Abstract
The COVID-19 pandemic, caused by a novel zoonotic coronavirus (CoV), SARS-CoV-2, has infected 46,182 million people, resulting in 1,197,026 deaths (as of 1 November 2020), with devastating and far-reaching impacts on economies and societies worldwide. The complex origin, extended human-to-human transmission, pathogenesis, host immune responses, and various clinical presentations of SARS-CoV-2 have presented serious challenges in understanding and combating the pandemic situation. Human CoVs gained attention only after the SARS-CoV outbreak of 2002–2003. On the other hand, animal CoVs have been studied extensively for many decades, providing a plethora of important information on their genetic diversity, transmission, tissue tropism and pathology, host immunity, and therapeutic and prophylactic strategies, some of which have striking resemblance to those seen with SARS-CoV-2. Moreover, the evolution of human CoVs, including SARS-CoV-2, is intermingled with those of animal CoVs. In this comprehensive review, attempts have been made to compare the current knowledge on evolution, transmission, pathogenesis, immunopathology, therapeutics, and prophylaxis of SARS-CoV-2 with those of various animal CoVs. Information on animal CoVs might enhance our understanding of SARS-CoV-2, and accordingly, benefit the development of effective control and prevention strategies against COVID-19.
1. Introduction
Since its first detection in cases with atypical pneumonia from the city of Wuhan, China, in December of 2019, the novel coronavirus (CoV), designated as SARS-CoV-2, has infected 46,182 million people, resulting in 1,197,026 deaths worldwide (as of 1 November 2020) [1,2,3,4]. The ensuing pandemic, known as COVID-19, is one of unprecedented proportions that has crippled the global economy and exerted far-reaching, detrimental impacts on societies [5,6]. The complex origin, extended human-to-human transmission, pathogenesis including immunopathology, and the various clinical presentations of SARS-CoV-2 have presented serious challenges to the scientific and medical communities towards properly understanding and combating the pandemic situation [3,7,8,9,10,11]. Data available so far strongly indicated that SARS-CoV-2 is a zoonotic virus that might have been derived from bats with, or without, the involvement of another animal species (pangolins?) as intermediate host/s [11]. SARS-CoV-2 is the 7th CoV to be reported in humans, and with respect to novelty, differs from the other important human respiratory CoVs: SARS-CoV and MERS-CoV [2,3,11,12].
Animal CoVs have been known to cause important diseases in livestock and companion animals [13,14,15,16,17,18,19,20,21,22,23]. Although human CoVs received attention only after the SARS-CoV outbreak of 2002–2003 [24], animal CoVs have been studied extensively for many decades, providing a plethora of important information on their complex genetic diversity, transmission, tissue tropism and pathology, host immune responses, therapeutics, and prophylaxis, some of which have striking resemblance to those seen with SARS-CoV-2. Moreover, the evolution of human CoVs including SARS-CoV-2 is intermingled with those of animal CoVs. Information on animal CoVs might enhance our understanding of SARS-CoV-2, and accordingly, benefit the development of effective control and prevention strategies against COVID-19. Since January 2020, many review articles have been written on the COVID-19 pandemic. However, these were mostly focused on various aspects of SARS-CoV-2 in humans. In this comprehensive review, attempts have been made to compare the current knowledge on evolution, transmission, pathogenesis, therapeutics, and prophylaxis of SARS-CoV-2 with those of CoVs in various animals.
9. Conclusions
Satirically, the crown-like morphology of SARS-CoV-2 indeed befits the virus, for SARS-CoV-2 would surely be crowned as one of the most devastating viruses of recent times with regards to its impact on health, economies, and societies worldwide [2,3,5,6]. Although the current SARS-CoV-2 pandemic has triggered a global wave of research studies on CoVs at an unprecedented scale, human CoVs attracted attention only after the SARS outbreak of 2002–2003. On the other hand, animal CoVs have long been identified as major causes of mortality and morbidity in livestock and companion animals. As a result, animal CoVs have been studied extensively, yielding a plethora of information on CoV evolution, transmission including interspecies transmission events, pathogenesis including immunopathology, therapeutics, and prophylaxis, evaluation of which might allow for a better understanding of CoV disease in humans.
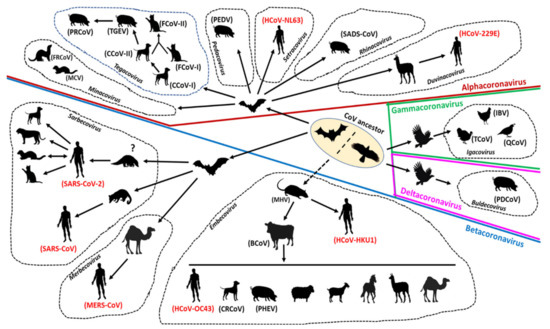
Surveillance and genome sequencing of CoVs from various animals species, including wildlife, were extremely crucial in tracing the possible origin and transmission of SARS-CoV-2 (and that of SARS-CoV and MERS), suggesting that these viruses were most likely derived from bats with/without an animal intermediate host (Figure 3) [11,24,139]. Antibodies against SARS-CoV have been detected in humans living near bat caves in China [89]. Recently, SARS-COV-2 has been reported in cats, dogs, minks, and captive wildlife, indicating possible human-to-animal transmission events [129]. Taken together, these observations underscored the significance of continuous monitoring of zoonotic and zooanthroponotic pathogens in animals, especially wildlife, and in humans residing near wildlife habitats. Once again, evolutionary analysis of SARS-CoV-2 reiterated the importance of reducing contact potential between humans and wildlife by addressing climate change and deforestation, and banning wildlife trade/wet animal markets [11,266,267].
Compared to the other human CoVs and many other zoonotic viruses, the rapidity and extent of human-to-human spread of SARS-CoV-2 has intrigued many researchers [1,3,7]. Although remarkable, this observation might not appear surprising, at least to animal virologists, for studies on animal CoVs have previously revealed the capability of CoVs to jump the species barrier, and successfully adapt and spread in a new host species. Widely prevalent animal CoVs, such as FCoV-II, PEDV, and TGEV, have been proposed to be derived from heterologous hosts including unrelated animal species (Figure 3) [13,15,16,17,18,19].
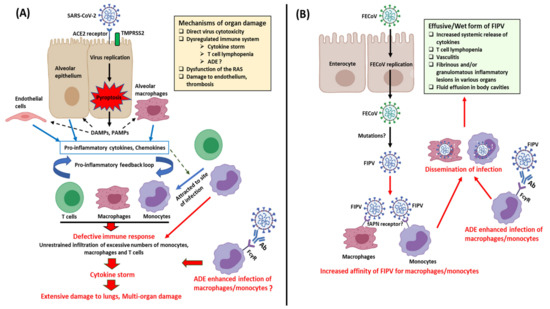
Studies on animal CoVs have revealed the enigmatic nature of CoVs, such as alterations in tissue tropism (pantropic CCoV-IIa, FIPV, and PRCoV), multi-organ tropism (BCoV, pantropic CCoV-IIa, FIPV, IBV, MHV, and PHEV), and/or immunopathology (FIPV) [14,16,17,18,19,20,21,22,23,37]. Although changes in the CoV genome have been proposed to induce alterations in tissue tropism, except for that of PRCoV, the molecular basis for such events remain to be clearly elucidated. Interestingly, SARS-CoV-2, considered as a respiratory pathogen, has been associated with extra-clinical manifestations, and viral RNA has been detected in various organs and fecal samples [8,9,137]. Furthermore, the cytokine storm observed in SARS-CoV-2 patients mirrors that in cats with FIP [9,17] (Figure 4). Considering the above, it would be interesting to compare the genetic variations and pathogenesis of SARS-CoV-2 strains from various organs.
The development of an effective and safe vaccine against SARS-CoV-2 is of paramount importance. Although there are no licensed vaccines against CoVs in humans so far, the various issues (lack of complete protection, short duration of immunity, correlates of protection against CoVs, emergence of multiple serotypes/subtypes including virulent recombinants between vaccine and field strains, and/or ADE enhanced disease) observed with the veterinary CoV vaccines should be taken into consideration while devising immunization strategies against SARS-CoV-2 [17,18,22,30,32,33,254,255].
SARS-CoV-2 has challenged the very basic tenet of human existence: “United We Stand, Divided We Fall”. Measures aimed at reducing human-to-human transmission have resulted in a crisis situation, where, unfortunately, “Divided We Stand, United We Fall” has become the motto for survival. However, pandemics, especially those caused by zoonotic pathogens such as SARS-CoV-2, could be avoided, or effectively controlled, by adopting a One Health approach that unites the various streams of animal, human, and environmental health, thereby reinforcing the concept of “United We Stand, Divided We Fall”.
Author Contributions
Conceptualization, S.G.; methodology, S.G., Y.S.M.; resources, S.G., Y.S.M.; data curation, S.G.; writing—original draft preparation, S.G.; writing—review and editing, S.G., Y.S.M.; visualization, S.G.; funding acquisition, S.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding The APC was funded by the One Health Center for Zoonoses and Tropical Veterinary Medicine, Ross University School of Veterinary Medicine, St. Kitts and Nevis.
Acknowledgments
The authors would like to thank Subhankar Sircar, PhD student, Indian Veterinary Research Institute, India, for assisting with the phylogenetic analysis.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Ortiz-Prado, E.; Simbaña-Rivera, K.; Gómez-Barreno, L.; Rubio-Neira, M.; Guaman, L.P.; Kyriakidis, N.C.; Muslin, C.; Jaramillo, A.M.G.; Barba-Ostria, C.; Cevallos-Robalino, D.; et al. Clinical, molecular, and epidemiological characterization of the SARS-CoV-2 virus and the Coronavirus Disease 2019 (COVID-19), a comprehensive literature review. Diagn. Microbiol. Infect. Dis. 2020, 98. [Google Scholar] [CrossRef] [PubMed]
- Petersen, E.; Koopmans, M.; Go, U.; Hamer, D.H.; Petrosillo, N.; Castelli, F.; Storgaard, M.; Al Khalili, S.; Simonsen, L. Comparing SARS-CoV-2 with SARS-CoV and influenza pandemics. Lancet Infect. Dis. 2020. [Google Scholar] [CrossRef]
- Dhama, K.; Khan, S.; Tiwari, R.; Sircar, S.; Bhat, S.; Malik, Y.S.; Singh, K.P.; Chaicumpa, W.; Bonilla-Aldana, D.K.; Rodriguez-Morales, A.J. Coronavirus disease 2019–COVID-19. Clin. Microbiol. Rev. 2020, 33, 1–48. [Google Scholar] [CrossRef] [PubMed]
- Coronavirus Resource Center, Johns Hopkins University School of Medicine. Available online: https://coronavirus.jhu.edu/ (accessed on 1 November 2020).
