The Molecular Mechanism by Which miR-211-5p Regulates the Proliferation and Differentiation of Preadipocytes in Meat Rabbits by Targeting TPK1
Simple Summary
Abstract
1. Introduction
2. Results
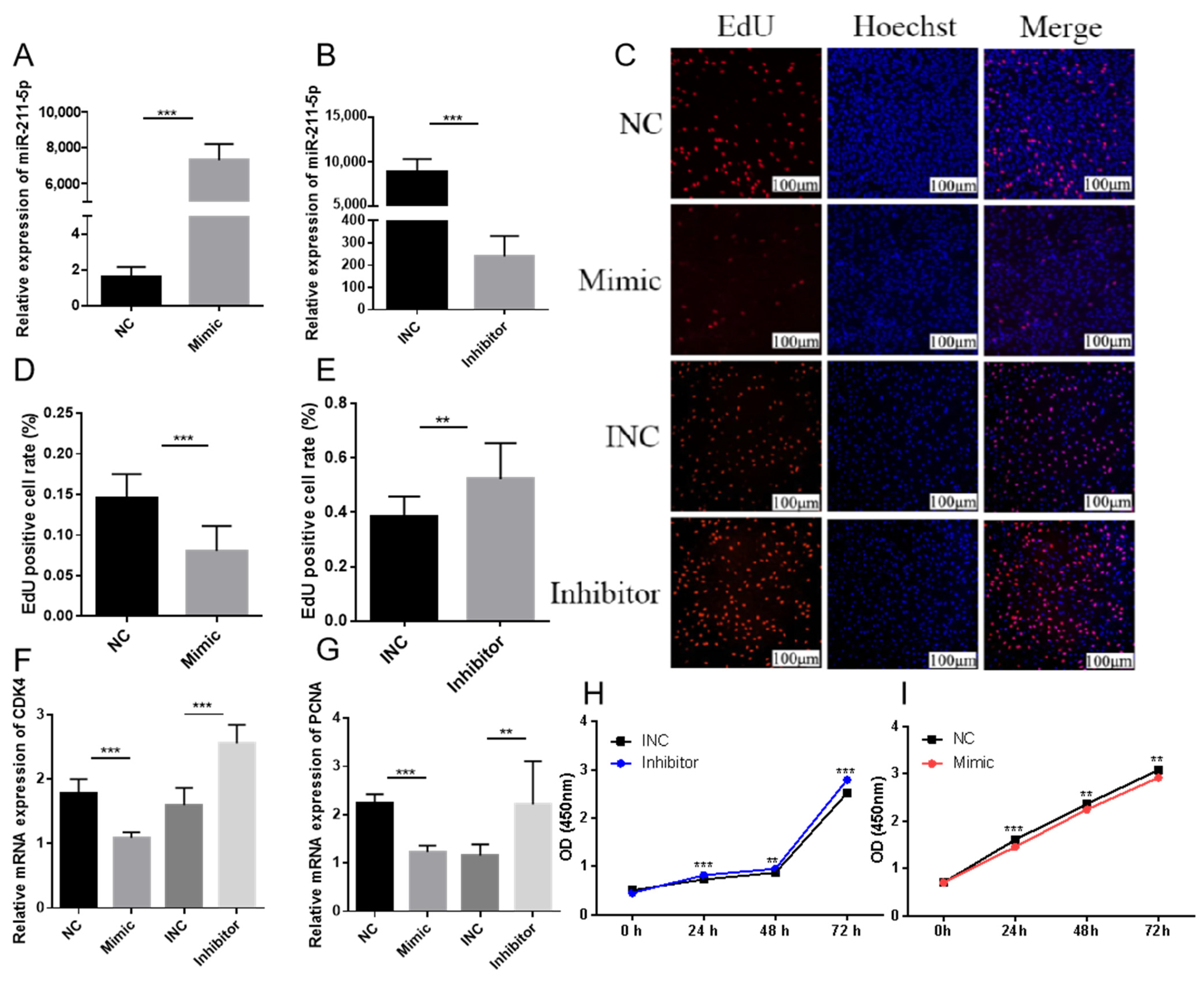
2.1. miR-211-5p Inhibits the Proliferation of Preadipocytes
2.2. miR-211-5p Promotes the Differentiation of Rabbit Preadipocytes
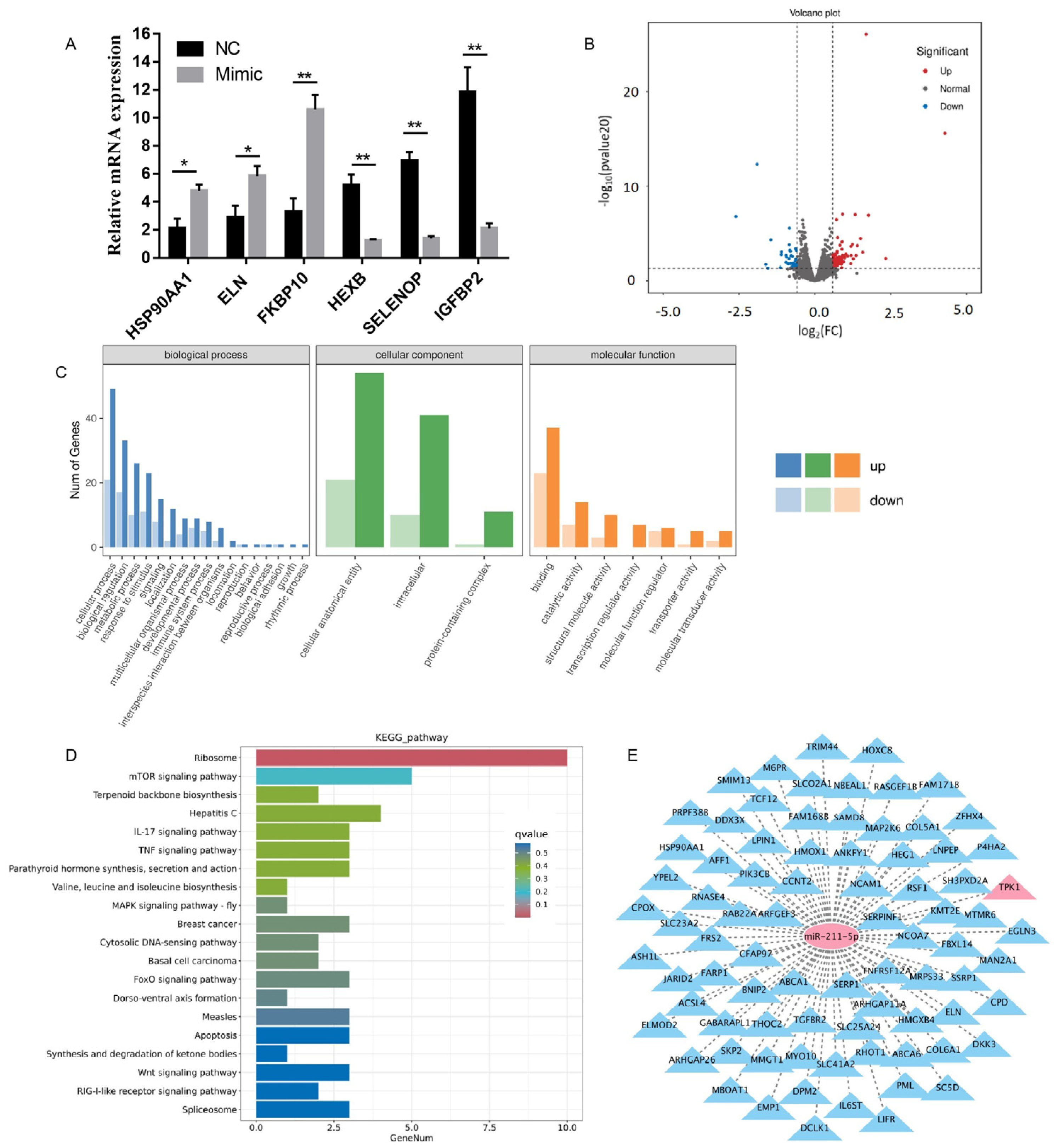
2.3. Analysis of Sequencing Results
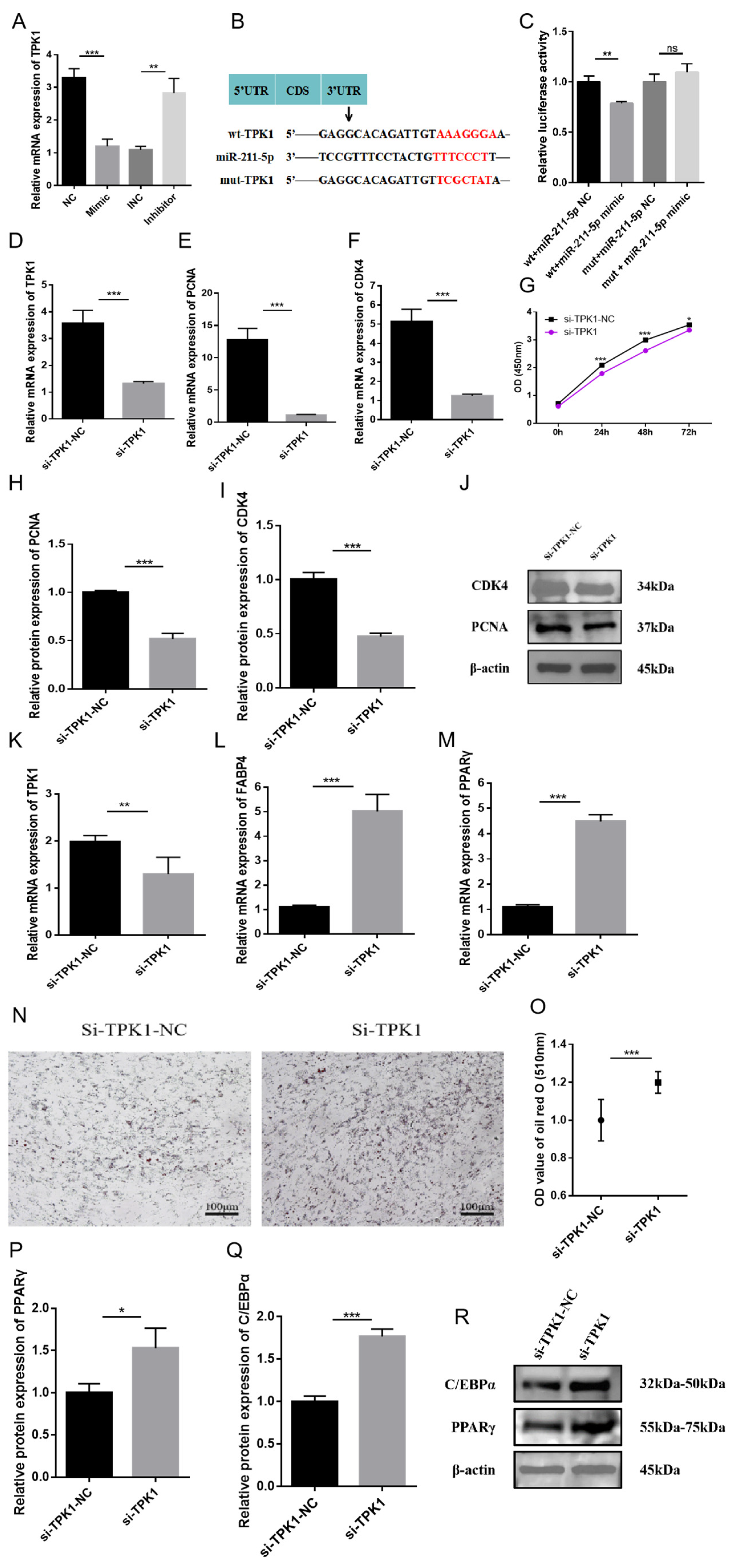
2.4. The Role of TPK1 in Regulating the Proliferation and Differentiation of Rabbit Preadipocytes
2.5. miR-211-5p Regulates the Proliferation and Differentiation of Rabbit Preadipocytes by Targeting TPK1
3. Discussion
4. Materials and Methods
4.1. Ethical Statement
4.2. Animal Cell Collection and Culture
4.3. Bioinformatic Analysis of Sequencing Data
4.4. RNA Extraction and Quantitative Real-Time PCR (RT-qPCR)
4.5. Cell Induced Differentiation
4.6. Transfection
4.7. Western Blotting (WB)
4.8. Cell Counting Kit 8 (CCK-8) Assay
4.9. EdU Proliferation Assay
4.10. Oil Red O Staining
4.11. Dual-Luciferase Reporter Assay
4.12. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Koenen, M.; Hill, M.A.; Cohen, P.; Sowers, J.R. Obesity, Adipose Tissue and Vascular Dysfunction. Circ. Res. 2021, 128, 951–968. [Google Scholar] [CrossRef] [PubMed]
- Hausman, G.J.; Bergen, W.G.; Etherton, T.D.; Smith, S.B. The history of adipocyte and adipose tissue research in meat animals. J. Anim. Sci. 2018, 96, 473–486. [Google Scholar] [CrossRef]
- Tang, Q.Q.; Lane, M.D. Adipogenesis: From stem cell to adipocyte. Annu. Rev. Biochem. 2012, 81, 715–736. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.X.; Rothenberg, M.E. MicroRNA. J. Allergy Clin. Immunol. 2018, 141, 1202–1207. [Google Scholar] [CrossRef]
