Abstract
Cronobacter species are a group of foodborne pathogenic bacteria that cause both intestinal and systemic human disease in individuals of all age groups. Little is known about the mechanisms that Cronobacter employ to survive and persist in foods and other environments. Toxin–antitoxin (TA) genes are thought to play a role in bacterial stress physiology, as well as in the stabilization of horizontally-acquired re-combinatorial elements such as plasmids, phage, and transposons. TA systems have been implicated in the formation of a persistence phenotype in some bacterial species including Escherichia coli and Salmonella. This project’s goal was to understand the phylogenetic relatedness among TA genes present in Cronobacter. Preliminary studies showed that two typical toxin genes, fic and hipA followed species evolutionary lines. A local database of 22 TA homologs was created for Cronobacter sakazakii and a Python version 3 shell script was generated to extract TA FASTA sequences present in 234 C. sakazakii genomes previously sequenced as part of Center for Food Safety and Applied Nutrition’s (CFSAN) GenomeTrakr project. BLAST analysis showed that not every C. sakazakii strain possessed all twenty-two TA loci. Interestingly, some strains contained either a toxin or an antitoxin component, but not both. Five common toxin genes: ESA_00258 (parDE toxin-antitoxin family), ESA_00804 (relBE family), ESA_01887 (relBE family), ESA_03838 (relBE family), and ESA_04273 (YhfG-Fic family) were selected for PCR analysis and the primers were designed to detect these genes. PCR analysis showed that 55 of 63 strains possessed three of these genes Sequence analysis identified homologs of the target genes and some of the strains were PCR-negative for one or more of the genes, pointing to potential nucleotide polymorphisms in those loci or that these toxin genes were absent. Phylogenetic studies using a Cronobacter pan genomic microarray showed that for the most part TAs follow species evolutionary lines except for a few toxin genes possessed by some C. malonaticus and C. universalis strains; this demonstrates that some TA orthologues share a common phylogeny. Within the C. sakazakii strains, the prevalence and distribution of these TA homologs by C. sakazakii strain BAA-894 (a powdered infant formula isolate) followed sequence-type evolutionary lineages. Understanding the phylogeny of TAs among the Cronobacter species is essential to design future studies to realize the physiological mechanisms and roles for TAs in stress adaptation and persistence of Cronobacter within food matrices and food processing environments.
1. Introduction
Toxin–antitoxin (TA) systems are small genetic elements found on plasmids, phage genomes, and chromosomes of many bacterial and archaeal species [1,2,3,4]. They are usually observed as bi-cistronic gene pairs with the antitoxin gene preceding the toxin gene. In general, the toxin component is a stable protein that inhibits cellular processes, and the antitoxin is an unstable protein that directly binds to its associated toxin or mRNA transcript, which allows for physiological processes to continue by homeostatic means. When bacteria encounter a stressful environment, the antitoxin gets degraded by host proteases, such as Lon and ClpXP; which releases the toxin to bind to mRNA transcripts, halting essential transcription of key genes involved in the physiological processes like transcription/translation of critical cellular processes, cell division, DNA replication, ATP synthesis, mRNA stability, or cell wall synthesis [5].
Bacterial TA systems were initially discovered through their involvement in maintaining plasmids [4], and subsequently these plasmids’ role in antibiotic resistance. It has been posited that plasmidborne toxin/antitoxin loci serve only to maintain plasmid DNA at the expense of the host organism, while studies reported by other authors suggest that plasmidborne TAs maintain plasmids for host competitive advantages [6]. Furthermore, like plasmidborne TAs, chromosomal TAs might play a role in stabilizing genomic and pathogenicity islands and prophages, which are similarly obtained through horizontal gene transfer mechanisms [6,7]. In fact, it is thought that the prophage P1 constitutes a low copy number plasmid through its lysogeny into the host cell’s chromosome [7]. It has been conjectured that during antibiotic therapy, a small population of cells are able to survive antibiotic treatment by entering a transient state of dormancy or persistence. However, a recent report by Goormaghtigh et al. [8] showed that the direct link between induction of type II TA systems and persistence brought on by antibiotics might not be as straightforward as was once thought in these cells.
