Simple Summary
Ligilactobcillus salivarius is a Gram-positive bacterium that commonly colonizes the mucous membranes of the digestive tracts of humans and animals, including birds. It belongs to the group of lactic acid bacteria which, by producing lactic acid, acidify the intestinal environment and limit the development of undesirable intestinal microflora. In addition, L. salivarius can produce other antimicrobial substances, such as bacteriocins and hydrogen peroxide. Due to limiting the development of unfavourable microflora and other health-promoting effects, L. salivarius bacteria are considered as potential probiotics that may increase animal health, and thus animal production indicators. In this work, we undertook research on the characteristics of L. salivarius strains from chickens, geese, turkeys and domestic pigeons. We showed great variation in phenotypic and genotypic traits between strains and the evolutionary adaptation of L. salivarius strains to the colonization of a specific host. The results of the study contribute to knowledge of the characteristics of the species L. salivarius and may be useful in the selection of probiotic strains.
Abstract
Ligilactobacillus salivarius is an important member of the human and animal gut microbiota, and selected strains are promising probiotics, but knowledge of the characteristics of avian isolates is still limited. In this study, we examined selected phenotypic and genotypic traits of 33 L. salivarius strains from geese, chickens, turkeys and pigeons. The strains varied in terms of cell size, colony morphology, broth growth characteristics, biofilm formation, tolerance to bile, hydrophobicity and phenotypic and genotypic antibiotic resistance profiles. Large variation among strains was noted for the utilization of sorbitol, salicin, trehalose, rhamnose, inulin and N-acetyl-D-glucosamine. The presence of genes related to sugar metabolism, i.e., mipB, tktA, rhaB and LSL_1894, was not always correlated with the biochemical phenotypic profile. Correlations were recorded between the host and utilization of certain sugars as well as tolerance to bile. The repA-type megaplasmid and genes coding for Abp118 bacteriocin were detected in 94% and 51.5% of L. salivarius strains, respectively. Phylogeny based on groEL gene sequences was partly correlated with the origin of the strains and revealed an evolutionary distance between L. salivarius strains from humans and birds. The results of the study contribute to knowledge of the characteristics of the species L. salivarius. Intraspecies variations of L. salivarius strains may affect their ability to colonize specific niches and utilize nutrients and reveal potential strain-dependent effects on host health.
1. Introduction
The species Ligilactobacillus salivarius was originally described in 1953 by Rogosa et al. [1] as Lactobacillus salivarius. This species name was used for nearly 70 years, but in accordance with the new taxonomic classification of Lactobacillaceae and Leuconostocaceae, the genus Lactobacillus, comprising about 260 species until March 2020, was divided into 25 genera, and the name L. salivarius was changed to Ligilactobacillus salivarius [2].
L. salivarius bacteria are Gram-positive, non-motile, non-spore-forming, catalase-negative, aerotolerant anaerobic rods commonly isolated from the intestines or faeces of birds and mammals, including geese [3], chickens [4], turkeys [5], pigeons [6], ducks [7], pigs [8] and cattle [9,10]. The presence of L. salivarius has also been confirmed in the oral cavity [11] and vagina of humans [12], in human breast milk [13] and in the gut of honeybees [14], as well as in grape wine [15], meat [16] and St. Ivel cheese [17].
The species L. salivarius mainly comprises homofermentative strains that ferment hexoses to lactic acid in the Embden–Meyerhof–Parnas (EMP) pathway, but heterofermentative strains of L. salivarius (e.g., UCC118) are also known. The latter have been shown to be able to degrade ribose via an inducible phosphoketolase, an enzyme of the pentose phosphate pathway, and to produce lactic acid, acetic acid and ethanol from hexoses. Some strains have D-lactate dehydrogenase, meaning that in addition to L(+) lactic acid, they can produce the D(−)-isomer [18,19]. The average genome size of L. salivarius strains is 2.14 ± 0.14 Mbp, with 2062.74 ± 134.26 genes and a GC content of 32.84 ± 0.12% [8]. A characteristic feature of L. salivarius is the presence of 100–380 kbp repA-type megaplasmids and small plasmids in the cells. Extra linear and circular megaplasmids are less common [20]. Studies conducted thus far have shown significant genotypic differentiation among L. salivarius strains, in both chromosomal and plasmid sequences. The greatest variability has been noted in genes encoding glycosyl hydrolases, bacteriocins and proteases, as well as genes responsible for exopolysaccharide synthesis [20].
L. salivarius has the Qualified Presumption of Safety (QPS) status granted by the European Food Safety Authority (EFSA) to biological agents considered safe for use [21], and several authors have demonstrated the antimicrobial activity of avian L. salivarius strains against bacterial pathogens, including Salmonella enterica, Clostridium perfringens, Staphylococcus aureus, Pasteurella multocida, Riemerella anatipestifer and Campylobacter spp. The competitive exclusion of unfavorable microflora by L. salivarius strains depends on the production of lactic acid, hydrogen peroxide and bacteriocins, as well as the ability of the L. salivarius strains to permanently colonize the intestine [4,22,23,24,25]. A wide range of bacteriocins produced by L. salivarius strains include class II salivaricins, i.e., salivaricins P, T, L, CRL 1328, LS1, LS2 and Abp118, which have been found in human and porcine strains and salivaricins SMXD5, FK22, OR7 and L-1077, which can be secreted by L. salivarius strains from poultry [22]. Despite the documented antimicrobial activity of avian L. salivarius strains, they are not often used as feed additives for poultry, and the best described probiotic strain to date is UCC118 from resected human terminal ileum. It shows tolerance to gastric acid, resistance to bile, enhanced adhesion to the human intestinal epithelial cells, antimicrobial and anti-inflammatory activity, and its genome is used as a reference point in various genetic analyses [26].
The aim of this study was to determine selected phenotypic and genotypic traits of L. salivarius strains from chickens, geese, turkeys and domestic pigeons, including growth characteristics, cell size, the ability to utilize various carbon sources, biofilm formation, bile tolerance, hydrophobicity and antibiotic susceptibility, as well as the presence of resistance genes, repA-type megaplasmids and genes involved in sugar metabolism. The determination of the intraspecies diversity of L. salivarius strains will contribute to a better understanding of the biological compatibility of these bacteria with the host, their biological activity and may be helpful in the selection of probiotic strains.
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
A total of 33 wild-type strains of Lactobacillus salivarius were used in the study. They were isolated from the fresh faeces of Green-legged Partridge hens (3 strains, Ch4b-Ch9b), broilers (7 strains, Ch10a-Ch50d), turkeys (9 strains, T2-T31a), domestic geese (7 strains, G2K-G50b) and domestic pigeons (7 strains, P2a-P23a) from lofts and farms located in Poland. Faecal samples were obtained from asymptomatic adult birds and were collected between 2013 and 2017 by veterinarians who supervised individual poultry flocks. Chickens were reared in a standard litter system, geese and turkeys in a semi-open system and pigeons in two-pen lofts. The diet of the birds did not contain microbiological feed additives and was adequate for the age, species and utility breed of the birds and the season. Bacteria were cultured in Man–Rogosa–Sharpe (MRS) medium supplemented with L-cysteine (0.05% w/v) at 37 °C, 5% CO2, and stored at −80 °C in MRS broth containing ~20% glycerol. Reference strains of L. salivarius, i.e., LMG 9476 (formerly L. salivarius subsp. Salicinus) and LMG 9477 (formerly L. salivarius subsp. Salivarius), as well as L. agilis LMG 9186 and L. saerimneri LMG 22875, were purchased from the BCCMTM/LMG Bacteria Collection (Gent, Belgium).
2.2. Identification of L. salivarius
L. salivarius strains were identified by MALDI-TOF mass spectrometry [27] and by sequence analysis of the groEL gene [18]. Additionally, the taxonomic affiliation of some of the isolates tested in this work (Ch8b, Ch10d, Ch24b, Ch50b, G2K, G19a, G24a, G24b, G31a, G39a and G50b) was determined in our previous studies based on the analysis of 16S-23S rDNA regions [3] and/or 16S rDNA [28].
A standard ethanol/formic acid extraction procedure was used to identify the strains in an UltrafleXtreme MALDI-TOF mass spectrometer (Bruker, Germany). Briefly, the 2–3 bacterial colonies were suspended in 300 µL of ultrapure water, and then 900 mL of absolute ethanol (≥99.8%) was added. In this form, the samples were kept at −20 °C up to 1 month. On the day of analysis, the mixture was centrifuged (13,000× g, 2.5 min), the supernatant removed and the bacterial pellet was resuspended in 50 µL of 70% formic acid. After vortexing (1 min), 50 µL of ≥99.9% acetonitrile was added and the resulting mixture was vortexed (1 min) and centrifuged (13,000× g for 2.5 min). Aliquots of 1 µL of the supernatant of each sample were spotted onto three positions of the 384 MTP AnchorChip TF stainless steel MALDI target plate (Bruker, Germany), overlaid with 1 μL matrix solution (HCCA a-cyano-4-hydroxycinnamic acid, Bruker Germany, dissolved in 50% acetonitrile/0.1% trifluoroacetic acid, 10 mg/mL) and allowed to co-crystalize for 1 min. Calibration was preceded with a bacterial test standard (E. coli DH5 alpha, Bruker Daltonics). Microbial mass spectra were analysed using MALDI Biotyper 3.1 software (Bruker, Germany) containing 5989 reference spectra, including 3 strains of L. salivarius [27]. The identification score criteria used were those recommended by the manufacturer: score ≥2.300 indicates a reliable species-level identification; 2.00–2.299, probable species; 1.700–1.999, a genus-level identification; and a score <1.700 indicates no reliable identification. The relationship among MSPs obtained from each strain was visualized in a score-oriented dendrogram using the unweighted pair group method with arithmetic mean (UPGMA) implemented in the MALDI Biotyper 3.1 software package.
The groEL gene was amplified using primers LSL_1212 groS_F2 (59-AAACCATTAGGAGATCGCGTT) and LSL_1211groL_R2 (59-ATCATACCGCCCATACCTG) [18]. The PCR product of ~1900 bp was sequenced using the Sanger method (Genomed, Warsaw, Poland) and compared to the sequences available in the GenBank database using the National Center for Biotechnology Information (NCBI) Basic Local Alignment Search Tool (BLAST). DNA sequences were deposited in GenBank (Accession Nos. MT862775-MT862808 and MT920907, Table A1 in Appendix A).
2.3. Morphology and Growth Features
L. salivarius cultures were Gram-stained, and cell size was assessed using an Olympus DP72 microscope and Cell˄F 3.1 software. The morphology of the colonies and the ability of L. salivarius strains to grow at 45 °C in an atmosphere containing 5% CO2 and at 37 °C under aerobic conditions were also assessed (the growth intensity was visually compared with respect to cultivation under standard conditions, i.e., 37 °C, 5% CO2). Suspension formation during the growth of the strains on MRS broth was visually assessed according to the following scale: − growth in the form of sediment; + low suspension; ++ high suspension. To assess the ability of L. salivarius bacteria to self-aggregate, tubes with 24-h MRS broth cultures were vortexed vigorously for 90 s, and the presence of bacterial aggregates in the broth was visually assessed (−, +, ++).
