar, P.; Loi, S.M.; Syeda, W.T.; Van Rheenen, T.E.; Bush, A.I.; Desmond, P.; Cropley, V.L.; Lane, D.J.R.; Opazo, C.M.; Moffat, B.A.; et al. Systematic Review: Quantitative Susceptibility Mapping (QSM) of Brain Iron Profile in Neurodegenerative Diseases. Front. Neurosci. 2021, 15. [Google Scholar] [CrossRef]
- Mittal, S.; Wu, Z.; Neelavalli, J.; Haacke, E. Susceptibility-Weighted Imaging: Technical Aspects and Clinical Applications, Part 2. Am. J. Neuroradiol. 2009, 30, 232–252. [Google Scholar] [CrossRef]
- Haacke, E.M.; Mittal, S.; Wu, Z.; Neelavalli, J.; Cheng, Y.-C. Susceptibility-Weighted Imaging: Technical Aspects and Clinical Applications, Part 1. Am. J. Neuroradiol. 2009, 30, 19–30. [Google Scholar] [CrossRef]
- Wu, B.; Li, W.; Guidon, A.; Liu, C. Whole brain susceptibility mapping using compressed sensing. Magn. Reson. Med. 2011, 67, 137–147. [Google Scholar] [CrossRef] [PubMed]
- Fan, A.P.; A Khalil, A.; Fiebach, J.B.; Zaharchuk, G.; Villringer, A.; Villringer, K.; Gauthier, C.J. Elevated brain oxygen extraction fraction measured by MRI susceptibility relates to perfusion status in acute ischemic stroke. J. Cereb. Blood Flow Metab. 2019, 40, 539–551. [Google Scholar] [CrossRef] [PubMed]
- Berberat, J.; Grobholz, R.; Boxheimer, L.; Rogers, S.; Remonda, L.; Roelcke, U. Differentiation Between Calcification and Hemorrhage in Brain Tumors Using Susceptibility-Weighted Imaging: A Pilot Study. Am. J. Roentgenol. 2014, 202, 847–850. [Google Scholar] [CrossRef] [PubMed]
- Eskreis-Winkler, S.; Zhang, Y.; Zhang, J.; Liu, Z.; Dimov, A.; Gupta, A.; Wang, Y. The clinical utility of QSM: Disease diagnosis, medical management, and surgical planning. NMR Biomed. 2016, 30. [Google Scholar] [CrossRef]
- Duyn, J.H.; Schenck, J. Contributions to magnetic susceptibility of brain tissue. NMR Biomed. 2016, 30. [Google Scholar] [CrossRef]
- Azuma, M.; Maekawa, K.; Yamashita, A.; Yokogami, K.; Enzaki, M.; Khant, Z.; Takeshima, H.; Asada, Y.; Wang, Y.; Hirai, T. Characterization of Carotid Plaque Components by Quantitative Susceptibility Mapping. Am. J. Neuroradiol. 2019, 41, 310–317. [Google Scholar] [CrossRef]
- Clarke, M.A.; Witt, A.A.; Robison, R.K.; Fleishman, S.; Combes, A.J.; Houston, D.; Prock, L.E.; Sweeney, G.; O’GRady, K.P.; McKnight, C.D.; et al. Cervical spinal cord susceptibility-weighted MRI at 7T: Application to multiple sclerosis. NeuroImage 2023, 284, 120460. [Google Scholar] [CrossRef]
- Marques, J.P.; Meineke, J.; Milovic, C.; Bilgic, B.; Chan, K.; Hedouin, R.; van der Zwaag, W.; Langkammer, C.; Schweser, F. QSM reconstruction challenge 2.0: A realistic in silico head phantom for MRI data simulation and evaluation of susceptibility mapping procedures. Magn. Reson. Med. 2021, 86, 526–542. [Google Scholar] [CrossRef]
