The Root Nodule Microbiome of Cultivated and Wild Halophytic Legumes Showed Similar Diversity but Distinct Community Structure in Yellow River Delta Saline Soils
Abstract
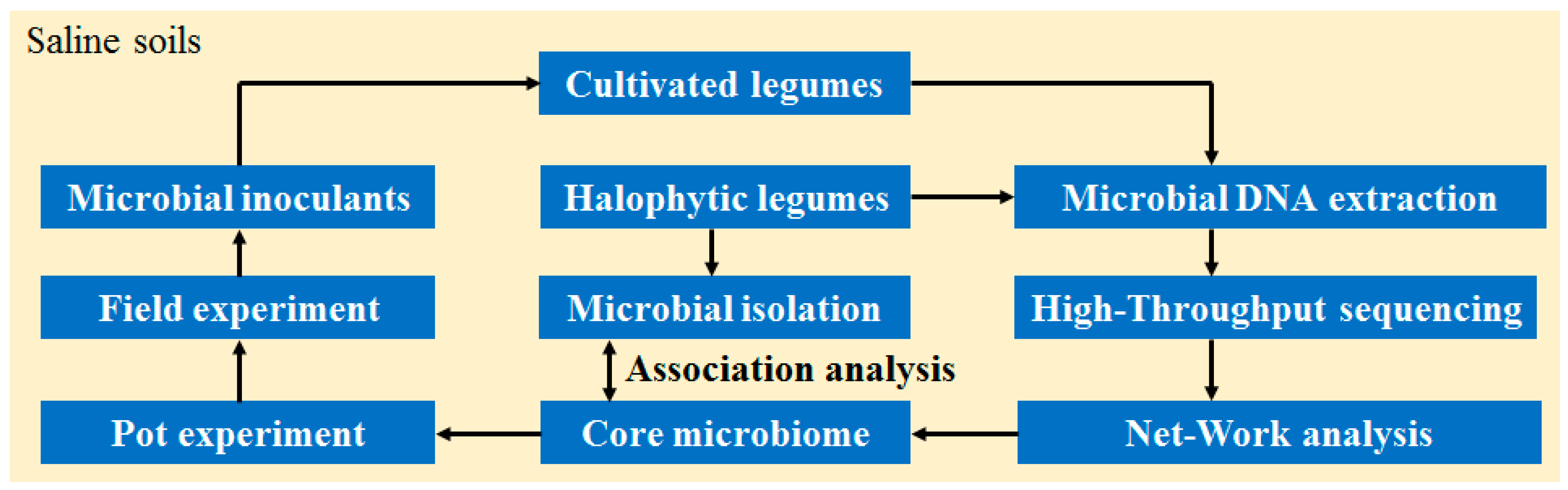
1. Introduction
2. Materials and Methods
2.1. Nodule Samples
2.2. Observation of Nodulation Characteristics
2.3. Determination of Nitrogenase Activity
2.4. Microbial DNA Extraction
2.5. Quantitative PCR (qPCR) of the Bacterial Community
2.6. 16S rRNA Gene Amplification and PacBio Sequencing
2.7. Sequence Data Analyses
3. Results
3.1. Natural Field Nodulation
3.2. Characteristics of Sample Sequence Tags
3.3. Microbial Community Richness and Diversity
3.4. Microbial Taxonomic Analysis at the Phylum and Genus Level
3.5. Microbial Taxonomic Analysis at the Species Levels
3.6. Comparative Analysis of Bacteria in Different Sample Groups
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Pennock, D.; McKenzie, N.; Montanarella, L. Status of the World’s Soil Resources; Technical Summary FAO: Rome, Italy, 2015. [Google Scholar]
- Li, J.; Pu, L.; Zhu, M.; Zhang, R. The present situation and hot issues in the salt-affected soil. Acta Geol. Sin. 2012, 67, 1233–1245. [Google Scholar]
- Lu, Z.; Yang, J.; Liu, G.; Li, J.; Liu, H.; Li, B. Relationship between soil salinization and groundwater characteristics in the Yellow River Delta. Acta Pedolocica Sin. 2017, 54, 1377–1385. [Google Scholar]
- Abiala, M.A.; Abdelrahman, M.; Burritt, D.; Tran, L.P. Salt stress tolerance mechanisms and potential applications of legumes for sustainable reclamation of salt-degraded soils. Land Degrad. Dev. 2018, 29, 3812–3822. [Google Scholar] [CrossRef]