- Nicola, M.; Alsafi, Z.; Sohrabi, C.; Kerwan, A.; Al-Jabir, A.; Iosifidis, C.; Agha, M.; Agha, R. The socio-economic implications of the coronavirus pandemic (COVID-19): A review. Int. J. Surg. 2020, 78, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Osofsky, J.D.; Osofsky, H.J.; Mamon, L.Y. Psychological and Social Impact of COVID-19. Psychol. Trauma Theory Res. Pract. Policy 2020. [Google Scholar] [CrossRef]
- Day, T.; Gandon, S.; Lion, S.; Otto, S.P. On the evolutionary epidemiology of SARS-CoV-2. Curr. Biol. 2020, 30, R849–R857. [Google Scholar] [CrossRef]
- Patel, K.P.; Vunnam, S.R.; Patel, P.A.; Krill, K.L.; Korbitz, P.M.; Gallagher, J.P.; Suh, J.E.; Vunnam, R.R. Transmission of SARS-CoV-2: An update of current literature. Eur. J. Clin. Microbiol. Infect. Dis. 2020. [Google Scholar] [CrossRef]
- Gupta, A.; Madhavan, M.V.; Sehgal, K.; Nair, N.; Mahajan, S.; Sehrawat, T.S.; Bikdeli, B.; Ahluwalia, N.; Ausiello, J.C.; Wan, E.Y.; et al. Extrapulmonary manifestations of COVID-19. Nat. Med. 2020, 26, 1017–1032. [Google Scholar] [CrossRef]
- Jeyanathan, M.; Afkhami, S.; Smaill, F.; Miller, M.S.; Lichty, B.D.; Xing, Z. Immunological considerations for COVID-19 vaccine strategies. Nat. Rev. Immunol. 2019, 4, 1–18. [Google Scholar] [CrossRef]
- Zhang, Y.Z.; Holmes, E.C. A Genomic Perspective on the Origin and Emergence of SARS-CoV-2. Cell 2020, 181, 223–227. [Google Scholar] [CrossRef]
- Liu, J.; Xie, W.; Wang, Y.; Xiong, Y.; Chen, S.; Han, J.; Wu, Q. A comparative overview of COVID-19, MERS and SARS: Review article. Int. J. Surg. 2020, 81, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Decaro, N.; Lorusso, A. Novel human coronavirus (SARS-CoV-2): A lesson from animal coronaviruses. Vet. Microbiol. 2020, 244. [Google Scholar] [CrossRef] [PubMed]
- Decaro, N.; Buonavoglia, C. Canine Coronavirus: Not Only an Enteric Pathogen. Vet. Clin. N. Am. Small Anim. Pract. 2011, 41, 1121–1132. [Google Scholar] [CrossRef] [PubMed]
- Perlman, S.; Netland, J. Coronaviruses post-SARS: Update on replication and pathogenesis. Nat. Rev. Microbiol. 2009, 7, 439–450. [Google Scholar] [CrossRef]
- Perlman, S.; Dandekar, A.A. Immunopathogenesis of coronavirus infections: Implications for SARS. Nat. Rev. Immunol. 2005, 5, 917–927. [Google Scholar] [CrossRef]
- Pedersen, N.C. An update on feline infectious peritonitis: Virology and immunopathogenesis. Vet. J. 2014, 201, 123–132. [Google Scholar] [CrossRef]
- Wang, Q.; Vlasova, A.N.; Kenney, S.P.; Saif, L.J. Emerging and re-emerging coronaviruses in pigs. Curr. Opin. Virol. 2019, 34, 39–49. [Google Scholar] [CrossRef]
- Vlasova, A.N.; Wang, Q.; Jung, K.; Langel, S.N.; Malik, Y.S.; Saif, L.J. Porcine Coronaviruses. In Emerging and Transboundary Animal Viruses; Malik, Y.S., Singh, R.K., Yadav, M.P., Eds.; Springer: Singapore, 2020; pp. 79–110. [Google Scholar] [CrossRef]
- Saif, L.J.; Jung, K. Comparative pathogenesis of bovine and porcine respiratory coronaviruses in the animal host species and sars-cov-2 in humans. J. Clin. Microbiol. 2020, 58, e01355. [Google Scholar] [CrossRef]
- Bande, F.; Arshad, S.S.; Omar, A.R.; Hair-Bejo, M.; Mahmuda, A.; Nair, V. Global distributions and strain diversity of avian infectious bronchitis virus: A review. Anim. Health Res. Rev. 2017, 18, 70–83. [Google Scholar] [CrossRef]
- Rohaim, M.A.; El Naggar, R.F.; Abdelsabour, M.A.; Mohamed, M.H.A.; El-Sabagh, I.M.; Munir, M. Evolutionary analysis of infectious bronchitis virus reveals marked genetic diversity and recombination events. Genes 2020, 11, 605. [Google Scholar] [CrossRef] [PubMed]
- Körner, R.W.; Majjouti, M.; Alcazar, M.A.A.; Mahabir, E. Of Mice and Men: The Coronavirus MHV and Mouse Models as a Translational Approach to Understand SARS-CoV-2. Viruses 2020, 12, 880. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Li, F.; Shi, Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019, 17, 181–192. [Google Scholar] [CrossRef] [PubMed]
- Gorbalenya, A.; Baker, S.; Baric, R.; de Groot, R.; Drosten, C.; Gulyaeva, A.; Haagmans, B.; Lauber, C.; Leontovich, A.; Neuman, B.; et al. Severe acute respiratory syndrome-related coronavirus: The species and its viruses—A statement of the Coronavirus Study Group. Nat. Microbiol. 2020, 5, 536–544. [Google Scholar] [CrossRef]
- Coronaviridae—International Committee on Taxonomy of Viruses (ICTV). Available online: https://talk.ictvonline.org/taxonomy/ (accessed on 18 September 2020).
- Mihindukulasuriya, K.A.; Wu, G.; St. Leger, J.; Nordhausen, R.W.; Wang, D. Identification of a Novel Coronavirus from a Beluga Whale by Using a Panviral Microarray. J. Virol. 2008, 82, 5084–5088. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef]
- Abdel-Moneim, A.S.; Abdelwhab, E.M. Evidence for SARS-COV-2 infection of animal hosts. Pathogens 2020, 9, 529. [Google Scholar] [CrossRef]
- Tizard, I.R. Vaccination against coronaviruses in domestic animals. Vaccine 2020, 38, 5123–5130. [Google Scholar] [CrossRef]
- Szczepanski, A.; Owczarek, K.; Bzowska, M.; Gula, K.; Drebot, I.; Ochman, M.; Maksym, B.; Rajfur, Z.; Mitchell, J.A.; Pyrc, K. Canine Respiratory Coronavirus, Bovine Coronavirus, and Human Coronavirus OC43: Receptors and Attachment Factors. Viruses 2019, 11, 328. [Google Scholar] [CrossRef]
- Gerdts, V.; Zakhartchouk, A. Vaccines for porcine epidemic diarrhea virus and other swine coronaviruses. Vet. Microbiol. 2017, 206, 45–51. [Google Scholar] [CrossRef]
- Saif, L.J. Vaccines for covid-19: Perspectives, prospects, and challenges based on candidate sars, mers, and animal coronavirus vaccines. Eur. Med. J. 2020. [Google Scholar] [CrossRef]
- Zhou, L.; Li, Q.N.; Su, J.N.; Chen, G.H.; Wu, Z.X.; Luo, Y.; Wu, R.T.; Sun, Y.; Lan, T.; Ma, J.Y. The re-emerging of SADS-CoV infection in pig herds in Southern China. Transbound. Emerg. Dis. 2019, 66, 2180–2183. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.L.; Yu, J.Q.; Huang, Y.W. Swine enteric alphacoronavirus (swine acute diarrhea syndrome coronavirus): An update three years after its discovery. Virus Res. 2020, 285. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Chen, J.; Liu, Y.; Da, S.; Shi, H.; Zhang, X.; Liu, J.; Cao, L.; Zhu, X.; Wang, X.; et al. Pathogenicity of porcine deltacoronavirus (PDCoV) strain NH and immunization of pregnant sows with an inactivated PDCoV vaccine protects 5-day-old neonatal piglets from virulent challenge. Transbound. Emerg. Dis. 2020, 67, 572–583. [Google Scholar] [CrossRef] [PubMed]
- Mora-Díaz, J.C.; Piñeyro, P.E.; Houston, E.; Zimmerman, J.; Giménez-Lirola, L.G. Porcine hemagglutinating encephalomyelitis virus: A review. Front. Vet. Sci. 2019, 6, 53. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Zhao, K.; He, W.; Gao, W.; Zhao, C.; Wang, L.; Pan, W.; Song, D.; Wang, C.; Gao, F. Comparative evaluation of two hemagglutinating encephalomyelitis coronavirus vaccine candidates in mice. Clin. Vaccine Immunol. 2012, 19, 1102–1109. [Google Scholar] [CrossRef]
- Legnardi, M.; Tucciarone, C.M.; Franzo, G.; Cecchinato, M. Infectious bronchitis virus evolution, diagnosis and control. Vet. Sci. 2020, 7, 79. [Google Scholar] [CrossRef] [PubMed]
- Bande, F.; Arshad, S.S.; Hair Bejo, M.; Moeini, H.; Omar, A.R. Progress and challenges toward the development of vaccines against avian infectious bronchitis. J. Immunol. Res. 2015, 424860. [Google Scholar] [CrossRef]
- Scherk, M.; Ford, R.B.; Gaskell, R.M.; Hartmann, K.; Hurley, K.F.; Lappin, M.R.; Levy, J.K.; Little, S.E.; Nordone, S.; Sparkes, A.H. 2013 AAFP Feline Vaccination. J. Feline Med. Surg. 2016, 15, 785–808. [Google Scholar] [CrossRef]
- Erles, K.; Brownlie, J. Canine Respiratory Coronavirus: An Emerging Pathogen in the Canine Infectious Respiratory Disease Complex. Vet. Clin. N. Am. Small Anim. Pract. 2008, 38, 815–825. [Google Scholar] [CrossRef]
- Tyrrel, D.A.J.; Almedia, J.D.; Berry, D.M.; Cunningham, C.H.; Hamre, D.; Hofstad, M.S.; Malluci, L.; McIntosh, K. Coronavirus. Nature 1968, 220, 650. [Google Scholar]
- Weiss, S.R.; Navas-Martin, S. Coronavirus Pathogenesis and the Emerging Pathogen Severe Acute Respiratory Syndrome Coronavirus. Microbiol. Mol. Biol. Rev. 2005, 69, 635–664. [Google Scholar] [CrossRef] [PubMed]
- Artika, I.M.; Dewantari, A.K.; Wiyatno, A. Molecular biology of coronaviruses: Current knowledge. Heliyon 2020, 6, e04743. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Grunewald, M.; Perlman, S. Coronaviruses: An Updated Overview of Their Replication and Pathogenesis. Methods Mol. Biol. 2020, 2203, 1–29. [Google Scholar] [CrossRef] [PubMed]
- Fehr, A.R.; Perlman, S. Coronaviruses: An overview of their replication and pathogenesis. In Coronaviruses: Methods and Protocols; Humana Press: Valley Stream, NY, USA, 2015; Volume 1282, pp. 1–23. [Google Scholar] [CrossRef]
- De Wit, E.; Van Doremalen, N.; Falzarano, D.; Munster, V.J. SARS and MERS: Recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016, 14, 523–534. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, Q.; Guo, D. Emerging coronaviruses: Genome structure, replication, and pathogenesis. J. Med. Virol. 2020, 92, 418–423. [Google Scholar] [CrossRef]