- Naji, N.; Lauzon, M.L.; Seres, P.; Stolz, E.; Frayne, R.; Lebel, C.; Beaulieu, C.; Wilman, A.H. Multisite reproducibility of quantitative susceptibility mapping and effective transverse relaxation rate in deep gray matter at 3 T using locally optimized sequences in 24 traveling heads. NMR Biomed. 2022, 35, e4788. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Clingman, C.; Golay, X.; van Zijl, P.C. Determining the longitudinal relaxation time (T1) of blood at 3.0 Tesla. Magn. Reson. Med. 2004, 52, 679–682. [Google Scholar] [CrossRef] [PubMed]
- Østergaard, L.; Weisskoff, R.M.; Chesler, D.A.; Gyldensted, C.; Rosen, B.R. High resolution measurement of cerebral blood flow using intravascular tracer bolus passages. Part I: Mathematical approach and statistical analysis. Magn. Reson. Med. 1996, 36, 715–725. [Google Scholar] [CrossRef] [PubMed]
- Boxerman, J.; Schmainda, K.; Weisskoff, R. Relative Cerebral Blood Volume Maps Corrected for Contrast Agent Extravasation Significantly Correlate with Glioma Tumor Grade, Whereas Uncorrected Maps Do Not. Am. J. Neuroradiol. 2006, 27, 859–867. [Google Scholar]
- Hua, J.; Qin, Q.; Donahue, M.J.; Zhou, J.; Pekar, J.J.; van Zijl, P.C. Inflow-based vascular-space-occupancy (iVASO) MRI. Magn. Reson. Med. 2011, 66, 40–56. [Google Scholar] [CrossRef]
- Tofts, P.S.; Kermode, A.G. Measurement of the blood-brain barrier permeability and leakage space using dynamic MR imaging. 1. Fundamental concepts. Magn. Reson. Med. 1991, 17, 357–367. [Google Scholar] [CrossRef]
- Sourbron, S.P.; Buckley, D.L. Classic models for dynamic contrast-enhanced MRI. NMR Biomed. 2013, 26, 1004–1027. [Google Scholar] [CrossRef]
- Lupo, J.M.; Cha, S.; Chang, S.M.; Nelson, S.J. Dynamic Susceptibility-Weighted Perfusion Imaging of High-Grade Gliomas: Characterization of Spatial Heterogeneity. Am. J. Neuroradiol. 2005, 26, 1446–1454. [Google Scholar]
- Mouridsen, K.; Friston, K.; Hjort, N.; Gyldensted, L.; Østergaard, L.; Kiebel, S. Bayesian estimation of cerebral perfusion using a physiological model of microvasculature. NeuroImage 2006, 33, 570–579. [Google Scholar] [CrossRef]
- Withey, S.B.; MacPherson, L.; Oates, A.; Powell, S.; Novak, J.; Abernethy, L.; Pizer, B.; Grundy, R.; Morgan, P.S.; Bailey, S.; et al. Dynamic susceptibility-contrast magnetic resonance imaging with contrast agent leakage correction aids in predicting grade in pediatric brain tumours: A multicenter study. Pediatr. Radiol. 2022, 52, 1134–1149. [Google Scholar] [CrossRef]
- Jackson, A.; Jayson, G.C.; Li, K.L.; Zhu, X.P.; Checkley, D.R.; Tessier, J.J.L.; Waterton, J.C. Reproducibility of quantitative dynamic contrast-enhanced MRI in newly presenting glioma. Br. J. Radiol. 2003, 76, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Sourbron, S.; Luypaert, R.; Morhard, D.; Seelos, K.; Reiser, M.; Peller, M. Deconvolution of bolus-tracking data: A comparison of discretization methods. Phys. Med. Biol. 2007, 52, 6761–6778. [Google Scholar] [CrossRef] [PubMed]
- Conte, G.M.; Altabella, L.; Castellano, A.; Cuccarini, V.; Bizzi, A.; Grimaldi, M.; Costa, A.; Caulo, M.; Falini, A.; Anzalone, N. Comparison of T1 mapping and fixed T1 method for dynamic contrast-enhanced MRI perfusion in brain gliomas. Eur. Radiol. 2019, 29, 3467–3479. [Google Scholar] [CrossRef] [PubMed]