- Zhao, K.; Li, F.; Fan, S.; Feng, L. Halophytes in China. Chin. Bull. Bot. 1999, 16, 10–16. [Google Scholar]
- Tian, B.; Zhang, C.; Ye, Y.; Wen, J.; Wu, Y.; Wang, H.; Li, L.; Cai, S.; Cai, W.; Cheng, Z.; et al. Beneficial traits of bacterial endophytes belonging to the core communities of the tomato root microbiome. Agric. Ecosyst. Environ. 2017, 247, 149–156. [Google Scholar] [CrossRef]
- Tian, X.; Zhang, C. Illumina-based analysis of endophytic and rhizosphere bacterial diversity of the coastal halophyte Messerschmidia sibirica. Front. Microbiol. 2017, 8, 2288. [Google Scholar] [CrossRef]
- Gao, D.; Wang, X.; Fu, S.; Zhao, J. Legume plants enhance the resistance of soil to ecosystem disturbance. Front. Plant. Sci. 2017, 8, 1295. [Google Scholar] [CrossRef]
- Larrainzar, E.; Wienkoop, S. A proteomic view on the role of legume symbiotic interactions. Front. Plant. Sci. 2017, 8, 1267. [Google Scholar] [CrossRef]
- Wang, X.; Teng, Y.; Zhang, N.; Christie, P.; Li, Z.; Luo, Y.; Wang, J. Rhizobial symbiosis alleviates polychlorinated biphenyls-induced systematic oxidative stress via brassinosteroids signaling in alfalfa. Sci. Total Environ. 2017, 592, 68–77. [Google Scholar] [CrossRef]
- Zhang, C.; Xin, H.; Zou, P. Plants at beach in Shandong; Agricultural science and Technology Press: Beijing, China, 2017. [Google Scholar]
- Li, Y.; Li, X.; Liu, Y.; Wang, E.; Ren, C.; Liu, W.; Xu, H.; Wu, H.; Jiang, N.; Li, Y.; et al. Genetic diversity and community structure of rhizobia nodulating Sesbania cannabina in saline–alkaline soils. Syst. Appl. Microbiol. 2016, 39, 195–202. [Google Scholar] [CrossRef]
- Zhao, L.; Fan, M.; Zhang, D.; Yang, R.; Zhang, F.; Lin, X.; Wei, X.; Shen, Y.; Wei, G. Distribution and diversity of rhizobia associated with wild soybean (Glycine soja Sieb. & Zucc.) in Northwest China. Syst. Appl. Microbiol. 2014, 37, 449–456. [Google Scholar]
