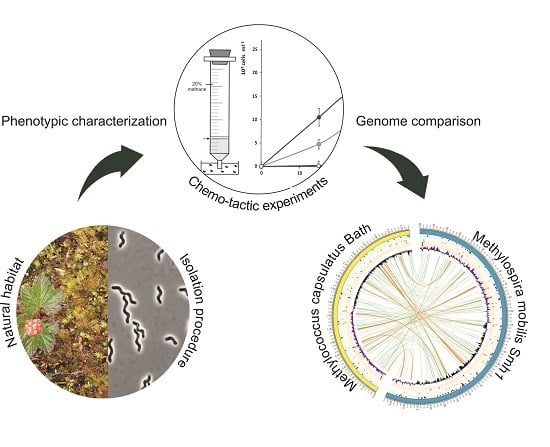
Thriving in Wetlands: Ecophysiology of the Spiral-Shaped Methanotroph Methylospira mobilis as Revealed by the Complete Genome Sequence
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation Procedure
2.2. Cultivation in a Range of Oxygen Concentrations and Analysis of Chemotactic Properties
2.3. Genome Sequencing and Assembly
2.4. Annotation
2.5. Comparative Genomics and Phylogenetic Analysis
3. Results
3.1. Isolation of Methylospira Mobilis Shm1
3.2. Phenotypic Characterization
3.3. General Genome Features of Strain Shm1 and Genome-Based Phylogeny
3.4. Genome-to-GENOME comparison of Strain Shm1 and Mc. capsulatus Bath
3.5. Genome-Encoded Adaptations to Oxygen Limitation
3.6. Nitrogen Metabolism
3.7. Motility
3.8. Helical Cell Shape
3.9. Insertion Sequences and Prophages
3.10. Signaling Systems
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hanson, R.; Hanson, T. Methanotrophic bacteria. Microbiol. Rev. 1996, 60, 439–471. [Google Scholar] [PubMed]
- Trotsenko, Y.A.; Murrell, J.C. Metabolic aspects of aerobic obligate methanotrophy. Adv. Appl. Microbiol. 2008, 63, 183–229. [Google Scholar] [PubMed]
- Chistoserdova, L.; Lidstrom, M.E. The prokaryotes: Prokaryotic physiology and biochemistry. In The Prokaryotes: Prokaryotic Physiology and Biochemistry; Rosenberg, E., Delong, E., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer-Verlag: Berlin/Heidelberg, Germany, 2013; pp. 267–285. ISBN 9-78364-230-1414. [Google Scholar]
- Khmelenina, V.N.; Murrell, J.C.; Smith, T.J.; Trotsenko, Y.A. Physiology and Biochemistry of the Aerobic Methanotrophs. In Aerobic Utilization of Hydrocarbons, Oils and Lipids. Handbook of Hydrocarbon and Lipid Microbiology; Rojo, F., Ed.; Springer: Cham, Switzerland, 2018; pp. 1–25. ISBN 9-78331-939-7825. [Google Scholar]
- Lieberman, R.L.; Shrestha, D.B.; Doan, P.E.; Hoffman, B.M.; Stemmler, T.L.; Rosenzweig, A.C. Purified particulate methane monooxygenase from Methylococcus capsulatus (Bath) is a dimer with both mononuclear copper and a copper-containing cluster. Proc. Natl. Acad. Sci. USA 2003, 100, 3820–3825. [Google Scholar] [CrossRef] [PubMed]
- Hakemian, A.S.; Rosenzweig, A.C. The Biochemistry of Methane Oxidation. Annu. Rev. Biochem. 2007, 76, 223–241. [Google Scholar] [CrossRef]
- Dedysh, S.N.; Knief, C. Diversity and phylogeny of described aerobic methanotrophs. In Methane Biocatalysis: Paving the Way to Sustainability; Kalyuzhnaya, M., Xing, X., Eds.; Springer: Cham, Switzerland, 2018; pp. 17–42. ISBN 9-78331-974-8665. [Google Scholar]
- Ettwig, K.F.; Butler, M.K.; Le Paslier, D.; Pelletier, E.; Mangenot, S.; Kuypers, M.M.M.; Schreiber, F.; Dutilh, B.E.; Zedelius, J.; de Beer, D.; et al. Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature 2010, 464, 543–548. [Google Scholar] [CrossRef]
- Sharp, C.E.; Smirnova, A.V.; Graham, J.M.; Stott, M.B.; Khadka, R.; Moore, T.R.; Grasby, S.E.; Strack, M.; Dunfield, P.F. Distribution and diversity of Verrucomicrobia methanotrophs in geothermal and acidic environments. Environ. Microbiol. 2014, 16, 1867–1878. [Google Scholar] [CrossRef]
