Simple Summary
In this paper, a new Pareas species of the P. hamptoni complex is described from southern Yunnan Province, China, based on morphological and molecular evidence. Genetically, the new species is most closely related to P. hamptoni sensu stricto, for which we confirm the distribution in China. Morphologically, the new species can be distinguished from P. hamptoni sensu stricto and all other congeners by a combination of morphological characteristics. The discovery of the new species brings the total number of recognized species of the genus Pareas to 31, of which 25 occur in China.
Abstract
We describe a new species of the genus Pareas, based on three specimens collected from Guanyinshan Provincial Nature Reserve in Yuanyang County, Honghe Prefecture, Yunnan Province, China. The new species is distinguished from its congeners by one preocular, one postocular or postocular fused with subocular, loreal not bordering the orbit, one row enlarged vertebral scales, five rows keeled mid-dorsal scales at the middle of the body, 189–192 ventral scales and 72–89 subcaudal scales. The dorsal surfaces of the head and body are yellowish red or yellowish brown, and the belly and ventral surfaces of the head and tail are pinkish yellow or yellow with more or less small black spots. Phylogenetic analyses of mitochondrial DNA recovered the new species being the sister taxon to P. hamptoni sensu stricto. The genetic divergences between the new species and P. hamptoni sensu stricto were 4.2% in the Cyt b sequences and 5.0% in the ND4 sequences. In addition, based on specimens collected from Honghe and Wenshan prefectures, we confirmed that P. hamptoni sensu stricto is distributed in China.
1. Introduction
The family Pareidae Romer, 1956 is an ancient lineage, but there is as yet practically no fossil record known for this group [1]. Zheng and Wiens [2] and Deepak et al. [3] estimated that Pareidae split from other caenophidians during the Paleocene, while Zaher et al. [4] inferred that this happened during the Eocene. Deepak et al. [3] and Poyarkov et al. [5] estimated that Pareidae divided into two subfamilies, namely, Pareatinae Romer, 1956 and Xylophiinae Deepak, Ruane and Gower, 2019, during the middle Eocene. Xylophiinae encompasses one genus, namely, Xylophis Beddome, 1878, while Pareatinae encompasses three genera, namely, Aplopeltura Duméril, 1853, Asthenodipsas Peters, 1864, and Pareas Wagler, 1830 [3,5,6].
The genus Pareas split from its sister genus Aplopeltura during the Oligocene [3,5]. Pareas covers a group of highly specialized snakes that only feed on snails or slugs [7,8,9,10,11]. This special habit also gives them a significant advantage in the niche, so that they occupied a vast area from Sundaland to northeastern India and southern China and evolved into a large number of different species [5,6,10,12,13,14,15,16].
Hampton’s slug-eating snake, Pareas hamptoni (Boulenger, 1905), was originally described based on a single specimen from Mogok, Myanmar [17]. Subsequently, this species was thought to be widely distributed, ranging across mainland Southeast Asia and southern China [18,19,20,21]. Thereafter, Wang et al. [6] demonstrated that the populations previously identified as P. hamptoni in southeast China belong to P. formosensis (Van Denburgh, 1909). Ding et al. [11] described the population that had previously been identified as P. hamptoni in south central Yunnan, China as a new species, P. geminatus Ding, Chen, Suwannapoom, Nguyen, Poyarkov & Vogel, 2020. They demonstrated that the populations previously identified as P. hamptoni in northeast, central, and central south Vietnam belong to P. formosensis, whereas the populations in northwest Vietnam belong to P. hamptoni sensu stricto. Liu and Rao [12] described the population that had previously been considered P. hamptoni from southwestern Yunnan, China, as a new species, P. xuelinensis Liu & Rao, 2021. Until now, whether P. hamptoni sensu stricto is distributed in China has been a matter of uncertainty.
During our field research in southern Yunnan Province, China, from 2019 to 2023, we collected a series of Pareas specimens that closely resemble P. hamptoni. However, further comprehensive analyses of their molecular and morphological characteristics have demonstrated that these specimens do not belong to the same species but are from two separate taxa. One corresponds to P. hamptoni sensu stricto, while the other is different from all known congeners. Therefore, we herein confirm the distribution of P. hamptoni sensu stricto in China and describe the unique taxon as a new species.
2. Materials and Methods
2.1. Sampling
Specimens were caught by hand at night. Photographs were taken in situ before the specimens were collected. Liver tissues were cut and stored in analytical pure ethanol and the snakes were preserved in a 75% concentration of ethanol. Specimens were deposited at Kunming Natural History Museum of Zoology, Kunming Institute of Zoology, Chinese Academy of Sciences (KIZ).