To date, there are six known classes, or types of TA systems [5]. Type I and Type III TA systems have antitoxins that are small, non-coding RNAs while Type II, IV, V, and VI TA systems have antitoxins that are proteins. Type I toxin–antitoxin systems depend on sequence homology of the complementary antitoxin RNA with the toxin mRNA. Translation of the mRNA is then prevented either through degradation by means of RNase III activity or by obstructing the Shine-Dalgarno sequence (or ribosome binding site) of the toxin mRNA. Type III antitoxins inhibit their cognate toxins through protein-RNA interactions. Type II antitoxins neutralize the harmful effects of their toxins by directly binding to them. Type IV antitoxins prevent their toxin from binding to cytoskeleton proteins. The Type V antitoxin is an endoribonuclease, which cleaves the mRNA of the toxin to halt its expression. Type VI TAs only occur in microorganisms rich in AT complexes, and its toxin blocks the beta sliding clamp, preventing DNA synthesis, which can be selectively targeted by proteases [2,9].
Even though the role of TAs in persistence of cells during antibiotic therapy is now controversial, these genes have not been well-characterized in the Cronobacter species [10,11]. Cronobacter species are Gram-negative bacteria that are opportunistic foodborne pathogens that cause intestinal and systemic human disease [12,13]. The genus Cronobacter taxonomically consists of seven species including Cronobacter sakazakii, Cronobacter malonaticus, Cronobacter turicensis, Cronobacter muytjensii, Cronobacter universalis, Cronobacter dublinensis (with three subspecies—C. dublinensis subsp. dublinensis, C. dublinensis subsp. lausannensis, and C. dublinensis subsp. lactaridi) and Cronobacter condimenti [12,13,14]. Cronobacter species were once thought to only infect neonates [15,16,17]. Cronobacter are now known to infect individuals of all ages [18,19,20,21]. The species considered most pathogenic include C. sakazakii, C. malonaticus, and C. turicensis, causing necrotizing enterocolitis, bacteremia, and meningitis in neonates and infants and pneumonia, septicemia, and urinary tract infections in adults [16,18,19,21,22,23,24]. However, all species of Cronobacter, except for C. condimenti have been associated with infections. Infantile infections are thought to occur through consumption of intrinsic or extrinsic contamination of reconstituted, temperature-abused powdered infant formula (PIF) [23,25]. Jason reported surveillance information on over 80 infant Cronobacter cases (which occurred between 1958 and 2010 and is defined here as a confirmed culture-positive case of septicemia or meningitis) where infants exclusively ingested breast milk, without consumption of a commercially manufactured PIF product, prior to illness onset [17]. Friedemann had also reported similar earlier observations [26]. More recently, Bowen et al. [27] and McMullan et al. [28] described cases involving infantile C. sakazakii septicemia/meningitis infections where infants had only consumed expressed maternal milk (EMM) during the first weeks after birth. Contaminated personal breast pumps were found to be the source of contamination. Further epidemiological investigations using whole genome sequencing (WGS) analysis determined that the clinical isolates obtained during the investigation were indistinguishable from those cultured from a contaminated breast pump and a home kitchen sink drain in the first case and a breast pump in the latter case. Of equal importance is that Cronobacter species are now known to be largely more ecologically prevalent than was once believed, and many researchers have reported their isolation from many types of foods besides PIF and follow-up-formulas, including other low water activity foods, such as dried milk protein products, cereals, cheeses (and cheese powders), licorice, candies, spices, teas, nuts, herbs, and pastas [13,29,30,31,32]. It has also been found to be associated with a variety of ready-to-eat vegetables, body surfaces, and intestinal contents of filth and stable flies, environments of PIF or dairy powder production facilities and households, household sink drains, and water, which might help explain how infections occur in adults [33,34,35,36,37,38,39,40,41,42]. Recently, Yong et al. reported an epidemiological investigation of an acute gastroenteritis outbreak in a high school caused by a mixture of sequence types (ST) ST73 and ST4 C. sakazakii strains and ST567 C. malonaticus strains. The clinical, implicated food, and environmental samples included rectal swabs, a bean curd braised pork dish, and food delivery boxes [43]. This is the first report of a foodborne gastroenteritis outbreak caused by Cronobacter strains involving young teens and adults.