- Li, F. Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu. Rev. Virol. 2016, 3, 237–261. [Google Scholar] [CrossRef]
- Hulswit, R.J.G.; de Haan, C.A.M.; Bosch, B.J. Coronavirus Spike Protein and Tropism Changes. In Advances in Virus Research; Academic Press: San Diego, CA, USA, 2016; Volume 96, pp. 29–57. [Google Scholar] [CrossRef]
- Huang, Y.; Yang, C.; Xu, X.F.; Xu, W.; Liu, S.W. Structural and functional properties of SARS-CoV-2 spike protein: Potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020. [Google Scholar] [CrossRef]
- Schoeman, D.; Fielding, B.C. Coronavirus envelope protein: Current knowledge. Virol. J. 2019, 16, 69. [Google Scholar] [CrossRef]
- DeDiego, M.L.; Álvarez, E.; Almazán, F.; Rejas, M.T.; Lamirande, E.; Roberts, A.; Shieh, W.-J.; Zaki, S.R.; Subbarao, K.; Enjuanes, L. A Severe Acute Respiratory Syndrome Coronavirus That Lacks the E Gene Is Attenuated In Vitro and In Vivo. J. Virol. 2007, 81, 1701–1713. [Google Scholar] [CrossRef]
- Kuo, L.; Masters, P.S. The Small Envelope Protein E Is Not Essential for Murine Coronavirus Replication. J. Virol. 2003, 77, 4597–4608. [Google Scholar] [CrossRef]
- Ortego, J.; Ceriani, J.E.; Patiño, C.; Plana, J.; Enjuanes, L. Absence of E protein arrests transmissible gastroenteritis coronavirus maturation in the secretory pathway. Virology 2007, 368, 296–308. [Google Scholar] [CrossRef]
- Almazán, F.; Dediego, M.L.; Sola, I.; Zuñiga, S.; Nieto-Torres, J.L.; Marquez-Jurado, S.; Andrés, G.; Enjuanes, L. Engineering a replication-competent, propagation-defective middle east respiratory syndrome coronavirus as a vaccine candidate. MBio 2013, 4. [Google Scholar] [CrossRef] [PubMed]
- Neuman, B.W.; Kiss, G.; Kunding, A.H.; Bhella, D.; Baksh, M.F.; Connelly, S.; Droese, B.; Klaus, J.P.; Makino, S.; Sawicki, S.G.; et al. A structural analysis of M protein in coronavirus assembly and morphology. J. Struct. Biol. 2011, 174, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Masters, P.S. The Molecular Biology of Coronaviruses. Adv. Virus Res. 2006, 65, 193–292. [Google Scholar] [CrossRef]
- Nal, B.; Chan, C.; Kien, F.; Siu, L.; Tse, J.; Chu, K.; Kam, J.; Staropoli, S.; Crescenzo-Chaigne, B.; Escriou, N.; et al. Differential maturation and subcellular localization of severe acute respiratory syndrome coronavirus surface proteins S, M and E. J. Gen. Virol. 2005, 86, 1423–1434. [Google Scholar] [CrossRef]
- McBride, R.; van Zyl, M.; Fielding, B.C. The coronavirus nucleocapsid is a multifunctional protein. Viruses 2014, 6, 2991. [Google Scholar] [CrossRef]
- Cui, L.; Wang, H.; Ji, Y.; Yang, J.; Xu, S.; Huang, X.; Wang, Z.; Qin, L.; Tien, P.; Zhou, X.; et al. The Nucleocapsid Protein of Coronaviruses Acts as a Viral Suppressor of RNA Silencing in Mammalian Cells. J. Virol. 2015, 89, 9029–9043. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Q.; Langereis, M.A.; Van Vliet, A.L.W.; Huizinga, E.G.; De Groot, R.J. Structure of coronavirus hemagglutinin-esterase offers insight into corona and influenza virus evolution. Proc. Natl. Acad. Sci. USA 2008, 105, 9065–9069. [Google Scholar] [CrossRef]
- Michel, C.J.; Mayer, C.; Poch, O.; Thompson, J.D. Characterization of accessory genes in coronavirus genomes. Virol. J. 2020, 17, 131. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.X.; Fung, T.S.; Chong, K.K.L.; Shukla, A.; Hilgenfeld, R. Accessory proteins of SARS-CoV and other coronaviruses. Antivir. Res. 2014, 109, 97–109. [Google Scholar] [CrossRef]
- Hartenian, E.; Nandakumar, D.; Lari, A.; Ly, M.; Tucker, J.M.; Glaunsinger, B.A. The molecular virology of Coronaviruses. J. Biol. Chem. 2020, 295, 12910–12934. [Google Scholar] [CrossRef] [PubMed]
- De Haan, C.A.M.; Masters, P.S.; Shen, X.; Weiss, S.; Rottier, P.J.M. The group-specific murine coronavirus genes are not essential, but their deletion, by reverse genetics, is attenuating in the natural host. Virology 2002, 296, 177–189. [Google Scholar] [CrossRef]
- Shen, S.; Wen, Z.L.; Liu, D.X. Emergence of a coronavirus infectious bronchitis virus mutant with a truncated 3b gene: Functional characterization of the 3b protein in pathogenesis and replication. Virology 2003, 311, 16–27. [Google Scholar] [CrossRef]
- Narayanan, K.; Huang, C.; Makino, S. SARS coronavirus accessory proteins. Virus Res. 2008, 133, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Su, S.; Wong, G.; Shi, W.; Liu, J.; Lai, A.C.K.; Zhou, J.; Liu, W.; Bi, Y.; Gao, G.F. Epidemiology, Genetic Recombination, and Pathogenesis of Coronaviruses. Trends Microbiol. 2016, 24, 490–502. [Google Scholar] [CrossRef] [PubMed]
- Forni, D.; Cagliani, R.; Clerici, M.; Sironi, M. Molecular Evolution of Human Coronavirus Genomes. Trends Microbiol. 2017, 25, 35–48. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.Y.; Huang, Y.; Lau, S.K.P.; Yuen, K.Y. Coronavirus genomics and bioinformatics analysis. Viruses 2010, 2, 1805–1820. [Google Scholar] [CrossRef]
- Phan, T. Genetic diversity and evolution of SARS-CoV-2. Infect. Genet. Evol. 2020, 81. [Google Scholar] [CrossRef]
- Robson, F.; Khan, K.S.; Le, T.K.; Paris, C.; Demirbag, S.; Barfuss, P.; Rocchi, P.; Ng, W.-L. Coronavirus RNA proofreading: Molecular basis and therapeutic targeting. Mol. Cell 2020. [Google Scholar] [CrossRef]
- Denison, M.R.; Graham, R.L.; Donaldson, E.F.; Eckerle, L.D.; Baric, R.S. Coronaviruses: An RNA proofreading machine regulates replication fidelity and diversity. RNA Biol. 2011, 8. [Google Scholar] [CrossRef]
- Graham, R.L.; Baric, R.S. Recombination, Reservoirs, and the Modular Spike: Mechanisms of Coronavirus Cross-Species Transmission. J. Virol. 2010, 84, 3134–3146. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Liu, W.J.; Xu, W.; Jin, T.; Zhao, Y.; Song, J.; Shi, Y.; Ji, W.; Jia, H.; Zhou, Y.; et al. A Bat-Derived Putative Cross-Family Recombinant Coronavirus with a Reovirus Gene. PLoS Pathog. 2016, 12. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Wu, C.; Li, X.; Song, Y.; Yao, X.; Wu, X.; Duan, Y.; Zhang, H.; Wang, Y.; Qian, Z.; et al. On the origin and continuing evolution of SARS-CoV-2. Natl. Sci. Rev. 2020, 7, 1012–1023. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef]
- Lam, T.T.Y.; Jia, N.; Zhang, Y.W.; Shum, M.H.H.; Jiang, J.F.; Zhu, H.C.; Tong, Y.G.; Shi, Y.X.; Ni, X.B.; Liao, Y.S.; et al. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature 2020, 583, 282–285. [Google Scholar] [CrossRef]
- Korber, B.; Fischer, W.M.; Gnanakaran, S.; Yoon, H.; Theiler, J.; Abfalterer, W.; Hengartner, N.; Giorgi, E.E.; Bhattacharya, T.; Foley, B.; et al. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell 2020, 182, 812–827. [Google Scholar] [CrossRef]
- Young, B.E.; Fong, S.-W.; Chan, Y.-H.; Mak, T.-M.; Ang, L.W.; Anderson, D.E.; Lee, C.Y.-P.; Amrun, S.N.; Lee, B.; Goh, Y.S.; et al. Effects of a major deletion in the SARS-CoV-2 genome on the severity of infection and the inflammatory response: An observational cohort study. Lancet 2020. [Google Scholar] [CrossRef]
- Grubaugh, N.D.; Hanage, W.P.; Rasmussen, A.L. Making Sense of Mutation: What D614G Means for the COVID-19 Pandemic Remains Unclear. Cell 2020, 182, 794–795. [Google Scholar] [CrossRef]
- Song, H.D.; Tu, C.C.; Zhang, G.W.; Wang, S.Y.; Zheng, K.; Lei, L.C.; Chen, Q.X.; Gao, Y.W.; Zhou, H.Q.; Xiang, H.; et al. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc. Natl. Acad. Sci. USA 2005, 102, 2430–2435. [Google Scholar] [CrossRef]
- He, J.F.; Peng, G.W.; Min, J.; Yu, D.W.; Liang, W.J.; Zhang, S.Y.; Xu, R.H.; Zheng, H.Y.; Wu, X.W.; Xu, J.; et al. Molecular Evolution of the SARS Coronavirus, during the Course of the SARS Epidemic in China. Science 2004, 303, 1666–1669. [Google Scholar] [CrossRef]
- Zhang, Z.; Shen, L.; Gu, X. Evolutionary Dynamics of MERS-CoV: Potential Recombination, Positive Selection and Transmission. Sci. Rep. 2016, 6, 25049. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.X.; Liang, J.Q.; Fung, T.S. Human Coronavirus-229E, -OC43, -NL63, and -HKU1. Ref. Modul. Life Sci. 2020. [CrossRef]
- Corman, V.M.; Muth, D.; Niemeyer, D.; Drosten, C. Hosts and Sources of Endemic Human Coronaviruses. Adv. Virus Res. 2018, 100, 163–188. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Li, S.Y.; Yang, X.L.; Huang, H.M.; Zhang, Y.J.; Guo, H.; Luo, C.M.; Miller, M.; Zhu, G.; Chmura, A.A.; et al. Serological Evidence of Bat SARS-Related Coronavirus Infection in Humans, China. Virol. Sin. 2018, 33, 104–107. [Google Scholar] [CrossRef]
- Ge, X.Y.; Li, J.L.; Yang, X.L.; Chmura, A.A.; Zhu, G.; Epstein, J.H.; Mazet, J.K.; Hu, B.; Zhang, W.; Peng, C.; et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature 2013, 503, 535–538. [Google Scholar] [CrossRef] [PubMed]
- Wong, A.C.P.; Li, X.; Lau, S.K.P.; Woo, P.C.Y. Global epidemiology of bat coronaviruses. Viruses 2019, 11, 174. [Google Scholar] [CrossRef]