- Stokes, A.M.; Bergamino, M.; Alhilali, L.; Hu, L.S.; Karis, J.P.; Baxter, L.C.; Bell, L.C.; Quarles, C.C. Evaluation of single bolus, dual-echo dynamic susceptibility contrast MRI protocols in brain tumor patients. J. Cereb. Blood Flow Metab. 2021, 41, 3378–3390. [Google Scholar] [CrossRef]
- E Emblem, K.; Mouridsen, K.; Bjornerud, A.; Farrar, C.T.; Jennings, D.; Borra, R.J.H.; Wen, P.Y.; Ivy, P.; Batchelor, T.T.; Rosen, B.R.; et al. Vessel architectural imaging identifies cancer patient responders to anti-angiogenic therapy. Nat. Med. 2013, 19, 1178–1183. [Google Scholar] [CrossRef]
- Barajas, R.F.; Phillips, J.J.; Parvataneni, R.; Molinaro, A.; Essock-Burns, E.; Bourne, G.; Parsa, A.T.; Aghi, M.K.; McDermott, M.W.; Berger, M.S.; et al. Regional variation in histopathologic features of tumor specimens from treatment-naive glioblastoma correlates with anatomic and physiologic MR Imaging. Neuro-Oncology 2012, 14, 942–954. [Google Scholar] [CrossRef]
- Willats, L.; Connelly, A.; Calamante, F. Improved deconvolution of perfusion MRI data in the presence of bolus delay and dispersion. Magn. Reson. Med. 2006, 56, 146–156. [Google Scholar] [CrossRef]
- Schmainda, K.M.; Prah, M.; Connelly, J.; Rand, S.D.; Hoffman, R.G.; Mueller, W.; Malkin, M.G. Dynamic-susceptibility contrast agent MRI measures of relative cerebral blood volume predict response to bevacizumab in recurrent high-grade glioma. Neuro-Oncology 2014, 16, 880–888. [Google Scholar] [CrossRef]
- Law, M.; Young, R.J.; Babb, J.S.; Peccerelli, N.; Chheang, S.; Gruber, M.L.; Miller, D.C.; Golfinos, J.G.; Zagzag, D.; Johnson, G. Gliomas: Predicting Time to Progression or Survival with Cerebral Blood Volume Measurements at Dynamic Susceptibility-weighted Contrast-enhanced Perfusion MR Imaging. Radiology 2008, 247, 490–498. [Google Scholar] [CrossRef]
| Disease/Condition | Most Relevant qMRI Biomarkers (Examples) | Typical Readouts (Units) | Representative Uses |
|---|---|---|---|
| Multiple sclerosis (MS) | Myelin/MT (MWF, MTsat), T1/R1, DTI (FA/RD), QSM (rim-positive), volumetry (GM/thal) | MWF (%), MTsat (p.u.), T1 (ms), R1 (s−1), FA/RD (–/mm2/s), χ (ppm), volumes (cm3) | Demyelination vs. repair; lesion staging; disability risk; progressive disease monitoring |
| Dementia/AD | Volumetry (hippocampus/cortex), ASL-CBF, QSM (deep nuclei iron) | cm3; cortical thickness (mm); CBF (mL/100 g/min); χ (ppm) | Early diagnosis; subtype patterns; progression tracking |
| Neuro-oncology (gliomas/metastases) | DSC rCBV, DCE Ktrans/Ve, ADC, QSM (calcification vs. hemorrhage) | rCBV (ratio), Ktrans (min−1), Ve (–), ADC (mm2/s), χ (ppm) | Grading; pseudo-progression vs. progression; early response (SRS/anti-angiogenic) |
| Ischemic stroke | DWI/ADC, DKI MK, ASL-CBF, DSC delay/MTT | ADC (mm2/s); MK (–); CBF (mL/100 g/min); MTT (s) | Core/penumbra; tissue-at-risk delineation |
| TBI | SWI/QSM (microbleeds), DTI (FA/RD), volumetry | χ (ppm); FA/RD; cm3 | Diffuse axonal injury; microhemorrhage burden; prognosis |