2.2. Morphometrics
Measurements were taken to the nearest 1 mm with a ruler. Paired meristic characteristics are given as left/right. The methodology of measurements and meristic counts is the same as that used by Liu et al. [22]. The abbreviations are as follows: anterior temporals (ATem); dorsal scale rows (DS), counted at one head length behind the head-mid-body-one head length before the vent; infralabials (InfL); loreal bordering orbit (LoBO); maxillary teeth (Max); the number of enlarged dorsal scale rows at mid-body (NED); the number of keeled dorsal scale rows (NKD), counted at one head length behind the head-mid-body one head length before the vent; postoculars (PosO); precloacal plate (Prec); preoculars (PreO); prefrontal bordering orbit (PrFBO); posterior temporals (PTem); subcaudals (Sc); subocular-postocular fused (SPOF); suboculars (SubO); supralabials (SupL); snout–vent length (SVL); tail length (TL); and ventrals (Vs).
2.3. Molecular Analyses
Molecular data were generated for nine specimens collected from Honghe and Wenshan prefectures, Yunnan Province, China. Other sequences used in the phylogenetic analyses were obtained from GenBank. All new sequences have been deposited in GenBank. Aplopeltura boa (Boie, 1828) was selected as the outgroup according to Liu et al. [23]. Details of all the sequences used in this study can be found in Table 1.

Table 1.
Samples used for molecular phylogenetic analysis in this study.
Total genomic DNA was extracted from liver tissue samples. The sequences of the mitochondrial genes, cytochrome b (Cyt b) and NADH dehydrogenase subunit 4 (ND4), were amplified by a polymerase chain reaction (PCR) using the primers L14910/H16064 [24] and ND4F/ND4LEUR [25], respectively. The amplification products were purified and sequenced by Tsingke Biotechnology Co., Ltd. (Beijing, China). The sequences were stitched using SeqMan in Lasergene 7.1 [26].
The sequences were aligned using ClustalW [27] that is integrated in MEGA 11 [28] with the default parameters. The genetic divergences (uncorrected p-distance) between species were calculated using MEGA 11 [28]. The best substitution models were selected with ModelFinder [29], using the Bayesian information criterion (BIC). Phylogenetic analyses were constructed based on the concatenated sequences of the Cyt b and ND4 gene sequences. Bayesian inference was performed with MrBayes 3.2.6 [30], using the GTR+F+I+G4 model for the first and second codon positions of Cyt b and ND4 and the GTR+F+G4 model for the third codon positions of Cyt b and ND4. Maximum likelihood analysis was performed with IQ-TREE 1.6.12 [31], using the GTR+F+I+G4 model for the first and second codon positions of Cyt b and ND4 and the TN+F+R3 model for the third codon positions of Cyt b and ND4.
3. Results
3.1. Morphological Results
Meristic and mensural characteristics were noted for all newly collected specimens (Table 2 and Table 3). The morphological characteristics of the specimens collected from Malipo County in Wenshan Prefecture and Hekou and Jianshui counties in Honghe Prefecture agree with the morphological characteristics of Pareas hamptoni sensu stricto given by Ding et al. [11]. However, the morphological characteristics of the specimens collected from Yuanyang County, Honghe Prefecture, can be distinguished from those of the specimens collected from Malipo, Hekou, and Jianshui, and all other named species of Pareas.

Table 2.
Measurements (in mm) and the scalation data of the type series of Pareas guanyinshanensis sp. nov. For abbreviations, see the Materials and Methods section.

Table 3.
Measurements (in mm) and scalation data of the specimens of Pareas hamptoni sensu stricto from China. For abbreviations, see the Materials and Methods section.
3.2. Molecular Results
Bayesian inference and maximum likelihood analysis resulted in similar topologies; the specimens from Malipo, Hekou, and Jianshui were clustered with Pareas hamptoni sensu stricto from Myanmar and Vietnam with strong supports, which should also be assigned to P. hamptoni sensu stricto, while the specimens from Yuanyang formed a distinct lineage sister to P. hamptoni sensu stricto with strong supports (Figure 1). The genetic divergences (uncorrected p-distance) between the specimens from Yuanyang and P. hamptoni sensu stricto were 4.2% and 5.0% in the Cyt b and ND4 gene sequences, respectively (Table 4 and Table 5).
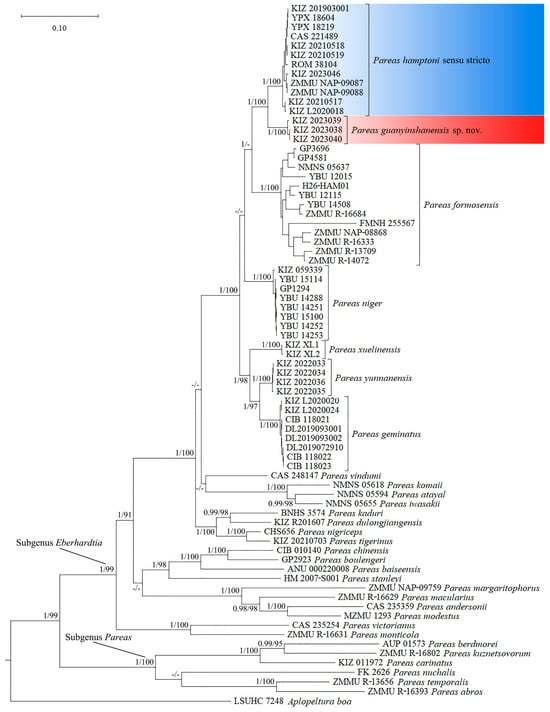
Figure 1.