- Knief, C. Diversity and habitat preferences of cultivated and uncultivated aerobic methanotrophic bacteria evaluated based on pmoA as molecular marker. Front. Microbiol. 2015, 6, 1346. [Google Scholar] [CrossRef]
- Danilova, O.V.; Suzina, N.E.; Van De Kamp, J.; Svenning, M.M.; Bodrossy, L.; Dedysh, S.N. A new cell morphotype among methane oxidizers: A spiral-shaped obligately microaerophilic methanotroph from northern low-oxygen environments. ISME J. 2016, 10, 2734–2743. [Google Scholar] [CrossRef]
- Murrell, J.C. Genomics of Methylococcus capsulatus. In Handbook of Hydrocarbon and Lipid Microbiology; Timmis, K.N., Ed.; Springer: Berlin/Heidelberg, Germany, 2010; ISBN 9-78354-077-5874. [Google Scholar]
- Eccleston, M.; Kelly, D.P. Assimilation and Toxicity of Some Exogenous. J. Gen. Microbiol. 1973, 75, 211–221. [Google Scholar] [CrossRef][Green Version]
- Kao, W.C.; Chen, Y.R.; Yi, E.C.; Lee, H.; Tian, Q.; Wu, K.M.; Tsai, S.F.; Yu, S.S.F.; Chen, Y.J.; Aebersold, R.; et al. Quantitative proteomic analysis of metabolic regulation by copper ions in Methylococcus capsulatus (Bath). J. Biol. Chem. 2004, 279, 51554–51560. [Google Scholar] [CrossRef]
- Ward, N.; Larsen, Ø.; Sakwa, J.; Bruseth, L.; Khouri, H.; Durkin, A.S.; Dimitrov, G.; Jiang, L.; Scanlan, D.; Kang, K.H.; et al. Genomic insights into methanotrophy: The complete genome sequence of Methylococcus capsulatus (Bath). PLoS Biol. 2004, 2, 707–713. [Google Scholar] [CrossRef]
- Lieven, C.; Petersen, L.A.H.; Jørgensen, S.B.; Gernaey, K.V.; Herrgard, M.J.; Sonnenschein, N. A genome-scale metabolic model for Methylococcus capsulatus predicts reduced efficiency uphill electron transfer to pMMO. BioRxiv 2018, 329714. [Google Scholar] [CrossRef]
- Gupta, A.; Ahmad, A.; Chothwe, D.; Madhu, M.K.; Srivastava, S.; Sharma, V.K. Genome-scale metabolic reconstruction and metabolic versatility of an obligate methanotroph Methylococcus capsulatus str. Bath. PeerJ 2018, 7, e6685. [Google Scholar] [CrossRef] [PubMed]
- Lieberman, R.L.; Rosenzweig, A.C. Crystal structure of a membrane-bound metalloenzyme that catalyses the biological oxidation of methane. Nature 2005, 434, 177–182. [Google Scholar] [CrossRef]
- Larmola, T.; Leppanen, S.M.; Tuittila, E.-S.; Aarva, M.; Merila, P.; Fritze, H.; Tiirola, M. Methanotrophy induces nitrogen fixation during peatland development. Proc. Natl. Acad. Sci. USA 2014, 111, 734–739. [Google Scholar] [CrossRef]
- Larsen, Ø.; Karlsen, O.A. Transcriptomic profiling of Methylococcus capsulatus (Bath)during growth with two different methane monooxygenases. Microbiologyopen 2016, 5, 254–267. [Google Scholar] [CrossRef]
- Frindte, K.; Maarastawi, S.A.; Lipski, A.; Hamacher, J.; Knief, C. Characterization of the first rice paddy cluster I isolate, Methyloterricola oryzae gen. nov., sp. nov. and amended description of Methylomagnum ishizawai. Int. J. Syst. Evol. Microbiol. 2017, 67, 4507–4514. [Google Scholar] [CrossRef]
- Frindte, K.; Kalyuzhnaya, M.G.; Bringel, F.; Dunfield, P.F.; Jetten, M.S.M.; Khmelenina, V.N.; Klotz, M.G.; Murrell, J.C.; Op den Camp, H.; Sakai, Y.; et al. Draft Genome Sequences of Two Gammaproteobacterial Methanotrophs Isolated from Rice Ecosystems. Genome Announc. 2017, 5, e00526-1. [Google Scholar] [CrossRef]