- Liu, J.; Che, Y.; Cai, K.; Zhao, B.; Qiao, L.; Pan, Y.; Yang, K.; Liu, W. miR-136 Regulates the Proliferation and Adipogenic Differentiation of Adipose-Derived Stromal Vascular Fractions by Targeting HSD17B12. Int. J. Mol. Sci. 2023, 24, 14892. [Google Scholar] [CrossRef]
- Chen, F.F.; Xiong, Y.; Peng, Y.; Gao, Y.; Qin, J.; Chu, G.Y.; Pang, W.J.; Yang, G.S. miR-425-5p Inhibits Differentiation and Proliferation in Porcine Intramuscular Preadipocytes. Int. J. Mol. Sci. 2017, 18, 2101. [Google Scholar] [CrossRef]
- Xu, Y.; Du, J.; Zhang, P.; Zhao, X.; Li, Q.; Jiang, A.; Jiang, D.; Tang, G.; Jiang, Y.; Wang, J.; et al. MicroRNA-125a-5p Mediates 3T3-L1 Preadipocyte Proliferation and Differentiation. Molecules 2018, 23, 317. [Google Scholar] [CrossRef]
- Yu, Y.; Chen, Y.; Wang, L.; Cheng, J.; Du, M.; Pan, S. Rno-miR-130b Attenuates Lipid Accumulation Through Promoting Apoptosis and Inhibiting Differentiation in Rat Intramuscular Adipocytes. Int. J. Molexular Sci. 2025, 26, 1399. [Google Scholar] [CrossRef]
- Wang, L.; Shen, Y.F.; Shi, Z.M.; Shang, X.J.; Jin, D.L.; Xi, F. Overexpression miR-211-5p hinders the proliferation, migration, and invasion of thyroid tumor cells by downregulating SOX11. J. Clin. Labratory Anal. 2018, 32, e22293. [Google Scholar] [CrossRef]
- Wang, K.; Jin, W.; Jin, P.; Fei, X.; Wang, X.; Chen, X. miR-211-5p Suppresses Metastatic Behavior by Targeting SNAI1 in Renal Cancer. Mol. Cancer Res. 2017, 15, 448–456. [Google Scholar] [CrossRef]