Bayesian phylogeny tree of Pareas, based on concatenated Cyt b and ND4 fragments. Node numbers before “/” indicate Bayesian posterior probabilities (values below 0.90 are not shown) and numbers after “/” indicate ultrafast bootstrap support for the maximum likelihood analyses (values below 90 are not shown).

Table 4.
Uncorrected p-distances (%), as calculated from Cyt b gene sequences. (1) Pareas guanyinshanensis sp. nov., (2) P. abros, (3) P. andersonii, (4) P. atayal, (5) P. baiseensis, (6) P. berdmorei, (7) P. boulengeri, (8) P. carinatus, (9) P. chinensis, (10) P. dulongjiangensis, (11) P. formosensis, (12) P. geminatus, (13) P. hamptoni sensu stricto, (14) P. iwasakii, (15) P. kaduri, (16) P. komaii, (17) P. kuznetsovorum, (18) P. macularius, (19) P. margaritophorus, (20) P. modestus, (21) P. monticola, (22) P. niger, (23) P. nigriceps, (24) P. nuchalis, (25) P. stanleyi, (26) P. temporalis, (27) P. tigerinus, (28) P. victorianus, (29) P. vindumi, (30) P. xuelinensis, and (31) P. yunnanensis.

Table 5.
Uncorrected p-distances (%), as calculated from the ND4 gene sequences. (1) Pareas guanyinshanensis sp. nov., (2) Pareas abros, (3) P. andersonii, (4) P. atayal, (5) P. baiseensis, (6) P. berdmorei, (7) P. boulengeri, (8) P. carinatus, (9) P. chinensis, (10) P. formosensis, (11) P. hamptoni sensu stricto, (12) P. komaii, (13) P. kuznetsovorum, (14) P. macularius, (15) P. margaritophorus, (16) P. monticola, (17) P. niger, (18) P. nuchalis, (19) P. stanleyi, (20) P. temporalis, (21) P. victorianus, and (22) P. vindumi.
3.3. Systematics
Class Reptilia Laurenti, 1768
Order Squamata Oppel, 1811
Suborder Serpentes Linnaeus, 1758
Infraorder Caenophidia Hoffstetter, 1939
Family Pareidae Romer, 1956
Subfamily Pareinae Romer, 1956
Genus Pareas Wagler, 1830
Subgenus Eberhardtia Angel, 1920
Pareas guanyinshanensis sp. nov.
urn:lsid:zoobank.org:act:AC079EB5-1E12-4AB6-BB55-666CD66D3C0F
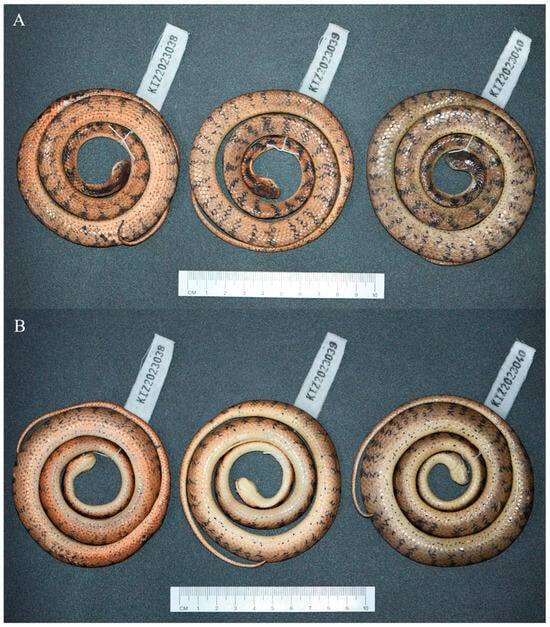
Figure 2.
The type series of Pareas guanyinshanensis sp. nov. in preservative. (A) Dorsal view; (B) ventral view.
