Transcriptomic Analysis of the Antiviral Responses in Ovine Type II Alveolar Epithelial Cells During Early Stage of Bluetongue Virus Infection
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Considerations
2.2. Cells and Viruses
2.3. Cell Culture and BTV Infection
2.4. RNA Extraction and Qualification
2.5. Library Construction and Sequencing
2.6. Bioinformatics Analyses
2.7. IPA Analysis of Differentially Expressed Genes
2.8. Protein–Protein Interaction (PPI) Analysis for Key Genes in OAECIIs
2.9. Real-Time Quantitative PCR
2.10. Statistical Analysis
3. Results
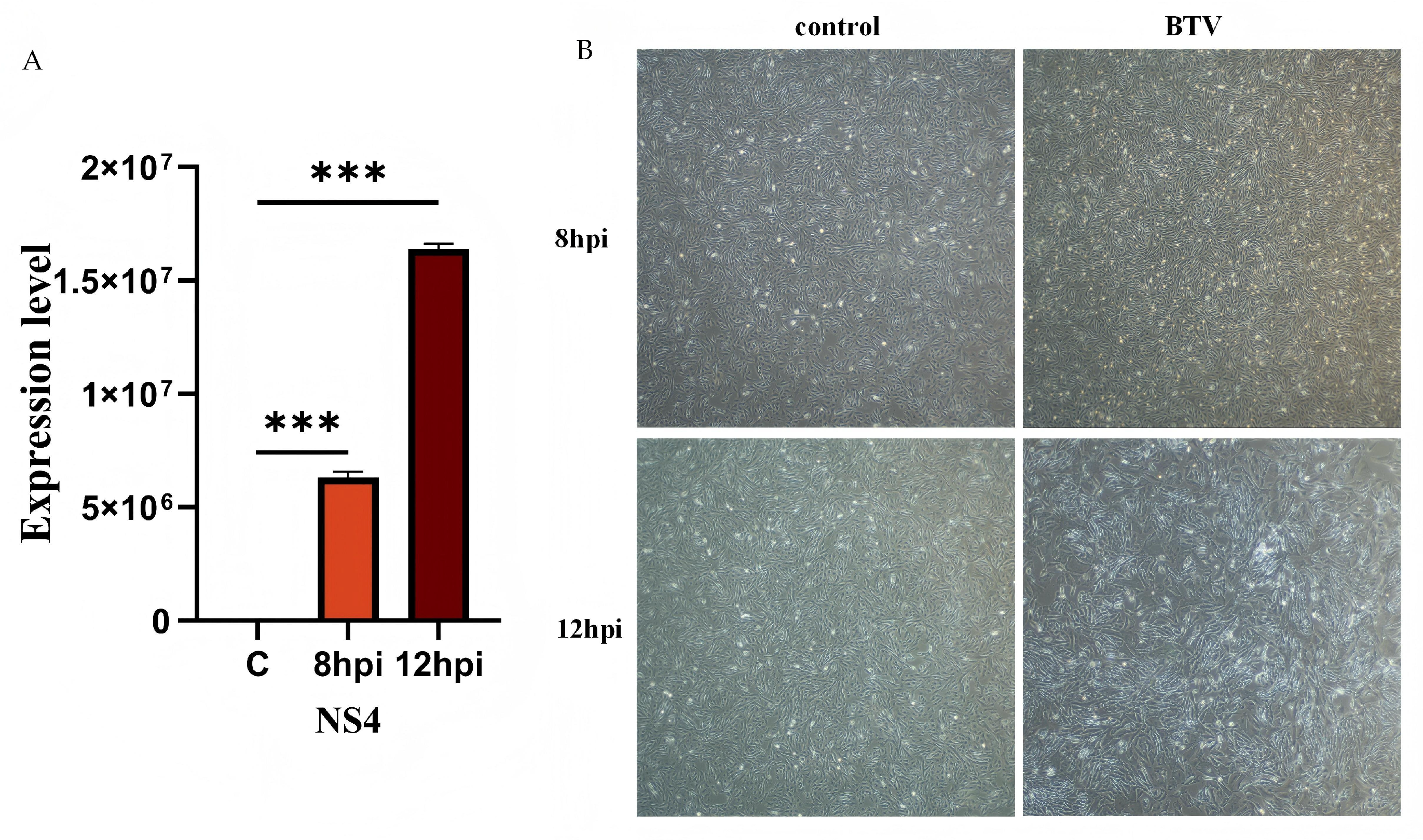
3.1. Viral Infection and Verification
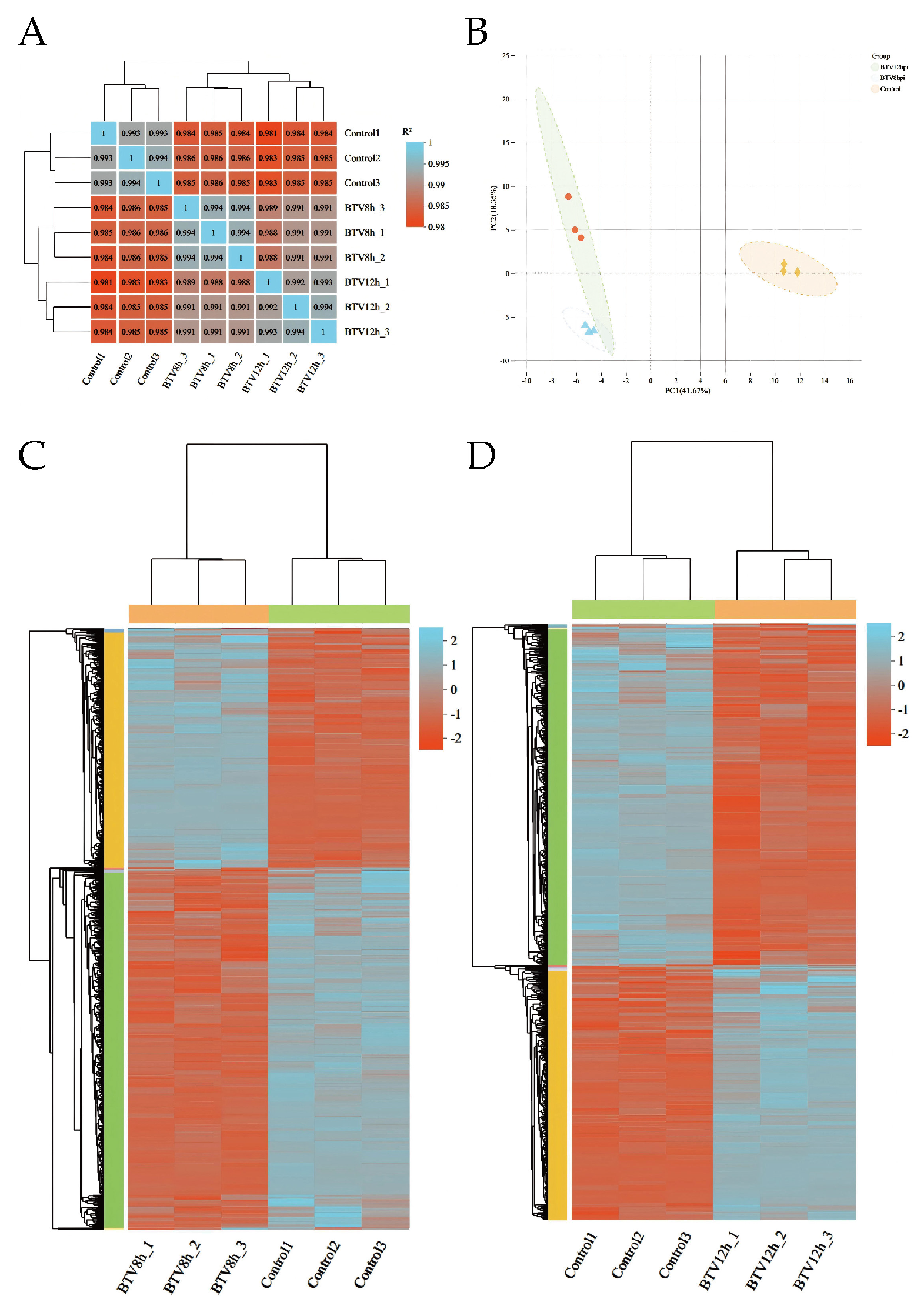
3.2. Inter-Sample Correlation and Gene Overlap Analysis
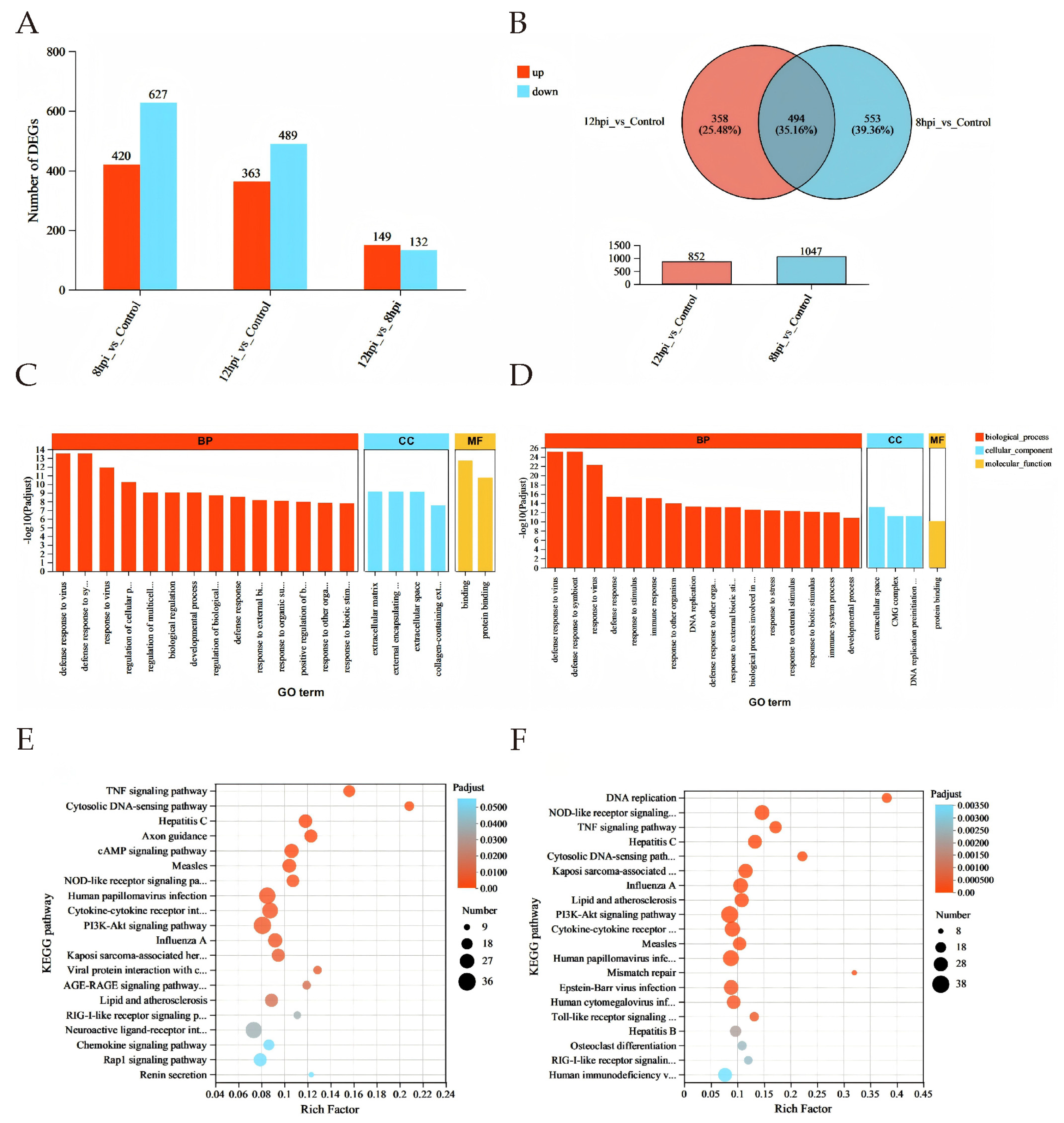
3.3. Differential Gene Expression Analysis
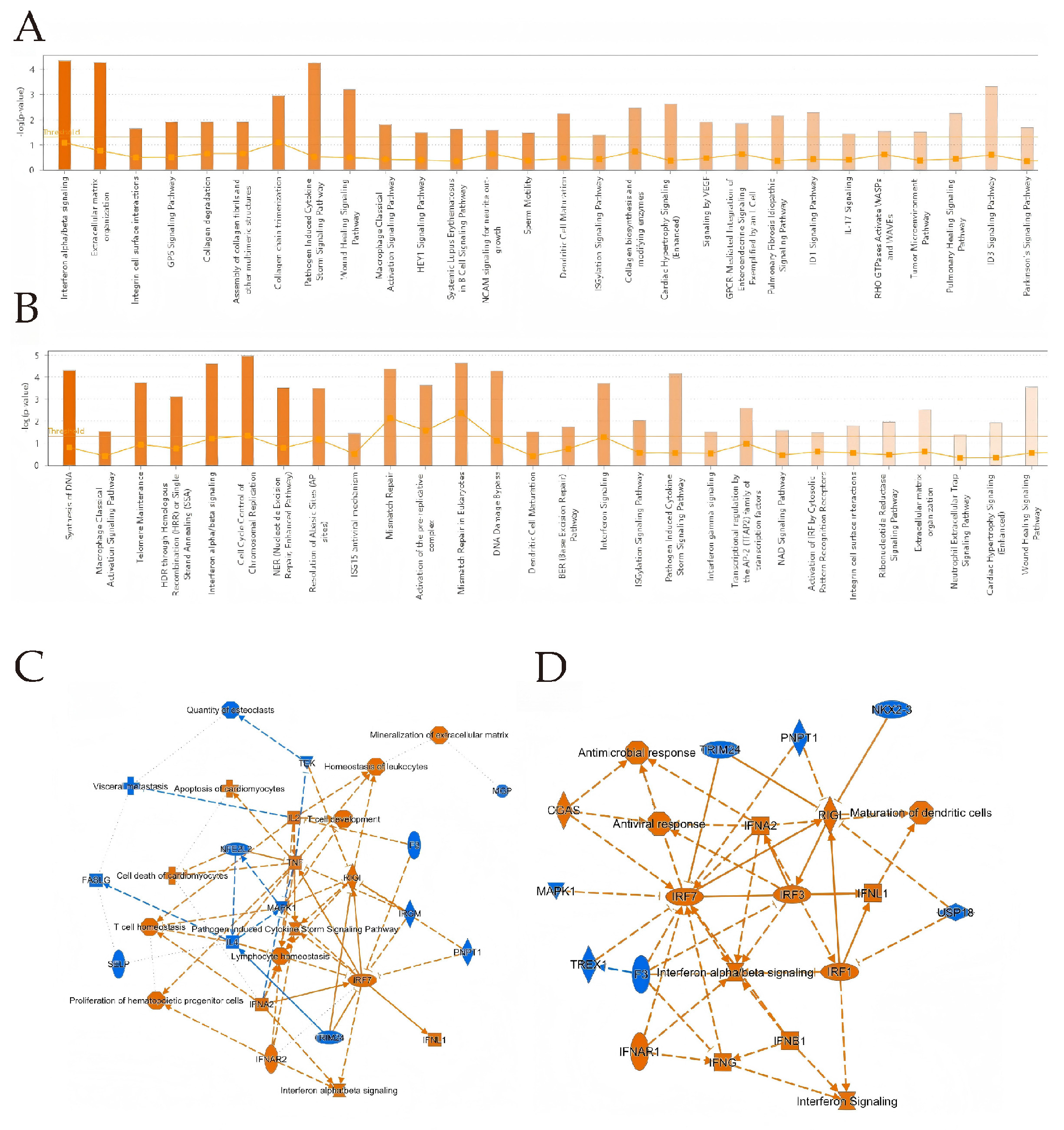
3.4. IPA Analysis
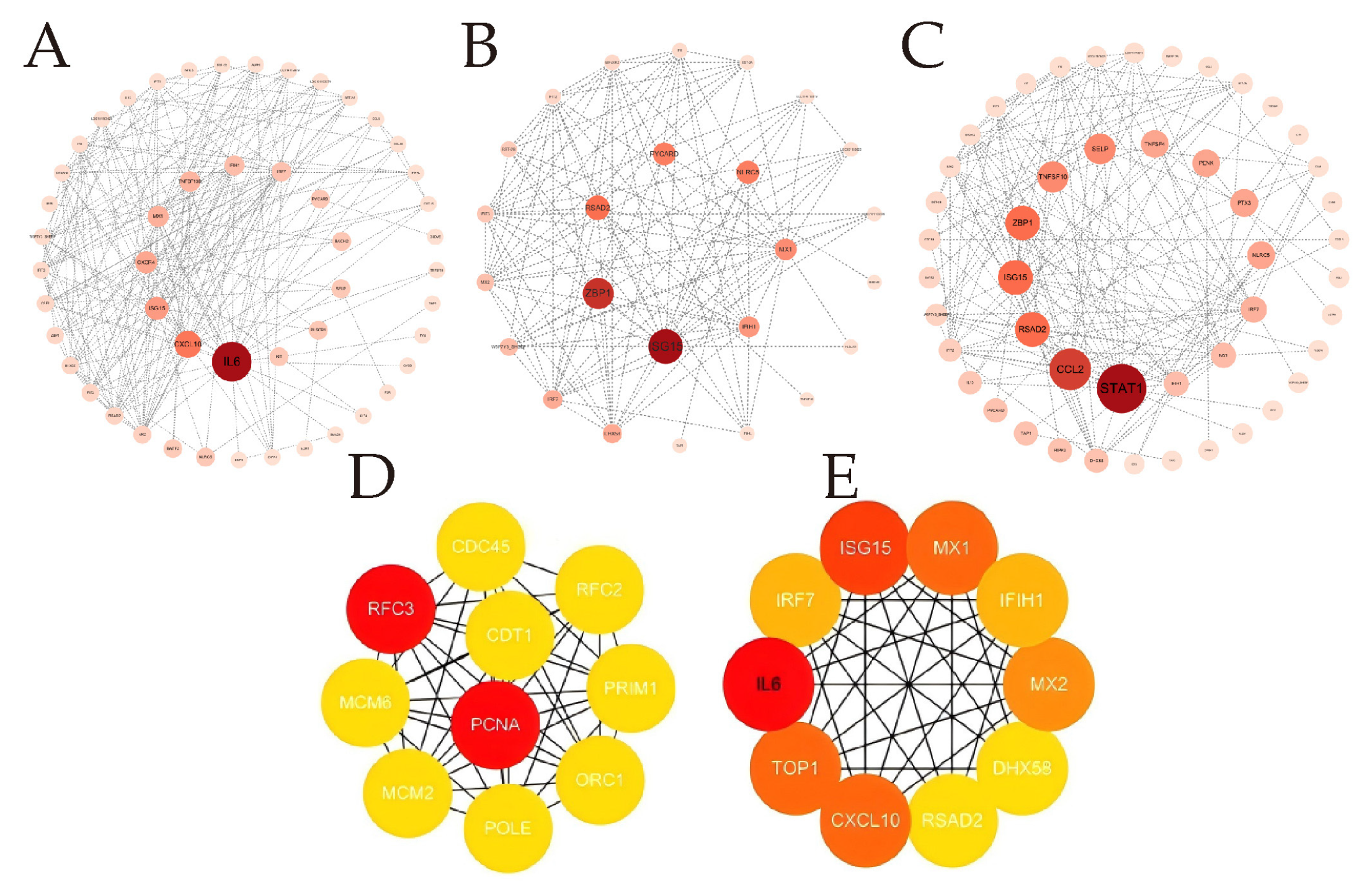
3.5. PPI Delineates Critical Genes in OAECIIs That Are Implicated in BTV Infection
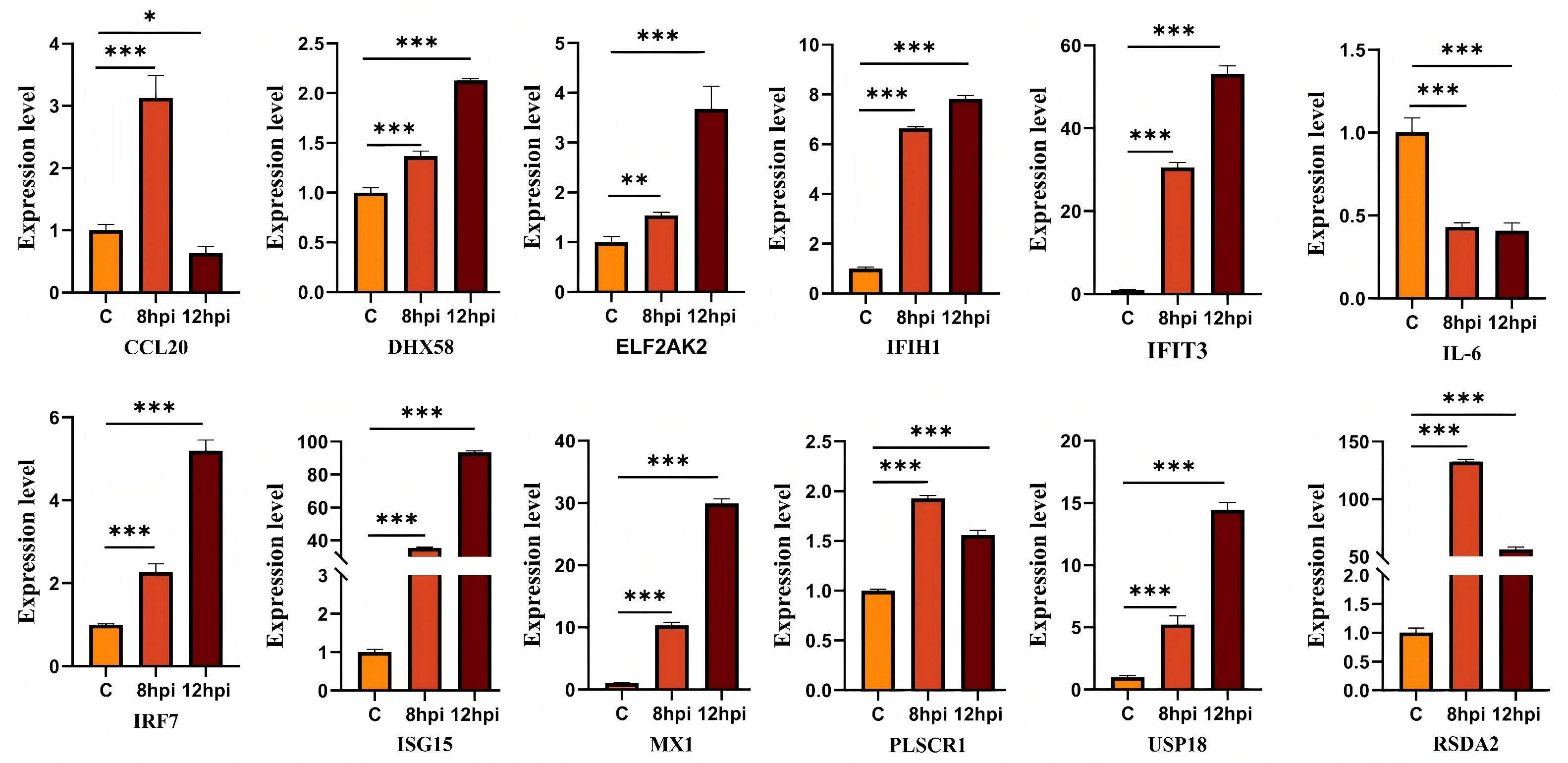
3.6. Validation of Dif-mRNAs Using RT-qPCR
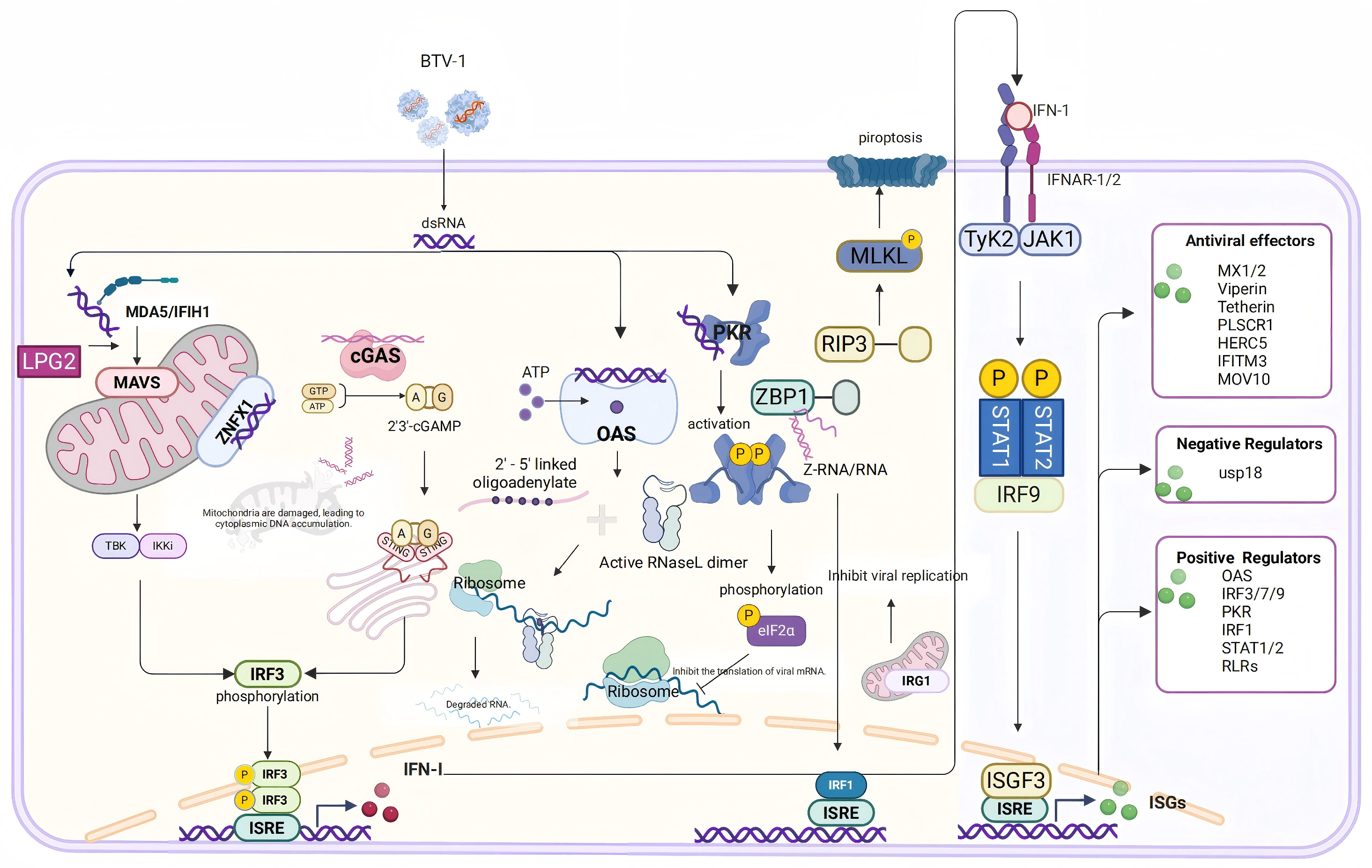
4. Discussion
4.1. Innate Immune Response and Interferon-Stimulated Gene Functions
4.2. Inflammation Response
4.3. Disruption of Cellular Structural Integrity
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Roy, P. Bluetongue Virus Structure and Assembly. Curr. Opin. Virol. 2017, 24, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Moulin, V.; Noordegraaf, C.V.; Makoschey, B.; van der Sluijs, M.; Veronesi, E.; Darpel, K.; Mertens, P.P.C.; de Smit, H. Clinical Disease in Sheep Caused by Bluetongue Virus Serotype 8, and Prevention by an Inactivated Vaccine. Vaccine 2012, 30, 2228–2235. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Gao, S.; Tian, Z.; Guo, Y.; Kang, D.; Xing, S.; Zhang, G.; Liu, G.; Luo, J.; Chang, H.; et al. Transcriptome Analysis of Responses to Bluetongue Virus Infection in Aedes Albopictus Cells. BMC Microbiol. 2019, 19, 121. [Google Scholar] [CrossRef] [PubMed]
- Hardy, A.; Bakshi, S.; Furnon, W.; MacLean, O.; Gu, Q.; Varjak, M.; Varela, M.; Aziz, M.A.; Shaw, A.E.; Pinto, R.M.; et al. The Timing and Magnitude of the Type I Interferon Response Are Correlated with Disease Tolerance in Arbovirus Infection. mBio 2023, 14, e00101-23. [Google Scholar] [CrossRef]
- Lu, D.; Li, Z.; Zhu, P.; Yang, Z.; Yang, H.; Li, Z.; Li, H.; Li, Z. Whole-Transcriptome Analyses of Sheep Embryonic Testicular Cells Infected with the Bluetongue Virus. Front. Immunol. 2022, 13, 1053059. [Google Scholar] [CrossRef]
- Pini, A. Study on the Pathogenesis of Bluetongue: Replication of the Virus in the Organs of Infected Sheep. Onderstepoort J. Vet. Res. 1976, 43, 159–164. [Google Scholar]
- Coen, M.L.; Ellis, J.A.; O’Toole, D.T.; Wilson, W.C. Cytokine Modulation of the Interaction between Bluetongue Virus and Endothelial Cells in Vitro. Vet. Pathol. 1991, 28, 524–532. [Google Scholar] [CrossRef]