Identification of TA genes is critical for a reliable estimation of gene function. Understanding the genomic landscape including the phylogenetic relatedness of TA genes among Cronobacter spp. is essential for designing future studies to identify mechanisms used for survival and persistence in foods. Furthermore, more information about the mechanisms of Cronobacter adaptation in a wide range of environments, could eventually lead to development of methods to control the contamination of foods by these organisms. This study establishes the first report describing the phylogeny, prevalence, and distribution of various type two toxin–antitoxin genes possessed by Cronobacter species in general, and specifically focuses on the prevalence, distribution, and phylogeny of type II TAs possessed by C. sakazakii strains. The predictive results described in this report lay the foundation for future functional genetic studies designed to understand the role of these genes in the control of the contamination of foods.
2. Materials and Methods
2.1. Bacterial Strains
The strains described in this report are shown in Table 1 and in Table S1. These strains were obtained through various surveillance studies reported by Restaino et al. [44], Jaradat et al. [45], Chon et al. [46], Yan et al. [47], Gopinath et al. [32], Chase et al. [48], and Jang et al. [40,41]. All strains, except for those whose genomes were obtained from the National Center for Biotechnology Information (NCBI) GenBank, were identified as Cronobacter using species-specific (rpoB and cgcA) PCR assays [49,50,51]. Multi-locus sequence typing (MLST) analysis of the strains was carried out following the protocol described by Joseph et al. [52]. Genome assemblies (FASTA) were submitted to the Cronobacter MLST website (http://www.pubmlst.org/cronobacter) where the MLST database assigned allelic, sequence type (ST), and clonal complex (CC) profiles for each strain based on a seven-loci MLST scheme [52,53]. All genomes were also submitted to Center for Food Safety and Applied Nutrition’s (CFSAN) Galaxy GenomeTrakr website’s MLST tool (https://galaxytrakr.org/?tool_id=toolshed.g2.bx.psu.edu%2Frepos%2Fiuc%2Fmlst%2Fmlst%2F2.15.1&version=2.15.1&__identifer=b7m1wdw5jjt) for analysis. Taken together, both approaches confirmed the identity of the strains.

Table 1.
Strain information of Cronobacter, Siccibacter, and Franconibacter isolates used for the phylogenetic microarray analysis in this study.
2.2. DNA Extraction for PCR Assay, Microarray, and Whole Genome Sequencing (WGS)
Frozen stocks of each strain were plated onto Enterobacter sakazakii Chromogenic Plating Medium (ESPM; R&F Products, Downers Grove, IL, USA) and were cultured overnight at 37 °C. A single colony of each Cronobacter strain displaying the typically blue-black to blue-gray colony on ESPM was inoculated into 5 mL of Trypticase soy broth (BBL, Cockeysville, MD, USA), supplemented with 1% NaCl (TSBS) and then incubated at 37 °C for 18 h with shaking at 150 rpm. Bacterial DNA extraction and purification were performed using a Qiagen QIACube instrument and its automated technology (QIAGEN Sciences, Germantown, MD, USA), as described previously and according to the manufacturer’s instructions [32,41,48,54]. The 109 TA orthologs identified on the pan genomic microarray are described in Table S2.