2.4. Fermentation Assay
Twenty-four carbohydrates, including pentoses (D/L-arabinose, ribose and xylose), hexoses (glucose, galactose, fructose and mannose), 6-deoxyhexoses (rhamnose and fucose), disaccharides (cellobiose, maltose, lactose, melibiose, saccharose and trehalose), trisaccharides (D-melezitose and D-raffinose), polysaccharides (inulin), sugar alcohols (mannitol, sorbitol and xylitol), alcoholic β-glucoside (salicin), N-acetylglucosamine (GlcNAc, an amide derivative of the monosaccharide glucose) and amygdalin (cyanogenic glycoside), were used in the experiment. All sugars were purchased in powdered form from Sigma-Aldrich (Poznań, Poland). The fermentation assay was performed according to the protocol developed by Hedberd et al. [29]. Two-percent aqueous carbohydrate solutions were made, filtered using 0.45 µm syringe filters and stored for no longer than 2 weeks at 4 °C. Freshly grown bacterial cultures were suspended in 0.85% NaCl so that the optical density was 1.0 on the McFarland scale. A 50 µL volume of modified MRS broth (pH 6.7) without carbohydrates was mixed with 50 µL each of 24 carbohydrates, 5 µL of sterile filtered 2% solution of bromocresol purple and 5 µL of bacterial suspension in the wells of microtiter plates. In the negative control, 0.85% NaCl was added instead of bacteria. Plates were incubated at 37 °C, 5% CO2, and examined for colour changes after 24, 48 and 72 h. Purple (pH > 6.8) was considered a negative result (−), yellow (pH < 5.2) indicated a positive reaction (+) and an intermediate colour (pH between 5.2 and 6.8) was considered poor fermentation (+/−). The experiment was performed in two independent repetitions.
2.5. Detection of repE, Abp118 Bacteriocin Genes and Genes Associated with Sugar Metabolism
The repE gene, which is a molecular marker of the repA-type megaplasmid characteristic of L. salivarius; the abp118β + abp118α gene coding for Abp118 bacteriocin (beta + alpha peptide); and the genes involved in the metabolism of rhamnose, sorbitol and pentoses, i.e., rhaB (coding for rhamnulokinase), LSL-1894 (sorbitol-6-phosphate 2-dehydrogenase), mipB (transaldolase) and tktA (transketolase), were detected using primers previously developed by Li et al. [30] (Table 1).

Table 1.
Primers used in this study.
Due to the heterogeneity of the size of the abp118β + abo118α amplicons, those selected were sequenced using the Sanger method and compared to reference sequences available in the GenBank database using the BLAST algorithm.
2.6. Phylogenetic Analysis
The groEL gene of the 33 wild-type and 2 reference L. salivarius strains were amplified and sequenced as described above (Section 2.2). The amino acid sequences were predicted using the NCBI translate tool ORF finder [31]. With MEGA X software, the groEL sequences were aligned using the ClustalW program, and the maximum likelihood method was used to create phylogenetic trees with a bootstrap support value of 500. The analysis included 30 additional L. salivarius strains, mainly of human origin [18], whose groEL sequences were retrieved from the NCBI genome database [32].
2.7. Bile Tolerance Test
The tolerance of L. salivarius isolates to bile salts was determined in a microplate assay. MRS medium (200 µL) containing 1% or 2% ox gall (BTL, Łódź, Poland) was inoculated with 0.5 µL of fresh broth cultures of lactobacilli. Following 24 and 48 h incubation at 37 °C, 5% CO2, the optical density of the bacterial cultures was measured at 620 nm. Positive controls were bacterial cultures grown without ox gall. The growth of each strain was expressed as a percentage of the OD620 value of the control samples [4]. To check the bacterial viability in the presence of bile, after 24 h of incubation, 10 µL was pipetted from each well and added to fresh MRS broth. Bacterial growth was assessed after 48 h.
2.8. Biofilm Formation
MRS broth was dispensed in 200 mL volumes into the wells of Nunc MaxiSorpTM 96-well flat-bottom plates (Biokom, Janki, Poland) and inoculated with 1 µL fresh bacterial culture. Following 48 h incubation at 37 °C, 5% CO2, the wells were emptied and washed 3 times with 0.85% NaCl. Adherent cells were stained with crystal violet (1% w/v, 50 µL) for 15 min. Unbound dye was washed off with water, and cell-bound dye was dissolved in 20% acetone in ethanol for 10 min; the absorbance (A570) was measured using a Microplate Reader 680 (Bio-Rad, Warszawa, Poland). Isolates were classified as follows: no biofilm producer (−), weak biofilm producer (+), moderate biofilm producer (++) and strong biofilm producer (+++), based on the absorbance value [23].
2.9. Measurement of Bacterial Hydrophobicity
The hydrophobicity of the bacteria was determined on the basis of microbial adhesion to xylene, as described previously [4]. Strains with hydrophobicity equal to or greater than 50% were considered hydrophobic.
2.10. Phenotypic and Genotypic Profiles of Antimicrobial Resistance
The antimicrobial susceptibility of L. salivarius strains was determined by the broth microdilution method, as described previously [6]. The following antimicrobial agents were included in the study: ampicillin, tetracycline, streptomycin, gentamicin, kanamycin, erythromycin, lincomycin, chloramphenicol and enrofloxacin. The categorization of strains into susceptible or resistant was based on the MIC cut offs recommended by EFSA [33]. For enrofloxacin and lincomycin that are not included in the EFSA guidelines, strains with MIC values above 32 µg/mL were considered resistant [6].
In order to determine the genotypic profiles of antimicrobial resistance in the tested strains, a number of resistance genes, i.e., tetK, tetL, tetM, tetO, ermA, ermB, ermC, mefA/E, lnuA, aph(3′)-IIIa, aac(6′)-Ie-aph(2″)-Ia, aph(2″)-Ib, aph(2″)-Ic, aph(2″)-Id, ant(4′)-Ia, ant(6)-Ia, cat, lsaE and the Tn916 integrase gene int-Tn, were detected using the primers and the methodology described in our previous papers [5,6].
2.11. Statistical Analysis
Yates’s chi-squared test was used to compare the frequency of occurrence of strains utilizing individual carbohydrates among the pools of strains from pigeons, chickens, turkeys and geese. The same test was also used to determine the relationship between the host and the ability of the strains to grow in bile-supplemented medium. The power of the relationships was assessed with contingency coefficients (C), which ranged from 0.000 to 0.707 for the tests. The level of significance was set as p < 0.05. The statistical analysis was performed using Microsoft Excel 2019.
3. Results
3.1. Identification of L. salivarius Strains
By processing the mass spectra with MALDI Biotyper software, an identification value of >2.000 was obtained for all isolates, indicating probability (2.000–2.299, 24 isolates) or high probability (2.300–3.000, nine isolates) that they belonged to the species Lactobacillus salivarius (Table A1). The similarity among the L. salivarius strains based on their MALDI-TOF mass spectra is shown in the dendrogram derived from UPGMA cluster analysis (Figure 1). At a level of similarity of ca. 60%, examined strains formed three main clusters. There was no correlation between the host and the mass profiles of the strains, although it should be noted that 5 out of 7 goose strains were grouped in cluster A. The pigeon strains were clustered in clades B and C, and the strains from chickens and turkeys were scattered across all three clades. Interestingly, the reference strains formed a separate cluster within clade A.
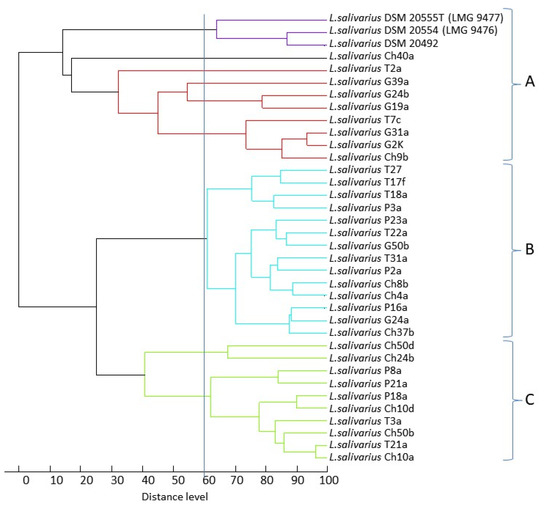
Figure 1.
Dendrogram derived from UPGMA cluster analysis of MALDI-TOF mass spectra obtained from the 33 wild-type L. salivarius strains. The relative distance between isolates is displayed as arbitrary units. One hundred indicates complete similarity and 0 indicates maximum dissimilarity. The vertical line corresponds to the level of similarity 60%.
Analysis using the BLAST algorithm showed that the groEL sequences (1572 nk, GenBank Acc. No. MT862777-MT862808 and MT920907) of all strains tested were homologous to the sequences of L. salivarius strains deposited in the GenBank database. At least the first 30 matches (query cover > 99%) indicated L. salivarius species for all groEL sequences analysed. For 32 of the 33 tested strains, the two best matches showed an identity of ≥99.49%, and only for the Ch10a strain, a slightly lower homology, i.e., ≥98.66%, was obtained (Table A1).
3.2. Morphology and Growth Characteristics
Most of the L. salivarius strains tested grew on MRS agar as large (1.2–2.2 mm), cream-colored, smooth-surfaced and brittle colonies; less frequently, the colonies were umbonate or rough with a wavy edge; several strains grew as shiny, sticky colonies (Figure 2, Table 2).

Figure 2.
Phenotypes of selected L. salivarius strains on MRS agar (strains T2a, G2K, T17f) and MRS broth (strains Ch4a, T3a, G2K, Ch9b).

Table 2.
L. salivarius strains used in this study and their colony morphology, broth growth characteristics, biofilm formation capacity, hydrophobicity and tolerance to 2% bile; capability of the strains to utilize arabinose (ARA), ribose (RIB) and xylose (XYL), rhamnose (RHA) and sorbitol (SOR). Distribution of the repE, bacteriocin Abp118 genes and genes related to sugar metabolism (mipB, tktA, rhaB and LSL_1894). ND*—the structure of the colonies was not determined as they were neither clearly brittle nor clearly sticky; the meaning of the signs “−”, “+”, “++” and “+++” are explained in the Section 2.