- DeMaula, C.D.; Leutenegger, C.M.; Jutila, M.A.; MacLachlan, N.J. Bluetongue Virus-Induced Activation of Primary Bovine Lung Microvascular Endothelial Cells. Vet. Immunol. Immunopathol. 2002, 86, 147–157. [Google Scholar] [CrossRef]
- Luo, S.; Chen, Y.; Ma, X.; Miao, H.; Jia, H.; Yi, H. Whole-Transcriptome Analyses of Ovine Lung Microvascular Endothelial Cells Infected with Bluetongue Virus. Vet. Res. 2024, 55, 122. [Google Scholar] [CrossRef]
- Kása, A.; Csortos, C.; Verin, A.D. Cytoskeletal Mechanisms Regulating Vascular Endothelial Barrier Function in Response to Acute Lung Injury. Tissue Barriers 2015, 3, e974448. [Google Scholar] [CrossRef]
- Makanya, A.; Anagnostopoulou, A.; Djonov, V. Development and Remodeling of the Vertebrate Blood-Gas Barrier. Biomed. Res. Int. 2013, 2013, 101597. [Google Scholar] [CrossRef] [PubMed]
- Fehrenbach, H. Alveolar Epithelial Type II Cell: Defender of the Alveolus Revisited. Respir. Res. 2001, 2, 33–46. [Google Scholar] [CrossRef] [PubMed]
- Sisson, T.H.; Mendez, M.; Choi, K.; Subbotina, N.; Courey, A.; Cunningham, A.; Dave, A.; Engelhardt, J.F.; Liu, X.; White, E.S.; et al. Targeted Injury of Type II Alveolar Epithelial Cells Induces Pulmonary Fibrosis. Am. J. Respir. Crit. Care Med. 2010, 181, 254–263. [Google Scholar] [CrossRef] [PubMed]
- Mason, R.J. Biology of Alveolar Type II Cells. Respirology 2006, 11, S12–S15. [Google Scholar] [CrossRef]
- Lowery, S.A.; Sariol, A.; Perlman, S. Innate Immune and Inflammatory Responses to SARS-CoV-2: Implications for COVID-19. Cell Host Microbe 2021, 29, 1052–1062. [Google Scholar] [CrossRef]
- Zhou, B.; Li, J.; Liang, X.; Yang, Z.; Jiang, Z. Transcriptome Profiling of Influenza a Virus-Infected Lung Epithelial (A549) Cells with Lariciresinol-4-β-D-Glucopyranoside Treatment. PLoS ONE 2017, 12, e0173058. [Google Scholar] [CrossRef]
- Onufer, A.P.; Mell, J.C.; Cort, L.; Rao, A.; Mdluli, N.V.; Carey, A.J. Influenza Virus-Induced Type I Interferons Disrupt Alveolar Epithelial Repair and Tight Junction Integrity in the Developing Lung. Mucosal Immunol. 2025, 18, 607–619. [Google Scholar] [CrossRef]
- González-Navajas, J.M.; Lee, J.; David, M.; Raz, E. Immunomodulatory Functions of Type I Interferons. Nat. Rev. Immunol. 2012, 12, 125–135. [Google Scholar] [CrossRef]
- Rojas, J.M.; Avia, M.; Martín, V.; Sevilla, N. Inhibition of the IFN Response by Bluetongue Virus: The Story so Far. Front. Microbiol. 2021, 12, 692069. [Google Scholar] [CrossRef]
- Li, Q.; Cai, X.; Luo, S.; Chen, Y.; Yi, H.; Ma, X. Advances in the anti-host interferon immune response of bluetongue virus. Chin. J. Biotechnol. 2024, 40, 4439–4451. [Google Scholar] [CrossRef]
- Avia, M.; Rojas, J.M.; Miorin, L.; Pascual, E.; Van Rijn, P.A.; Martín, V.; García-Sastre, A.; Sevilla, N. Virus-Induced Autophagic Degradation of STAT2 as a Mechanism for Interferon Signaling Blockade. EMBO Rep. 2019, 20, e48766. [Google Scholar] [CrossRef] [PubMed]