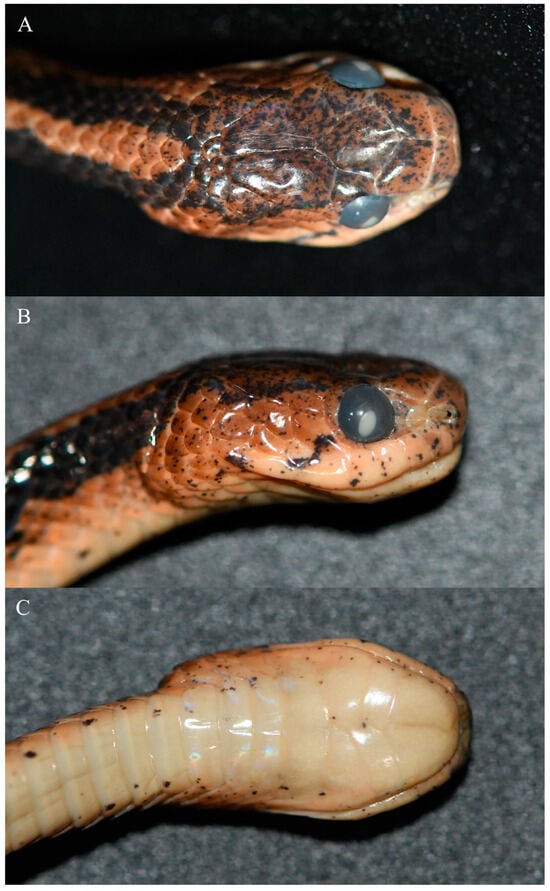
Figure 3.
Close-up views of the head of the holotype (KIZ 2023038) in preservative. (A) Dorsal view; (B) right side view; (C) ventral view.

Figure 4.
Pareas guanyinshanensis sp. nov. in life. (A) The holotype (KIZ 2023038); (B) the paratype (KIZ 2023040).
Holotype. KIZ 2023038, adult female, collected from Guanyinshan Provincial Nature Reserve, Shuijingwan Village, Ganiang Township, Yuanyang County, Honghe Prefecture, Yunnan Province, China (23°3′5″ N, 102°51′54″ E; 1750 m a.s.l.) on 17 May 2023 by Shuo Liu.
Paratypes. KIZ 2023039–2023040, two adult females, with the same collection data as the holotype.
Diagnosis. SVL 482–540 mm, TL/SVL 0.26–0.30; prefrontal bordering orbit; loreal not bordering orbit; 1 preocular; 1 postocular or postocular fused with subocular; 7–8 supralabials; 6–8 infralabials; infralabial not fused with chin-shield; 3 chin-shield pairs; dorsal scales in 15 rows throughout; 1 row of vertebral scales enlarged; scales not keeled at anterior part of body, 5 rows of mid-dorsal scales keeled at middle part of body, 5–7 rows of mid-dorsal scales keeled at posterior part of body; ventral scales 189–192; subcaudals 72–89, paired; precloacal plate undivided; maxillary teeth 4–5. Dorsal surface of head dark yellowish red or yellowish brown, with dense small black spots; dorsal surface of body yellowish red or yellowish brown; belly and ventral surfaces of head and tail pinkish yellow or yellow, with more or less small black spots; iris reddish yellow or yellow.
Description of the holotype. Adult female, SVL 488 mm, TL 146 mm, TL/SVL 0.30, TL/total length 0.23; body elongated, laterally compressed; head distinct from neck; snout wide and blunt, projecting beyond lower jaw; rostral approximately as wide as high, slightly visible from above; nasal undivided; internasal elongated; prefrontal approximately trapezoidal, bordering orbits; frontal shield-shaped, slightly longer than wide; parietals large, longer than wide, gradually narrower posteriorly, median suture approximately equal to length of frontal; single loreal, not bordering orbit; 1 supraocular, approximately triangular; 1 preocular; 1 postocular and 1 elongated crescent-shaped subocular; 2 anterior temporals, 3 posterior temporals; 7 supralabials on each side, separated from eyes; 7 infralabials on left side and 8 infralabials on right side, anterior-most in contact with its opposite between mental and anterior chin-shields, first 4 in contact with anterior chin-shield on left side and first 5 in contact with anterior chin-shield on right side; 3 chin-shields pairs, first pair and third pair triangle and large, second pair small and elongate, chin-shields interlaced, no mental groove under chin and throat; ventral scales 190; precloacal plate undivided; subcaudals 89, paired; dorsal scales in 15 rows throughout, 1 row of vertebral scales distinctly enlarged, scales not keeled at anterior body, 5 rows of mid-dorsal scales keeled at middle and posterior body; 5 maxillary teeth on left side and 4 maxillary teeth on right side.
Coloration of the holotype in life. Dorsal surface of head dark yellowish red, scattered with dense, small black spots; dorsal surface of body yellowish red; 2 wide black stripes on dorsal neck from occipital region; a black stripe from lower posterior orbit downwards and backwards to junction of last 2 supralabials and a black dot on last supralabial on each side of head; approximately 47 vertical black stripes on each side of body, most stripes on different sides connected to each other on vertebrals; some irregular black stripes on each side of tail; belly and ventral surfaces of head and tail pinkish yellow with many small black spots; iris reddish yellow, pupil black.
Coloration of the holotype in preservative. Yellowish red dorsal surfaces of head and body faded to pinkish brown; black stripes on sides of body and tail still distinct; pinkish-yellow belly and ventral surfaces of head and tail faded to pinkish white; iris changed to grayish black and pupil changed to white.