- Jiang, Y.; Wang, Z.; Ying, C.; Hu, J.; Zeng, T.; Gao, L. FMR1/circCHAF1A/miR-211-5p/HOXC8 feedback loop regulates proliferation and tumorigenesis via MDM2-dependent p53 signaling in GSCs. Oncogene 2021, 40, 4094–4110. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Shao, J.; Li, Y.; Elzo, M.A.; Jia, X.; Lai, S. Genome-wide identification and characterization of perirenal adipose tissue microRNAs in rabbits fed a high-fat diet. Biosci. Rep. 2021, 41, BSR20204297. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Wang, H.; Mu, J.; Wang, H.; Peng, Y.; Li, Q.; Mao, D.; Guo, L. MiRNA-211 triggers an autophagy-dependent apoptosis in cervical cancer cells: Regulation of Bcl-2. Naunyn-Schmiedeberg’s Arch. Pharmacol. 2020, 393, 359–370. [Google Scholar] [CrossRef]
- Peng, Y.; Chen, F.F.; Ge, J.; Zhu, J.Y.; Shi, X.E.; Li, X.; Yu, T.Y.; Chu, G.Y.; Yang, G.S. miR-429 Inhibits Differentiation and Promotes Proliferation in Porcine Preadipocytes. Int. J. Mol. Sci. 2016, 17, 2047. [Google Scholar] [CrossRef]
- Wang, S.; Pan, C.; Ma, X.; Yang, C.; Tang, L.; Huang, J.; Wei, X.; Li, H.; Ma, Y. Identification and Functional Verification Reveals that miR-195 Inhibiting THRSP to Affect Fat Deposition in Xinyang Buffalo. Front. Genet. 2021, 12, 736441. [Google Scholar] [CrossRef]
- Tang, T.; Jiang, G.; Shao, J.; Wang, M.; Zhang, X.; Xia, S.; Sun, W.; Jia, X.; Wang, J.; Lai, S. lncRNA MSTRG4710 Promotes the Proliferation and Differentiation of Preadipocytes through miR-29b-3p/IGF1 Axis. Int. J. Mol. Sci. 2023, 24, 15715. [Google Scholar] [CrossRef]
- Luo, G.; Hu, S.; Lai, T.; Wang, J.; Wang, L.; Lai, S. MiR-9-5p promotes rabbit preadipocyte differentiation by suppressing leptin gene expression. Lipids Health Dis. 2020, 19, 126. [Google Scholar] [CrossRef]
- Mir, B.A.; Albrecht, E.; Ali, A.; Hansson, O.; Maak, S. MicroRNA-100 Reduced Fetal Bovine Muscle Satellite Cell Myogenesis and Augmented Intramuscular Lipid Deposition by Modulating IGF1R. Cells 2022, 11, 451. [Google Scholar] [CrossRef]