- Forzan, M.; Marsh, M.; Roy, P. Bluetongue Virus Entry into Cells. J. Virol. 2007, 81, 4819–4827. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Crawford, S.E.; Hyser, J.M.; Estes, M.K.; Prasad, B.V. Rotavirus Non-Structural Proteins: Structure and Function. Curr. Opin. Virol. 2012, 2, 380–388. [Google Scholar] [CrossRef] [PubMed]
- Ratinier, M.; Caporale, M.; Golder, M.; Franzoni, G.; Allan, K.; Nunes, S.F.; Armezzani, A.; Bayoumy, A.; Rixon, F.; Shaw, A.; et al. Identification and Characterization of a Novel Non-Structural Protein of Bluetongue Virus. PLoS Pathog. 2011, 7, e1002477. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A Fast Spliced Aligner with Low Memory Requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie Enables Improved Reconstruction of a Transcriptome from RNA-Seq Reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Desmet, C.J.; Ishii, K.J. Nucleic Acid Sensing at the Interface between Innate and Adaptive Immunity in Vaccination. Nat. Rev. Immunol. 2012, 12, 479–491. [Google Scholar] [CrossRef]
- Chauveau, E.; Doceul, V.; Lara, E.; Adam, M.; Breard, E.; Sailleau, C.; Viarouge, C.; Desprat, A.; Meyer, G.; Schwartz-Cornil, I.; et al. Sensing and Control of Bluetongue Virus Infection in Epithelial Cells via RIG-I and MDA5 Helicases. J. Virol. 2012, 86, 11789–11799. [Google Scholar] [CrossRef]
- Stower, H. CARs for Macrophages. Nat. Med. 2020, 26, 649. [Google Scholar] [CrossRef]
- Satoh, T.; Kato, H.; Kumagai, Y.; Yoneyama, M.; Sato, S.; Matsushita, K.; Tsujimura, T.; Fujita, T.; Akira, S.; Takeuchi, O. LGP2 Is a Positive Regulator of RIG-I- and MDA5-Mediated Antiviral Responses. Proc. Natl. Acad. Sci. USA 2010, 107, 1512–1517. [Google Scholar] [CrossRef]
- Hornung, V.; Hartmann, R.; Ablasser, A.; Hopfner, K.-P. OAS Proteins and cGAS: Unifying Concepts in Sensing and Responding to Cytosolic Nucleic Acids. Nat. Rev. Immunol. 2014, 14, 521–528. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Dong, B.; Asthana, A.; Silverman, R.H.; Yan, N. RNA Helicase SKIV2L Limits Antiviral Defense and Autoinflammation Elicited by the OAS-RNase L Pathway. EMBO J. 2024, 43, 3876–3894. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Lee, J.H.; Park, J.-E.; Cho, J.; Yi, H.; Kim, V.N. PKR Is Activated by Cellular dsRNAs during Mitosis and Acts as a Mitotic Regulator. Genes. Dev. 2014, 28, 1310–1322. [Google Scholar] [CrossRef] [PubMed]
- Silverman, R.H. Viral Encounters with 2’,5’-Oligoadenylate Synthetase and RNase L during the Interferon Antiviral Response. J. Virol. 2007, 81, 12720–12729. [Google Scholar] [CrossRef]
- Zevini, A.; Olagnier, D.; Hiscott, J. Crosstalk between Cytoplasmic RIG-I and STING Sensing Pathways. Trends Immunol. 2017, 38, 194–205. [Google Scholar] [CrossRef]
- Louloudes-Lázaro, A.; Nogales-Altozano, P.; Rojas, J.M.; Veloz, J.; Carlón, A.B.; Van Rijn, P.A.; Martín, V.; Fernández-Sesma, A.; Sevilla, N. Double-Stranded RNA Orbivirus Disrupts the DNA-Sensing cGAS-Sting Axis to Prevent Type I IFN Induction. Cell Mol. Life Sci. 2025, 82, 55. [Google Scholar] [CrossRef]