- Lau, S.K.P.; Woo, P.C.Y.; Li, K.S.M.; Tsang, A.K.L.; Fan, R.Y.Y.; Luk, H.K.H.; Cai, J.-P.; Chan, K.-H.; Zheng, B.-J.; Wang, M.; et al. Discovery of a Novel Coronavirus, China Rattus Coronavirus HKU24, from Norway Rats Supports the Murine Origin of Betacoronavirus 1 and Has Implications for the Ancestor of Betacoronavirus Lineage A. J. Virol. 2015, 89, 3076–3092. [Google Scholar] [CrossRef]
- Wille, M.; Holmes, E.C. Wild birds as reservoirs for diverse and abundant gamma- and deltacoronaviruses. FEMS Microbiol. Rev. 2020. [Google Scholar] [CrossRef]
- Lorusso, A.; Decaro, N.; Schellen, P.; Rottier, P.J.M.; Buonavoglia, C.; Haijema, B.-J.; de Groot, R.J. Gain, Preservation, and Loss of a Group 1a Coronavirus Accessory Glycoprotein. J. Virol. 2008, 82, 10312–10317. [Google Scholar] [CrossRef]
- van Nguyen, D.; Terada, Y.; Minami, S.; Yonemitsu, K.; Nagata, N.; Le, T.D.H.; Kuwata, R.; Shimoda, H.; Maeda, K. Characterization of canine coronavirus spread among domestic dogs in Vietnam. J. Vet. Med. Sci. 2017, 79, 343–349. [Google Scholar] [CrossRef]
- Wang, X.; Li, C.; Guo, D.; Wei, S.; Geng, Y.; Wang, E.; Wang, Z.; Zhao, X.; Su, M.; Liu, Q.; et al. Co-circulation of canine coronavirus I and IIa/b with high prevalence and genetic diversity in Heilongjiang Province, Northeast China. PLoS ONE 2016, 11. [Google Scholar] [CrossRef] [PubMed]
- Moutinho Costa, E.; Xavier de Castro, T.; de Oliveira Bottino, F.; Nasser Cubel Garcia, R.d.C. Molecular characterization of canine coronavirus strains circulating in Brazil. Vet. Microbiol. 2014, 168, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Decaro, N.; Mari, V.; Elia, G.; Addie, D.D.; Camero, M.; Lucente, M.S.; Martella, V.; Buonavoglia, C. Recombinant canine coronaviruses in dogs, Europe. Emerg. Infect. Dis. 2010, 16, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Santana-Clavijo, N.F.; Reyes Romero, D.P.; Arango Fajardo, D.F.; Velandia Muñoz, A.; Taniwaki, S.A.; de Souza Silva, S.O.; Brandão, P.E. Molecular diversity of Alphacoronavirus 1 in dogs and cats in Colombia. Heliyon 2020, 6. [Google Scholar] [CrossRef] [PubMed]
- Terada, Y.; Matsui, N.; Noguchi, K.; Kuwata, R.; Shimoda, H.; Soma, T.; Mochizuki, M.; Maeda, K. Emergence of pathogenic coronaviruses in cats by homologous recombination between feline and canine coronaviruses. PLoS ONE 2014, 9, e0106534. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, M.A. Feline Infectious Peritonitis: Update on Pathogenesis, Diagnostics, and Treatment. Vet. Clin. N. Am. Small Anim. Pract. 2020, 50, 1001–1011. [Google Scholar] [CrossRef]
- Barker, E.N.; Stranieri, A.; Helps, C.R.; Porter, E.L.; Davidson, A.D.; Day, M.J.; Knowles, T.; Kipar, A.; Tasker, S. Limitations of using feline coronavirus spike protein gene mutations to diagnose feline infectious peritonitis. Vet. Res. 2017, 48. [Google Scholar] [CrossRef]
- Kipar, A.; Meli, M.L. Feline Infectious Peritonitis: Still an Enigma? Vet. Pathol. 2014, 51, 505–526. [Google Scholar] [CrossRef]
- Wise, A.G.; Kiupel, M.; Garner, M.M.; Clark, A.K.; Maes, R.K. Comparative sequence analysis of the distal one-third of the genomes of a systemic and an enteric ferret coronavirus. Virus Res. 2010, 149, 42–50. [Google Scholar] [CrossRef]
- Stout, A.E.; André, N.M.; Jaimes, J.A.; Millet, J.K.; Whittaker, G.R. Coronaviruses in cats and other companion animals: Where does SARS-CoV-2/COVID-19 fit? Vet. Microbiol. 2020, 247, 108777. [Google Scholar] [CrossRef]
- Lamers, M.M.; Smits, S.L.; Hundie, G.B.; Provacia, L.B.; Koopmans, M.; Osterhaus, A.D.M.E.; Haagmans, B.L.; Raj, V.S. Naturally occurring recombination in ferret coronaviruses revealed by complete genome characterization. J. Gen. Virol. 2016, 97, 2180–2186. [Google Scholar] [CrossRef] [PubMed]
- Minami, S.; Kuroda, Y.; Terada, Y.; Yonemitsu, K.; Van Nguyen, D.; Kuwata, R.; Shimoda, H.; Takano, A.; Maeda, K. Detection of novel ferret coronaviruses and evidence of recombination among ferret coronaviruses. Virus Genes 2016, 52, 858–862. [Google Scholar] [CrossRef] [PubMed]
- Jung, K.; Saif, L.J.; Wang, Q. Porcine epidemic diarrhea virus (PEDV): An update on etiology, transmission, pathogenesis, and prevention and control. Virus Res. 2020, 286. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.; Kulcsar, K.; Misra, V.; Frieman, M.; Mossman, K. Bats and coronaviruses. Viruses 2019, 11, 41. [Google Scholar] [CrossRef]
- Zhou, P.; Fan, H.; Lan, T.; Yang, X.L.; Shi, W.F.; Zhang, W.; Zhu, Y.; Zhang, Y.W.; Xie, Q.M.; Mani, S.; et al. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature 2018, 556, 255–259. [Google Scholar] [CrossRef]
- Lin, S.Y.; Chen, H.W. Infectious bronchitis virus variants: Molecular analysis and pathogenicity investigation. Int. J. Mol. Sci. 2017, 18, 2030. [Google Scholar] [CrossRef]
- Brown, P.A.; Touzain, F.; Briand, F.X.; Gouilh, A.M.; Courtillon, C.; Allée, C.; Lemaitre, E.; De Boisséson, C.; Blanchard, Y.; Eterradossi, N. First complete genome sequence of European Turkey coronavirus suggests complex recombination history related with US Turkey and guinea fowl coronaviruses. J. Gen. Virol. 2016, 97, 110–120. [Google Scholar] [CrossRef]
- Wang, Y.; Cui, X.; Chen, X.; Yang, S.; Ling, Y.; Song, Q.; Zhu, S.; Sun, L.; Li, C.; Li, Y.; et al. A recombinant infectious bronchitis virus from a chicken with a spike gene closely related to that of a turkey coronavirus. Arch. Virol. 2020, 165, 703–707. [Google Scholar] [CrossRef]
- Arslan, M.; Xu, B.; Gamal El-Din, M. Transmission of SARS-CoV-2 via fecal-oral and aerosols–borne routes: Environmental dynamics and implications for wastewater management in underprivileged societies. Sci. Total Environ. 2020, 743. [Google Scholar] [CrossRef]
- Jiang, X.; Luo, M.; Zou, Z.; Wang, X.; Chen, C.; Qiu, J. Asymptomatic SARS-CoV-2 infected case with viral detection positive in stool but negative in nasopharyngeal samples lasts for 42 days. J. Med. Virol. 2020. [Google Scholar] [CrossRef]
- Liu, J.; Xiao, Y.; Shen, Y.; Shi, C.; Chen, Y.; Shi, P.; Gao, Y.; Wang, Y.; Lu, B. Detection of SARS-CoV-2 by RT-PCR in anal from patients who have recovered from coronavirus disease 2019. J. Med. Virol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Hindson, J. COVID-19: Faecal–oral transmission? Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 259. [Google Scholar] [CrossRef] [PubMed]
- Conticini, E.; Frediani, B.; Caro, D. Can atmospheric pollution be considered a co-factor in extremely high level of SARS-CoV-2 lethality in Northern Italy? Environ. Pollut. 2020, 261, 114465. [Google Scholar] [CrossRef] [PubMed]
- Coccia, M. Factors determining the diffusion of COVID-19 and suggested strategy to prevent future accelerated viral infectivity similar to COVID. Sci. Total Environ. 2020, 729, 138474. [Google Scholar] [CrossRef] [PubMed]
- Van Doremalen, N.; Bushmaker, T.; Morris, D.H.; Holbrook, M.G.; Gamble, A.; Williamson, B.N.; Tamin, A.; Harcourt, J.L.; Thornburg, N.J.; Gerber, S.I.; et al. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N. Engl. J. Med. 2020, 382, 1564–1567. [Google Scholar] [CrossRef]
- Niederwerder, M.C.; Hesse, R.A. Swine enteric coronavirus disease: A review of 4 years with porcine epidemic diarrhoea virus and porcine deltacoronavirus in the United States and Canada. Transbound. Emerg. Dis. 2018, 65, 660–675. [Google Scholar] [CrossRef]
- Priestnall, S.L. Canine Respiratory Coronavirus: A Naturally Occurring Model of COVID-19? Vet. Pathol. 2020, 57, 467–471. [Google Scholar] [CrossRef]
- Ramakrishnan, S.; Kappala, D. Avian infectious bronchitis virus. In Recent Advances in Animal Virology; Malik, Y.S., Singh, R.K., Yadav, M.P., Eds.; Springer: Singapore, 2020; pp. 301–319. [Google Scholar] [CrossRef]
- Ignjatović, J.; Sapats, S. Avian infectious bronchitis virus. OIE Rev. Sci. Tech. 2000, 19, 493–508. [Google Scholar] [CrossRef]
- Decaro, N.; Campolo, M.; Desario, C.; Cirone, F.; D’Abramo, M.; Lorusso, E.; Greco, G.; Mari, V.; Colaianni, M.L.; Elia, G.; et al. Respiratory disease associated with bovine coronavirus infection in cattle herds in Southern Italy. J. Vet. Diagn. Investig. 2008, 20, 28–32. [Google Scholar] [CrossRef]
- Barthold, S.W.; Beck, D.S.; Smith, A.L. Mouse hepatitis virus and host determinants of vertical transmission and maternally-derived passive immunity in mice. Arch. Virol. 1988, 100, 171–183. [Google Scholar] [CrossRef]
- Hartmann, K. Feline infectious peritonitis. Vet. Clin. N. Am. Small Anim. Pract. 2005, 35, 39–79. [Google Scholar] [CrossRef] [PubMed]
- Pereira, C.G.; Saraiva, G.L.; Vidigal, P.M.P.; Fietto, J.L.R.; Bressan, G.C.; Moreira, M.A.S.; de Almeida, M.R.; Júnior, A.S. Distribution of infectious bronchitis virus strains in different organs and evidence of vertical transmission in natural infection. Arch. Virol. 2016, 161, 3355–3363. [Google Scholar] [CrossRef] [PubMed]
- OIE. Infection with SARS-Cov-2 in Animals. Available online: https://www.oie.int/fileadmin/Home/MM/A_Factsheet_SARS-CoV-2__1_.pdf (accessed on 18 September 2020).