- Lin, X.; Du, Y.; Lu, W.; Gui, W.; Sun, S.; Zhu, Y.; Wang, G.; Eserberg, D.T.; Zheng, F.; Zhou, J.; et al. CircRNF111 Protects Against Insulin Resistance and Lipid Deposition via Regulating miR-143-3p/IGF2R Axis in Metabolic Syndrome. Front. Cell Dev. Biol. 2021, 9, 663148. [Google Scholar] [CrossRef]
- Wang, L.; Xie, Y.; Chen, W.; Zhang, Y.; Zeng, Y. miR-34a Regulates Lipid Droplet Deposition in 3T3-L1 and C2C12 Cells by Targeting LEF1. Cells 2022, 12, 167. [Google Scholar] [CrossRef]
- Guo, L.; Cui, H.; Zhao, G.; Liu, R.; Li, Q.; Zheng, M.; Guo, Y.; Wen, J. Intramuscular preadipocytes impede differentiation and promote lipid deposition of muscle satellite cells in chickens. BMC Genom. 2018, 19, 838. [Google Scholar] [CrossRef] [PubMed]
- Ye, L.; Wang, F.; Wang, J.; Wu, H.; Yang, H.; Yang, Z.; Huang, H. Role and mechanism of miR-211 in human cancer. J. Cancer 2022, 13, 2933–2944. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Yu, X.; Li, B.; Gao, C.; Chen, Y.; Zhang, X.; Li, W.; Yang, L.; Fan, Z. miR-211-5p targeting MMP9 regulates the expressions of AQP4 in traumatic brain injury. Acta Neurol. Belg. 2023, 123, 1321–1329. [Google Scholar] [CrossRef] [PubMed]
- Zeng, B.; Chen, Y.; Chen, H.; Zhao, Q.; Sun, Z.; Liu, D.; Li, X.; Zhang, Y.; Wang, J.; Xing, H.R. Exosomal miR-211-5p regulates glucose metabolism, pyroptosis, and immune microenvironment of melanoma through GNA15. Pharmacol. Res. 2023, 188, 106660. [Google Scholar] [CrossRef]
- Qin, X.; Zhang, J.; Lin, Y.; Sun, X.M.; Zhang, J.N.; Cheng, Z.Q. Identification of MiR-211-5p as a tumor suppressor by targeting ACSL4 in Hepatocellular Carcinoma. J. Transl. Med. 2020, 18, 326. [Google Scholar] [CrossRef]