- Takaoka, A.; Wang, Z.; Choi, M.K.; Yanai, H.; Negishi, H.; Ban, T.; Lu, Y.; Miyagishi, M.; Kodama, T.; Honda, K.; et al. DAI (DLM-1/ZBP1) Is a Cytosolic DNA Sensor and an Activator of Innate Immune Response. Nature 2007, 448, 501–505. [Google Scholar] [CrossRef]
- Zhang, T.; Yin, C.; Boyd, D.F.; Quarato, G.; Ingram, J.P.; Shubina, M.; Ragan, K.B.; Ishizuka, T.; Crawford, J.C.; Tummers, B.; et al. Influenza Virus Z-RNAs Induce ZBP1-Mediated Necroptosis. Cell 2020, 180, 1115–1129.e13. [Google Scholar] [CrossRef]
- Li, H.; Zheng, T.; Chen, M.; Lei, X.; Li, S.; Chen, X.; Wang, S.; Ning, Z. ZBP1 Inhibits the Replication of Senecavirus a by Enhancing NF-κB Signaling Pathway Mediated Antiviral Response in Porcine Alveolar Macrophage 3D4/21 Cells. Cell. Mol. Biol. Lett. 2024, 29, 83. [Google Scholar] [CrossRef]
- Mohd Jaafar, F.; Monsion, B.; Mertens, P.P.C.; Attoui, H. Identification of Orbivirus Non-Structural Protein 5 (NS5), Its Role and Interaction with RNA/DNA in Infected Cells. Int. J. Mol. Sci. 2023, 24, 6845. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yuan, S.; Jia, X.; Ge, Y.; Ling, T.; Nie, M.; Lan, X.; Chen, S.; Xu, A. Mitochondria-Localised ZNFX1 Functions as a dsRNA Sensor to Initiate Antiviral Responses through MAVS. Nat. Cell Biol. 2019, 21, 1346–1356. [Google Scholar] [CrossRef] [PubMed]
- Xing, J.; Zhang, A.; Du, Y.; Fang, M.; Minze, L.J.; Liu, Y.-J.; Li, X.C.; Zhang, Z. Identification of Poly(ADP-Ribose) Polymerase 9 (PARP9) as a Noncanonical Sensor for RNA Virus in Dendritic Cells. Nat. Commun. 2021, 12, 2681. [Google Scholar] [CrossRef] [PubMed]
- Ooi, E.L.; Chan, S.T.; Cho, N.E.; Wilkins, C.; Woodward, J.; Li, M.; Kikkawa, U.; Tellinghuisen, T.; Gale, M.; Saito, T. Novel Antiviral Host Factor, TNK1, Regulates IFN Signaling through Serine Phosphorylation of STAT1. Proc. Natl. Acad. Sci. USA 2014, 111, 1909–1914. [Google Scholar] [CrossRef]
- Gomes, M.T.R.; Guimarães, E.S.; Oliveira, S.C. ZBP1 Senses Brucella Abortus DNA Triggering Type I Interferon Signaling Pathway and Unfolded Protein Response Activation. Front. Immunol. 2025, 15, 1511949. [Google Scholar] [CrossRef]
- Doceul, V.; Chauveau, E.; Lara, E.; Bréard, E.; Sailleau, C.; Zientara, S.; Vitour, D. Dual Modulation of Type I Interferon Response by Bluetongue Virus. J. Virol. 2014, 88, 10792–10802. [Google Scholar] [CrossRef]
- Schneider, W.M.; Chevillotte, M.D.; Rice, C.M. Interferon-Stimulated Genes: A Complex Web of Host Defenses. Annu. Rev. Immunol. 2014, 32, 513–545. [Google Scholar] [CrossRef]
- Park, J.M.; Yang, S.W.; Yu, K.R.; Ka, S.H.; Lee, S.W.; Seol, J.H.; Jeon, Y.J.; Chung, C.H. Modification of PCNA by ISG15 Plays a Crucial Role in Termination of Error-Prone Translesion DNA Synthesis. Mol. Cell 2014, 54, 626–638. [Google Scholar] [CrossRef]
- Gao, S.; von der Malsburg, A.; Paeschke, S.; Behlke, J.; Haller, O.; Kochs, G.; Daumke, O. Structural Basis of Oligomerization in the Stalk Region of Dynamin-like MxA. Nature 2010, 465, 502–506. [Google Scholar] [CrossRef]