- Dunfield, P.F.; Khmelenina, V.N.; Suzina, N.E.; Trotsenko, Y.A.; Dedysh, S.N. Methylocella silvestris sp. nov., a novel methonotroph isolated from an acidic forest cambisol. Int. J. Syst. Evol. Microbiol. 2003, 53, 1231–1239. [Google Scholar] [CrossRef]
- Berk, S.G.; Guerry, P.; Colwell, R.R. Separation of small ciliate protozoa from bacteria by sucrose gradient centrifugation. Appl. Environ. Microbiol. 1976, 31, 450–452. [Google Scholar]
- Yamamura, H.; Hayakawa, M.; Iimura, Y. Application of sucrose-gradient centrifugation for selective isolation of Nocardia spp. from soil. J. Appl. Microbiol. 2003, 95, 677–685. [Google Scholar] [CrossRef] [PubMed]
- Bussmann, I.; Rahalkar, M.; Schink, B. Cultivation of methanotrophic bacteria in opposing gradients of methane and oxygen. FEMS Microbiol. Ecol. 2006, 56, 331–344. [Google Scholar] [CrossRef] [PubMed]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M.; et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2014, 42, 206–214. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG Tools for Functional Characterization of Genome and Metagenome Sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef]
- Lowe, T.M.; Eddy, S.R. TRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1996, 25, 955–964. [Google Scholar] [CrossRef]
- Lagesen, K.; Hallin, P.; Rødland, E.A.; Stærfeldt, H.H.; Rognes, T.; Ussery, D.W. RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007, 35, 3100–3108. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S.; Sato, Y.; Kawashima, M. Data, information, knowledge and principle: Back to metabolism in KEGG. Nucleic Acids Res. 2014, 42, 199–205. [Google Scholar] [CrossRef]
- Varani, A.M.; Siguier, P.; Gourbeyre, E.; Charneau, V.; Chandler, M. ISsaga is an ensemble of web-based methods for high throughput identification and semi-automatic annotation of insertion sequences in prokaryotic genomes. Genome Biol. 2011, 12, R30. [Google Scholar] [CrossRef]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinf. 2013, 14, 60. [Google Scholar] [CrossRef] [PubMed]
- Parks, D.H.; Chuvochina, M.; Waite, D.W.; Rinke, C.; Skarshewski, A.; Chaumeil, P.A.; Hugenholtz, P. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol. 2018, 36, 996. [Google Scholar] [CrossRef] [PubMed]
- Harris, R.S. Improved Pairwise Alignment of Genomic DNA. Ph.D. Thesis, Pennsylvania State University, State College, PA, USA, 2007. [Google Scholar]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018, 46, D493–D496. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. Fasttree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. [Google Scholar] [CrossRef]
- Rissanen, A.J.; Saarenheimo, J.; Tiirola, M.; Peura, S.; Aalto, S.L.; Karvinen, A.; Nykänen, H. Gammaproteobacterial methanotrophs dominate methanotrophy in aerobic and anaerobic layers of boreal lake waters. Aquat. Microb. Ecol. 2018, 81, 257–276. [Google Scholar] [CrossRef]