- Singer, E.; Bushnell, B.; Coleman-Derr, D.; Bowman, B.; Bowers, R.M.; Levy, A.; Gies, E.A.; Cheng, J.-F.; Copeland, A.; Klenk, H.-P.; et al. High-resolution phylogenetic microbial community profiling. ISME J. 2016, 10, 2020. [Google Scholar] [CrossRef]
- Motooka, D.; Fujimoto, K.; Tanaka, R.; Yaguchi, T.; Gotoh, K.; Maeda, Y.; Furuta, Y.; Kurakawa, T.; Goto, N.; Yasunaga, T.; et al. Fungal ITS1 deep-sequencing strategies to reconstruct the composition of a 26-species community and evaluation of the gut mycobiota of healthy Japanese individuals. Front. Microbiol. 2017, 8, 238. [Google Scholar] [CrossRef]
- Zaied, K.A.; Kosba, Z.A.; Nassef, M.A.; EI-saied, A.I. Induction of Rhizobium inoculants harboring salicylic acid gene. Aust. J. Bas. 2009, 3, 1386–1411. [Google Scholar]
- Fierer, N.; Jackson, J.; Vilgalys, R.; Jackson, R. Assessment of soil microbial community structure by use of taxon-specific quantitative PCR assays. Appl. Environ. Microbiol. 2005, 71, 4117–4120. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microb. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucl. Acids Res. 2013, 41, 590–596. [Google Scholar] [CrossRef]
- Clark, K.R.; Gorley, R.N. PRIMER v6: User Manual/Tutorial. PRIMER-E: Plymouth; Plymouth Marine Laboratory: Plymouth, UK, 2006. [Google Scholar]
- Dang, X.; Xie, Z.; Liu, W.; Sun, Y.; Liu, X.; Zhu, Y.; Staehelin, C. The genome of Ensifer alkalisoli YIC4027 provides insights for host specificity and environmental adaptations. BMC Genom. 2019, 20, 643. [Google Scholar] [CrossRef]