- Kane, M.; Yadav, S.S.; Bitzegeio, J.; Kutluay, S.B.; Zang, T.; Wilson, S.J.; Schoggins, J.W.; Rice, C.M.; Yamashita, M.; Hatziioannou, T.; et al. MX2 Is an Interferon-Induced Inhibitor of HIV-1 Infection. Nature 2013, 502, 563–566. [Google Scholar] [CrossRef]
- Helbig, K.J.; Carr, J.M.; Calvert, J.K.; Wati, S.; Clarke, J.N.; Eyre, N.S.; Narayana, S.K.; Fiches, G.N.; McCartney, E.M.; Beard, M.R. Viperin Is Induced Following Dengue Virus Type-2 (DENV-2) Infection and Has Anti-Viral Actions Requiring the C-Terminal End of Viperin. PLoS Neglected Trop. Dis. 2013, 7, e2178. [Google Scholar] [CrossRef] [PubMed]
- Arnaud, F.; Black, S.G.; Murphy, L.; Griffiths, D.J.; Neil, S.J.; Spencer, T.E.; Palmarini, M. Interplay between Ovine Bone Marrow Stromal Cell Antigen 2/Tetherin and Endogenous Retroviruses. J. Virol. 2010, 84, 4415–4425. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Jiang, W.; Wu, L.; Gaudet, R.G.; Park, E.-S.; Su, M.; Cheppali, S.K.; Cheemarla, N.R.; Kumar, P.; Uchil, P.D.; et al. PLSCR1 Is a Cell-Autonomous Defence Factor against SARS-CoV-2 Infection. Nature 2023, 619, 819–827. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.H.; Lei, H.Y.; Liu, H.S.; Lin, Y.S.; Liu, C.C.; Yeh, T.M. Dengue Virus Infects Human Endothelial Cells and Induces IL-6 and IL-8 Production. Am. J. Trop. Med. Hyg. 2000, 63, 71–75. [Google Scholar] [CrossRef]
- Sinha, S.; Singh, K.; Ravi Kumar, Y.S.; Roy, R.; Phadnis, S.; Meena, V.; Bhattacharyya, S.; Verma, B. Dengue Virus Pathogenesis and Host Molecular Machineries. J. Biomed. Sci. 2024, 31, 43. [Google Scholar] [CrossRef]
- Songprakhon, P.; Panya, A.; Choomee, K.; Limjindaporn, T.; Noisakran, S.; Tarasuk, M.; Yenchitsomanus, P.-T. Cordycepin Exhibits Both Antiviral and Anti-Inflammatory Effects against Dengue Virus Infection. iScience 2024, 27, 110711. [Google Scholar] [CrossRef]
- Li, Z.; Lu, D.; Yang, H.; Li, Z.; Zhu, P.; Xie, J.; Liao, D.; Zheng, Y.; Li, H. Bluetongue Virus Non-Structural Protein 3 (NS3) and NS4 Coordinatively Antagonize Type Ⅰ Interferon Signaling by Targeting STAT1. Vet. Microbiol. 2021, 254, 108986. [Google Scholar] [CrossRef]
- Fu, Z.; Chen, K.; Zheng, F.; Gong, W.; Chao, D.; Lu, C. FCGBP Promotes Ovarian Cancer Progression via Activation of IL-6/JAK-STAT Signaling Pathway. J. Transl. Med. 2025, 23, 827. [Google Scholar] [CrossRef]
- Xu, H.-Y.; Jiang, M.-T.; Yang, Y.-F.; Huang, Y.; Yang, W.-D.; Li, H.-Y.; Wang, X. Microalgae-Based Fucoxanthin Attenuates Rheumatoid Arthritis by Targeting the JAK-STAT Signaling Pathway and Gut Microbiota. J. Agric. Food Chem. 2025, 73, 11708–11719. [Google Scholar] [CrossRef]
- Pourcelot, M.; da Silva Moraes, R.A.; Lacour, S.; Fablet, A.; Caignard, G.; Vitour, D. Activation of Inflammasome during Bluetongue Virus Infection. Pathogens 2023, 12, 801. [Google Scholar] [CrossRef]
- Garlanda, C.; Bottazzi, B.; Magrini, E.; Inforzato, A.; Mantovani, A. PTX3, a Humoral Pattern Recognition Molecule, in Innate Immunity, Tissue Repair, and Cancer. Physiol. Rev. 2018, 98, 623–639. [Google Scholar] [CrossRef] [PubMed]