All L. salivarius strains, except G2K, showed the ability to grow at 45 °C, 5% CO2. All strains also grew under aerobic conditions at 37 °C, but the most abundant growth was recorded at 37 °C, 5% CO2. The growth of the strains on the broth was varied; most grew as sediment, some formed a suspension, and the cells of several strains adhered to the walls of polystyrene tubes (Figure 2). Many of the L. salivarius strains that grew on MRS agar as brittle colonies showed autoaggregation when grown on MRS broth (Table 2).
In the Gram method, cells of some L. salivarius strains stained purple, and others were Gram-labile. The cells had the form of straight or slightly bent rods varying in length from 1.8 to 7.0 µM; their thickness was 0.5 to 0.8 µM (Figure 3).
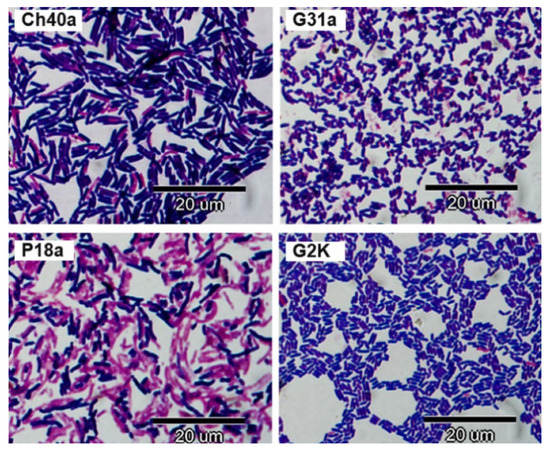
Figure 3.
Gram staining results for selected avian L. salivarius strains from chicken (Ch40a), geese (G31a, G2K) and pigeon (P18a).
3.3. Fermentation Assay
All L. salivarius strains were able to utilize hexoses, i.e., glucose, galactose, fructose and mannose, and disaccharides, i.e., sucrose and melibiose. As many as 97% (32/33) of the isolates grew on the medium with mannitol (except T21a) and lactose (except G2K), and 94% (31/33) of strains fermented raffinose (the exceptions were T22a and G2K). None of the strains fermented pentoses, i.e., arabinose, ribose and xylose and non-fermented xylitol or fucose. Half of the strains (16/33) showed the ability to utilize sorbitol. Substantial variation was noted among the strains for the utilization of sorbitol (48% of isolates), salicin (12%), trehalose (61%), rhamnose (24%), inulin (15%) and N-acetylglucosamine (82%) (Figure 4, Table A2).
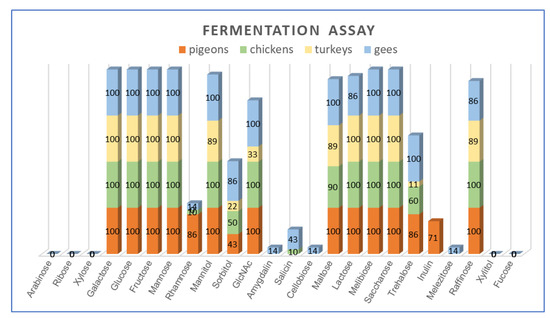
Figure 4.
Percentage of L. salivarius strains capable of utilizing individual carbohydrates.
Statistical analysis showed a significant (p < 0.05) relationship between the ability of L. salivarius strains to utilize several carbohydrates and the host. The fermentation of rhamnose and inulin was significantly more frequent in the pool of pigeon isolates compared to strains from other bird species (power of observed relations was at an average level, as indicated contingency coefficient ranged from 0.315 to 0.417). A significantly lower frequency of trehalose and N-Acetylglucosamine utilization was noted in strains from turkeys compared to isolates from pigeons (C = 0.338), chickens (C = 0.187) and geese (C = 0.420). The utilization of salicin was more frequent in geese strains compared to turkey isolates (C = 0.203).
In terms of carbohydrate utilization, the strain G2K derived from geese stood out from the rest. It was the only strain that did not ferment lactose and grew in broth supplemented with amygdalin, cellobiose and melezitose. It was also one of the two strains unable to use raffinose (Table A2). Despite this biochemical difference, the sequence analysis of 16S rDNA of the G2K strain (GenBank Acc. No. MW642195) confirmed its affiliation to the species of L. salivarius (identity > 99%).
3.4. Presence of repE, abp118β + abp118α and Genes Determining Carbohydrate Utilization
The repE gene, which in addition to the repA gene is a recognized marker of repA-type megaplasmids commonly found in L. salivarius strains [30], was detected in 31 (94%) of 33 wild-type isolates and in reference strain L. salivarius LMG 9477.
In the PCR with the use of primers specific for the gene encoding of the peptide alfa and beta of Abp118 bacteriocin, the PCR product was obtained for 18 (54.5%) L. salivarius strains, including seven isolates from turkeys, six isolates from domestic pigeons, three isolates from chicken and two isolates from geese (Table 2). The amplicon size was 410 bp for 16 strains, 390 bp for isolate T18a and 430 bp for isolate P3a (Table 3).

Table 3.
BLAST results of sequence analysis of abp118 a + abp118 β bacteriocin amplicons.
Sequence analysis showed that the 410 and 390 bp amplicons corresponded to the gene encoding the Abp118 bacteriocin (identity ≥ 98%) or salivaricin P (identity ≥ 98%); however, in the case of the T18a strain (390 bp), there was a 24 consecutive nucleotide deletion in the gene encoding Abp118 alpha subunits (compared to the sequence of strain UCC118, Acc. No. AF408405.1). Within the DNA sequence of the P81, T27 and T18a amplicons, eight variable sites were detected, but they did not translate into changes in the putative amino acid sequence of bacteriocin (Table A3). The sequence (387 nk) of the 430 bp amplicon obtained for strain P3a showed 99% similarity to the gene of ABC transporter permease and bacteriocin and 98% similarity to the blp1 and bimlp genes encoding putative bacteriocin subunit a and putative bacteriocin immunity protein (Table 3).
Genes associated with sugar metabolism were noted in strains containing megaplasmids as well as in one isolate (G24b) in which the repE gene was not detected. Despite the inability of the L. salivarius strains to ferment pentoses, 15% of them contained the tktA gene coding for transketolase, and 36% had the mipB gene encoding transaldolase; two strains from pigeons (P18a and P21a) had both genes which complete the pentose phosphate pathway. All rhamnose-fermenting strains contained the rhaB gene encoding rhamnulokinase, but it was also found in six isolates unable to grow on rhamnose-supplemented medium. Similarly, all L. salivarius strains utilizing sorbitol (48%), as well as three isolates (from pigeons) unable to ferment this carbohydrate, harboured the LSL_1894 gene encoding sorbitol-6-phosphate2-dehydrogenase (Table 2).
3.5. Phylogenetic Analysis
In the dendrogram resulting from the comparative analysis of the groEL sequence of wild-type isolates tested in this study and 30 additional L. salivarius strains (21 strains from humans and nine strains from animals or other sources), previously characterized by Li et al. [17], two reference strains, LMG 9476 (formerly subspecies salicinus) and LMG 9477 (formerly subspecies salivarius), were separated into distinct branches, each of which comprised a mixture of strains (Figure 5A).
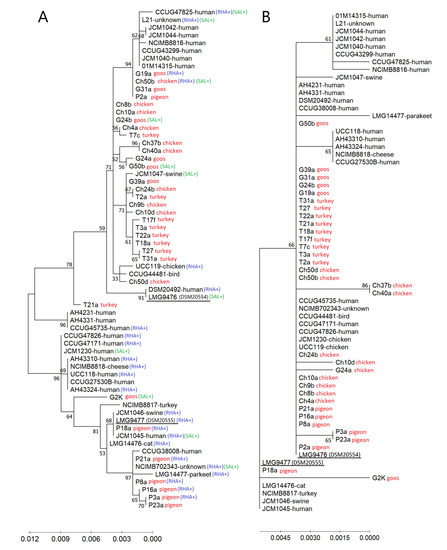
Figure 5.
(A) Phylogenetic trees based on the groEL gene sequences of 33 wild-type avian L. salivarius strains tested in this study and 30 L. salivarius strains (mainly of human origin) previously described by Li et al. [18] built by the maximum likelihood method. The percentage of replicate trees in which the associated taxa were clustered together in the bootstrap test (500 replicates) is shown next to the branches. Scale bars show genetic distance; RHA+ and SAL+ indicate the ability of strains to utilize rhamnose and salicin, respectively. (B) Dendrogram showing the similarity among the predicted amino acid sequences of the GroEL chaperonin built by the maximum likelihood method.
The LMG 9477 strain formed a common clade together with the pigeon-derived L. salivarius strains (except for the P2a strain) and with the goose-derived G2K strain. Most pigeon strains and human strains of this clade showed the ability to ferment rhamnose. However, several other strains (including wild-type strains Ch50b and G19a) with the ability to utilize this carbon source were clustered in a second major clade containing the LMG 9476 strain. Strains capable of utilizing salicin were scattered in both clades. The clade containing L. salivarius LMG 9476 was further split into several clusters, one of which grouped the majority (6 of 9) of turkey isolates (T3a, T17f, T18a, T22a, T27 and T31a). Human strains were largely in separate clusters, within both the LMG 9476 clade and the LMG 9477 clade, while single reference animal strains (from birds and swine) were generally grouped with the wild-type isolates tested in this study (Figure 5A). This indicates a phylogenetic distance between L. salivarius strains from humans and birds.
The significant differences within the groEL gene sequence did not translate into differences in the amino acid sequence. The predicted amino acid sequences of GroEL chaperonin were identical for 25 out of 33 isolates tested in this study and 11 other strains (mainly of human origin); they clustered together in the dendrogram with the L. salivarius LMG 9476 strain (formerly subsp. salicinus) (Figure 5B). Interestingly, in the groEL gene of the Ch10d strain (Acc. No. MT862788), a deletion of as many as 18 consecutive nucleotides was noted, corresponding to eight amino acids.
The G + C content in the groEL gene of avian strains grouped with the LMG 9477 strain ranged from 36.22 (P3a and P23a) to 36.64 mol% (G2K), and the average content of G + C in this clade was 36.36 mol%. In the avian strains forming the second main clade (containing the LMG 9476 strain), the mean G + C content was slightly lower, i.e., 36.21 mol%, and ranged from 35.96 (strain Ch24b) to 36.47 mol% (T21a).
3.6. Tolerance to Bile
All isolates tested were able to survive for 24 h in the presence of 2% bile, and 73% of them even grew in the MRS broth supplemented with 1% or 2% ox gall. However, the growth of the vast majority of isolates was considered poor (3.5%–17% compared to the positive control), and only two strains, G2K (100% growth) and Ch40a (24%–38%), showed intensive growth on bile medium (Table 2). There were no significant differences in growth intensity in broth supplemented with 1% and 2% bile (Table A4), but most strains showed more intense growth on bile medium after 48 h compared to the 24-h culture (Table 2). A significantly higher frequency of strains capable of growing in the presence of 2% bile was recorded in the pool of pigeon and goose isolates compared to strains from chickens and turkeys (C 0.205–2.39) after 48 h of incubation. After 24 h of incubation, a significantly higher frequency of growth was recorded only in the pool of pigeon isolates compared to turkey strains (C = 0.264).