Variations. Morphometric and meristic data for the type series of the new species are provided in Table 2. The paratypes resemble the holotype, except that the subocular and the postocular are fused on one side of the head. The posterior temporals vary in number from two to four. The dorsal surfaces of the head and body vary from light yellowish red to yellowish brown, the belly and ventral surfaces of the head and tail vary from yellowish white to yellow, the number of vertical black stripes on each side of the body vary from 46 to 53, and the iris varies from reddish yellow to yellow in the paratypes. In addition, the small black spots on the belly are much fewer in one paratype.
Etymology. The specific epithet guanyinshanensis refers to Guanyinshan Provincial Nature Reserve, where the new species was found.
Distribution. This new species is currently known only from Guanyinshan Provincial Nature Reserve in Yuanyang County, Honghe Prefecture, Yunnan Province, China (Figure 5).
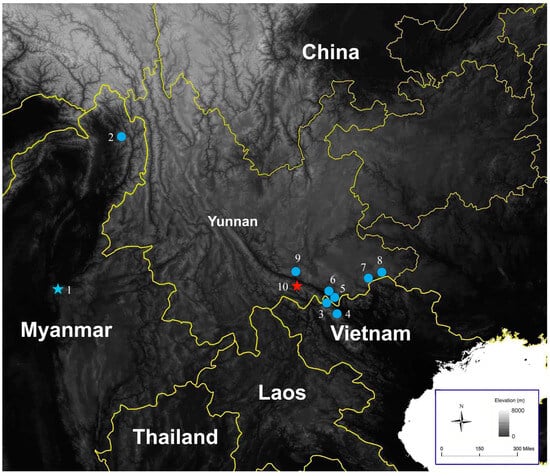
Figure 5.
Map showing the type locality (shown by a blue star) of Pareas hamptoni sensu stricto, other confirmed distributions (blue dots) of P. hamptoni sensu stricto, and the type locality (indicated by a red star) of Pareas guanyinshanensis sp. nov. (1) Mogok, Mandalay, Myanmar; (2) Naung Mon, Putao, Kachin, Myanmar; (3) Bat Xat, Lao Cai, Vietnam; (4) Sa Pa, Lao Cai, Vietnam; (5,6) Hekou, Yunnan, China; (7,8) Malipo, Yunnan, China; (9) Jianshui, Yunnan, China; (10) Yuanyang, Yunnan, China.
Habitat. All specimens of the new species were found on small branches or on the ground beside a stream at night, with forest and farmland nearby (Figure 6).

Figure 6.
The habitat of Pareas guanyinshanensis sp. nov. at the type locality.
Comparison. Pareas guanyinshanensis sp. nov. can be distinguished from P. abros Poyarkov, Nguyen, Vogel & Orlov, 2022, P. baiseensis Wu, Gong, Huang & Xu, 2023, P. berdmorei Theobald, 1868, P. carinatus Wagler, 1830, P. kuznetsovorum Poyarkov, Yushchenko & Nguyen, 2022, P. nuchalis (Boulenger, 1900), and P. temporalis Le, Tran, Hoang & Stuart, 2021 by having one elongated crescent-shaped subocular or postocular and subocular that is fused into one semicircular scale (vs. 1–3 relatively short suboculars, with the subocular and postocular not fused).
Pareas guanyinshanensis sp. nov. can be distinguished from P. andersonii (Boulenger, 1888), P. modestus Theobald, 1868, P. macularius Theobald, 1868, and P. margaritophorus (Jan 1866) by its yellowish-red or yellowish-brown body background color (vs. body background colors of grey, dark grey, brownish gray, or completely black).
Pareas guanyinshanensis sp. nov. can be distinguished from P. atayal You, Poyarkov & Lin, 2015, P. chinensis (Barbour, 1912), P. formosensis, and P. komaii (Maki, 1931) by one row of vertebral scales being enlarged (vs. three rows that are enlarged).
Pareas guanyinshanensis sp. nov. can be distinguished from P. boulengeri (Angel, 1920), P. stanleyi (Boulenger, 1914), P. vindumi Vogel, 2015, and P. xuelinensis by the vertebral scales being enlarged (vs. not enlarged).
Pareas guanyinshanensis sp. nov. can be distinguished from P. dulongjiangensis Liu, Yang, Rao, Guo & Rao, 2023, P. monticola (Cantor, 1839), and P. victorianus Vogel, Nguyen & Poyarkov, 2021 by the loreal not being in contact with the orbit (vs. in contact with the orbit).
Pareas guanyinshanensis sp. nov. can be distinguished from P. geminatus by having more ventrals (189–192 vs. 170–188). Pareas guanyinshanensis sp. nov. can be distinguished from P. iwasakii (Maki, 1937) by having no distinct black stripe on the lateral head (vs. two clear thin black stripes on each side of the head).
Pareas guanyinshanensis sp. nov. can be distinguished from P. kaduri Bhosale, Phansalkar, Sawant, Gowande, Patel & Mirza, 2020, P. niger (Pope, 1928), P. nigriceps Guo & Deng, 2009, P. tigerinus Liu, Zhang, Poyarkov, Hou, Wu, Rao, Nguyen & Vogel, 2023, and P. yunnanensis (Vogt, 1922) by the dorsal surface of the head being yellowish red or yellowish brown with small black spots (vs. a large black blotch on the dorsal surface of the head or the dorsal surface of the head being solid black).