- Halfmann, P.J.; Hatta, M.; Chiba, S.; Maemura, T.; Fan, S.; Takeda, M.; Kinoshita, N.; Hattori, S.I.; Sakai-Tagawa, Y.; Iwatsuki-Horimoto, K.; et al. Transmission of SARS-CoV-2 in Domestic Cats. N. Engl. J. Med. 2020, 383, 592–594. [Google Scholar] [CrossRef] [PubMed]
- Gaudreault, N.N.; Trujillo, J.D.; Carossino, M.; Meekins, D.A.; Madden, D.W.; Indran, S.V.; Bold, D.; Balaraman, V.; Kwon, T.; Libanori Artiaga, B.; et al. SARS-CoV-2 infection, disease and transmission in domestic cats Running title: SARS-CoV-2 in domestic cats. Emerg Microbes Infect. 2020, 9, 2322–2332. [Google Scholar] [CrossRef] [PubMed]
- Schlottau, K.; Rissmann, M.; Graaf, A.; Schön, J.; Sehl, J.; Wylezich, C.; Höper, D.; Mettenleiter, T.C.; Balkema-Buschmann, A.; Harder, T.; et al. SARS-CoV-2 in fruit bats, ferrets, pigs, and chickens: An experimental transmission study. Lancet Microbe 2020, 1, e218–e225. [Google Scholar] [CrossRef]
- Shi, J.; Wen, Z.; Zhong, G.; Yang, H.; Wang, C.; Liu, R.; He, X.; Shuai, L.; Sun, Z.; Zhao, Y.; et al. Susceptibility of ferrets, cats, dogs, and different domestic animals to SARS-coronavirus-2. Science 2020, 368, 1016–1020. [Google Scholar] [CrossRef] [PubMed]
- Olival, K.J.; Cryan, P.M.; Amman, B.R.; Baric, R.S.; Blehert, D.S.; Brook, C.E.; Calisher, C.H.; Castle, K.T.; Coleman, J.T.H.; Daszak, P.; et al. Possibility for reverse zoonotic transmission of SARS-CoV-2 to free-ranging wildlife: A case study of bats. PLoS Pathog. 2020, 16, e1008758. [Google Scholar] [CrossRef]
- Oreshkova, N.; Molenaar, R.J.; Vreman, S.; Harders, F.; Oude Munnink, B.B.; Van Der Honing, R.W.H.; Gerhards, N.; Tolsma, P.; Bouwstra, R.; Sikkema, R.S.; et al. SARS-CoV-2 infection in farmed minks, the Netherlands, April and May 2020. Eurosurveillance 2020, 25, 2001005. [Google Scholar] [CrossRef]
- Temmam, S.; Barbarino, A.; Maso, D.; Behillil, S.; Enouf, V.; Huon, C.; Jaraud, A.; Chevallier, L.; Backovic, M.; Pérot, P.; et al. Absence of SARS-CoV-2 infection in cats and dogs in close contact with a cluster of COVID-19 patients in a veterinary campus. One Health 2020, 10, 100164. [Google Scholar] [CrossRef]
- Tay, M.Z.; Poh, C.M.; Rénia, L.; MacAry, P.A.; Ng, L.F.P. The trinity of COVID-19: Immunity, inflammation and intervention. Nat. Rev. Immunol. 2020, 20, 363–374. [Google Scholar] [CrossRef]
- Lai, C.C.; Ko, W.C.; Lee, P.I.; Jean, S.S.; Hsueh, P.R. Extra-respiratory manifestations of COVID-19. Int. J. Antimicrob. Agents 2020, 56, 106024. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Guo, H.; Zhou, P.; Shi, Z.L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2020, 6, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Shang, J.; Ye, G.; Shi, K.; Wan, Y.; Luo, C.; Aihara, H.; Geng, Q.; Auerbach, A.; Li, F. Structural basis of receptor recognition by SARS-CoV-2. Nature 2020, 581, 221–224. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Zhang, Y.; Wu, L.; Niu, S.; Song, C.; Zhang, Z.; Lu, G.; Qiao, C.; Hu, Y.; Yuen, K.Y.; et al. Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell 2020, 181, 894–904. [Google Scholar] [CrossRef] [PubMed]
- Moreira, R.A.; Chwastyk, M.; Baker, J.L.; Guzman, H.V.; Poma, A.B. Quantitative determination of mechanical stability in the novel coronavirus spike protein. Nanoscale 2020, 12, 16409–16413. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wang, R.; Wang, M.; Wei, G.W. Mutations Strengthened SARS-CoV-2 Infectivity. J. Mol. Biol. 2020, 432, 5212–5226. [Google Scholar] [CrossRef] [PubMed]
- Qiao, B.; De La Cruz, M.O. Enhanced binding of SARS-CoV-2 spike protein to receptor by distal polybasic cleavage sites. ACS Nano 2020, 14, 10616–10623. [Google Scholar] [CrossRef]
- Plante, J.A.; Liu, Y.; Liu, J.; Xia, H.; Johnson, B.A.; Lokugamage, K.G.; Zhang, X.; Muruato, A.E.; Zou, J.; Fontes-Garfias, C.R.; et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature 2020. [Google Scholar] [CrossRef]
- Domingo, P.; Mur, I.; Pomar, V.; Corominas, H.; Casademont, J.; de Benito, N. The four horsemen of a viral Apocalypse: The pathogenesis of SARS-CoV-2 infection (COVID-19). EBioMedicine 2020, 58, 102887. [Google Scholar] [CrossRef] [PubMed]
- Tekes, G.; Thiel, H.J. Feline Coronaviruses: Pathogenesis of Feline Infectious Peritonitis. Adv. Virus Res. 2016, 96, 193–218. [Google Scholar] [CrossRef]
- Rissi, D.R. A retrospective study of the neuropathology and diagnosis of naturally occurring feline infectious peritonitis. J. Vet. Diagn. Investig. 2018, 30, 392–399. [Google Scholar] [CrossRef] [PubMed]
- Malbon, A.J.; Fonfara, S.; Meli, M.L.; Hahn, S.; Egberink, H.; Kipar, A. Feline infectious peritonitis as a systemic inflammatory disease: Contribution of liver and heart to the pathogenesis. Viruses 2019, 11, 1144. [Google Scholar] [CrossRef] [PubMed]
- Goitsuka, R.; Ohashi, T.; Ono, K.; Yasukawa, K.; Koishibara, Y.; Fukui, H.; Ohsugi, Y.; Hasegawa, A. IL-6 activity in feline infectious peritonitis. J. Immunol. 1990, 144, 2599–2603. [Google Scholar] [PubMed]
- Gubernatorova, E.O.; Gorshkova, E.A.; Polinova, A.I.; Drutskaya, M.S. IL-6: Relevance for immunopathology of SARS-CoV-2. Cytokine Growth Factor Rev. 2020, 53, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Schultze, B.; Krempl, C.; Ballesteros, M.L.; Shaw, L.; Schauer, R.; Enjuanes, L.; Herrler, G. Transmissible gastroenteritis coronavirus, but not the related porcine respiratory coronavirus, has a sialic acid (N-glycolylneuraminic acid) binding activity. J. Virol. 1996, 70, 5634–5637. [Google Scholar] [CrossRef]
- Schwegmann-Weßels, C.; Herrler, G. Transmissible gastroenteritis virus infection: A vanishing specter. Dtsch. Tierarztl. Wochenschr. 2006, 113, 157–159. [Google Scholar]
- Alfano, F.; Fusco, G.; Mari, V.; Occhiogrosso, L.; Miletti, G.; Brunetti, R.; Galiero, G.; Desario, C.; Cirilli, M.; Decaro, N. Circulation of pantropic canine coronavirus in autochthonous and imported dogs, Italy. Transbound. Emerg. Dis. 2020. [Google Scholar] [CrossRef]
- Pinto, L.D.; Barros, I.N.; Budaszewski, R.F.; Weber, M.N.; Mata, H.; Antunes, J.R.; Boabaid, F.M.; Wouters, A.T.B.; Driemeier, D.; Brandão, P.E.; et al. Characterization of pantropic canine coronavirus from Brazil. Vet. J. 2014, 202, 659–662. [Google Scholar] [CrossRef]
- Decaro, N.; Cordonnier, N.; Demeter, Z.; Egberink, H.; Elia, G.; Grellet, A.; Poder, S.L.; Mari, V.; Martella, V.; Ntafis, V.; et al. European surveillance for pantropic canine coronavirus. J. Clin. Microbiol. 2013, 51, 83–88. [Google Scholar] [CrossRef]
- Marinaro, M.; Mari, V.; Bellacicco, A.L.; Tarsitano, E.; Elia, G.; Losurdo, M.; Rezza, G.; Buonavoglia, C.; Decaro, N. Prolonged depletion of circulating CD4+ T lymphocytes and acute monocytosis after pantropic canine coronavirus infection in dogs. Virus Res. 2010, 152, 73–78. [Google Scholar] [CrossRef]
- Zhang, G.; Li, B.; Yoo, D.; Qin, T.; Zhang, X.; Jia, Y.; Cui, S. Animal coronaviruses and SARS-CoV-2. Transbound. Emerg. Dis. 2020, 16. [Google Scholar] [CrossRef] [PubMed]
- Ellis, J. What is the evidence that bovine coronavirus is a biologically significant respiratory pathogen in cattle? Can. Vet. J. 2019, 60, 147–152. [Google Scholar] [PubMed]
- Zhang, X.; Hasoksuz, M.; Spiro, D.; Halpin, R.; Wang, S.; Vlasova, A.; Janies, D.; Jones, L.R.; Ghedin, E.; Saif, L.J. Quasispecies of bovine enteric and respiratory coronaviruses based on complete genome sequences and genetic changes after tissue culture adaptation. Virology 2007, 363, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.T.; Chen, T.C.; Lin, S.Y.; Mase, M.; Murakami, S.; Horimoto, T.; Chen, H.W. Emerging lethal infectious bronchitis coronavirus variants with multiorgan tropism. Transbound. Emerg. Dis. 2020, 67, 884–893. [Google Scholar] [CrossRef] [PubMed]
- Cavanagh, D. Coronavirus avian infectious bronchitis virus. Vet. Res. 2007, 38, 281–297. [Google Scholar] [CrossRef] [PubMed]
- Compton, S.R.; Ball-Goodrich, L.J.; Johnson, L.K.; Johnson, E.A.; Paturzo, F.X.; Macy, J.D. Pathogenesis of enterotropic mouse hepatitis virus in immunocompetent and immunodeficient mice. Comp. Med. 2004, 54, 681–689. [Google Scholar] [PubMed]
- Kaddoura, M.; AlIbrahim, M.; Hijazi, G.; Soudani, N.; Audi, A.; Alkalamouni, H.; Haddad, S.; Eid, A.; Zaraket, H. COVID-19 Therapeutic Options Under Investigation. Front. Pharmacol. 2020, 11, 1196. [Google Scholar] [CrossRef]
- Magro, G. COVID-19: Review on latest available drugs and therapies against SARS-CoV-2. Coagulation and inflammation cross-talking. Virus Res. 2020, 286, 1980970. [Google Scholar] [CrossRef]