3.7. Hydrophobicity
The vast majority (82%) of the L. salivarius strains tested were hydrophobic, and for as many as 68% of the strains, including all turkey isolates, the %H value was >95%. Six isolates, including two (28%) strains from pigeons, one (10%) strain from chickens and three (43%) strains from geese, were classified as hydrophilic; the extremely low value of %H = 0 was recorded for two strains, i.e., Ch50b and G2K. It is worth noting that all strains showing low or moderate hydrophobicity, i.e., 0%–55%, were able to grow in broth containing 1% or 2% bile (Table 2).
3.8. Phenotypic and Genotypic Profiles of Antimicrobial Resistance
The phenotypic and genotypic drug resistance profiles of some L. salivarius strains tested in this work, i.e., those from pigeons and turkeys, were taken from our previous papers [5,6] (Table 4).

Table 4.
Phenotypic and genotypic antibiotic resistance profiles of Ligilactobacillus salivarius strains.
In the pool of studied isolates, antibiotic-resistant strains were found with high frequency, and as many as 51% of them were multiresistant (resistance to at least three groups of antimicrobial agents). Resistance to streptomycin, kanamycin, enrofloxacin and tetracycline was reported most frequently (>40% of strains), and high MIC values for ampicillin were confirmed in several isolates from turkeys. Strains from pigeons, chickens and turkeys showed the highest frequency of resistance, while goose isolates were susceptible to all antibiotics, except for aminoglycosides. Only one strain, i.e., G24a, was susceptible to all antimicrobial agents (Table 4).
Phenotypic antibiotic resistance was correlated with the presence of resistance genes. TetL and tetM, ermB and lnuA genes were widespread in pigeon, chicken and turkey strains, and other resistance genes were less frequently detected. None of the considered resistance genes were detected in 12 L. salivarius strains, including all strains from geese (Table 4).
4. Discussion
Among the main goals of this research was to evaluate the fermentation profiles of avian L. salivarius strains. Historically, the ability to ferment rhamnose or salicin was considered to be a criterion distinguishing L. salivarius subsp. salivarius from L. salivarius subsp. salicinius. However, subsequent polyphasic analyses by Li et al. [18] showed that the initially proposed taxonomic criteria were insufficient, and ultimately, these two subspecies were unified into a single species. The validity of this unification was also confirmed by the results of this study, as most of the avian L. salivarius strains were unable to utilize rhamnose (utilized by 24% strains) and salicin (utilized by 12% strains). Moreover, one isolate (Ch50b) showed the ability to utilize both of these carbohydrates. Our results differ significantly from those of Li et al. [18], who showed the fermentation of rhamnose and salicin by 57% and 27% of L. salivarius strains (mainly of human origin), respectively. The ability of all L. salivarius isolates in the present study to ferment glucose, galactose, fructose, mannose and sucrose, as well as their inability to utilize arabinose and xylose, is fully consistent with the previous study by Li et al. [18]. However, contrary to our results, these authors reported the utilization of D-ribose by several (8/30) L. salivarius strains. Although sorbitol-fermenting ability was originally considered a characteristic feature of the species L. salivarius [1], more than half of the avian strains were unable to utilize this carbohydrate. Our results differ significantly from the reports of Li et al. [18], who noted sorbitol fermentation in 88% of strains in a pool of human isolates. Even higher values are indicated by the API 50CHL test guidelines [34], according to which as many as 98% of L. salivarius strains are able to utilize sorbitol.
The correlation recorded in the present study among the utilization of certain carbohydrates (rhamnose, inulin, NAG, trehalose and salicin) by L. salivarius strains and the host indicates the evolutionary variability of L. salivarius bacteria. The phenomenon of the evolution of commensal bacteria in accordance with the nutritional conditions of the host environment has long been widely recognized [35,36], and Lee et al. [8] demonstrated that among the host-specific genes detected in L. salivarius strains, several are associated with nutrient utilization. The diversity in the biochemical profiles across tested L. salivarius strains may be the result of the variability of the genome within the repA-type megaplasmids [20], where several genes related to sugar metabolism (mipB, tktA, rhaB and LSL_1894) are located [30]. The presence of the rhamnulokinase gene was detected not only in L. salivarius strains fermenting rhamnose but also in strains without this ability, which has previously been demonstrated by Li et al. [30] and points to gene silencing or a lack of other genes that are essential for completing the pathway. The same authors also reported the prevalence of genes encoding transaldolase (mipB) and transketolase (tktA) in human L. salivarius strains lacking ribose utilization. This study is the first report indicating the possibility of the utilization of cellobiose and melezitose as carbon sources by L. salivarius strains [1,18]. It is also worth emphasizing that inulin, which is utilized by L. salivarius strains from pigeons, is a long-chain polysaccharide that acts as a prebiotic by selectively stimulating the growth and activity of beneficial intestinal microbiota [37]. The utilization of inulin-type fructans is common in bifidobacteria, while in lactobacilli, the degradation of this polysaccharide is rare [38].
We showed that groEL sequences can be used not only for phylogenetic analyses but also for the identification of L. salivarius strains. Several authors have demonstrated that the groEL gene can be successfully used to identify a variety of Gram-positive bacteria, including lactobacilli [39], enterococci [40] and bifidobacteria [41]. Contrary to earlier reports [18], we showed a certain correlation between groEL-based phylogenesis and the host and biochemical profiles of the isolates tested. The slightly higher mean G + C content in the groEL sequences of L. salivarius strains grouped with the reference strain LMG 9477 (DSM 20555T subs. salivarius) than the G + C content in the LMG9477 clade is consistent with the earlier results of Li et al. [18].
In this study, we demonstrated large diversity among avian strains of L. salivarius in terms of colony morphology, growth characteristics on broth (sediment, suspension formation and autoaggregation ability) and biofilm formation, as well as hydrophobicity and bile resistance. These features may be dependent on the production of extracellular polysaccharides (EPS) by bacteria [42,43,44,45]. EPS are thought to protect bacterial cells against extreme conditions (temperature, light intensity, pH or osmotic stress) or biotic stress [43], and they can be either weakly or strongly bound to the bacterial cell surface [46]. EPS of lactobacilli are involved in cell adhesion/recognition mechanisms and could confer a range of local and systemic health benefits to the host, including immune modulation, lowering blood cholesterol or stimulation of the growth of beneficial gut microbiota [47,48].
Several authors have reported that EPS-rich Lactobacillus strains grow as slimy colonies on agar and form a suspension on broth [43,44]. EPS-poor strains form sediment at the bottom of the tube as they grow on the broth. It, therefore, appears that the growth characteristics of L. salivarius bacteria on both agar and broth allow for the initial differentiation of EPS-rich and EPS-poor strains.
Biofilm formation by probiotic bacteria is considered a beneficial property, as it may allow them to colonize the intestinal epithelium while displacing unfavourable microflora. The positive correlation recorded in this study between the ability to self-aggregate and biofilm formation has previously been noted for other Lactobacillus species [49]. Autoaggregation is mediated by surface autoagglutinins, mainly proteins, and is often among the first steps in forming biofilms [50]. Strong biofilm formation by strains with brittle colonies, which are likely to have poor EPS production, is consistent with the results of Tsuneda et al. [45], who showed that EPS-poor strains adhere more strongly to a glass surface than EPS-rich strains. Moreover, they showed that the EPS of EPS-poor strains contain twice as much protein as the EPS of EPS-rich strains.
Several researchers have reported a positive correlation between the hydrophobicity of Lactobacillus strains and their adhesion to epithelial cells [7,51]. In our study, we did not find a clear relationship between the hydrophobicity of L. salivarius strains and their biofilm formation capacity or colony morphology. This is in line with reports by Tsuneda et al. [45], who found no correlation between hydrophobicity and cell adhesiveness for either EPS-rich or EPS-poor strains.
Bile tolerance is among the most crucial properties for probiotic bacteria, as it determines their ability to survive transit through the duodenum. Bacterial resistance to bile is mediated by bile salt hydrolysing enzymes (BSH) and by EPS, which acts as a protective coating against unfavourable environmental factors [42,52]. A positive correlation between EPS production and bile tolerance was recorded in bifidobacteria [42], but in the case of lactobacilli, the relationship between EPS and bile resistance remains unclear [53]. We found no relationship between colony structure or broth growth characteristics (which are dependent on EPS production) and bile tolerance, and, therefore, it seems that EPS do not play a leading role in determining bile resistance in L. salivarius strains. The tendency observed in this study of increased bile tolerance of L. salivarius strains over time is consistent with several previous reports [53,54].
Bacteriocinogenic non-pathogenic strains are desirable probiotics. The production of the bacteriocin Abp118, the gene of which we detected in most strains tested in this study, has previously been confirmed in many human L. salivarius strains [30]. It is a class IIb heat-stable dipeptide bacteriocin that shows a high homology to salivaricin P (they differ only in two amino acids) [55]. Both bacteriocins inhibit the growth of sensitive strains of Listeria monocytogenes, Enterococcus sp., Lactobacillus sp., Pediococcus sp., Lecuconostoc sp. and Streptococcus sp. [55]. Bases on these results, it can be assumed that abp118-positive L. salivarius strains inhabiting the gastrointestinal tract of birds contribute to competitive exclusion and modulate the intestinal microbiota. It should be noted, however, that not all L. salivarius strains containing genes encoding the Abp118 bacteriocin show antibacterial activity in vitro [30]. Therefore, further studies are needed to determine the expression of the abp118alpha and abp118beta genes in avian L. salivarius strains and the related antimicrobial activity.
The issue of antibiotic resistance of Lactobacillus bacteria from poultry and domestic pigeons, including L. salivarius strains, was widely discussed in our previous papers [5,6,56,57]. Due to drug resistance, most L. salivarius strains from poultry and domestic pigeons are disqualified as potential probiotics. Only strains showing drug sensitivity and no resistance genes can be used as microbial feed additives [33]. Despite the beneficial effects of L. salivarius in birds [25], consideration should also be given to the risk of possible transmission of resistance genes between them and other members of the intestinal microbiota, including potentially pathogenic bacteria. Many resistance genes detected in this study have previously been found in lactobacilli and other lactic acid bacteria on mobile elements [5,6], and the conjugative Tn916/Tn1545-like transposon has been identified in one L. salivarius strain (T22a).
5. Conclusions
The results of the phenotypic and genotypic analyses carried out in this study broaden the knowledge on the species L. salivarius and may be useful both in determining taxonomic affiliation and in the selection of probiotic strains. Due to the phenotypic diversity of avian L. salivarius, including the presumed production of Abp118 bacteriocin and differentiation in EPS production, individual strains can be assumed to have varying abilities to colonize distinct niches and different effects on the host. The host-specific traits of the L. salivarius strains indicate their evolutionary adaptation and confirm the principle promoted by scientists that, due to biological compatibility, probiotics intended for a given host species should be derived from its natural microflora.