- Murayama, T.; Kikuchi, M.; Miita, T.; Yamada, R.; Matsubara, K.; Sadanari, H.; Mukaida, N. Human Cytomegalovirus Replication Supported by Virus-Induced Activation of CCL2-CCR2 Interactions. Biochem. Biophys. Res. Commun. 2014, 453, 321–325. [Google Scholar] [CrossRef] [PubMed]
- Held, K.S.; Chen, B.P.; Kuziel, W.A.; Rollins, B.J.; Lane, T.E. Differential Roles of CCL2 and CCR2 in Host Defense to Coronavirus Infection. Virology 2004, 329, 251–260. [Google Scholar] [CrossRef]
- Kong, Y.; Ma, J. Dynamic Mechanisms of the Membrane Water Channel Aquaporin-1 (AQP1). Proc. Natl. Acad. Sci. USA 2001, 98, 14345–14349. [Google Scholar] [CrossRef] [PubMed]
- Shabanian, K.; Shabanian, T.; Karsai, G.; Pontiggia, L.; Paneni, F.; Ruschitzka, F.; Beer, J.H.; Saeedi Saravi, S.S. AQP1 Differentially Orchestrates Endothelial Cell Senescence. Redox Biol. 2024, 76, 103317. [Google Scholar] [CrossRef]
- Tesse, A.; Gena, P.; Rützler, M.; Calamita, G. Ablation of Aquaporin-9 Ameliorates the Systemic Inflammatory Response of LPS-Induced Endotoxic Shock in Mouse. Cells 2021, 10, 435. [Google Scholar] [CrossRef]
- Karimi, N.; Ahmadi, V. Aquaporin Channels in Skin Physiology and Aging Pathophysiology: Investigating Their Role in Skin Function and the Hallmarks of Aging. Biology 2024, 13, 862. [Google Scholar] [CrossRef]
- Mineta, K.; Yamamoto, Y.; Yamazaki, Y.; Tanaka, H.; Tada, Y.; Saito, K.; Tamura, A.; Igarashi, M.; Endo, T.; Takeuchi, K.; et al. Predicted Expansion of the Claudin Multigene Family. FEBS Lett. 2011, 585, 606–612. [Google Scholar] [CrossRef]
- Furuse, M. Molecular Basis of the Core Structure of Tight Junctions. Cold Spring Harb. Perspect. Biol. 2010, 2, a002907. [Google Scholar] [CrossRef]
- Maginnis, M.S. Virus-Receptor Interactions: The Key to Cellular Invasion. J. Mol. Biol. 2018, 430, 2590–2611. [Google Scholar] [CrossRef]
- Looi, K.; Buckley, A.G.; Rigby, P.J.; Garratt, L.W.; Iosifidis, T.; Zosky, G.R.; Larcombe, A.N.; Lannigan, F.J.; Ling, K.-M.; Martinovich, K.M.; et al. Effects of Human Rhinovirus on Epithelial Barrier Integrity and Function in Children with Asthma. Clin. Exp. Allergy 2018, 48, 513–524. [Google Scholar] [CrossRef]
| GenBank | Target Genes | Full Name of Gene | Primer Sequences (5′-3′) | |
|---|---|---|---|---|
| NM_007393.5 | β-Actin | Forward | CCACTGTCGAGTCGCGTCC | |
| Reverse | ATTCCCACCATCACACCCTGG | |||
| XM_004004655 | IFIH1 | Interferon Induced with Helicase C Domain 1 | Forward | GGTCAGCACGAGGAATAA |
| Reverse | CTGTGGTAGCGATAAGCA | |||
| NC_056056.1 | RSAD2 | radical S-adenosyl methionine domain containing 2 | Forward | GGTCTGCTGATGCTGAAGGAA |
| Reverse | TCACAGGAGATGGCGAGGAT | |||
| XM_027960293 | IFIT3 | Interferon Induced Protein with Tetratricopeptide Repeats 3 | Forward | GGCGGCTGAATGCTATGAGA |
| Reverse | CTTCATGCTCAGTTGCTGGC | |||
| NC_056055.1 | CCL20 | C-C motif chemokine ligand 20 | Forward | TGCTCTTGCTCCACCTCTG |
| Reverse | GCTTGCTTCACCCACTTCTTC | |||
| NC_056065.1 | ISG15 | Ovis aries ISG15 ubiquitin like modifier | Forward | AGACTGTGGCTGTGCTCAAG |