- Quan, J.; Pan, X.; He, T.; Lin, C.; Lai, Y.; Chen, P.; Zhang, Z.; Yang, S.; Wang, T.; Lai, Y. Tumor suppressor miR-211-5p is associated with cellular migration, proliferation and apoptosis in renal cell carcinoma. Exp. Ther. Med. 2018, 15, 4019–4028. [Google Scholar] [CrossRef]
- Qin, X.; He, X.; Chen, L.; Han, Y.; Yun, Y.; Wu, J.; Sha, L.; Borjigin, G. Transcriptome analysis of adipose tissue in grazing cattle: Identifying key regulators of fat metabolism. Open Life Sci. 2024, 19, 20220843. [Google Scholar] [CrossRef]
- Ke, N.; Chen, L.; Liu, Q.; Xiong, H.; Chen, X.; Zhou, X. Downregulation of miR-211-5p Promotes Carboplatin Resistance in Human Retinoblastoma Y79 Cells by Affecting the GDNF-LIF Interaction. Front. Oncol. 2022, 12, 848733. [Google Scholar] [CrossRef]
- Bai, Y.; Li, X. hsa_circ_0008285 Facilitates the Progression of Cervical Cancer by Targeting miR-211-5p/SOX4 Axis. Cancer Manag. Res. 2020, 12, 3927–3936. [Google Scholar] [CrossRef]
- Tao, D.; Liu, Z.; Wang, L.; Li, C.; Zhang, R.; Ni, N. CircPAG1 interacts with miR-211-5p to promote the E2F3 expression and inhibit the high glucose-induced cell apoptosis and oxidative stress in diabetic cataract. Cell Cycle 2022, 21, 708–719. [Google Scholar] [CrossRef]
- Jiang, G.; Wen, L.; Deng, W.; Jian, Z.; Zheng, H. Regulatory role of miR-211-5p in hepatocellular carcinoma metastasis by targeting ZEB2. Biomed. Pharmacother. Biomed. Pharmacother. 2017, 90, 806–812. [Google Scholar] [CrossRef] [PubMed]
- Sahu, U.; Villa, E.; Reczek, C.R.; Zhao, Z.; O’Hara, B.P.; Torno, M.D.; Mishra, R.; Shannon, W.D.; Asara, J.M.; Gao, P.; et al. Pyrimidines maintain mitochondrial pyruvate oxidation to support de novo lipogenesis. Science 2024, 383, 1484–1492. [Google Scholar] [CrossRef] [PubMed]