- Brown, A.J.; Won, J.J.; Graham, R.L.; Dinnon, K.H.; Sims, A.C.; Feng, J.Y.; Cihlar, T.; Denison, M.R.; Baric, R.S.; Sheahan, T.P. Broad spectrum antiviral remdesivir inhibits human endemic and zoonotic deltacoronaviruses with a highly divergent RNA dependent RNA polymerase. Antivir. Res. 2019, 169, 104541. [Google Scholar] [CrossRef]
- Agostini, M.L.; Andres, E.L.; Sims, A.C.; Graham, R.L.; Sheahan, T.P.; Lu, X.; Smith, E.C.; Case, J.B.; Feng, J.Y.; Jordan, R.; et al. Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. MBio 2018, 9, e00221-18. [Google Scholar] [CrossRef]
- Amirian, E.S.; Levy, J.K. Current knowledge about the antivirals remdesivir (GS-5734) and GS-441524 as therapeutic options for coronaviruses. One Health 2020, 9, 100128. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, N.C.; Perron, M.; Bannasch, M.; Montgomery, E.; Murakami, E.; Liepnieks, M.; Liu, H. Efficacy and safety of the nucleoside analog GS-441524 for treatment of cats with naturally occurring feline infectious peritonitis. J. Feline Med. Surg. 2019, 21, 271–281. [Google Scholar] [CrossRef]
- Murphy, B.G.; Perron, M.; Murakami, E.; Bauer, K.; Park, Y.; Eckstrand, C.; Liepnieks, M.; Pedersen, N.C. The nucleoside analog GS-441524 strongly inhibits feline infectious peritonitis (FIP) virus in tissue culture and experimental cat infection studies. Vet. Microbiol. 2018, 219, 226–233. [Google Scholar] [CrossRef]
- Dickinson, P.J.; Bannasch, M.; Thomasy, S.M.; Murthy, V.D.; Vernau, K.M.; Liepnieks, M.; Montgomery, E.; Knickelbein, K.E.; Murphy, B.; Pedersen, N.C. Antiviral treatment using the adenosine nucleoside analogue GS-441524 in cats with clinically diagnosed neurological feline infectious peritonitis. J. Vet. Intern. Med. 2020, 34, 1587–1593. [Google Scholar] [CrossRef] [PubMed]
- Yan, V.C.; Muller, F.L. Advantages of the Parent Nucleoside GS-441524 over Remdesivir for Covid-19 Treatment. ACS Med. Chem. Lett. 2020, 11, 1361–1366. [Google Scholar] [CrossRef] [PubMed]
- Tu, Y.F.; Chien, C.S.; Yarmishyn, A.A.; Lin, Y.Y.; Luo, Y.H.; Lin, Y.T.; Lai, W.Y.; Yang, D.M.; Chou, S.J.; Yang, Y.P.; et al. A review of sars-cov-2 and the ongoing clinical trials. Int. J. Mol. Sci. 2020, 21, 2657. [Google Scholar] [CrossRef]
- Vuong, W.; Khan, M.B.; Fischer, C.; Arutyunova, E.; Lamer, T.; Shields, J.; Saffran, H.A.; McKay, R.T.; van Belkum, M.J.; Joyce, M.; et al. Feline coronavirus drug inhibits the main protease of SARS-CoV-2 and blocks virus replication. Nat. Commun. 2020, 11, 4282. [Google Scholar] [CrossRef]
- Pedersen, N.C.; Kim, Y.; Liu, H.; Galasiti Kankanamalage, A.C.; Eckstrand, C.; Groutas, W.C.; Bannasch, M.; Meadows, J.M.; Chang, K.O. Efficacy of a 3C-like protease inhibitor in treating various forms of acquired feline infectious peritonitis. J. Feline Med. Surg. 2018, 20, 378–392. [Google Scholar] [CrossRef]
- Kim, Y.; Liu, H.; Galasiti Kankanamalage, A.C.; Weerasekara, S.; Hua, D.H.; Groutas, W.C.; Chang, K.O.; Pedersen, N.C. Reversal of the Progression of Fatal Coronavirus Infection in Cats by a Broad-Spectrum Coronavirus Protease Inhibitor. PLoS Pathog. 2016, 12, e1005531. [Google Scholar] [CrossRef]
- Ye, G.; Wang, X.; Tong, X.; Shi, Y.; Fu, Z.F.; Peng, G. Structural basis for inhibiting porcine epidemic diarrhea virus replication with the 3C-Like protease inhibitor GC376. Viruses 2020, 12, 240. [Google Scholar] [CrossRef]
- Perera, K.D.; Galasiti Kankanamalage, A.C.; Rathnayake, A.D.; Honeyfield, A.; Groutas, W.; Chang, K.O.; Kim, Y. Protease inhibitors broadly effective against feline, ferret and mink coronaviruses. Antivir. Res. 2018, 160, 79–86. [Google Scholar] [CrossRef]
- Hsieh, L.E.; Lin, C.N.; Su, B.L.; Jan, T.R.; Chen, C.M.; Wang, C.H.; Lin, D.S.; Lin, C.T.; Chueh, L.L. Synergistic antiviral effect of Galanthus nivalis agglutinin and nelfinavir against feline coronavirus. Antivir. Res. 2010, 88, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Chu, H.F.; Chen, C.C.; Moses, D.C.; Chen, Y.H.; Lin, C.H.; Tsai, Y.C.; Chou, C.Y. Porcine epidemic diarrhea virus papain-like protease 2 can be noncompetitively inhibited by 6-thioguanine. Antivir. Res. 2018, 158, 199–205. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; StJohn, S.E.; Osswald, H.L.; O’Brien, A.; Banach, B.S.; Sleeman, K.; Ghosh, A.K.; Mesecar, A.D.; Baker, S.C. Coronaviruses Resistant to a 3C-Like Protease Inhibitor Are Attenuated for Replication and Pathogenesis, Revealing a Low Genetic Barrier but High Fitness Cost of Resistance. J. Virol. 2014, 88, 11886–11898. [Google Scholar] [CrossRef] [PubMed]
- Hung, I.F.N.; Lung, K.C.; Tso, E.Y.K.; Liu, R.; Chung, T.W.H.; Chu, M.Y.; Ng, Y.Y.; Lo, J.; Chan, J.; Tam, A.R.; et al. Triple combination of interferon beta-1b, lopinavir–ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: An open-label, randomised, phase 2 trial. Lancet 2020, 395, 1695–1704. [Google Scholar] [CrossRef]
- Weiss, R.C.; Oostrom-Ram, T. Inhibitory effects of ribavirin alone or combined with human alpha interferon on feline infectious peritonitis virus replication in vitro. Vet. Microbiol. 1989, 20, 255–265. [Google Scholar] [CrossRef]
- Weiss, R.C.; Cox, N.R.; Martinez, M.L. Evaluation of free or liposome-encapsulated ribavirin for antiviral therapy of experimentally induced feline infectious peritonitis. Res. Vet. Sci. 1993, 55, 162–172. [Google Scholar] [CrossRef]
- Kim, Y.; Lee, C. Ribavirin efficiently suppresses porcine nidovirus replication. Virus Res. 2013, 171, 44–53. [Google Scholar] [CrossRef]
- Hu, T.Y.; Frieman, M.; Wolfram, J. Insights from nanomedicine into chloroquine efficacy against COVID-19. Nat. Nanotechnol. 2020, 15, 247–249. [Google Scholar] [CrossRef]
- Takano, T.; Katoh, Y.; Doki, T.; Hohdatsu, T. Effect of chloroquine on feline infectious peritonitis virus infection in vitro and in vivo. Antivir. Res. 2013, 99, 100–107. [Google Scholar] [CrossRef]
- Takano, T.; Satoh, K.; Doki, T.; Tanabe, T.; Hohdatsu, T. Antiviral effects of hydroxychloroquine and type I interferon on in vitro fatal feline coronavirus infection. Viruses 2020, 12, 576. [Google Scholar] [CrossRef] [PubMed]
- Li, C.C.; Wang, X.J. Three kinds of treatment with Homoharringtonine, Hydroxychloroquine or shRNA and their combination against coronavirus PEDV in vitro. Virol. J. 2020, 17, 71. [Google Scholar] [CrossRef] [PubMed]
- Maisonnasse, P.; Guedj, J.; Contreras, V.; Behillil, S.; Solas, C.; Marlin, R.; Naninck, T.; Pizzorno, A.; Lemaitre, J.; Gonçalves, A.; et al. Hydroxychloroquine use against SARS-CoV-2 infection in non-human primates. Nature 2020. [Google Scholar] [CrossRef] [PubMed]
- Sharun, K.; Dhama, K.; Patel, S.K.; Pathak, M.; Tiwari, R.; Singh, B.R.; Sah, R.; Bonilla-Aldana, D.K.; Rodriguez-Morales, A.J.; Leblebicioglu, H. Ivermectin, a new candidate therapeutic against SARS-CoV-2/COVID-19. Ann. Clin. Microbiol. Antimicrob. 2020, 19, 23. [Google Scholar] [CrossRef]
- Heidary, F.; Gharebaghi, R. Ivermectin: A systematic review from antiviral effects to COVID-19 complementary regimen. J. Antibiot. 2020, 73, 593–602. [Google Scholar] [CrossRef] [PubMed]
- Caly, L.; Druce, J.D.; Catton, M.G.; Jans, D.A.; Wagstaff, K.M. The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antivir. Res. 2020, 178, 104787. [Google Scholar] [CrossRef] [PubMed]
- Glowacka, P.; Rudnicka, L.; Warszawik-Hendzel, O.; Sikora, M.; Goldust, M.; Gajda, P.; Stochmal, A.; Blicharz, L.; Rakowska, A.; Olszewska, M. The antiviral properties of cyclosporine. Focus on coronavirus, hepatitis C virus, influenza virus, and human immunodeficiency virus infections. Biology 2020, 9, 192. [Google Scholar] [CrossRef]
- Pawlotsky, J.-M. COVID-19 Pandemic: Time to Revive the Cyclophilin Inhibitor Alisporivir. Clin. Infect. Dis. 2020, 15, ciaa587. [Google Scholar] [CrossRef]
- Softic, L.; Brillet, R.; Berry, F.; Ahnou, N.; Nevers, Q.; Morin-Dewaele, M.; Hamadat, S.; Bruscella, P.; Fourati, S.; Pawlotsky, J.M.; et al. Inhibition of SARS-CoV-2 infection by the cyclophilin inhibitor alisporivir (Debio 025). Antimicrob. Agents Chemother. 2020, 64, e00876-20. [Google Scholar] [CrossRef]
- Tanaka, Y.; Sato, Y.; Sasaki, T. Feline coronavirus replication is affected by both cyclophilin A and cyclophilin B. J. Gen. Virol. 2017, 98, 190–200. [Google Scholar] [CrossRef]
- Mahendran, A.S.K.; Lim, Y.S.; Fang, C.M.; Loh, H.S.; Le, C.F. The Potential of Antiviral Peptides as COVID-19 Therapeutics. Front. Pharmacol. 2020, 11, 575444. [Google Scholar] [CrossRef] [PubMed]
- Outlaw, V.K.; Bovier, F.T.; Mears, M.C.; Cajimat, M.N.; Zhu, Y.; Lin, M.J.; Addetia, A.; Lieberman, N.A.P.; Peddu, V.; Xie, X.; et al. Inhibition of Coronavirus Entry In Vitro and Ex Vivo by a Lipid-Conjugated Peptide Derived from the SARS-CoV-2 Spike Glycoprotein HRC Domain. MBio 2020, 11, e01935-20. [Google Scholar] [CrossRef] [PubMed]