- Esposti, M.D.; Mentel, M.; Martin, W.; Sousa, F.L. Oxygen reductases in alphaproteobacterial genomes: Physiological evolution from low to high oxygen environments. Front. Microbiol. 2019, 10, 1–15. [Google Scholar] [CrossRef]
- Morris, R.L.; Schmidt, T.M. Shallow breathing: Bacterial life at low O2. Nat. Rev. Microbiol. 2013, 11, 205–212. [Google Scholar] [CrossRef]
- D’Mello, R.; Hill, S.; Poole, R.K. The cytochrome bd quinol oxidase in Escherichia coli has an extremely high oxygen affinity and two oxygen-binding haems: Implications for regulation of activity in vivo by oxygen inhibition. Microbiology 1996, 142, 755–763. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, J.; Handa, Y.; Sone, N. A novel cytochrome b(o/a)3-type oxidase from Bacillus stearothermophilus catalyzes cytochrome c-551 oxidation. J. Biochem. 1997, 122, 764–771. [Google Scholar] [CrossRef] [PubMed]
- Charon, N.W.; Goldstein, S.F.; Curci, K.; Limberger, R.J. The bent-end morphology of Treponema phagedenis is associated with short, left-handed, periplasmic flagella. J. Bacteriol. 1991, 173, 4820–4826. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Motaleb, M.A.; Corum, L.; Bono, J.L.; Elias, A.F.; Rosa, P.; Samuels, D.S.; Charon, N.W. Borrelia burgdorferi periplasmic flagella have both skeletal and motility functions. Proc. Natl. Acad. Sci. USA 2000, 97, 10899–10904. [Google Scholar] [CrossRef]
- Karlinsey, J.E.; Tanaka, S.; Bettenworth, V.; Yamaguchi, S.; Boos, W.; Aizawa, S.-I.; Hughes, K.T. Completion of the hook–basal body complex of the Salmonella typhimurium flagellum is coupled to FlgM secretion and fliC transcription. Mol. Microbiol. 2000, 37, 1220–1231. [Google Scholar] [CrossRef]
- Kutsukake, K.; Iino, T. Role of the FliA-FlgM regulatory system on the transcriptional control of the flagellar regulon and flagellar formation in Salmonella typhimurium. J. Bacteriol. 1994, 176, 3598–3605. [Google Scholar] [CrossRef]
- Caccamo, P.D.; Brun, Y.V. The Molecular Basis of Noncanonical Bacterial Morphology. Trends Microbiol. 2018, 26, 191–208. [Google Scholar] [CrossRef]
- Yang, D.C.; Blair, K.M.; Salama, N.R. Staying in Shape: The Impact of Cell Shape on Bacterial Survival in Diverse Environments. Microbiol. Mol. Biol. Rev. 2016, 80, 187–203. [Google Scholar] [CrossRef]
- Kofoid, E.C.; Parkinson, J.S. Transmitter and receiver modules in bacterial signaling proteins. Proc. Natl. Acad. Sci. USA 1988, 85, 4981–4985. [Google Scholar] [CrossRef]
- Kim, D.J.; Forst, S. Genomic analysis of the histidine kinase family in bacteria and archaea. Microbiology 2001, 147, 1197–1212. [Google Scholar] [CrossRef]
- Galperin, M.Y. Systems Biology and Biotechnology of Escherichia coli; Lee, S.Y., Ed.; Springer: New York, NY, USA, 2009. [Google Scholar]
- Bilwes, A.M.; Alex, L.A.; Crane, B.R.; Simon, M.I. Structure of CheA, a signal-transducing histidine kinase. Cell 1999, 96, 131–141. [Google Scholar] [CrossRef]
- Wuichet, K.; Alexander, R.P.; Zhulin, I.B. Comparative Genomic and Protein Sequence Analyses of a Complex System Controlling Bacterial Chemotaxis. Methods Enzymol. 2007, 422, 3. [Google Scholar]