Pareas guanyinshanensis sp. nov. is phylogenetically closely related to P. hamptoni sensu stricto. However, Pareas guanyinshanensis sp. nov. can be distinguished from P. hamptoni sensu stricto by having fewer subcaudals (72–89 vs. 91–99) and a shorter tail (TL/SVL 0.26–0.30 vs. 0.32–0.37) (Table 6).

Table 6.
Comparison between Pareas guanyinshanensis sp. nov. and P. hamptoni sensu stricto. Measurements are in mm. The data for P. hamptoni sensu stricto were obtained by combining those of the original description, Ding et al. [11], and this study.
4. Discussion
Poyarkov et al. [5] divided the genus Pareas into two subgenera, namely, Pareas and Eberhardtia, and partitioned the subgenus Eberhardtia into four species groups, namely, the P. chinensis, P. hamptoni, P. monticola, and P. margaritophorus species groups. Currently, the P. chinensis species group consists of P. baiseensis, P. boulengeri, P. chinensis, and P. stanleyi; the P. monticola species group consists of only P. monticola and P. victorianus; and the P. margaritophorus species group consists of P. andersonii, P. modestus, P. macularius, and P. margaritophorus. The new species obviously belongs to the P. hamptoni species group. Therefore, the P. hamptoni species group should currently contain P. atayal, P. dulongjiangensis, P. formosensis, P. geminatus, Pareas guanyinshanensis sp. nov., P. hamptoni, P. iwasakii, P. kaduri, P. komaii, P. niger, P. nigriceps, P. tigerinus, P. vindumi, P. xuelinensis, and P. yunnanensis.
As the new species is phylogenetically the sister to and most resembles Pareas hamptoni sensu stricto, in order to compare the new species with P. hamptoni sensu stricto, we rely on the morphological data given by Ding et al. [11], combined with the original description [17] of this species and the new data in this paper, to obtain a relatively reliable morphological characterization of P. hamptoni sensu stricto. Although the morphological data of P. hamptoni sensu stricto given by Ding et al. [11] are slightly different from the original description [17] of this species, Ding et al. examined five specimens of P. hamptoni sensu stricto, which included the holotype of this species. Therefore, when the morphological data of P. hamptoni sensu stricto given by Ding et al. [11] are inconsistent with the original description [17] of this species, we adopt those given by Ding et al. [11].
Pareas hamptoni was once considered to be widely distributed, from Myanmar, Thailand, Laos, and Vietnam to southern China [18]. Wang et al. [6] restricted the distribution of P. hamptoni sensu stricto to Myanmar. Ding et al. [11] demonstrated that P. hamptoni sensu stricto is also distributed in northern Vietnam. Based on the specimens collected from Honghe and Wenshan prefectures in Yunnan Province, we confirmed the distribution of P. hamptoni sensu stricto in China (Figure 5) and describe a new species that is closely related to P. hamptoni sensu stricto. The distribution areas of these two species do not overlap, as the new species is from the southwest of the Red River, while all specimens of P. hamptoni sensu stricto in China were from the northeast of the Red River. But further downstream of the Red River, the situation is different. The record of P. hamptoni sensu stricto in Vietnam is from Lao Cai, which is located southwest of the Red River. In this way, P. hamptoni sensu stricto is distributed on both sides of the lower reaches of the Red River. However, in the relative upstream, P. hamptoni sensu stricto and the new species are distributed on different sides of the Red River, respectively.
5. Conclusions
A new Pareas species of the P. hamptoni complex (Figure 7) is described in this paper, based on three specimens collected from Guanyinshan Provincial Nature Reserve in Yuanyang County, Honghe Prefecture, Yunnan Province, China. Currently, the new species is known only from its type locality. The local ecological environment is relatively well maintained, and this species is less threatened at present.

Figure 7.
Species of the Pareas hamptoni complex in life. (A) Pareas guanyinshanensis sp. nov., from its type locality in Yuanyang County, Honghe Prefecture, Yunnan Province, China; (B) P. hamptoni sensu stricto, from Hekou County, Honghe Prefecture, Yunnan Province, China; (C) P. formosensis, from Nanping City, Fujian Province, China; (D) P. niger, from its type locality in Kunming City, Yunnan Province, China; (E) P. xuelinensis, from its type locality in Xuelin Township, Lancang County, Puer City, Yunnan Province, China; (F) P. geminatus, from its type locality in Jiangcheng County, Puer City, Yunnan Province, China; (G) P. yunnanensis, from its type locality in Dali City, Dali Prefecture, Yunnan Province, China.