Author Contributions
Conceptualization, M.D.; Methodology, M.D. and D.S.-P.; Formal Analysis, M.D., T.H. and S.I.; Investigation, M.D., D.S-P., A.P., D.P.-O. and T.H.; Resources, M.D., R.U.-C. and A.P.; Data Curation, M.D., D.P.-O. and T.H.; Writing—Original Draft Preparation, M.D.; Writing—Review and Editing, R.U.-C. and S.I.; Visualization, M.D.; Funding Acquisition, M.D. and R.U.-C. All authors have read and agreed to the published version of the manuscript.
Funding
This research has been supported by the Polish National Science Centre (NCN), project no. 2017/01/X/NZ8/00478. The funds received were used to purchase the materials necessary to perform the tests. The financing body did not participate in the research design, analysis of results, or writing of the manuscript.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
Nucleotide sequences reported in this paper are available in the NCBI GenBank database under the following accession numbers: MT862775-MT862808 and MT920907 (groEL gene), MW478293-MW478294 and MW478296-MW478297 (genes coding for bacteriocin), MW642195 (16S rDNA of the G2K strain).
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A

Table A1.
Identification of L. salivarius strains by MALDI-TOF mass spectrometry, groEL sequence analysis and analysis of 16S rDNA or 16S-23S rDNA regions.
Table A1.
Identification of L. salivarius strains by MALDI-TOF mass spectrometry, groEL sequence analysis and analysis of 16S rDNA or 16S-23S rDNA regions.
| Strain | Identification by MALDI-TOF MS | Identification Based on Sequence Analysis of the groEL Gene | Analysis of 16S rDNA or 16S-23S rDNA | |||
|---|---|---|---|---|---|---|
| groEL Acc. No. | Two Best Matches; GenBank Accesion Number | Query Cover [%] | Identity [%] | |||
| P2a | L. salivarius 2.393 | MT862777 | Ligilactobacillus salivarius strain 2D; CP047412.1 | 100 | 99.94 | ND |
| Lactobacillus salivarius str. Ren Select; CP011403.1 | 100 | 99.87 | ||||
| P3a | L. salivarius 2.362 | MT862778 | Lactobacillus salivarius CECT 5713; CP002034.1 | 100 | 99.81 | ND |
| Lactobacillus salivarius strain NCIMB702343 groEL; DQ444339.1 | 99 | 99.81 | ||||
| P8a | L. salivarius 2.050 | MT862779 | Lactobacillus salivarius CECT 5713; CP002034.1 | 100 | 99.94 | ND |
| Lactobacillus salivarius strain NCIMB702343 groEL; DQ444339.1 | 99 | 99.94 | ||||
| P16a | L. salivarius 2.367 | MT862780 | Lactobacillus salivarius CECT 5713; CP002034.1 | 100 | 99.94 | ND |
| Lactobacillus salivarius strain NCIMB702343 groEL; DQ444339.1 | 100 | 99.94 | ||||
| P18a | L. salivarius 2.228 | MT862781 | Ligilactobacillus salivarius strain BCRC 14759; CP024067.1 | 100 | 99.94 | ND |
| Ligilactobacillus salivarius strain ZLS006; CP020858.1 | 100 | 99.94 | ||||
| P21a | L. salivarius 2.023 | MT862782 | Lactobacillus salivarius CECT 5713; CP002034.1 | 100 | 99.94 | ND |
| Lactobacillus salivarius strain NCIMB702343 groEL; DQ444339.1 | 99 | 99.94 | ||||
| P23a | L. salivarius 2.322 | MT862783 | Lactobacillus salivarius CECT 5713; CP002034.1 | 100 | 99.81 | ND |
| Lactobacillus salivarius strain NCIMB702343 groEL; DQ444339.1 | 99 | 99.81 | ||||
| Ch4a | L. salivarius 2.118 | MT862784 | Lactobacillus salivarius strain CCUG44481 groEL; DQ444352.1 | 100 | 99.75 | ND |
| Ligilactobacillus salivarius strain CICC 23174; CP017107.1 | 100 | 99.68 | ||||
| Ch8b | L. salivarius 2.168 | MT862785 | Lactobacillus salivarius strain CCUG44481 groEL; DQ444352.1 | 100 | 99.81 | L. salivarius [3,28] |
| Ligilactobacillus salivarius strain CICC 23174; CP017107.1 | 100 | 99.75 | ||||
| Ch9b | L. salivarius 2.264 | MT862786 | Lactobacillus salivarius strain CCUG44481 groEL; DQ444352.1 | 100 | 99.87 | ND |
| Ligilactobacillus salivarius strain CICC 23174; CP017107.1 | 100 | 99.81 | ||||
| Ch10a | L. salivarius 2.006 | MT862787 | Lactobacillus salivarius strain CCUG44481 groEL; DQ444352.1 | 100 | 99.87 | ND |
| Ligilactobacillus salivarius strain CICC 23174; CP017107.1 | 100 | 99.81 | ||||
| Ch10d | L. salivarius 2.249 | MT862788 | Lactobacillus salivarius strain CCUG44481 groEL; DQ444352.1 | 100 | 98.66 | L. salivarius [3,28] |
| Ligilactobacillus salivarius strain CICC 23174; CP017107.1 | 100 | 98.60 | ||||
| Ch24b | L. salivarius 2.080 | MT862789 | Lactobacillus salivarius strain CCUG44481 groEL; DQ444352.1 | 100 | 99.68 | L. salivarius [3,28] |
| Ligilactobacillus salivarius strain CICC 23174; CP017107.1 | 100 | 99.62 | ||||
| Ch37b | L. salivarius 2.324 | MT862790 | Lactobacillus salivarius strain CCUG44481 groEL; DQ444352.1 | 100 | 99.43 | ND |
| Ligilactobacillus salivarius strain CICC 23174; CP017107.1 | 100 | 99.43 | ||||
| Ch40a | L. salivarius 2.001 | MT920907 | Lactobacillus salivarius strain CCUG44481 groEL; DQ444352.1 | 100 | 99.62 | ND |
| Ligilactobacillus salivarius strain CICC 23174; CP017107.1 | 100 | 99.55 | ||||
| Ch50b | L. salivarius 2.241 | MT862791 | Ligilactobacillus salivarius strain 2D; CP047412.1 | 100 | 100 | L. salivarius [3,28] |
| Lactobacillus salivarius str. Ren Select; CP011403.1 | 100 | 99.94 | ||||
| Ch50d | L. salivarius 2.124 | MT862792 | Lactobacillus salivarius strain CCUG44481 groEL; DQ444352.1 | 100 | 99.75 | ND |
| Ligilactobacillus salivarius strain CICC 23174; CP017107.1 | 100 | 99.55 | ||||
| T2a | L. salivarius 2.010 | MT862793 | Lactobacillus salivarius strain CCUG44481 groEL; DQ444352.1 | 100 | 99.81 | ND |
| Ligilactobacillus salivarius strain CICC 23174; CP017107.1 | 100 | 99.75 | ||||
| T3a | L. salivarius 2.164 | MT862794 | Lactobacillus salivarius strain CCUG44481 groEL; DQ444352.1 | 100 | 99.81 | ND |
| Ligilactobacillus salivarius strain CICC 23174; CP017107.1 | 100 | 99.75 | ||||
| T7c | L. salivarius 2.002 | MT862795 | Lactobacillus salivarius strain CCUG44481 groEL; DQ444352.1 | 100 | 99.75 | ND |
| Ligilactobacillus salivarius strain CICC 23174; CP017107.1 | 100 | 99.68 | ||||
| T17f | L. salivarius 2.311 | MT862796 | Ligilactobacillus salivarius strain CICC 23174; CP017107.1 | 100 | 99.81 | ND |
| Lactobacillus salivarius strain CCUG44481 groEL; DQ444352.1 | 100 | 99.75 | ||||
| T18a | L. salivarius 2.390 | MT862797 | Lactobacillus salivarius strain CCUG44481 groEL; DQ444352.1 | 100 | 99.81 | ND |
| Ligilactobacillus salivarius strain CICC 23174; CP017107.1 | 100 | 99.75 | ||||
| T21a | L. salivarius 2.003 | MT862798 | Lactobacillus salivarius strain CCUG44481 groEL; DQ444352.1 | 100 | 99.49 | ND |
| Ligilactobacillus salivarius strain CICC 23174; CP017107.1 | 100 | 99.43 | ||||
| T22a | L. salivarius 2.136 | MT862799 | Lactobacillus salivarius strain CCUG44481 groEL; DQ444352.1 | 100 | 99.81 | ND |
| Ligilactobacillus salivarius strain CICC 23174; CP017107.1 | 100 | 99.75 | ||||
| T27 | L. salivarius 2.400 | MT862800 | Lactobacillus salivarius strain CCUG44481 groEL; DQ444352.1 | 100 | 99.75 | ND |
| Ligilactobacillus salivarius strain CICC 23174; CP017107.1 | 100 | 99.68 | ||||
| T31a | L. salivarius 2.241 | MT862801 | Lactobacillus salivarius strain CCUG44481 groEL; DQ444352.1 | 100 | 99.68 | ND |
| Ligilactobacillus salivarius strain CICC 23174; CP017107.1 | 100 | 99.62 | ||||
| G2K | L. salivarius 2.106 | MT862802 | Lactobacillus salivarius strain JCM1230 groEL; DQ444335.1 | 100 | 99.55 | L. salivarius [3,28] |
| Lactobacillus salivarius isolate LPM01 genome; LT604074.1 | 100 | 99.49 | ||||
| G19a | L. salivarius 2.139 | MT862803 | Ligilactobacillus salivarius strain 2D; CP047412.1 | 100 | 100 | L. salivarius [3,28] |
| Lactobacillus salivarius str. Ren Select; CP011403.1 | 100 | 99.94 | ||||
| G24a | L. salivarius 2.273 | MT862804 | Lactobacillus salivarius strain CCUG44481 groEL; DQ444352.1 | 100 | 99.68 | L. salivarius [3] |
| Ligilactobacillus salivarius strain CICC 23174; CP017107.1 | 100 | 99.62 | ||||
| G24b | L. salivarius 2.305 | MT862805 | Lactobacillus salivarius strain CCUG44481 groEL; DQ444352.1 | 100 | 99.87 | L. salivarius [3] |
| Ligilactobacillus salivarius strain CICC 23174; CP017107.1 | 100 | 99.81 | ||||
| G31a | L. salivarius 2.211 | MT862806 | Ligilactobacillus salivarius strain 2D; CP047412.1 | 100 | 100 | L. salivarius [3] |
| Lactobacillus salivarius str. Ren Select; CP011403.1 | 100 | 99.94 | ||||
| G39a | L. salivarius 2.029 | MT862807 | Lactobacillus salivarius strain CCUG44481 groEL; DQ444352.1 | 100 | 99.81 | L. salivarius [3] |
| Ligilactobacillus salivarius strain CICC 23174; CP017107.1 | 100 | 99.75 | ||||
| G50b | L. salivarius 2.135 | MT862808 | Lactobacillus salivarius strain CCUG44481 groEL; DQ444352.1 | 100 | 99.75 | L. salivarius [3] |
| Ligilactobacillus salivarius strain CICC 23174; CP017107.1 | 100 | 99.68 |

Table A2.