- Chai, W.; Stewart, V. NasR, a novel RNA-binding protein, mediates nitrate-responsive transcription antitermination of the Klebsiella oxytoca M5al nasF operon leader in vitro. J. Mol. Biol. 1998, 283, 339–351. [Google Scholar] [CrossRef] [PubMed]
- Shu, C.J.; Ulrich, L.E.; Zhulin, I.B. The NIT domain: A predicted nitrate-responsive module in bacterial sensory receptors. Trends Biochem. Sci. 2003, 28, 121–124. [Google Scholar] [CrossRef]
- Zhang, W.; Phillips, G.N. Structure of the oxygen sensor in Bacillus subtilis: Signal transduction of chemotaxis by control of symmetry. Structure 2003, 11, 1097–1110. [Google Scholar] [CrossRef]
- Sharma, G.; Parales, R.; Singer, M. In silico characterization of a novel putative aerotaxis chemosensory system in the myxobacterium, Corallococcus coralloides. BMC Genom. 2018, 19, 757. [Google Scholar] [CrossRef]
- Hengge, R. Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 2009, 7, 263–273. [Google Scholar] [CrossRef]
- Galperin, M.Y. What bacteria want. Environ. Microbiol. 2018, 20, 4221–4229. [Google Scholar] [CrossRef]
- Rahalkar, M.; Bussmann, I.; Schink, B. Methylosoma difficile gen. nov., sp. nov., a novel methanotroph enriched by gradient cultivation from littoral sediment of Lake Constance. Int. J. Syst. Evol. Microbiol. 2007, 57, 1073–1080. [Google Scholar] [CrossRef]
- Deutzmann, J.S.; Hoppert, M.; Schink, B. Characterization and phylogeny of a novel methanotroph, Methyloglobulus morosus gen. nov., spec. nov. Syst. Appl. Microbiol. 2014, 37, 165–169. [Google Scholar] [CrossRef]
- Giuffrè, A.; Borisov, V.B.; Arese, M.; Sarti, P.; Forte, E. Cytochrome bd oxidase and bacterial tolerance to oxidative and nitrosative stress. Biochim. Biophys. Acta Bioenerg. 2014, 1837, 1178–1187. [Google Scholar] [CrossRef] [PubMed]
- Borisov, V.B.; Forte, E.; Siletsky, S.A.; Arese, M.; Davletshin, A.I.; Sarti, P.; Giuffrè, A. Cytochrome bd protects bacteria against oxidative and nitrosative stress: A potential target for next-generation antimicrobial agents. Biochemistry 2015, 80, 565–575. [Google Scholar] [CrossRef] [PubMed]
- Galperin, M.Y. A census of membrane-bound and intracellular signal transduction proteins in bacteria: Bacterial IQ, extroverts and introverts. BMC Microbiol. 2005, 5, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Bi, S.; Sourjik, V. Stimulus sensing and signal processing in bacterial chemotaxis. Curr. Opin. Microbiol. 2018, 45, 22–29. [Google Scholar] [CrossRef]
- Ortega, Á.; Zhulin, I.B.; Krell, T. Sensory Repertoire of Bacterial Chemoreceptors. Microbiol. Mol. Biol. Rev. 2017, 81, 1–28. [Google Scholar] [CrossRef]
- Salah Ud-Din, A.I.M.; Roujeinikova, A. Methyl-accepting chemotaxis proteins: A core sensing element in prokaryotes and archaea. Cell. Mol. Life Sci. 2017, 74, 3293–3303. [Google Scholar] [CrossRef]
- Boyd, E.S.; Hamilton, T.L.; Peters, J.W. An alternative path for the evolution of biological nitrogen fixation. Front. Microbiol. 2011, 2, 1–11. [Google Scholar] [CrossRef]
- Han, D.; Dedysh, S.N.; Liesack, W. Unusual genomic traits suggest Methylocystis bryophila S285 to be well adapted for life in peatlands. Genome Biol. Evol. 2018, 10, 623–628. [Google Scholar] [CrossRef]