A key to the members of the Pareas hamptoni complex:
| 1 | Dorsal head solid black........................................................................................................................2 |
| Dorsal head yellowish brown to reddish brown.............................................................................3 | |
| 2 | Dorsal body almost solid black........................................................................................Pareas niger |
| Dorsal body yellow to brownish red with black bars......................................Pareas yunnanensis | |
| 3 | Vertebral scales not enlarged..................................................................................Pareas xuelinensis |
| Three rows of vertebral scales enlarged...............................................................Pareas formosensis | |
| One row of vertebral scales enlarged................................................................................................4 | |
| 4 | Subcaudals more than 91....................................................................Pareas hamptoni sensu stricto |
| Subcaudals less than 91.......................................................................................................................5 | |
| 5 | Ventrals less than 188................................................................................................Pareas geminatus |
| Ventrals more than 189..................................................................Pareas guanyinshanensis sp. nov. |
Author Contributions
Investigation, S.L. (Shuo Liu), M.M., M.L., B.L., X.L. and S.L. (Song Li); conceptualization, S.L. (Song Li) and D.R.; data curation, S.L (Shuo Liu) and S.L. (Song Li); formal analysis, all authors; methodology, all authors; writing—original draft, S.L. (Shuo Liu); writing—review and editing, D.R.; supervision, S.L. (Song Li) and M.M. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Science-Technology Basic Condition Platform from the Ministry of Science and Technology of the People’s Republic of China (grant no. 2005DKA21402), the project of Yuanyang Guanyin Mountains Provincial Nature Reserve Integrative Scientific Expedition (grant no. E2HX105B), and the project of the Ministry of Ecology and Environment of China, “Investigation and assessment of amphibians and reptiles in southern Yunnan”.
Institutional Review Board Statement
Specimen collection was performed in accordance with the Wild Animals Protection Law of the People’s Republic of China and approved by Guanyinshan Provincial Nature Reserve Management and Protection Bureau (project number E2HX105B). The study was approved by the Ethics Committee of Kunming Institute of Zoology, Chinese Academy of Sciences (protocol code IACUC-OE-2023-12-004).
Informed Consent Statement
Not applicable.
Data Availability Statement
All data are presented in this article.
Acknowledgments
We thank the forest rangers of Guanyinshan Provincial Nature Reserve for their assistance in the fieldwork. We would like to thank our workmates for their help and advice. We are also grateful to the editors and reviewers for their valuable comments on the manuscript.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Smith, K.T.; Georgalis, G.L. The Diversity and Distribution of Palaeogene Snakes—A Review, with Comments on Vertebral Sufficiency. In The Origin and Early Evolution of Snakes; Gower, D., Zaher, H., Eds.; Cambridge University Press: Cambridge, UK, 2022; pp. 55–84. [Google Scholar]
- Zheng, Y.; Wiens, J.J. Combining phylogenomic and supermatrix approaches, and a time-calibrated phylogeny for squamate reptiles (lizards and snakes) based on 52 genes and 4162 species. Mol. Phylogenet. Evol. 2016, 94, 537–547. [Google Scholar] [CrossRef]
- Deepak, V.; Ruane, S.; Gower, D.J. A New Subfamily of Fossorial Colubroid Snakes from the Western Ghats of Peninsular India. J. Nat. Hist. 2019, 52, 2919–2934. [Google Scholar] [CrossRef]
- Zaher, H.; Murphy, R.W.; Arredondo, J.C.; Graboski, R.; Machado-Filho, P.R.; Mahlow, K.; Montingelli, G.G.; Quadros, A.B.; Orlov, N.L.; Wilkinson, M.; et al. Large-Scale Molecular Phylogeny, Morphology, Divergence-Time Estimation, and the Fossil Record of Advanced Caenophidian Snakes (Squamata: Serpentes). PLoS ONE 2019, 14, e0216148. [Google Scholar] [CrossRef]