2.3. Microarray Analysis (MA)
The microarray used in this study was an Affymetrix MyGeneChip Custom Array (Affymetrix design number: FDACRONOa520845F), which utilizes the whole genome sequences of 15 Cronobacter strains, as well as 18 plasmids. These 15 strains encompassed all currently proposed species of Cronobacter. A ≥97% identity threshold level between gene homologs to positively predict allelic coverage was used to obtain genes that were represented on the array as described by Tall et al. [54]. Each gene (19,287 Cronobacter gene targets) was represented on the array by 22 unique 25-mer oligonucleotide probes, as described by Tall et al. [54].
2.4. Whole Genome Sequencing (WGS), Assemblies, and Annotation for Comparative Genomic Analysis
Two milliliters of an overnight TSBS (total 3 mL volume) culture of each C. sakazakii strain was taken, and genomic DNA was extracted using the automated Qiagen QIACube instrument (QIAGEN, Inc., Germantown, MD, USA) as described above. Typical yields of the purified genomic DNA were 5–50 µg from a final elution volume of 200 µL and these were used for the WGS and Microarray analyses. Each strain’s DNA was quantified using a Qubit dsDNA BR assay kit (Invitrogen, Thermo Fisher Scientific, Wilmington, DE, USA) and Qubit 2.0 fluorometer (Life Technologies, Grand Island, NY, USA). The DNA samples were prepared with nuclease-free deionized water (molecular biology grade, Thermo Fisher Scientific, Waltham, MA, USA) with a final concentration of 0.2 ng/μL. Genomic libraries were prepared using the Nextera library prep kit (Illumina, San Diego, CA, USA) and whole genome sequencing was conducted using an Illumina MiSeq platform utilizing either 500 or 600 cycles of paired-end reads.
Raw sequence reads (FASTQ datasets) from the Illumina sequencing were trimmed for removal of adaptor sequences and for quality control purposes, and then de novo assembled using the CLC Genomics Workbench version 9.0 (CLC bio, Aarhus, Denmark). For annotation, the FASTA files of the assemblies were uploaded onto the Rapid Annotation Subsystems Technology (RAST) server (online annotation; http://rast.theseed.org). For routine prokaryotic genome annotation, the files were uploaded using the PGAP pipeline at NCBI.
Nucleotide sequences of the strains were deposited into NCBI’s GenBank and were released to the public through submission to NCBI under the FDA-CFSAN bioproject Cronobacter GenomeTrkr Project (PRJNA258403), which is part of the FDA’s Center for Food Safety and Applied Nutrition (CFSAN) foodborne pathogen research umbrella project at NCBI (PRJNA186875) project. The accession numbers of the genomes used in the study are shown in Table S1.
2.5. PCR
Five type II toxin genes and represented TA families—ESA_00258 (parDE toxin-antitoxin family), ESA_00804 (relBE family), ESA_01887 (relBE family), ESA_03838 (relBE family), ESA_04273 (YhfG-Fic family) were found in the C. sakazakii strain BAA-894 using the PostgreSQL-based TAfinder tool: TADB2 (http://bioinfo-mml.sjtu.edu.cn/TADB2/) [55], and were run in NCBI’s Primer-BLAST so that the PCR primers could be designed to detect these five toxins in other Cronobacter strains. Primer sequences and PCR reaction parameters are given in Table 2.

Table 2.
Description of the PCR primers, amplicon sizes, and PCR reaction parameters # targeting five common C. sakazakii toxin genes used in this study.
2.6. Bioinformatics Analysis: A Database and Local BLAST Analysis
The Type II database located at http://bioinfo-mml.sjtu.edu.cn/TADB2/ was used for screening WGS assemblies. A local database of these assemblies was created and formatted for BLAST analysis. Nucleotide sequences of Type II TAs were obtained from TADB and were confirmed to be present in C. sakazakii strain BAA-894 using this local database. Reference genomes were obtained from NCBI and in-house python and Perl scripts were used to parse the BLAST output and identify homologous sequences of TA genes in 234 genomes of C. sakazakii strains.