| Spinal cord/DCM | DTI (FA/MD), MT/MTsat, MWF | FA/MD; MTsat; MWF (%) | Subclinical degeneration; severity; outcome prediction |
| Modality | Typical Acquisition | Approx. Time | Primary Outputs (Units) | Strengths | Common Limitations |
|---|---|---|---|---|---|
| T1 relaxometry | IR/MP2RAGE; VFA (B1-corrected); SyMRI | ~4–8 min | T1 (ms), R1 (s−1) | Myelin/sclerosis sensitivity; whole-brain maps | B1/MT bias; sequence heterogeneity |
| T2 relaxometry/MWF | MESE/GRASE; mcDESPOT | ~4–8 min | T2 (ms), MWF (%) | Myelin-related specificity | Stimulated echoes; ill-posed multi-component fits |
| MT (MTR/MTsat/qMT) | GRE with MT prep; multi-parametric MT | ~4–7 min | MTR (p.u.), MTsat (p.u.), qMT params | Myelin/macromolecule sensitivity | B1 dependence; vendor diversity |
| Diffusion (DWI/DTI/DKI/NODDI) | EPI with ≥30 dirs; multi-b shells | ~3–10 min | ADC/FA/MD; MK/NDI/ODI | Microstructure; tractography | EPI distortions; motion/eddy; model dependence |
| SWI/QSM | 3D multi-echo GRE | ~4–7 min | SWI (qual.), χ (ppm) | Veins/iron; calcification vs. hemorrhage | Ill-posed inversion; regularization trade-offs |
| Perfusion (DSC/DCE/ASL) | T2* EPI (DSC); 3D GRE (DCE); pCASL | ~2–3/5–15/4–6 min | rCBV/rCBF/MTT; Ktrans/Ve/Vp/Kep; CBF/ATT | Vascular density/permeability/flow | Leakage/AIF/ATT; SNR; model variance |
| Volumetry | 3D T1 (MPRAGE/SPGR) | ~4–6 min | Regional volumes (cm3), thickness (mm) | Objective atrophy metrics |
| Feature | DSC | DCE |
|---|---|---|
| Sequence | T2*-weighted EPI | T1-weighted 3D GRE |
| Key Parameters | rCBV, rCBF, MTT | Ktrans, Ve, Kep, Vp |
| Temporal Resolution | <2 s | ~4–6 s |
| Duration | ~2–3 min | ~5–15 min |
| Sensitivity | Microvascular density | Capillary permeability |
| Limitations | Leakage effects, susceptibility | AIF estimation, modeling variability |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saltarelli, G.; Di Cerbo, G.; Innocenzi, A.; De Felici, C.; Splendiani, A.; Di Cesare, E. Quantitative MRI in Neuroimaging: A Review of Techniques, Biomarkers, and Emerging Clinical Applications. Brain Sci. 2025, 15, 1088. https://doi.org/10.3390/brainsci15101088
Saltarelli G, Di Cerbo G, Innocenzi A, De Felici C, Splendiani A, Di Cesare E. Quantitative MRI in Neuroimaging: A Review of Techniques, Biomarkers, and Emerging Clinical Applications. Brain Sciences. 2025; 15(10):1088. https://doi.org/10.3390/brainsci15101088
Chicago/Turabian StyleSaltarelli, Gaspare, Giovanni Di Cerbo, Antonio Innocenzi, Claudia De Felici, Alessandra Splendiani, and Ernesto Di Cesare. 2025. "Quantitative MRI in Neuroimaging: A Review of Techniques, Biomarkers, and Emerging Clinical Applications" Brain Sciences 15, no. 10: 1088. https://doi.org/10.3390/brainsci15101088
APA StyleSaltarelli, G., Di Cerbo, G., Innocenzi, A., De Felici, C., Splendiani, A., & Di Cesare, E. (2025). Quantitative MRI in Neuroimaging: A Review of Techniques, Biomarkers, and Emerging Clinical Applications. Brain Sciences, 15(10), 1088. https://doi.org/10.3390/brainsci15101088