| Reverse | GCGGGTGCTCATCATCCAT | |||
| NC_056074.1 | IRF7 | interferon regulatory factor 7 | Forward | CCGCACTACACCATCTACCTG |
| Reverse | AGCCTGTTCCACCTCCATCA | |||
| NC_056054.1 | MXI | Ovis aries MX dynamin like GTPase 1 | Forward | TCGGTATCGTGGCAGAGAGT |
| Reverse | TGGCAGTTCGGTGGAGGTT | |||
| NC_056054.1 | PLSCR1 | phospholipid scramblase 1 | Forward | TGAGAGGCGAGAGGATGTACT |
| Reverse | ATGGGAACTGGATGCCAAAGT | |||
| NC_056064.1 | DHX58 | DExH-box helicase 58 | Forward | CGAAACTGGAGGTGCTGGAA |
| Reverse | TCTGAGTCTTCTGGCTGTTGT | |||
| NC_056057.1 | IL-6 | Ovis aries interleukin 6 | Forward | GGTTCAATCAGGCGATTTGCT |
| Reverse | GTGTGTGGCTGGAGTGGTTAT | |||
| NC_056056.1 | USP18 | ubl carboxyl-terminal hydrolase 18 | Forward | TGAGGAGCAGAGGAAGAGTGT |
| Reverse | TTCAAGCGGATGGTGTAGAGG | |||
| NC_056056.1 | eIF2aK2 | eukaryotic translation initiation factor 2 alpha kinase 2 | Forward | AGAAGGTAGAGCGTGAAGTGA |
| Reverse | TCGTCAATCCATTCCGCCAAT | |||
| KM099648.1 | NS4 | non-structural protein NS4 | Forward | ATGGTGAGGGGGCACAACAGAA |
| Reverse | CCCATCCTCCTCTGCTCGCT |
| Upstream Regulator | Molecule | Activation Z-Score | p-Value of Overlap | Target Molecules Number |
|---|---|---|---|---|
| TNF | Cytokine | 2.051 | 7.54 × 10−13 | 90 |
| IMMUNOGLOBULIN(complex) | complex | 1.960 | 3.01 × 10−12 | 71 |
| IRF7 | transcription regulator | 3.296 | 2.43 × 10−11 | 18 |
| tetradecanoylphorbol acetate | chemical drug | 1.737 | 5.82 × 10−11 | 60 |
| dexamethasone | chemical drug | −0.893 | 2.19 × 10−10 | 91 |
| vidutolimod | chemical drug | 2.673 | 5.35 × 10−10 | 10 |
| IFNAR2 | transcription regulator | 2.000 | 8.42 × 10−10 | 15 |
| SN-011 | chemical reagent | −1.722 | 1.24 × 10−9 | 13 |
| pyridostatin | chemical reagent | 3.426 | 1.25 × 10−9 | 12 |
| STAT1 | transcription regulator | 0.968 | 1.73 × 10−9 | 20 |
| TNF(family) | group | −0.458 | 2.54 × 10−9 | 23 |
| MAPK1 | kinase | −2.848 | 2.55 × 10−9 | 31 |
| TWISTI | transcription regulator | 0.600 | 2.84 × 10−9 | 24 |
| TREX1 | enzyme | −2.492 | 3.27 × 10−9 | 18 |
| MAP3K7 | kinase | 0.015 | 3.72 × 10−9 | 17 |
| TGFβ1 | growth factor | 0.367 | 3.91 × 10−9 | 82 |
| NONO | transcription regulator | 2.646 | 5.70 × 10−9 | 14 |
| IL1B | Cytokine | 1.496 | 6.83 × 10−9 | 56 |
| CHROMR | other | 2.755 | 7.59 × 10−9 | 17 |
| WNT3A | Cytokine | 0.362 | 9.29 × 10−9 | 26 |
| INTERFERON ALPHA(family) | group | 2.376 | 1.00 × 10−8 | 35 |
| Tretinoin | biological drug | 2.615 | 1.08 × 10−8 | 70 |
| Lipopolysaccharide | Cytokine | 1.953 | 1.09 × 10−8 | 96 |
| F3 | transcription regulator | −3.267 | 1.64 × 10−8 | 19 |
| IFNL1 | Cytokine | 2.436 | 1.87 × 10−8 | 11 |
| IFN BETA(family) | group | 2.507 | 2.73 × 10−8 | 21 |
| Stallimycin | biological drug | 1.632 | 2.91 × 10−8 | 10 |
| IL17A | Cytokine | −0.695 | 4.17 × 10−8 | 28 |
| IFNAR1 | transcription regulator | 2.741 | 4.42 × 10−8 | 17 |
| IRF3 | transcription regulator | 1.896 | 4.28 × 10−8 | 33 |
| Upstream Regulator | Molecule | Activation Z-Score | p-Value of Overlap | Target Molecules Number |