- Li, Q.Q.; Wang, E.T.; Zhang, Y.Z.; Zhang, Y.M.; Tian, C.F.; Sui, X.H.; Chen, W.F.; Chen, W.X. Diversity and biogeography of rhizobia isolated from root nodules of Glycine max grown in Hebei Province, China. Microb. Ecol. 2011, 61, 917–931. [Google Scholar] [CrossRef]
- Naamala, J.; Jaiswal, S.K.; Dakora, F.D. Microsymbiont diversity and phylogeny of native bradyrhizobia associated with soybean (Glycine max L. Merr.) nodulation in South African soils. Syst. Appl. Microbiol. 2016, 39, 336–344. [Google Scholar] [CrossRef]
- de Castro Pires, R.; dos Reis Junior, F.B.; Zilli, J.E.; Fischer, D.; Hofmann, A.; James, E.K.; Simon, M.F. Soil characteristics determine the rhizobia in association with different species of Mimosa in central Brazil. Plant. Soil 2018, 423, 411–428. [Google Scholar] [CrossRef]
- Xiao, X.; Chen, W.; Zong, L.; Yang, J.; Jiao, S.; Lin, Y.; Wang, E.; Wei, G. Two cultivated legume plants reveal the enrichment process of the microbiome in the rhizocompartments. Mol. Ecol. 2017, 26, 1641–1651. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Shi, F.; Hamid, M.I.; Zhu, Y. Endophytic bacterial diversity of wild soybean (Glycine soja) varieties with different resistance to soybean cyst nematode (Heterodera glycines). Acta Microbiol. Sin. 2014, 54, 926–935. [Google Scholar]
- Dong, X.; Liu, X.; Zhang, B.; Wang, Y. Diversity of rhizobia and endophytic bacteria isolated from nodules of Sesbania spp. Chin. J. Trop. Crop. 2010, 31, 88–92. [Google Scholar]
- Pootakham, W.; Mhuantong, W.; Yoocha, T.; Putchim, L.; Sonthirod, C.; Naktang, C.; Thongtham, N.; Tangphatsornruang, S. High resolution profiling of coral-associated bacterial communities using full-length 16S rRNA sequence data from PacBio SMRT sequencing system. Sci. Rep. 2017, 7, 2774. [Google Scholar] [CrossRef]