- Oshkin, I.Y.; Miroshnikov, K.K.; Dedysh, N. Draft Genome Sequence of Methylocystis heyeri H2T,a Methanotroph with Habitat-Specific Adaptations, Isolated from a Peatland Ecosystem. Microbiol. Resour. Announc. 2019, 8, e00454-19. [Google Scholar] [CrossRef]









| Genome Characteristics | Strain Shm1 | Mc. capsulatus Bath |
|---|---|---|
| Accession number | CP044205 | AE017282 |
| Size (MB) | 4.7 | 3.3 |
| Contigs | 1 | 1 |
| G+C content (mol %) | 54.0 | 63.6 |
| Coding sequences | 4858 | 3167 |
| rRNAs (5S, 16S, 23S) | 3, 3, 3 | 2, 2, 2 |
| tRNAs | 49 | 51 |
| CRISPR loci | 2 | 2 |
| pMMO operon | 2 | 2 |
| sMMO operon | 1 | 1 |
| IS elements | 211 | 61 |
| Prophages | 5 (incomplete) | 2 (complete) |
| Microorganism (Genome Accession Number) | Total Number of Terminal Oxidases | |||
|---|---|---|---|---|
| Low-Affinity | High-Affinity | High-/Low-Affinity | ||
| aa3 | bo3 | bd | b(o)a3/ba3 | |
| Strain Shm1 (CP044205) | 1 * | none | 3 | none |
| Methylococcus capsulatus Bath (AE017282.2) | 1 | none | 1 | 1 |
| Methylomonas methanica MC09 (NC_015572.1) | 2 | 1 | 2 | none |
| Methylomonas sp. LW13 (NZ_CP033381.1) | 1 | none | 2 | 1 |
| Methylobacter tundripaludum SV96 (AEGW00000000.2) | 2 | none | 2 | none |
| Methylomicrobium buryatense 5GB1C (NZ_CP035467.1) | 1 | 1 | none | 1 |
| Microorganism | Total Proteins | RRs | HisK | MCP | GGDEF | GGDEF +EAL | EAL |
|---|---|---|---|---|---|---|---|
| Strain Shm1 | 4043 | 73 | 43 | 29 | 21 | 18 | 4 |
| Mc. capsulatus Bath | 2959 | 24 | 16 | 2 | 5 | 11 | 1 |
| M. methanica MC09 | 4403 | 87 | 40 | 14 | 26 | 34 | 5 |
| Methylomonas sp. LW13 | 4609 | 101 | 51 | 20 | 23 | 30 | 4 |
| M. tundripaludum SV96 | 4080 | 75 | 46 | 22 | 20 | 26 | 4 |
| M. buryatense 5GB1C | 4254 | 102 | 50 | 18 | 21 | 37 | 4 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oshkin, I.Y.; Miroshnikov, K.K.; Danilova, O.V.; Hakobyan, A.; Liesack, W.; Dedysh, S.N. Thriving in Wetlands: Ecophysiology of the Spiral-Shaped Methanotroph Methylospira mobilis as Revealed by the Complete Genome Sequence. Microorganisms 2019, 7, 683. https://doi.org/10.3390/microorganisms7120683
Oshkin IY, Miroshnikov KK, Danilova OV, Hakobyan A, Liesack W, Dedysh SN. Thriving in Wetlands: Ecophysiology of the Spiral-Shaped Methanotroph Methylospira mobilis as Revealed by the Complete Genome Sequence. Microorganisms. 2019; 7(12):683. https://doi.org/10.3390/microorganisms7120683
Chicago/Turabian StyleOshkin, Igor Y., Kirill K. Miroshnikov, Olga V. Danilova, Anna Hakobyan, Werner Liesack, and Svetlana N. Dedysh. 2019. "Thriving in Wetlands: Ecophysiology of the Spiral-Shaped Methanotroph Methylospira mobilis as Revealed by the Complete Genome Sequence" Microorganisms 7, no. 12: 683. https://doi.org/10.3390/microorganisms7120683
APA StyleOshkin, I. Y., Miroshnikov, K. K., Danilova, O. V., Hakobyan, A., Liesack, W., & Dedysh, S. N. (2019). Thriving in Wetlands: Ecophysiology of the Spiral-Shaped Methanotroph Methylospira mobilis as Revealed by the Complete Genome Sequence. Microorganisms, 7(12), 683. https://doi.org/10.3390/microorganisms7120683