|---|---|---|---|---|
| TNF | cytokine | 1.43 | 1.2 × 10−13 | 87 |
| pyridostain | chemical reagent | 3.713 | 1.27 × 10−12 | 14 |
| TGFB1 | growth factor | −0.474 | 8.31 × 10−12 | 84 |
| NONO | transcription regulator | 2.97 | 1.41 × 10−11 | 16 |
| TWISTI | transcription regulator | 0.187 | 1.64 × 10−11 | 26 |
| etrogen | chemical drug | 1.043 | 2.64 × 10−11 | 29 |
| ETV6 | transcription regulator | −2.731 | 2.73 × 10−11 | 13 |
| TP53 | transcription regulator | −1.279 | 3.30 × 10−11 | 87 |
| PGR | ligand-dependent nuclear | −0.422 | 5.00 × 10−11 | 36 |
| ETV3 | transcription regulator | −3.317 | 5.65 × 10−11 | 11 |
| TNIK | kinase | 3.286 | 5.65 × 10−11 | 11 |
| F3 | transcription regulator | −3.946 | 9.37 × 10−11 | 21 |
| vidutolimod | chemical drug | 3.357 | 1.80 × 10−10 | 15 |
| STAT1 | transcription regulator | 1.858 | 2.62 × 10−10 | 30 |
| IRF7 | transcription regulator | 3.910 | 7.28 × 10−10 | 16 |
| IL18 | Cytokine | 1.104 | 2.85 × 10−9 | 54 |
| dexamethasone | chemical drug | −2.278 | 3.19 × 10−9 | 83 |
| IFNA2 | cytokine | 3.616 | 6.93 × 10−9 | 21 |
| CG(complex) | complex | −1.786 | 6.97 × 10−9 | 34 |
| CSF1 | cytokine | 0.373 | 9.13 × 10−9 | 27 |
| IFNAR(family) | group | 3.676 | 9.35 × 10−9 | 14 |
| IRGM | enzyme | −2.949 | 9.54 × 10−9 | 12 |
| IFNAR1 | transcription regulator | 3.539 | 1.34 × 10−8 | 17 |
| NTRK1 | kinase | −1.666 | 1.38 × 10−8 | 23 |
| IRF1 | transcription regulator | 2.842 | 1.94 × 10−8 | 19 |
| IL6 | Cytokine | 0.321 | 2.10 × 10−8 | 41 |
| doxoorubicin | chemical drug | 0.260 | 2.26 × 10−8 | 41 |
| FGF2 | growth factor | −0.402 | 2.29 × 10−8 | 32 |
| RIPK2 | kinase | 2.376 | 3.61 × 10−8 | 15 |
| CGAS | enzyme | 2.250 | 3.68 × 10−8 | 12 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Chen, Y.; Lei, N.; Ye, Z.; Pu, S.; Luo, S.; Ma, X.; Yang, S.; Wang, G.; Jia, H.; Yi, H. Transcriptomic Analysis of the Antiviral Responses in Ovine Type II Alveolar Epithelial Cells During Early Stage of Bluetongue Virus Infection. Animals 2026, 16, 243. https://doi.org/10.3390/ani16020243
Chen Y, Lei N, Ye Z, Pu S, Luo S, Ma X, Yang S, Wang G, Jia H, Yi H. Transcriptomic Analysis of the Antiviral Responses in Ovine Type II Alveolar Epithelial Cells During Early Stage of Bluetongue Virus Infection. Animals. 2026; 16(2):243. https://doi.org/10.3390/ani16020243
Chicago/Turabian StyleChen, Yunyi, Nijing Lei, Zhenghao Ye, Shaohua Pu, Shimei Luo, Xianping Ma, Shaoyu Yang, Guanghua Wang, Huaijie Jia, and Huashan Yi. 2026. "Transcriptomic Analysis of the Antiviral Responses in Ovine Type II Alveolar Epithelial Cells During Early Stage of Bluetongue Virus Infection" Animals 16, no. 2: 243. https://doi.org/10.3390/ani16020243
APA StyleChen, Y., Lei, N., Ye, Z., Pu, S., Luo, S., Ma, X., Yang, S., Wang, G., Jia, H., & Yi, H. (2026). Transcriptomic Analysis of the Antiviral Responses in Ovine Type II Alveolar Epithelial Cells During Early Stage of Bluetongue Virus Infection. Animals, 16(2), 243. https://doi.org/10.3390/ani16020243