3. Results and Discussion
3.1. Pan-Genomic Microarray Analysis Demonstrates That Cronobacter TA Allelic Sequence Divergence Aligned along Species Taxa Lines and Within C. sakazakii Aligned with ST Lineages
The ~19,000 probe sets comprising the FDA Cronobacter custom designed Affymetrix microarray represents the total Cronobacter pan genomic content, and for this study the probesets were narrowed down to 109, representing TA orthologues from the seven species [53]. C. universalis was only represented by two homologs [FIG00553297: hypothetical protein (ESA_00913) and a putative transcriptional regulator (ESA_01147)]. The twenty-two TA homologs possessed by the C. sakazakii strain BAA-894 were confirmed to be present on the Cronobacter pan genomic microarray by using BLAST analysis, which compared the TADB sequences with sequences contained in the microarray’s chip design file (Supplemental Table S2). Some of these homologs were represented by TAs from other C. sakazakii strains or by TA orthologs of the other species, e.g., TA genes ESA_00803 and ESA_01887 from C. turicensis; ESA_00912 from the C. sakazakii strain E764; ESA_01146 from the C. sakazakii strain Es35; ESA_04371 from C. sakazakii strain ES15; and ESA_04372 from C. condimenti strain LMG 26250T. Two plasmid TA homologs were represented on the microarray as FIG00554131: hypothetical protein (ESA_00912) and was from the C. sakazakii strain Es35 and ESA3_p05543 was from the C. universalis strain NCTC 9529T. There were two phage TA homologs [encoding for YeeV toxin protein and YkfI toxin protein, prophage TAs from the C. sakazakii strain 29544T] that were represented on the microarray and were from C. dublinensis and C. malonaticus strain LMG2326T. The presence–absence calls generated by the Affymetrix algorithm were brought into MEGA7 [56] to create a phylogenetic tree showing relatedness. Figure 1 represents the results of this analysis and demonstrated that in some but not all species, the TA allelic sequence divergence aligned along the species taxa lines. For example, microarray analysis (MA) showed that the two C. malonaticus strains [Md99g, CI825 (LMG2326T)] and the C. universalis strain NCTC 9529T (797) clustered with some of the C. sakazakii strains, signifying that some TAs between these species share a common phylogenetic history. Supplementary Table S2 shows which toxin or antitoxin were present or absent in these strains. Many of these TAs, e.g., the parDE family of TA, were also shared by other members of the Enterobacteriaceae [5,57].
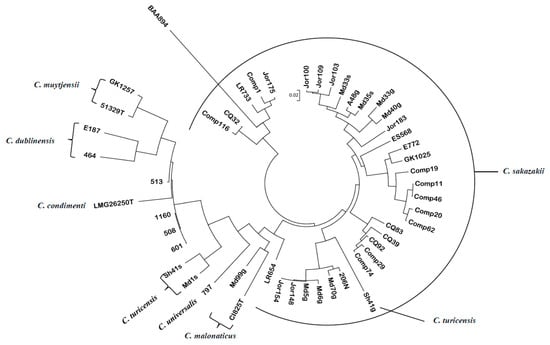
Figure 1.
Evolutionary relationships of 50 Cronobacter and phylogenetically related strains, which were generated from a gene-difference matrix using just the 109 toxin–antitoxin (TA) probeset homologs shown in Supplementary Table S2. The evolutionary history was inferred using the neighbor-joining method [55]. The microarray experimental protocol, as described by Tall et al. [53], was used for the interrogation of the strains and for the analysis. The optimal tree with the sum of branch length = 2.25204503 is shown. The tree was drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the p-distance method [56] and are in the units (0.02) of the number of base differences per site. Evolutionary analyses were conducted in MEGA5 [54]. The Cronobacter microarray demonstrated that for the most part, the TA allelic sequence divergence aligned along species taxa lines and within C. sakazakii, it aligned with ST lineages.
Preliminary analysis of fic-like and hipA homologs, two common toxin genes obtained by BLAST analysis from 42 strains representative of the seven species supports the findings obtained by MA. The dendrogram trees shown in Figure 2 demonstrate the relatedness of fic-like and hipA genes among these Cronobacter strains.