- Vuong, H.B.; Thrall, P.H.; Barrett, L.G. Host species and environmental variation can influence rhizobial community composition. J. Ecol. 2017, 105, 540–548. [Google Scholar] [CrossRef]
- Lorite, M.J.; Estrella, M.J.; Escaray, F.J.; Sannazzaro, A.; Videira e Castro, I.M.; Monza, J.; Sanjuán, J.; León-Barrios, M. The rhizobia-lotus symbioses: Deeply specific and widely diverse. Front. Microbiol. 2018, 9, 2055. [Google Scholar] [CrossRef]
- Bertrand, A.; Dhont, C.; Bipfubusa, M.; Chalifour, F.P.; Drouin, P.; Beauchamp, C.J. Improving salt stress responses of the symbiosis in alfalfa using salt-tolerant cultivar and rhizobial strain. Appl. Soil Ecol. 2015, 87, 108–117. [Google Scholar] [CrossRef]
- Qu, L.; Huang, Y.; Zhu, C.; Zeng, H.; Shen, C.; Liu, C.; Zhao, Y.; Pi, E. Rhizobia-inoculation enhances the soybean’s tolerance to salt stress. Plant. Soil 2016, 400, 209–222. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, Z.; Zhang, P.; Cao, Y.; Hu, T.; Yang, P. Rhizobium symbiosis contribution to short-term salt stress tolerance in alfalfa (Medicago sativa L. ). Plant Soil 2016, 402, 247–261. [Google Scholar] [CrossRef]
- Sgroy, V.; Cassán, F.; Masciarelli, O.; Del Papa, M.F.; Lagares, A.; Luna, V. Isolation and characterization of endophytic plant growth-promoting (PGPB) or stress homeostasis-regulating (PSHB) bacteria associated to the halophyte Prosopis strombulifera. Appl. Microbiol. Biotechnol. 2009, 85, 371–381. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Zhou, N.; Zhao, Z.; Zhang, K.; Wu, G.; Tian, C. Isolation of endophytic plant growth-promoting bacteria associated with the halophyte Salicornia europaea and evaluation of their promoting activity under salt stress. Curr. Microbiol. 2016, 73, 574–581. [Google Scholar] [CrossRef] [PubMed]