- Doki, T.; Takano, T.; Koyama, Y.; Hohdatsu, T. Identification of the peptide derived from S1 domain that inhibits type I and type II feline infectious peritonitis virus infection. Virus Res. 2015, 204, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Liu, I.J.; Tsai, W.T.; Hsieh, L.E.; Chueh, L.L. Peptides corresponding to the predicted heptad repeat 2 domain of the feline coronavirus spike protein are potent inhibitors of viral infection. PLoS ONE 2013, 8, e82081. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Wang, B.; Ji, C.M.; Cong, X.; Wang, M.; Huang, Y.W. Identification of a peptide derived from the heptad repeat 2 region of the porcine epidemic diarrhea virus (PEDV) spike glycoprotein that is capable of suppressing PEDV entry and inducing neutralizing antibodies. Antivir. Res. 2018, 150, 1–8. [Google Scholar] [CrossRef]
- Wang, C.; Wang, S.; Li, D.; Wei, D.Q.; Zhao, J.; Wang, J. Human Intestinal Defensin 5 Inhibits SARS-CoV-2 Invasion by Cloaking ACE2. Gastroenterology 2020, 159, 1145–1147. [Google Scholar] [CrossRef]
- Abu-Farha, M.; Thanaraj, T.A.; Qaddoumi, M.G.; Hashem, A.; Abubaker, J.; Al-Mulla, F. The role of lipid metabolism in COVID-19 virus infection and as a drug target. Int. J. Mol. Sci. 2020, 21, 3544. [Google Scholar] [CrossRef]
- Yan, B.; Chu, H.; Yang, D.; Sze, K.H.; Lai, P.M.; Yuan, S.; Shuai, H.; Wang, Y.; Kao, R.Y.T.; Chan, J.F.W.; et al. Characterization of the lipidomic profile of human coronavirus-infected cells: Implications for lipid metabolism remodeling upon coronavirus replication. Viruses 2019, 11, 73. [Google Scholar] [CrossRef]
- Radenkovic, D.; Chawla, S.; Pirro, M.; Sahebkar, A.; Banach, M. Cholesterol in Relation to COVID-19: Should We Care about It? J. Clin. Med. 2020, 9, 1909. [Google Scholar] [CrossRef]
- Wang, S.; Li, W.; Hui, H.; Tiwari, S.K.; Zhang, Q.; Croker, B.A.; Rawlings, S.; Smith, D.; Carlin, A.F.; Rana, T.M. Cholesterol 25-Hydroxylase inhibits SARS -CoV-2 and other coronaviruses by depleting membrane cholesterol. EMBO J. 2020, e106057. [Google Scholar] [CrossRef]
- Takano, T.; Endoh, M.; Fukatsu, H.; Sakurada, H.; Doki, T.; Hohdatsu, T. The cholesterol transport inhibitor U18666A inhibits type I feline coronavirus infection. Antivir. Res. 2017, 145, 96–102. [Google Scholar] [CrossRef] [PubMed]
- Jeon, J.H.; Lee, C. Cholesterol is important for the entry process of porcine deltacoronavirus. Arch. Virol. 2018, 163, 3119–3124. [Google Scholar] [CrossRef]
- Toelzer, C.; Gupta, K.; Yadav, S.K.N.; Borucu, U.; Davidson, A.D.; Kavanagh Williamson, M.; Shoemark, D.K.; Garzoni, F.; Staufer, O.; Milligan, R.; et al. Free fatty acid binding pocket in the locked structure of SARS-CoV-2 spike protein. Science 2020, eabd3255. [Google Scholar] [CrossRef] [PubMed]
- Marovich, M.; Mascola, J.R.; Cohen, M.S. Monoclonal Antibodies for Prevention and Treatment of COVID-19. JAMA J. Am. Med. Assoc. 2020, 324, 131–132. [Google Scholar] [CrossRef] [PubMed]
- Sewell, H.F.; Agius, R.M.; Kendrick, D.; Stewart, M. Vaccines, convalescent plasma, and monoclonal antibodies for covid-19. BMJ 2020, 370, m2722. [Google Scholar] [CrossRef]
- NIH: Clinical Trials of Monoclonal Antibodies to Prevent COVID-19 Now Enrolling. Available online: https://www.nih.gov/news-events/news-releases/clinical-trials-monoclonal-antibodies-prevent-covid-19-now-enrolling (accessed on 20 September 2020).
- Chang, C.Y.; Cheng, I.C.; Chang, Y.C.; Tsai, P.S.; Lai, S.Y.; Huang, Y.L.; Jeng, C.R.; Pang, V.F.; Chang, H.W. Identification of Neutralizing Monoclonal Antibodies Targeting Novel Conformational Epitopes of the Porcine Epidemic Diarrhoea Virus Spike Protein. Sci. Rep. 2019, 9, 2529. [Google Scholar] [CrossRef]
- Zhang, F.; Chen, Y.; Ke, Y.; Zhang, L.; Zhang, B.; Yang, L.; Zhu, J. Single chain fragment variable (scFv) antibodies targeting the spike protein of porcine epidemic diarrhea virus provide protection against viral infection in piglets. Viruses 2019, 11, 58. [Google Scholar] [CrossRef]
- Gong, L.; Lin, Y.; Qin, J.; Li, Q.; Xue, C.; Cao, Y. Neutralizing antibodies against porcine epidemic diarrhea virus block virus attachment and internalization. Virol. J. 2018, 15, 133. [Google Scholar] [CrossRef]
- Lee, D.H.; Jeon, Y.S.; Park, C.K.; Kim, S.; Lee, D.S.; Lee, C. Immunoprophylactic effect of chicken egg yolk antibody (IgY) against a recombinant S1 domain of the porcine epidemic diarrhea virus spike protein in piglets. Arch. Virol. 2015, 160, 2197–2207. [Google Scholar] [CrossRef]
- Zhang, F.; Chen, Y.; Yang, L.; Zhu, J. Construction and characterization of porcine single-chain fragment variable antibodies that neutralize transmissible gastroenteritis virus in vitro. Arch. Virol. 2019, 164, 983–994. [Google Scholar] [CrossRef]
- Vautherot, J.F.; Madelaine, M.F.; Boireau, P.; Laporte, J. Bovine coronavirus peplomer glycoproteins: Detailed antigenic analyses of S1, S2 and HE. J. Gen. Virol. 1992, 73, 1725–1737. [Google Scholar] [CrossRef] [PubMed]
- Zou, N.; Wang, F.; Duan, Z.; Xia, J.; Wen, X.; Yan, Q.; Liu, P.; Cao, S.; Huang, Y. Development and Characterization of Neutralizing Monoclonal Antibodies against the S1 Subunit Protein of QX-like Avian Infectious Bronchitis Virus Strain Sczy3. Monoclon. Antib. Immunodiagn. Immunother. 2015, 34, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.Q.; Li, Y.F.; Guo, J.Q.; Shen, H.G.; Zhang, D.Y.; Zhou, J.Y. Production and characterization of monoclonal antibodies to poly100S1 protein of avian infectious bronchitis virus. Zoonoses Public Health 2007, 54, 69–77. [Google Scholar] [CrossRef]
- Deregt, D.; Gifford, G.A.; Ijaz, M.K.; Watts, T.C.; Gilchrist, J.E.; Haines, D.M.; Babiuk, L.A. Monoclonal antibodies to bovine coronavirus glycoproteins E2 and E3: Demonstration of in vivo virus-neutralizing activity. J. Gen. Virol. 1989, 70 Pt 4, 993–998. [Google Scholar] [CrossRef]
- Doki, T.; Takano, T.; Kawagoe, K.; Kito, A.; Hohdatsu, T. Therapeutic effect of anti-feline TNF-alpha monoclonal antibody for feline infectious peritonitis. Res. Vet. Sci. 2016, 104, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Qureshi, A.; Tantray, V.G.; Kirmani, A.R.; Ahangar, A.G. A review on current status of antiviral siRNA. Rev. Med. Virol. 2018, 28, e1976. [Google Scholar] [CrossRef] [PubMed]
- de Andrade Santos, I.; Grosche, V.R.; Bergamini, F.R.G.; Sabino-Silva, R.; Jardim, A.C.G. Antivirals Against Coronaviruses: Candidate Drugs for SARS-CoV-2 Treatment? Front. Microbiol. 2020, 11, 1818. [Google Scholar] [CrossRef]
- Lundstrom, K. Coronavirus pandemic-Therapy and vaccines. Biomedicines 2020, 8, 109. [Google Scholar] [CrossRef]
- McDonagh, P.; Sheehy, P.A.; Norris, J.M. In vitro inhibition of feline coronavirus replication by small interfering RNAs. Vet. Microbiol. 2011, 150, 220–229. [Google Scholar] [CrossRef]
- Anis, E.A.; Wilkes, R.P.; Kania, S.A.; Legendre, A.M.; Kennedy, M.A. Effect of small interfering RNAs on in vitro replication and gene expression of feline coronavirus. Am. J. Vet. Res. 2014, 75, 828–834. [Google Scholar] [CrossRef]
- McDonagh, P.; Sheehy, P.A.; Norris, J.M. Combination siRNA therapy against feline coronavirus can delay the emergence of antiviral resistance in vitro. Vet. Microbiol. 2015, 176, 10–18. [Google Scholar] [CrossRef]
- Li, K.; Li, H.; Bi, Z.; Song, D.; Zhang, F.; Lei, D.; Luo, S.; Li, Z.; Gong, W.; Huang, D.; et al. Significant inhibition of re-emerged and emerging swine enteric coronavirus in vitro using the multiple shRNA expression vector. Antivir. Res. 2019, 166, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Gu, W.Y.; Li, Y.; Liu, B.J.; Wang, J.; Yuan, G.F.; Chen, S.J.; Zuo, Y.Z.; Fan, J.H. Short hairpin RNAs targeting M and N genes reduce replication of porcine deltacoronavirus in ST cells. Virus Genes 2019, 55, 795–801. [Google Scholar] [CrossRef] [PubMed]
- Lan, Y.; Lu, H.; Zhao, K.; He, W.; Chen, K.; Wang, G.; Song, D.; Gao, F. In vitro inhibition of porcine hemagglutinating encephalomyelitis virus replication with siRNAs targeting the spike glycoprotein and replicase polyprotein genes. Intervirology 2011, 55, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Yu, K.; Deng, S.; Wang, H.; Zhang, Y.; Chen, X.; Wang, K.; Hu, R.; Lian, Z.; Li, N. Small interfering RNA expression inhibits avian infectious bronchitis virus replication and inflammatory response. Antivir. Ther. 2016, 21, 469–479. [Google Scholar] [CrossRef]