Carbohydrate fermentation profiles of L. salivarius strains: ARA—arabinosel; RIB—ribose; XYL—xylose; GAL—galactose; GLU—glucose; FRU—fructose; MNE—mannose; RHA—rhamnose; MAN—mannitol; SOR—sorbitol; NAG-N—acethylglucosamine; AMY—amygdalin; SAL—salicin; CEL—cellobiose; MAL—maltose; LAC—lactose; MEL—melibiose; SAC—sacharose; TRE—trehalose; INU—inulin; MLZ—Melezitose; RAF—raffinose; XLT—xylitol; FUC—fucose. Symbols indicate the following: −, negative reaction; ±, weak rection; +, positive reaction after 24, 48 and 72 h, respectively.
Table A2.
Carbohydrate fermentation profiles of L. salivarius strains: ARA—arabinosel; RIB—ribose; XYL—xylose; GAL—galactose; GLU—glucose; FRU—fructose; MNE—mannose; RHA—rhamnose; MAN—mannitol; SOR—sorbitol; NAG-N—acethylglucosamine; AMY—amygdalin; SAL—salicin; CEL—cellobiose; MAL—maltose; LAC—lactose; MEL—melibiose; SAC—sacharose; TRE—trehalose; INU—inulin; MLZ—Melezitose; RAF—raffinose; XLT—xylitol; FUC—fucose. Symbols indicate the following: −, negative reaction; ±, weak rection; +, positive reaction after 24, 48 and 72 h, respectively.
| Pentoses | Hexoses | 6-Deoxy Hexoses | Sugar Alcohols | Disaccharides | Trisaccharides | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbohydrate► | ARA | RIB | XYL | GAL | GLU | FRU | MNE | FUC | RHA | XLT | MAN | SOR | NAG | AMY | SAL | CEL | MAL | LAC | MEL | SAC | TRE | INU | MLZ | RAF | |
| Ref. | LMG9476 | −−− | −−− | −−− | +++ | +++ | +++ | +++ | −−− | −−− | −−− | +++ | +++ | +++ | −−− | +++ | −−− | +++ | +++ | +++ | +++ | +++ | −−− | −−− | +++ |
| LMG9477 | −−− | −−− | −−− | +++ | +++ | +++ | +++ | −−− | +++ | −−− | +++ | +++ | +++ | −−− | −−− | −−− | +++ | +++ | +++ | +++ | +++ | −−− | −−− | +++ | |
| pigeons | P2a | −−− | −−− | −−− | +++ | +++ | +++ | +++ | −−− | +++ | −−− | +++ | −−− | +++ | −−− | −−− | −−− | +++ | +++ | +++ | +++ | +++ | −−− | −−− | +++ |
| P3a | −−− | −−− | −−− | +++ | +++ | +++ | +++ | −−− | +++ | −−− | +++ | −−− | +++ | −−− | −−− | −−− | +++ | +++ | +++ | +++ | +++ | +++ | −−− | +++ | |
| P 8a | −−− | −−− | −−− | +++ | +++ | +++ | +++ | −−− | ±++ | −−− | +++ | −−− | ±++ | −−− | −−− | −−− | +++ | +++ | +++ | +++ | −−− | +++ | −−− | +++ | |
| P 16a | −−− | −−− | −−− | +++ | +++ | +++ | +++ | −−− | ±++ | −−− | +++ | ±++ | +++ | −−− | −−− | −−− | +++ | +++ | +++ | +++ | −−+ | +++ | −−− | +++ | |
| P 18a | −−− | −−− | −−− | +++ | +++ | +++ | +++ | −−− | ±++ | −−− | +++ | ±++ | ±++ | −−− | −−− | −−− | +++ | +++ | +++ | +++ | +++ | −−− | −−− | +++ | |
| P 21a | −−− | −−− | −−− | +++ | +++ | +++ | +++ | −−− | ±++ | −−− | ±++ | −−− | ±++ | −−− | −−− | −−− | +++ | +++ | +++ | +++ | +++ | +++ | −−− | +++ | |
| P 23a | −−− | −−− | −−− | +++ | +++ | +++ | +++ | −−− | −−− | −−− | +++ | +++ | +++ | −−− | −−− | −−− | +++ | +++ | +++ | +++ | +++ | +++ | −−− | +++ | |
| Total [%] | 0 | 0 | 0 | 100 | 100 | 100 | 100 | 0 | 86 | 0 | 100 | 43 | 100 | 0 | 0 | 0 | 100 | 100 | 100 | 100 | 86 | 71 | 0 | 100 | |
| chicken | Ch 4a | −−− | −−− | −−− | +++ | +++ | +++ | +++ | −−− | −−− | −−− | −++ | −−− | −++ | −−− | −−− | −−− | −−− | −++ | −++ | −++ | −−− | −−− | −−− | −++ |
| Ch 8b | −−− | −−− | −−− | −++ | +++ | +++ | +++ | −−− | −−− | −−− | ±++ | −−− | +++ | −−− | −−− | −−− | +++ | +++ | +++ | +++ | −−− | −−− | −−− | +++ | |
| Ch 9b | −−− | −−− | −−− | +++ | +++ | +++ | +++ | −−− | −−− | −−− | +++ | −−− | +++ | −−− | −−− | −−− | +++ | +++ | +++ | +++ | +++ | −−− | −−− | +++ | |
| Ch 10a | −−− | −−− | −−− | +++ | +++ | +++ | +++ | −−− | −−− | −−− | +++ | +++ | +++ | −−− | −−− | −−− | +++ | +++ | +++ | +++ | −−− | −−− | −−− | +++ | |
| Ch10d | −−− | −−− | −−− | +++ | +++ | +++ | +++ | −−− | −−− | −−− | +++ | +++ | +++ | −−− | −−− | −−− | +++ | +++ | +++ | +++ | −−− | −−− | −−− | +++ | |
| Ch 24b | −−− | −−− | −−− | +++ | +++ | +++ | +++ | −−− | −−− | −−− | +++ | −−± | +++ | −−− | −−− | −−− | +++ | +++ | +++ | +++ | +++ | −−− | −−− | +++ | |
| Ch 37b | −−− | −−− | −−− | +++ | +++ | +++ | +++ | −−− | −−− | −−− | +++ | +++ | +++ | −−− | −−− | −−− | +++ | +++ | +++ | +++ | +++ | −−− | −−− | +++ | |
| Ch 40a | −−− | −−− | −−− | +++ | +++ | +++ | +++ | −−− | −−− | −−− | +++ | +++ | +++ | −−− | −−− | −−− | +++ | +++ | +++ | +++ | +++ | −−− | −−− | +++ | |
| Ch 50b | −−− | −−− | −−− | +++ | +++ | +++ | +++ | −−− | ±++ | −−− | +++ | −−− | +++ | −−− | +++ | −−− | +++ | +++ | +++ | +++ | +++ | −−− | −−− | +++ | |
| Ch 50d | −−− | −−− | −−− | +++ | +++ | +++ | +++ | −−− | −−− | −−− | +++ | −−± | +++ | −−− | −−− | −−− | +++ | +++ | +++ | +++ | +++ | −−− | −−− | +++ | |
| Total: [%] | 0 | 0 | 0 | 100 | 100 | 100 | 100 | 0 | 10 | 0 | 100 | 50 | 100 | 0 | 10 | 0 | 90 | 100 | 100 | 100 | 60 | 0 | 0 | 100 | |
| turkey | T 2a | −−− | −−− | −−− | +++ | +++ | +++ | +++ | −−− | −−− | −−− | +++ | −−± | +++ | −−− | −−− | −−− | +++ | +++ | +++ | +++ | +++ | −−− | −−− | +++ |
| T 3a | −−− | −−− | −−− | +++ | +++ | +++ | +++ | −−− | −−− | −−− | −++ | −−− | −++ | −−− | −−− | −−− | −++ | −++ | −++ | −++ | −−− | −−− | −−− | −++ | |
| T 7c | −−− | −−− | −−− | +++ | +++ | +++ | +++ | −−− | −−− | −−− | +++ | −−− | −−− | −−− | −−− | −−− | +++ | +++ | +++ | +++ | −−− | −−− | −−− | +++ | |
| T 17f | −−− | −−− | −−− | +++ | +++ | +++ | +++ | −−− | −−− | −−− | +++ | −−− | −−− | −−− | −−− | −−− | +++ | +++ | +++ | +++ | −−− | −−− | −−− | +++ | |
| T 18a | −−− | −−− | −−− | +++ | +++ | +++ | +++ | −−− | −−− | −−− | +++ | +++ | −−− | −−− | −−− | −−− | +++ | +++ | +++ | +++ | −−− | −−− | −−− | +++ | |
| T 21a | −−− | −−− | −−− | +++ | +++ | +++ | +++ | −−− | −−− | −−− | −−± | −−− | −−± | −−− | −−− | −−− | +++ | ±±+ | +++ | +++ | −−− | −−− | −−− | +++ | |
| T 22a | −−− | −−− | −−− | +++ | +++ | +++ | +++ | −−− | −−− | −−− | +++ | −−− | −−− | −−− | −−− | −−− | ±++ | −++ | −++ | +++ | −−− | −−− | −−− | −−− | |
| T 27 | −−− | −−− | −−− | +++ | +++ | +++ | +++ | −−− | −−− | −−− | +++ | −−− | +++ | −−− | −−− | −−− | +++ | +++ | +++ | +++ | −−− | −−− | −−− | +++ | |
| T 31a | −−− | −−− | −−− | +++ | +++ | +++ | +++ | −−− | −−− | −−− | +++ | +++ | −−− | −−− | −−− | −−− | −−± | +++ | +++ | +++ | −−− | −−− | −−− | +++ | |
| Total: [%] | 0 | 0 | 0 | 100 | 100 | 100 | 100 | 0 | 0 | 0 | 89 | 22 | 33 | 0 | 0 | 0 | 89 | 100 | 100 | 100 | 11 | 0 | 0 | 89 | |
| gees | G 2K | −−− | −−− | −−− | +++ | +++ | +++ | +++ | −−− | −−− | −−− | +++ | +++ | +++ | +++ | +++ | +++ | +++ | −−± | −++ | +++ | +++ | −−− | +++ | −−− |
| G 19a | −−− | −−− | −−− | +++ | +++ | +++ | +++ | −−− | +++ | −−− | +++ | −−± | +++ | −−− | −−− | −−− | +++ | +++ | ±++ | +++ | +++ | −−− | −−− | +++ | |
| G 24a | −−− | −−− | −−− | +++ | +++ | +++ | +++ | −−− | −−− | −−− | +++ | +++ | +++ | −−± | −−− | −−− | +++ | +++ | +++ | +++ | +++ | −−− | −−− | +++ | |
| G 24b | −−− | −−− | −−− | +++ | +++ | +++ | +++ | −−− | −−− | −−− | +++ | +++ | +++ | −−+ | +++ | −−− | +++ | +++ | +++ | +++ | +++ | −−− | −−− | +++ | |
| G 31a | −−− | −−− | −−− | +++ | +++ | +++ | +++ | −−− | −−− | −−− | +++ | +++ | +++ | −−− | −−− | −−− | +++ | +++ | +++ | +++ | +++ | −−− | −−− | +++ | |
| G 39a | −−− | −−− | −−− | +++ | +++ | +++ | +++ | −−− | −−− | −−− | +++ | +++ | +++ | −−− | −−− | −−− | +++ | +++ | +++ | +++ | +++ | −−− | −−− | +++ | |
| G 50b | −−− | −−− | −−− | +++ | +++ | +++ | +++ | −−− | −−− | −−− | +++ | +++ | +++ | −−− | +++ | −−− | +++ | +++ | +++ | +++ | +++ | −−− | −−− | +++ | |
| Total: [%] | 0 | 0 | 0 | 100 | 100 | 100 | 100 | 0 | 14 | 0 | 100 | 86 | 100 | 14 | 43 | 14 | 100 | 86 | 100 | 100 | 100 | 0 | 14 | 86 | |
| Total: [%] | 0 | 0 | 0 | 100 | 100 | 100 | 100 | 0 | 24 | 0 | 97 | 48 | 82 | 3 | 12 | 3 | 94 | 97 | 100 | 100 | 61 | 15 | 3 | 94 | |

Table A3.