- Torre, S.N.; Barcia-Piedras, J.M.; Mateos-Naranjo, E.; Redondo-Gómez, S.; Camacho, M.; Caviedes, M.A.; Pajuelo, E.; Rodríguez-Llorente, I.D. Assessing the role of endophytic bacteria in the halophyte Arthrocnemum macrostachyum salt tolerance. Plant. Biol. 2017, 19, 249–256. [Google Scholar] [CrossRef]
- Rejili, M.; Mahdhi, M.; Fterich, A.; Dhaoui, S.; Guefrachi, I.; Abdeddayem, R.; Mars, M. Symbiotic nitrogen fixation of wild legumes in Tunisia: Soil fertility dynamics, field nodulation and nodules effectiveness. Agric. Ecosyst. Environ. 2012, 157, 60–69. [Google Scholar] [CrossRef]

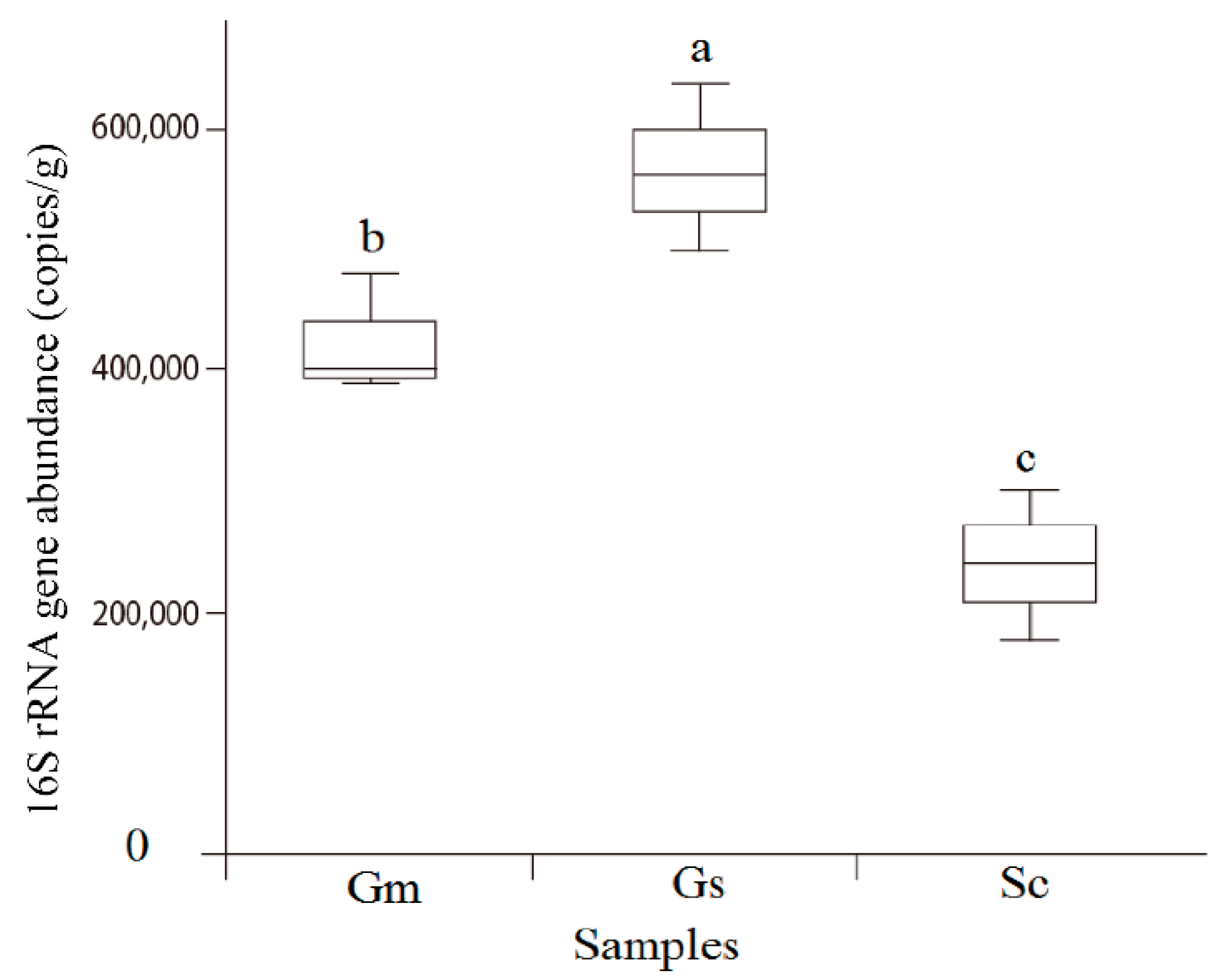
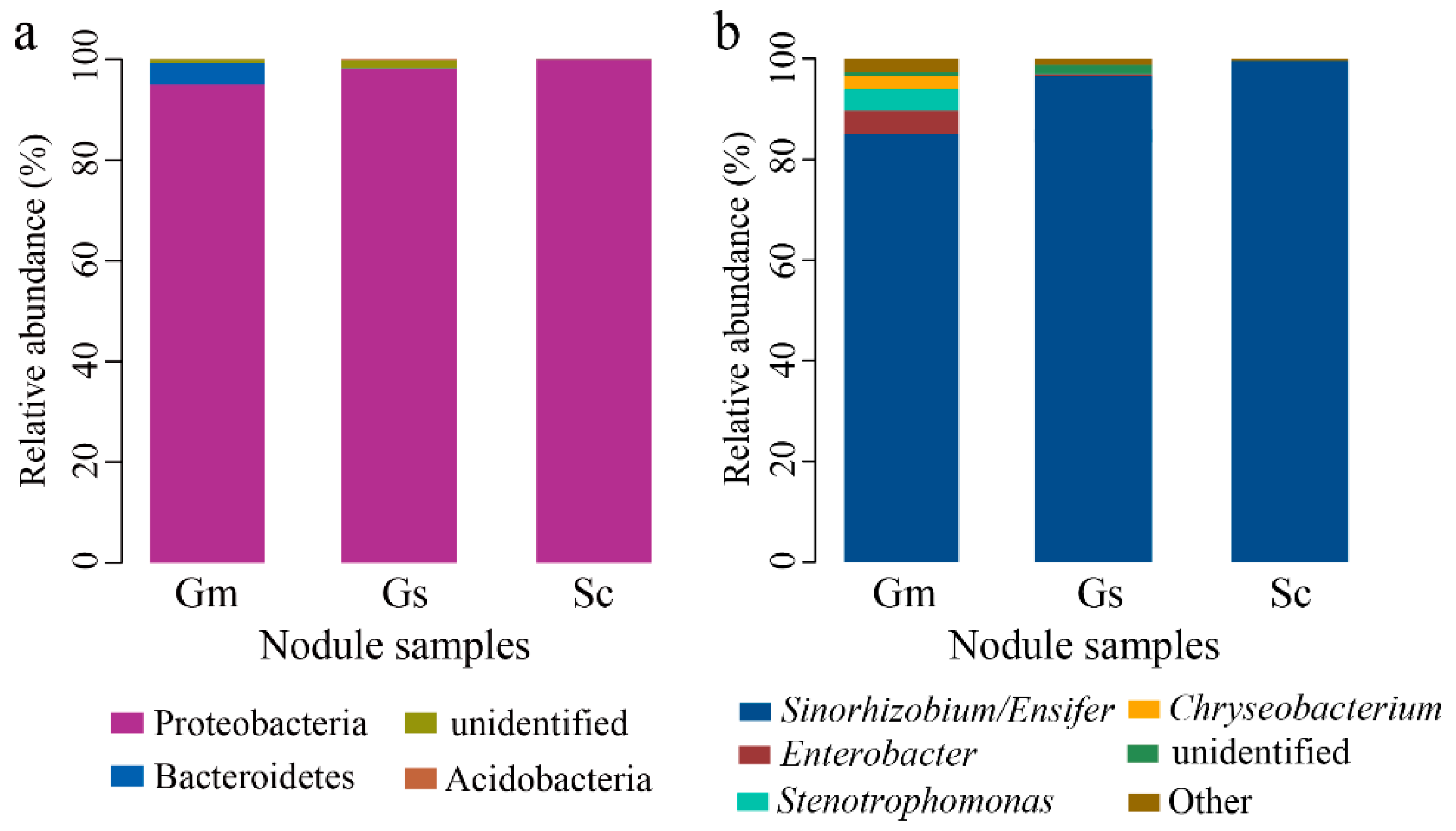
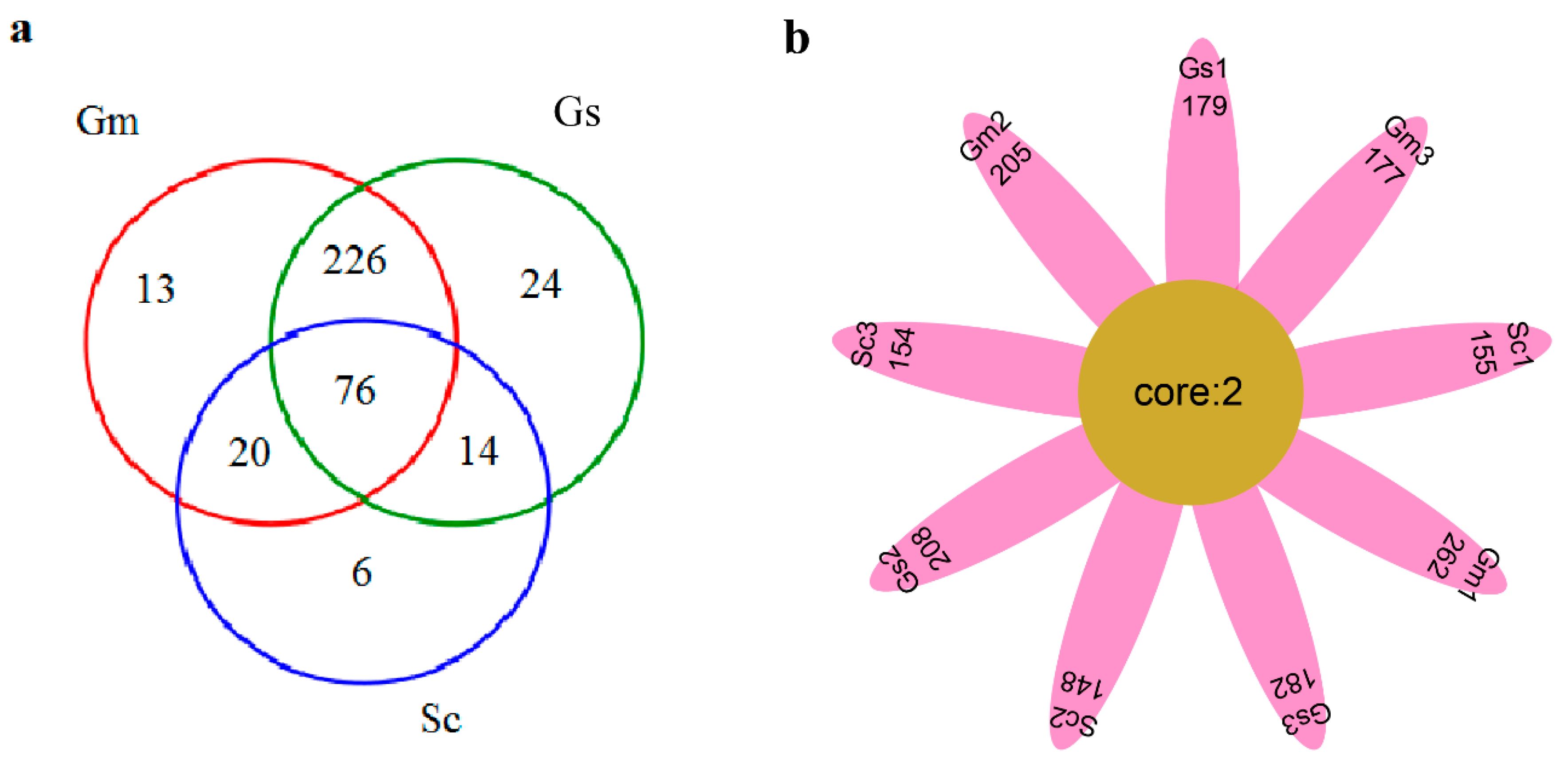

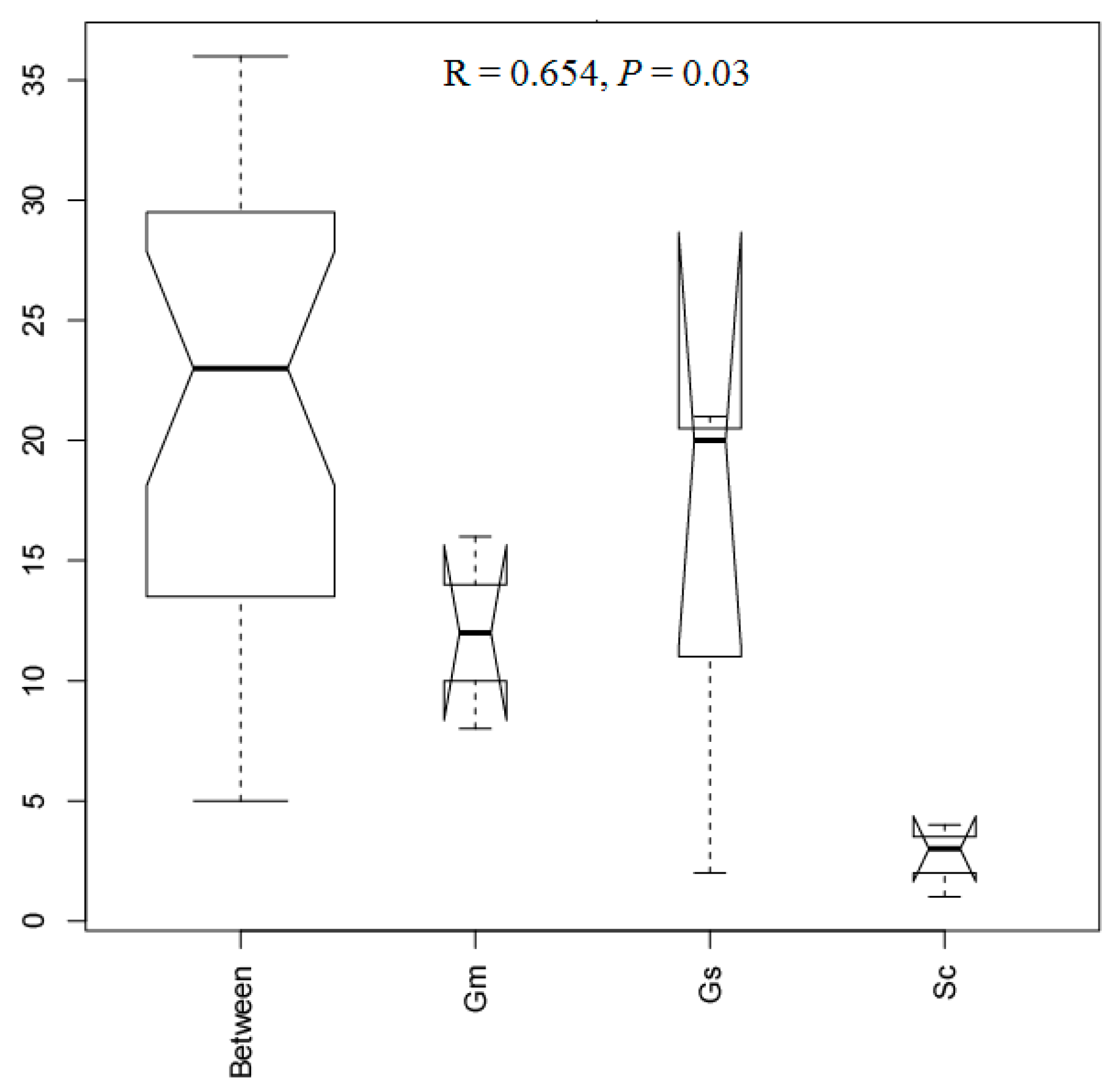
| Plant Species | External Color of Nodules | Shape of Nodule | Nodule Size (mm) | Effective Nodule Weight Per Plant (g) | Nodule Nitrogenase Activity [μmol (g h)−1] |
|---|---|---|---|---|---|
| G. max | white, pink, brown | round to massive | 3–9 | 0.39 ± 0.07 b | 3.52 ± 0.15 b |
| G. soja | white, pink, brown | round to massive | 3–11 | 0.57 ± 0.07 a | 4.37 ± 0.17 a |
| S. cannabina | white, pink, brown | round | 3–9 | 0.61 ± 0.05 a | 4.27 ± 0.11 a |
| Species | OTUs | Coverage (%) | Chao1 Richness | Shannon Diversity | PD Whole Tree |
|---|---|---|---|---|---|
| G. max | 217 ± 35 ab | 87.6 ± 2.1 b | 284 ± 41 a | 5.78 ± 0.55 a | 4.17 ± 0.77 a |
| G. soja | 191 ± 13 a | 90.2 ± 1.5 b | 228 ± 30 a | 5.88 ± 0.34 a | 3.33 ± 0.62 ab |
| S. cannabina | 154 ± 3 b | 96.1 ± 0.2 a | 112± 5 b | 5.28 ± 0.04 a | 1.82 ± 0.04 b |