- Lim, R.G.; Al-Dalahmah, O.; Wu, J.; Gold, M.P.; Reidling, J.C.; Tang, G.; Adam, M.; Dansu, D.K.; Park, H.J.; Casaccia, P.; et al. Huntington disease oligodendrocyte maturation deficits revealed by single-nucleus RNAseq are rescued by thiamine-biotin supplementation. Nat. Commun. 2022, 13, 7791. [Google Scholar] [CrossRef] [PubMed]
- You, S.; Wang, M.J.; Hou, Z.Y.; Wang, W.D.; Du, T.T.; Xue, N.N.; Ji, M.; Chen, X.G. Chlorogenic Acid Induced Neuroblastoma Cells Differentiation via the ACAT1-TPK1-PDH Pathway. Pharmaceuticals 2023, 16, 877. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Wang, M.; Tang, T.; Zhou, J.; Sun, W.; Jia, X.; Wang, J.; Yu, H.; Lai, S. The Molecular Mechanism by Which miR-211-5p Regulates the Proliferation and Differentiation of Preadipocytes in Meat Rabbits by Targeting TPK1. Animals 2025, 15, 1497. https://doi.org/10.3390/ani15101497
Zhang X, Wang M, Tang T, Zhou J, Sun W, Jia X, Wang J, Yu H, Lai S. The Molecular Mechanism by Which miR-211-5p Regulates the Proliferation and Differentiation of Preadipocytes in Meat Rabbits by Targeting TPK1. Animals. 2025; 15(10):1497. https://doi.org/10.3390/ani15101497
Chicago/Turabian StyleZhang, Xiaoxiao, Meigui Wang, Tao Tang, Jing Zhou, Wenqiang Sun, Xianbo Jia, Jie Wang, Hengwei Yu, and Songjia Lai. 2025. "The Molecular Mechanism by Which miR-211-5p Regulates the Proliferation and Differentiation of Preadipocytes in Meat Rabbits by Targeting TPK1" Animals 15, no. 10: 1497. https://doi.org/10.3390/ani15101497
APA StyleZhang, X., Wang, M., Tang, T., Zhou, J., Sun, W., Jia, X., Wang, J., Yu, H., & Lai, S. (2025). The Molecular Mechanism by Which miR-211-5p Regulates the Proliferation and Differentiation of Preadipocytes in Meat Rabbits by Targeting TPK1. Animals, 15(10), 1497. https://doi.org/10.3390/ani15101497