Analysis of DNA sequence and putative protein sequences of Abp118 bacteriocin.
Table A3.
Analysis of DNA sequence and putative protein sequences of Abp118 bacteriocin.
 |

Table A4.
The ability of avian L. salivarius strains to grow on MRS broth supplemented with 1% or 2% ox gall after 24 h and 48 h.
Table A4.
The ability of avian L. salivarius strains to grow on MRS broth supplemented with 1% or 2% ox gall after 24 h and 48 h.
| Isolate | Host Source | 1% Bile | 1% Bile | 2% Bile | 2% Bile |
|---|---|---|---|---|---|
| 24 h | 48 h | 24 h | 48 h | ||
| LMG 9476 | human | <3% | 5% | <3% | 6% |
| LMG 9477 | human | <3% | 5% | <3% | 4% |
| P2a | pigeon | 9% | 9% | 9% | 11% |
| P3a | pigeon | 8% | 9% | 8% | 9% |
| P8a | pigeon | 4% | 8% | <3% | 7% |
| P16a | pigeon | 7% | 8% | 4% | 12% |
| P18a | pigeon | 9% | 7% | 8% | 8% |
| P21a | pigeon | <3% | 6% | <3% | 6% |
| P23a | pigeon | 6% | 6% | 6% | 6% |
| Ch4a | chicken | 4% | 7% | <3% | 5% |
| Ch8b | chicken | <3% | <3% | <3% | <3% |
| Ch9b | chicken | <3% | <3% | <3% | <3% |
| Ch10a | chicken | <3% | <3% | <3% | <3% |
| Ch10d | chicken | <3% | <3% | <3% | <3% |
| Ch24b | chicken | <3% | <3% | <3% | <3% |
| Ch37b | chicken | 4.5% | 13% | 3.5% | 12% |
| Ch40a | chicken | 26% | 38% | 24% | 37% |
| Ch50b | chicken | <3% | 10% | 5% | 10% |
| Ch50d | chicken | 8% | 6% | 5% | 4.5% |
| T2a | turkey | 5% | 4.5% | 4% | 4% |
| T3a | turkey | 14% | 6% | 8% | 6% |
| T7c | turkey | <3% | <3% | <3% | <3% |
| T17f | turkey | <3% | 4% | <3% | <3% |
| T18a | turkey | <3% | <3% | <3% | <3% |
| T21a | turkey | <3% | <3% | <3% | <3% |
| T22a | turkey | <3% | <3% | <3% | <3% |
| T27 | turkey | <3% | 10% | <3% | 3.5% |
| T31a | turkey | 5.5% | 10% | <3% | 6% |
| G2K | goose | 100% | 100% | 100% | 100% |
| G19a | goose | 13% | 13% | 13% | 17% |
| G24a | goose | 13% | 15% | 13% | 16% |
| G24b | goose | 11% | 11% | 9% | 10% |
| G31a | goose | 14.5% | 17% | 13% | 17% |
| G39a | goose | 4.5% | 6% | <3% | 7% |
| G 50b | goose | 8% | 11% | 7% | 13% |
References
- Rogosa, M.; Wiseman, R.F.; Mitchell, J.A.; Disraely, M.N.; Beaman, A.J. Species differentiation of oral lactobacilli from man including description of Lactobacillus salivarius nov. spec. and Lactobacillus cellobiosus nov. spec. J. Bacteriol. 1953, 65, 681–699. [Google Scholar] [CrossRef]
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.M.A.P.; Harris, H.M.B.; Mattarelli, P.; O’Toole, P.W.; Pot, B.; Vandamme, P.; Walter, J.; et al. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef] [PubMed]
- Dec, M.; Urban-Chmiel, R.; Gnat, S.; Puchalski, A.; Wernicki, A. Identification of Lactobacillus strains of goose origin using MALDI-TOF mass spectrometry and 16S-23S rDNA intergenic spacer PCR analysis. Res. Microbiol. 2014, 165, 190–201. [Google Scholar] [CrossRef] [PubMed]
- Dec, M.; Puchalski, A.; Nowaczek, A.; Wernicki, A. Antimicrobial activity of Lactobacillus strains of chicken origin against bacterial pathogens. Int. Microbiol. 2016, 19, 57–67. [Google Scholar]
- Dec, M.; Nowaczek, A.; Stepień-Pyśniak, D.; Wawrzykowski, J.; Urban-Chmiel, R. Identification and antibiotic susceptibility of lactobacilli isolated from turkeys. BMC Microbiol. 2018, 18, 168. [Google Scholar] [CrossRef]
- Dec, M.; Stępień-Pyśniak, D.; Nowaczek, A.; Puchalski, A.; Urban-Chmiel, R. Phenotypic and genotypic antimicrobial resistance profiles of fecal lactobacilli from domesticated pigeons in Poland. Anaerobe 2020, 65, 102251. [Google Scholar] [CrossRef]
- Ehrmann, M.A.; Kurzak, P.; Bauer, J.; Vogel, R.F. Characterization of lactobacilli towards their use as probiotic adjuncts in poultry. J. Appl. Microbiol. 2002, 92, 966–975. [Google Scholar] [CrossRef]
- Lee, J.Y.; Han, G.G.; Kim, E.B.; Choi, Y.J. Comparative genomics of Lactobacillus salivarius strains focusing on their host adaptation. Microbiol. Res. 2017, 205, 48–58. [Google Scholar] [CrossRef]
- Adetoye, A.; Pinloche, E.; Adeniyi, B.A.; Ayeni, F.A. Characterization and anti-salmonella activities of lactic acid bacteria isolated from cattle faeces. BMC Microbiol. 2018, 18, 96. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.C.; Ptak, C.P.; Chang, C.Y.; Ian, M.K.; Chia, M.Y.; Chen, T.H.; Kuo, C.J. Autochthonous Lactic Acid Bacteria Isolated from Dairy Cow Feces Exhibiting Promising Probiotic Properties and in vitro Antibacterial Activity Against Foodborne Pathogens in Cattle. Front. Vet. Sci. 2020, 7, 239. [Google Scholar] [CrossRef]
- Obata, J.; Takeshita, T.; Shibata, Y.; Yamanaka, W.; Unemori, M.; Akamine, A.; Yamashita, Y. Identification of the microbiota in carious dentin lesions using 16S rRNA gene sequencing. PLoS ONE 2014, 9, e103712. [Google Scholar] [CrossRef] [PubMed]
- Pino, A.; Bartolo, E.; Caggia, C.; Cianci, A.; Randazzo, C.L. Detection of vaginal lactobacilli as probiotic candidates. Sci. Rep. 2019, 9, 3355. [Google Scholar] [CrossRef]
- Soto, A.; Martín, V.; Jiménez, E.; Mader, I.; Rodríguez, J.M.; Fernández, L. Lactobacilli and bifidobacteria in human breast milk: Influence of antibiotherapy and other host and clinical factors. J. Pediatr. Gastroenterol. Nutr. 2014, 59, 78–88. [Google Scholar] [CrossRef]
- Audisio, M.C.; Albarracín, L.; Torres, M.J.; Saavedra, L.; Hebert, E.M.; Villena, J. Draft genome sequences of Lactobacillus salivarius A3iob and Lactobacillus johnsonii CRL1647, novel potential probiotic strains for honeybees (Apis mellifera L.). Microbiol. Resour. Announc. 2018, 7, e00975-18. [Google Scholar] [CrossRef]
- Kačániová, M.; Hleba, L.; Pochop, J.; Kádasi-Horáková, M.; Fikselová, M.; Rovná, K. Determination of wine microbiota using classical method, polymerase chain method and Step One Real-Time PCR during fermentation process. J. Environ. Sci. Health B 2012, 47, 571–578. [Google Scholar] [CrossRef]
- Luo, Z.; Gasasira, V.; Huang, Y.; Liu, D.; Yang, X.; Jiang, S.; Hu, W. Effect of Lactobacillus salivarius H strain isolated from Chinese dry-cured ham on the color stability of fresh pork. Food Sci. Hum. Wellness 2013, 2, 139–145. [Google Scholar] [CrossRef]
- Public Databases for Molecular Typing and Microbial Genome Diversity (PubMLST). Available online: https://pubmlst.org/ (accessed on 3 March 2021).