| Species | Gm | Gs | Sc | Species | Gm | Gs | Sc |
|---|---|---|---|---|---|---|---|
| Rhizobia | Ensifer saheli | 0.21 | 0.18 | 0.00 | |||
| Ensifer americanum | 42.81 | 46.45 | 0.56 | Ensifer kostiense | 0.31 | 0.06 | 0.00 |
| Ensifer alkalisoli YIC4027 | 8.10 | 11.31 | 68.23 | Ensifer sp. T1Gs6 | 0.18 | 0.10 | 0.00 |
| Ensifer sp. | 13.57 | 17.23 | 16.46 | Ensifer sp. R7-568 | 0.13 | 0.14 | 0.00 |
| Ensifer sp. MSMC310 | 5.99 | 7.18 | 0.00 | Ensifer sp. 209 | 0.06 | 0.13 | 0.00 |
| Ensifer sp. C9 | 0.90 | 0.87 | 7.59 | Others | 0.33 | 0.25 | 0.00 |
| Ensifer meliloti | 0.74 | 1.15 | 3.99 | Total | 85.80 | 95.62 | 100 |
| Ensifer sp. CEQ1 | 2.84 | 2.64 | 0.00 | Non-rhizobial bacteria | |||
| Ensifer fredii | 1.94 | 1.34 | 0.00 | Enterobacter cloacae | 3.62 | 0.13 | 0.00 |
| Ensifer sp. ENCBTM 34 | 2.04 | 0.36 | 0.00 | Stenotrophomonas sp. CanR-75 | 2.79 | 0.00 | 0.00 |
| Ensifer adhaerens | 0.82 | 1.51 | 0.00 | Stenotrophomonas maltophilia | 2.41 | 0.00 | 0.00 |
| Ensifer sp. T2GRs3 | 0.89 | 0.75 | 0.53 | unidentified | 0.95 | 1.44 | 0.00 |
| Ensifer sp. ORS 1236 | 0.62 | 0.57 | 0.21 | Chryseobacterium wanjuense | 0.01 | 1.35 | 0.00 |
| Ensifer numidicus | 0.13 | 0.39 | 0.79 | Flavobacterium johnsoniae | 1.15 | 0.02 | 0.00 |
| Ensifer sp. L1GRs3 | 0.49 | 0.51 | 0.30 | Enterobacter asburiae | 0.83 | 0.00 | 0.00 |
| Ensifer sp. SEMIA 6161 | 0.09 | 0.18 | 0.89 | Chryseobacterium sp. KJ9C8 | 0.00 | 0.79 | 0.00 |
| Ensifer terangae | 1.00 | 0.13 | 0.00 | Sphingobacterium bambusae | 0.38 | 0.00 | 0.00 |
| Ensifer mexicanus | 0.41 | 0.60 | 0.00 | Xanthomonas pisi | 0.27 | 0.00 | 0.00 |
| Ensifer xinjiangense | 0.47 | 0.37 | 0.00 | Sphingobacterium sp. 1.3 | 0.23 | 0.02 | 0.00 |