- Poyarkov, N.A.; Nguyen, T.V.; Pawangkhanant, P.; Yushchenko, P.V.; Brakels, P.; Nguyen, L.H.; Nguyen, H.N.; Suwannapoom, C.; Orlov, N.; Vogel, G. An integrative taxonomic revision of slug-eating snakes (Squamata: Pareidae: Pareineae) reveals unprecedented diversity in Indochina. PeerJ 2022, 10, e12713. [Google Scholar] [CrossRef]
- Wang, P.; Che, J.; Liu, Q.; Li, K.; Jin, J.Q.; Jiang, K.; Shi, L.; Guo, P. A revised taxonomy of Asia snail-eating snakes Pareas (Squamata, Pareidae): Evidence from morphological comparison and molecular phylogeny. ZooKeys 2020, 939, 45–64. [Google Scholar] [CrossRef]
- Hoso, M.; Asami, T.; Hori, M. Right-handed snakes: Convergent evolution of asymmetry for functional specialization. Biol. Lett. 2007, 3, 169–172. [Google Scholar] [CrossRef]
- Wallach, V.; Williams, K.L.; Boundy, J. Snakes of the World. A Catalogue of Living and Extinct Species; CRC Press: Boca Raton, FL, USA, 2014. [Google Scholar]
- Vogel, G. A New Montane Species of the Genus Pareas Wagler, 1830 (Squamata: Pareatidae) from Northern Myanmar. Taprobanica 2015, 7, 1–7. [Google Scholar] [CrossRef]
- You, C.W.; Poyarkov, N.A.; Lin, S.M. Diversity of the snail-eating snakes Pareas (Serpentes, Pareatidae) from Taiwan. Zool. Scr. 2015, 44, 349–361. [Google Scholar] [CrossRef]
- Ding, L.; Chen, Z.; Suwannapoom, C.; Nguyen, T.V.; Poyarkov, N.A.; Vogel, G. A new species of the Pareas hamptoni complex (Squamata Serpentes: Pareidae) from the Golden Triangle. Taprobanica 2020, 9, 174–193. [Google Scholar] [CrossRef]
- Liu, S.; Rao, D.Q. A new species of the genus Pareas (Squamata, Pareidae) from Yunnan, China. ZooKeys 2021, 1011, 121–138. [Google Scholar] [CrossRef]
- Vogel, G.; Nguyen, T.V.; Zaw, T.; Poyarkov, N.A. A new species of the Pareas monticola complex (Squamata, Serpentes, Pareidae) from Chin Mountains with additions to the Pareas fauna of Myanmar. J. Nat. Hist. 2021, 54, 2577–2612. [Google Scholar] [CrossRef]
- Gong, Y.; Wu, J.; Huang, S.; Xu, Y.; Yang, D.; Liu, Y.; Liang, S.; Lee, P. A New Species of Pareas (Squamata, Pareidae) from Guangxi Province, China. Animals 2023, 13, 2233. [Google Scholar] [CrossRef] [PubMed]
- Uetz, P. The Reptile Database. Available online: http://www.reptile-database.org (accessed on 2 November 2023).
- Le, D.T.T.; Tran, T.G.; Hoang, H.D.; Stuart, B.L. A New Species of Pareas (Squamata, Pareidae) from Southern Vietnam. Vertebr. Zool. 2021, 71, 439–451. [Google Scholar] [CrossRef]
- Boulenger, G.A. Descriptions of Two New Snakes from Upper Burma. Bombay Nat. Hist. Soc. 1905, 16, 235–236. [Google Scholar] [CrossRef]
- Nguyen, S.V.; Ho, C.T.; Nguyen, T.Q. Herpetofauna of Vietnam; Edition Chimaira: Frankfurt, Germany, 2009. [Google Scholar]
- Zhao, E.M.; Huang, M.H.; Zong, Y.; Jiang, Y.M.; Huang, Q.Y.; Zhao, H.; Ma, J.F.; Zheng, J.; Huang, Z.J.; Wei, G.; et al. Fauna Sinica, Reptilia, Squamata. Serpentes; Science Press: Beijing, China, 1998. [Google Scholar]
- Zhao, E.M. Snakes of China; Anhui Science Technology Publishing House: Hefei, China, 2006. [Google Scholar]
- Yang, D.T.; Rao, D.Q. Amphibia and Reptilia of Yunnan; Yunnan Science and Technology Press: Kuming, China, 2008. [Google Scholar]
- Liu, S.; Zhang, D.R.; Poyarkov, N.A.; Hou, M.; Wu, L.; Rao, D.Q.; Nguyen, T.V.; Vogel, G. Resurrection of Pareas yunnanensis (Vogt, 1922) with Description of a New Species of Pareas from Yunnan Province, China (Squamata, Pareidae). Eur. J. Taxon. 2023, 860, 1–26. [Google Scholar] [CrossRef]
- Liu, S.; Yang, M.J.; Rao, J.Q.; Guo, Y.H.; Rao, D.Q. A New Species of Pareas Wagler, 1830 (Squamata, Pareidae) from Northwestern Yunnan, China. Taxonomy 2023, 3, 169–182. [Google Scholar] [CrossRef]
- Queiroz, A.D.; Lawson, R.; Lemos-Espinal, J.A. Phylogenetic relationships of North American garter snakes (Thamnophis) based on four mitochondrial genes: How much DNA is enough? Mol. Phylogenet. Evol. 2002, 22, 315–329. [Google Scholar] [CrossRef]
- Salvi, D.; Harris, D.J.; Kaliontzopoulou, A.; Carretero, M.A.; Pinho, C. Persistence across Pleistocene ice ages in Mediterranean and extra-Mediterranean refugia: Phylogeographic insights from the common wall lizard. BMC Evol. Biol. 2013, 13, 147. [Google Scholar] [CrossRef]
- Burland, T.G. DNASTAR’s Lasergene sequence analysis software. Methods Mol. Biol. 2000, 132, 71–91. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the Sensitivity of Progressive Multiple Sequence Alignment Through Sequence Weighting, Position-Specific Gap Penalties and Weight Matrix Choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast Model Selection for Accurate Phylogenetic Estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).