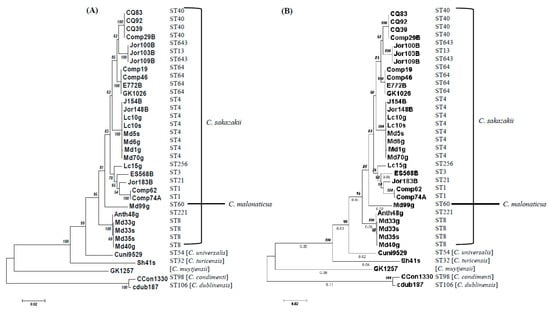
Figure 2.
The phylogeny of fic-like (A) and hipA (B) toxins were inferred using the neighbor-joining method [55]. The optimal trees with the sum of branch length of 0.38280022 (A) and 0.47999136 (B) are shown. The trees are drawn to scale, with branch lengths (next to the branches) in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the maximum composite likelihood method [58] and are in the units of the number of base substitutions per site. In both trees, the analysis involved 35 nucleotide sequences. All positions containing gaps and missing data were eliminated. There was a total of 603 positions in (A) and 1320 positions in (B) in the final dataset. Evolutionary analyses were conducted in MEGA7 [54]. The accession numbers for nucleotide sequences can be found in Table S1.
The phylogenies based on the sequence divergence of these TA genes, shown in Figure 1 and Figure 2, demonstrate that most strains cluster according to species lineages or in the case of C. sakazakii, the strains cluster according to the sequence type. This analysis involved fic-like and hipA type II toxin nucleotide sequences from C. sakazakii and single fic-like/hipA nucleotide sequences, each from C. malonaticus, C. universalis, C. turicensis, C. muytjensii, C. dublinensis, and C. condimenti strains, and each Cronobacter species clustered separately. Take note that the C. sakazakii strains clustered according to the assigned sequence type designations obtained from (https://pubmlst.org/cronobacter/), as maintained by the University of Oxford [59].
3.2. PCR and BLAST Analyses Showed That Not Every C. sakazakii Strain Possessed Similar Numbers or Types of TAs
PCR analysis also exemplified the phylogenetic relatedness of five representative type II TA homologs among C. sakazakii strains (the results are summarized in Table 3) and demonstrated that in some strains, TA alleles are shared. PCR results from 63 C. sakazakii strains obtained from environmental samples of USA dairy powder manufacturing facilities showed that 55 of the 63 strains (87%) were PCR-positive for these toxin genes (data not shown), suggesting that there were other sequence variations present among the strain’s genomes that were not captured by PCR or that these toxin genes were absent.

Table 3.
A summary table as an example of the toxin diversity that is among the representative C. sakazakii strains obtained from environmental samples of dairy powder manufacturing facilities that had at least one negative PCR result.
Further investigation using the TA sequences identified by local BLAST analysis showed that not every C. sakazakii strain possessed all twenty-two TA pairs. The results from this local BLAST analysis of these 22 TA genes against 234 C. sakazakii genomes gave an output organized by alleles (the results are summarized in Table 4) and the results of the BLAST analysis for each strain is shown in Supplementary Table S3. The BLAST results shown in Supplementary Table S3 demonstrate that for the strains analyzed by PCR they either possessed the alleles targeted by PCR, or that these alleles were absent. Interestingly, some strains contained either the toxin or the antitoxin component, but not both; and the prevalence and distribution of these TA homologs followed sequence type evolutionary lineages. Of the 234 strains analyzed for this experiment, toxin ESA_00804 (100%), antitoxin ESA_00803 (98%), and toxin ESA_01887 (99%) were the most conserved across the C. sakazakii strains of different sequence types. The only strains that contained all 22 TA homologs were C. sakazakii strains possessing ST1, but not every ST1 C. sakazakii contained all 22 TA pairs.

Table 4.