- Ritz, S.; Egberink, H.; Hartmann, K. Effect of Feline Interferon-Omega on the Survival Time and Quality of Life of Cats with Feline Infectious Peritonitis. J. Vet. Intern. Med. 2007, 21, 1193–1197. [Google Scholar] [CrossRef]
- Gil, S.; Leal, R.O.; Duarte, A.; McGahie, D.; Sepúlveda, N.; Siborro, I.; Cravo, J.; Cartaxeiro, C.; Tavares, L.M. Relevance of feline interferon omega for clinical improvement and reduction of concurrent viral excretion in retrovirus infected cats from a rescue shelter. Res. Vet. Sci. 2013, 94, 753–763. [Google Scholar] [CrossRef]
- Mochizuki, M.; Nakatani, H.; Yoshida, M. Inhibitory effects of recombinant feline interferon on the replication of feline enteropathogenic viruses in vitro. Vet. Microbiol. 1994, 39, 145–152. [Google Scholar] [CrossRef]
- Shen, H.; Zhang, C.; Guo, P.; Liu, Z.; Sun, M.; Sun, J.; Li, L.; Dong, J.; Zhang, J. Short communication: Antiviral activity of porcine IFN-λ3 against porcine epidemic diarrhea virus in vitro. Virus Genes 2016, 52, 877–882. [Google Scholar] [CrossRef]
- Shan, L.; Fu, F.; Xue, M.; Zhu, X.; Li, L.; Feng, L.; Liu, P. Interferon gamma inhibits transmissible gastroenteritis virus infection mediated by an IRF1 signaling pathway. Arch. Virol. 2019, 164, 2659–2669. [Google Scholar] [CrossRef]
- Pei, J.; Sekellick, M.J.; Marcus, P.I.; Choi, I.S.; Collisson, E.W. Chicken interferon type I inhibits infectious bronchitis virus replication and associated respiratory illness. J. Interferon Cytokine Res. 2001, 21, 1071–1077. [Google Scholar] [CrossRef] [PubMed]
- Jung, K.; Alekseev, K.P.; Zhang, X.; Cheon, D.-S.; Vlasova, A.N.; Saif, L.J. Altered Pathogenesis of Porcine Respiratory Coronavirus in Pigs due to Immunosuppressive Effects of Dexamethasone: Implications for Corticosteroid Use in Treatment of Severe Acute Respiratory Syndrome Coronavirus. J. Virol. 2007, 81, 13681–13693. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Alekseev, K.; Jung, K.; Vlasova, A.; Hadya, N.; Saif, L.J. Cytokine Responses in Porcine Respiratory Coronavirus-Infected Pigs Treated with Corticosteroids as a Model for Severe Acute Respiratory Syndrome. J. Virol. 2008, 82, 4420–4428. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Shimizu, M.; Shimizu, Y. Effects of ambient temperatures on clinical and immune responses of pigs infected with transmissible gastro-enteritis virus. Vet. Microbiol. 1979, 4, 109–116. [Google Scholar] [CrossRef]
- Tsunemitsu, H.; Smith, D.R.; Saif, L.J. Experimental inoculation of adult dairy cows with bovine coronavirus and detection of coronavirus in feces by RT-PCR. Arch. Virol. 1999, 144, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Balzarini, J.; Keyaerts, E.; Vijgen, L.; Egberink, H.; De Clercq, E.; Van Ranst, M.; Printsevskaya, S.S.; Olsufyeva, E.N.; Solovieva, S.E.; Preobrazhenskaya, M.N. Inhibition of feline (FIPV) and human (SARS) coronavirus by semisynthetic derivatives of glycopeptide antibiotics. Antivir. Res. 2006, 72, 20–33. [Google Scholar] [CrossRef] [PubMed]
- Balzarini, J.; Keyaerts, E.; Vijgen, L.; Vandermeer, F.; Stevens, M.; De Clercq, E.; Egberink, H.; Van Ranst, M. Pyridine N-oxide derivatives are inhibitory to the human SARS and feline infectious peritonitis coronavirus in cell culture. J. Antimicrob. Chemother. 2006, 57, 472–481. [Google Scholar] [CrossRef]
- Li, H.J.; Gao, D.S.; Li, Y.T.; Wang, Y.S.; Liu, H.Y.; Zhao, J. Antiviral effect of lithium chloride on porcine epidemic diarrhea virus in vitro. Res. Vet. Sci. 2018, 118, 288–294. [Google Scholar] [CrossRef]
- Takano, T.; Akiyama, M.; Doki, T.; Hohdatsu, T. Antiviral activity of itraconazole against type i feline coronavirus infection. Vet. Res. 2019, 50, 5. [Google Scholar] [CrossRef]
- Yang, C.W.; Lee, Y.Z.; Hsu, H.Y.; Shih, C.; Chao, Y.S.; Chang, H.Y.; Lee, S.J. Targeting Coronaviral Replication and Cellular JAK2 Mediated Dominant NF-κB Activation for Comprehensive and Ultimate Inhibition of Coronaviral Activity. Sci. Rep. 2017, 7, 4105. [Google Scholar] [CrossRef]
- Legendre, A.M.; Kuritz, T.; Galyon, G.; Baylor, V.M.; Heidel, R.E. Polyprenyl immunostimulant treatment of cats with presumptive non-effusive feline infectious peritonitis in a field study. Front. Vet. Sci. 2017, 4, 7. [Google Scholar] [CrossRef] [PubMed]
- Anis, E.A.; Dhar, M.; Legendre, A.M.; Wilkes, R.P. Transduction of hematopoietic stem cells to stimulate RNA interference against feline infectious peritonitis. J. Feline Med. Surg. 2017, 19, 680–686. [Google Scholar] [CrossRef] [PubMed]
- Mani, J.S.; Johnson, J.B.; Steel, J.C.; Broszczak, D.A.; Neilsen, P.M.; Walsh, K.B.; Naiker, M. Natural product-derived phytochemicals as potential agents against coronaviruses: A review. Virus Res. 2020, 284, 197989. [Google Scholar] [CrossRef] [PubMed]
- Decaro, N.; Martella, V.; Saif, L.J.; Buonavoglia, C. COVID-19 from veterinary medicine and one health perspectives: What animal coronaviruses have taught us. Res. Vet. Sci. 2020, 131, 21–23. [Google Scholar] [CrossRef] [PubMed]
- WHO Solidarity Trial Consortium; Pan, H.; Peto, R.; Abdool Karim, Q.; Alejandria, M.; Henao Restrepo, A.M.; Hernandez Garcia, C.; Kieny, M.P.; Malekzadeh, R.; Murthy, S.; et al. Repurposed antiviral drugs for COVID-19; interim WHO SOLIDARITY trial results. medRxiv 2020. [Google Scholar] [CrossRef]
- Jordan, B. Vaccination against infectious bronchitis virus: A continuous challenge. Vet. Microbiol. 2017, 206, 137–143. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, L.; Shang, Y.; Tan, R.; Ji, M.; Yue, X.; Wang, N.; Liu, J.; Wang, C.; Li, Y.; et al. Emergence and evolution of highly pathogenic porcine epidemic diarrhea virus by natural recombination of a low pathogenic vaccine isolate and a highly pathogenic strain in the spike gene. Virus Evol. 2020, 6, veaa049. [Google Scholar] [CrossRef]
- Iwasaki, A.; Yang, Y. The potential danger of suboptimal antibody responses in COVID-19. Nat. Rev. Immunol. 2020, 20, 339–341. [Google Scholar] [CrossRef]
- Screaton, G.; Mongkolsapaya, J.; Yacoub, S.; Roberts, C. New insights into the immunopathology and control of dengue virus infection. Nat. Rev. Immunol. 2015, 15, 745–759. [Google Scholar] [CrossRef]
- Weingartl, H.; Czub, M.; Czub, S.; Neufeld, J.; Marszal, P.; Gren, J.; Smith, G.; Jones, S.; Proulx, R.; Deschambault, Y.; et al. Immunization with Modified Vaccinia Virus Ankara-Based Recombinant Vaccine against Severe Acute Respiratory Syndrome Is Associated with Enhanced Hepatitis in Ferrets. J. Virol. 2004, 78, 12672–12676. [Google Scholar] [CrossRef]
- Eroshenko, N.; Gill, T.; Keaveney, M.K.; Church, G.M.; Trevejo, J.M.; Rajaniemi, H. Implications of antibody-dependent enhancement of infection for SARS-CoV-2 countermeasures. Nat. Biotechnol. 2020, 38, 789–791. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Wei, Q.; Lin, Q.; Fang, J.; Wang, H.; Kwok, H.; Tang, H.; Nishiura, K.; Peng, J.; Tan, Z.; et al. Anti-spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS-CoV infection. JCI Insight 2019, 4, e123158. [Google Scholar] [CrossRef] [PubMed]
- Bálint, Á.; Farsang, A.; Szeredi, L.; Zádori, Z.; Belák, S. Recombinant feline coronaviruses as vaccine candidates confer protection in SPF but not in conventional cats. Vet. Microbiol. 2014, 169, 154–162. [Google Scholar] [CrossRef]
- Weiss, R.C.; Scott, F.W. Antibody-mediated enhancement of disease in feline infectious peritonitis: Comparisons with dengue hemorrhagic fever. Comp. Immunol. Microbiol. Infect. Dis. 1981, 4, 175–189. [Google Scholar] [CrossRef]
- Vennema, H.; de Groot, R.J.; Harbour, D.A.; Dalderup, M.; Gruffydd-Jones, T.; Horzinek, M.C.; Spaan, W.J. Early death after feline infectious peritonitis virus challenge due to recombinant vaccinia virus immunization. J. Virol. 1990, 64, 1407–1409. [Google Scholar] [CrossRef]
- Takano, T.; Yamada, S.; Doki, T.; Hohdatsu, T.; Vopat, T. Pathogenesis of oral type I feline infectious peritonitis virus (FIPV) infection: Antibody dependent enhancement infection of cats with type I FIPV via the oral route. J. Vet. Med. Sci. 2019, 81, 911–915. [Google Scholar] [CrossRef]
- Orr-Burks, N.; Gulley, S.L.; Gallardo, R.A.; Toro, H.; Van Ginkel, F.W. Immunoglobulin a as an early humoral responder after mucosal avian coronavirus vaccination. Avian Dis. 2014, 58, 279–286. [Google Scholar] [CrossRef]
- Manzanedo, R.D.; Manning, P. COVID-19: Lessons for the climate change emergency. Sci. Total Environ. 2020, 742, 140563. [Google Scholar] [CrossRef]
- Afelt, A.; Frutos, R.; Devaux, C. Bats, coronaviruses, and deforestation: Toward the emergence of novel infectious diseases? Front. Microbiol. 2018, 9, 702. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).