| Mesorhizobium mediterraneum | 0.07 | 0.17 | 0.45 | Stenotrophomonas sp. 2TP1A | 0.03 | 0.21 | 0.00 |
| Ensifer sp. S39 | 0.33 | 0.33 | 0.00 | Enterobacter sp. B1-2 | 0.20 | 0.00 | 0.00 |
| Ensifer sp. ORS 1085 | 0.23 | 0.40 | 0.00 | Others | 1.33 | 0.42 | 0.00 |
| Ensifer kummerowiae | 0.10 | 0.32 | 0.00 | Total | 14.20 | 4.38 | 0.00 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, Y.; Liang, J.; Zhao, D.-L.; Meng, C.; Xu, Z.-C.; Xie, Z.-H.; Zhang, C.-S. The Root Nodule Microbiome of Cultivated and Wild Halophytic Legumes Showed Similar Diversity but Distinct Community Structure in Yellow River Delta Saline Soils. Microorganisms 2020, 8, 207. https://doi.org/10.3390/microorganisms8020207
Zheng Y, Liang J, Zhao D-L, Meng C, Xu Z-C, Xie Z-H, Zhang C-S. The Root Nodule Microbiome of Cultivated and Wild Halophytic Legumes Showed Similar Diversity but Distinct Community Structure in Yellow River Delta Saline Soils. Microorganisms. 2020; 8(2):207. https://doi.org/10.3390/microorganisms8020207
Chicago/Turabian StyleZheng, Yanfen, Jing Liang, Dong-Lin Zhao, Chen Meng, Zong-Chang Xu, Zhi-Hong Xie, and Cheng-Sheng Zhang. 2020. "The Root Nodule Microbiome of Cultivated and Wild Halophytic Legumes Showed Similar Diversity but Distinct Community Structure in Yellow River Delta Saline Soils" Microorganisms 8, no. 2: 207. https://doi.org/10.3390/microorganisms8020207
APA StyleZheng, Y., Liang, J., Zhao, D.-L., Meng, C., Xu, Z.-C., Xie, Z.-H., & Zhang, C.-S. (2020). The Root Nodule Microbiome of Cultivated and Wild Halophytic Legumes Showed Similar Diversity but Distinct Community Structure in Yellow River Delta Saline Soils. Microorganisms, 8(2), 207. https://doi.org/10.3390/microorganisms8020207