- Li, Y.; Raftis, E.; Canchaya, C.; Fitzgerald, G.F.; van Sinderen, D.; O’Toole, P.W. Polyphasic analysis indicates that Lactobacillus salivarius subsp. salivarius and Lactobacillus salivarius subsp. salicinius do not merit separate subspecies status. Int. J. Sys. Evol. Microbiol. 2006, 56, 2397–2403. [Google Scholar]
- Sun, Z.; Yu, J.; Dan, T.; Zhang, W.; Zhang, H. Phylogenesis and evolution of lactic acid bacteria. In Lactic Acid Bacteria. Fundamentals and Practice; Zhang, H., Cai, Y., Eds.; Springer Science + Business Media: Dordrecht, The Netherlands, 2014; pp. 1–101. [Google Scholar]
- Harris, H.M.B.; Bourin, M.J.B.; Claesson, M.J.; O’Toole, P.W. Phylogenomics and comparative genomics of Lactobacillus salivarius, a mammalian gut commensal. Microb. Genom. 2017, 3, e000115. [Google Scholar] [CrossRef] [PubMed]
- EFSA. Update of the list of QPS-recommended biological agents intentionally added to food or feed as notified to EFSA 10: Suitability of taxonomic units notified to EFSA until March. EFSA J. 2019, 17, 5753. [Google Scholar]
- Messaoudi, S.; Manai, M.; Kergourlay, G.; Prévost, H.; Connil, N.; Chobert, J.-M.; Dousset, X. Lactobacillus salivarius: Bacteriocin and probiotic activity. Food Microbiol. 2013, 36, 296–304. [Google Scholar] [PubMed]
- Dec, M.; Nowaczek, A.; Urban-Chmiel, R.; Stępień-Pyśniak, D.; Wernicki, A. Probiotic potential of Lactobacillus isolates of chicken origin with anti-Campylobacter activity. J. Vet. Med. Sci. 2018, 80, 1195–1203. [Google Scholar] [CrossRef]
- Dec, M.; Puchalski, A.; Urban-Chmiel, R.; Wernicki, A. Screening of Lactobacillus strains of domestic goose origin against bacterial poultry pathogens for use as probiotics. Poult. Sci. 2014, 93, 2464–2472. [Google Scholar] [CrossRef]
- Pascual, M.; Hugas, M.; Badiola, J.I.; Monfort, J.M.; Garriga, M. Lactobacillus salivarius CTC2197 prevents Salmonella enteritidis colonization in chickens. Appl. Environ. Microbiol. 1999, 65, 4981–4986. [Google Scholar] [CrossRef]
- Neville, B.A.; O’Toole, P.W. Probiotic properties of Lactobacillus salivarius and closely related Lactobacillus species. Future Microbiol. 2010, 5, 759–774. [Google Scholar] [CrossRef]
- Dudzic, A.; Urban-Chmiel, R.; Stępień-Pyśniak, D.; Dec, M.; Puchalski, A.; Wernicki, A. Isolation, identification and antibiotic resistance of Campylobacter strains isolated from domestic and free-living pigeons. Br. Poult. Sci. 2016, 57, 172–178. [Google Scholar] [CrossRef]
- Dec, M.; Puchalski, A.; Urban-Chmiel, R.; Wernicki, A. 16S-ARDRA and MALDI-TOF mass spectrometry as tools for identification of Lactobacillus bacteria isolated from poultry. BMC Microbiol. 2016, 16, 105. [Google Scholar] [CrossRef]
- Hedberg, M.; Hasslöf, P.; Sjöström, I.; Twetman, S.; Stecksén-Blicks, C. Sugar fermentation in probiotic bacteria—An in vitro study. Oral Microbiol. Immunol. 2008, 23, 482–485. [Google Scholar] [CrossRef]
- Li, Y.; Canchaya, C.; Fang, F.; Raftis, E.; Ryan, K.A.; van Pijkeren, J.P.; van Sinderen, D.; O’Toole, P.W. Distribution of megaplasmids in Lactobacillus salivarius and other lactobacilli. J. Bacteriol. 2007, 189, 6128–6139. [Google Scholar] [CrossRef]
- Open Reading Frame Finder (ORFinder). Available online: https://www.ncbi.nlm.nih.gov/orffinder/ (accessed on 3 March 2021).
- GenBank. Available online: https://www.ncbi.nlm.nih.gov/genbank/ (accessed on 3 March 2021).
- EFSA. Guidance on the characterisation of microorganisms used as feed additives or as production organisms. EFSA J. 2018, 16, 5206. [Google Scholar]
- Technical Library of bioMérieux. Available online: https://techlib.biomerieux.com/ (accessed on 3 March 2021).
- Alverdy, J.; Zaborina, O.; Wu, L. The impact of stress and nutrition on bacterial–host interactions at the intestinal epithelial surface. Curr. Opin. Clin. Nutr. Metab. Care 2005, 8, 205–209. [Google Scholar] [CrossRef] [PubMed]
- Hooper, L.V.; Midtvedt, T.; Gordon, J.I. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu. Rev. Nutr. 2002, 22, 283–307. [Google Scholar] [CrossRef]
- Teng, P.Y.; Kim, W.K. Review: Roles of Prebiotics in Intestinal Ecosystem of Broilers. Front. Vet. Sci. 2018, 5, 245. [Google Scholar] [CrossRef]
- Buntin, N.; Hongpattarakere, T.; Douillard, J.R.F.P.; Paulin, L.; Boeren, S.; Shetty, S.A.; de Vos, W.M. An inducible operon is involved in inulin utilization in Lactobacillus plantarum strains, as revealed by comparative proteogenomics and metabolic profiling. Appl. Environ. Microbiol. 2016, 83, e02402-16. [Google Scholar] [CrossRef]
- Xie, M.; Pan, M.; Jiang, Y.; Liu, X.; Lu, W.; Zhao, J.; Zhang, H.; Chen, W. groEL Gene-Based Phylogenetic Analysis of Lactobacillus Species by High-Throughput Sequencing. Genes 2019, 10, 530. [Google Scholar] [CrossRef]
- Hung, W.W.; Chen, Y.H.; Tseng, S.P.; Jao, Y.T.; Teng, L.J.; Hung, W.C. Using groEL as the target for identification of Enterococcus faecium clades and 7 clinically relevant Enterococcus species. J. Microbiol. Immunol. Infect. 2019, 52, 255–264. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Lu, W.; Wang, L.; Pan, M.; Zhang, H.; Zhao, J.; Chen, W. Assessment of Bifidobacterium Species Using groEL Gene on the Basis of Illumina MiSeq High-Throughput Sequencing. Genes 2017, 8, 336. [Google Scholar] [CrossRef]
- Alp, G.; Aslim, B. Relationship between the resistance to bile salts and low pH with exopolysaccharide (EPS) production of Bifidobacterium spp. isolated from infants feces and breast milk. Anaerobe 2010, 16, 101–105. [Google Scholar] [CrossRef]
- Caggianiello, G.; Kleerebezem, M.; Spano, G. Exopolysaccharides produced by lactic acid bacteria: From health-promoting benefits to stress tolerance mechanisms. Appl. Microbiol. Biotechnol. 2016, 100, 3877–3886. [Google Scholar] [CrossRef]
- Oleksy, M.; Klewicka, E. Exopolysaccharides produced by Lactobacillus sp.: Biosynthesis and applications. Crit. Rev. Food Sci. Nutr. 2018, 58, 450–462. [Google Scholar] [CrossRef]
- Tsuneda, S.; Aikawa, H.; Hayashi, H.; Yuasa, A.; Hirata, A. Extracellular polymeric substances responsible for bacterial adhesion onto solid surface. FEMS Microbiol. Lett. 2003, 223, 287–292. [Google Scholar] [CrossRef]
- Chapot-Chartier, M.P.; Vinogradov, E.; Sadovskaya, I.; Andre, G.; Mistou, M.Y.; Trieu-Cuot, P.; Sylviane Furlan, S.; Elena Bidnenko, E.; Courtin, P.; Péchoux, C.; et al. Cell Surface of Lactococcus lactis is covered by a protective polysaccharide pellicle. J. Biol. Chem. 2010, 285, 10464–10471. [Google Scholar] [CrossRef] [PubMed]
- Patten, D.A.; Laws, A.P. Lactobacillus-produced exopolysaccharides and their potential health benefits: A review. Benef. Microbes 2015, 6, 457–471. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, T.; Simsek, Ö. Potential health benefits of ropy exopolysaccharides produced by Lactobacillus plantarum. Molecules 2020, 25, 3293. [Google Scholar] [CrossRef]
- Terraf, M.C.; Juárez Tomás, M.S.; Nader-Macías, M.E.; Silva, C. Screening of biofilm formation by beneficial vaginal lactobacilli and influence of culture media components. J. Appl. Microbiol. 2012, 113, 1517–1529. [Google Scholar] [CrossRef]
- Trunk, T.; Khalil, H.S.; Leo, J.C. Bacterial autoaggregation. AIMS Microbiol. 2018, 4, 140–164. [Google Scholar] [CrossRef]
- Kos, B.; Susković, J.; Vuković, S.; Simpraga, M.; Frece, J.; Matosić, S. Adhesion and aggregation ability of probiotic strain Lactobacillus acidophilus M92. J. Appl. Microbiol. 2003, 94, 981–987. [Google Scholar] [CrossRef]
- Leivers, S.; Hidalgo-Cantabrana, C.; Robinson, G.; Margolles, A.; Ruas-Madiedo, P. Structure of the high molecular weight exopolysaccharide produced by Bifidobacterium animalis subsp. lactis IPLA-R1 and sequence analysis of its putative eps cluster. Carbohydr. Res. 2011, 346, 2710–2717. [Google Scholar] [CrossRef]
- Ruiz, L.; Margolles, A.; Sánchez, B. Bile resistance mechanisms in Lactobacillus and Bifidobacterium. Front. Microbiol. 2013, 4, 396. [Google Scholar] [CrossRef]
- Burns, P.; Sánchez, B.; Vinderola, G.; Ruas-Madiedo, P.; Ruiz, L.; Margolles, A.; Reinheimer, J.; de los Reyes-Gavilán, C.G. Inside the adaptation process of Lactobacillus delbrueckii subsp. lactis to bile. Int. J. Food Microbiol. 2010, 142, 132–141. [Google Scholar] [CrossRef] [PubMed]
- Barrett, E.; Hayes, M.; O’Connor, P.; Gardiner, G.; Fitzgerald, G.F.; Stanton, C.; Ross, R.P.; Hill, C. Salivaricin P, one of a family of two-component antilisterial bacteriocins produced by intestinal isolates of Lactobacillus salivarius. Appl. Environ. Microbiol. 2007, 73, 3719–3723. [Google Scholar] [CrossRef]
- Dec, M.; Urban-Chmiel, R.; Stępień-Pyśniak, D.; Wernicki, A. Assessment of antibiotic susceptibility in Lactobacillus isolates from chickens. Gut Pathog. 2017, 9, 54. [Google Scholar] [CrossRef] [PubMed]
- Dec, M.; Wernicki, A.; Puchalski, A.; Urban-Chmiel, R. Antibiotic susceptibility of Lactobacillus strains isolated from domestic geese. Br. Poult. Sci. 2015, 56, 416–424. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).