Summary table of percent distribution for each of the twenty-two TA genes in the 234 C. sakazakii strains analyzed using the local database. TA FASTA sequences downloaded from TADB were used as the query in the local database blasting against the C. sakazakii genomes sequenced by WGS.
Figure 3 shows a heat map with toxin and antitoxin homologs and the phylogeny among 34 C. sakazakii strains. This was created to give a visualization of the presence and absence of the TA homologs in these representative C. sakazakii strains. Manual overlaying of sequence type data obtained from the Cronobacter Pubmed MLST site (https://pubmlst.org/cronobacter/) again showed that most of the strains clustered according to sequence type. This figure also showed that not all toxin–antitoxin homologs were present in these representative C. sakazakii strains, which supported the previously reported BLAST, MA, and PCR results.
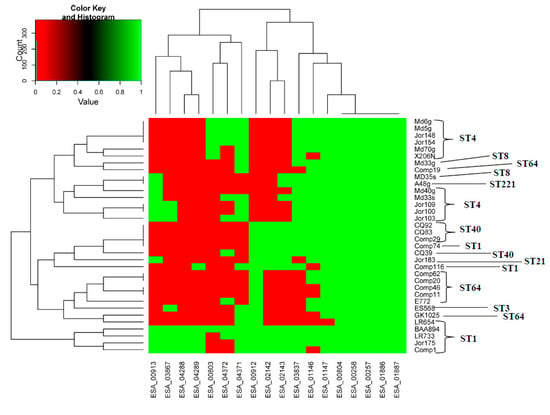
Figure 3.
Thirty-four C. sakazakii genomes were analyzed by MA and BLAST against 18 Type II toxin-antitoxin sequences. Using a 90% BLAST homology cut off, the presence or absence of the TA gene in these strains was determined. This information was converted to a binary matrix 1 (Green) = present, 0 (Red) = absent. This was used in a python version 3 script that resulted in the development of a heat map. NOTE: Overlaying sequence type (ST) information onto the phylogeny created by this analysis showed that most of the strains clustered according to ST, as expected. BLAST analysis showed that not every C. sakazakii strain possessed all 18 Type II TA loci. This figure was meant only to serve as an example of what TAs are present.
3.3. Acquisition of TAs Follow Different Evolutionary Incidences Which Draw a Parallel with Occurrence of ST Lineages
The molecular clock analysis shown in Figure 4 demonstrated that the C. sakazakii strains possibly originated from ancestral strain(s) that might have acquired TA genes at different evolutionary incidences; this draws a parallel with the occurrence of ST lineages. Recently Negrete et al. [60] reported similar evolutionary findings for efflux pumps possessed by C. sakazakii. We conjectured that the acquisition of these genes might have occurred either as independent evolutionary events through a horizontal gene transfer (such as through the attainment of plasmids, phage, or by transposition), or in certain situations, such as that of ST1, ST83, ST64, ST40, and ST4 strains, the TA genes could have been indiscriminately acquired through a robust microevolutionary selection process, also possibly involving horizontal gene transfer, some of which might have provided functional benefits. A more comprehensive analysis including candidates of closely related Enterobacteriaceae members might shed more light on how and when emergence of TA gene systems in Cronobacter occurred.
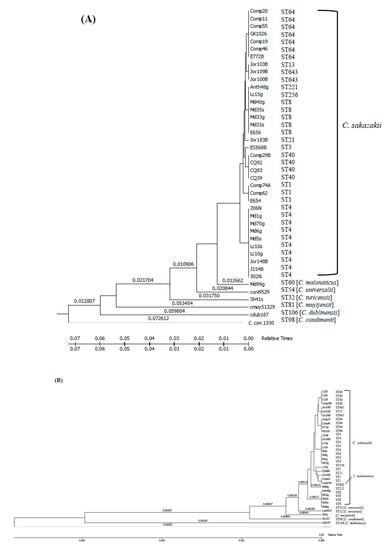
Figure 4.
A timetree analysis of evolutionary relationship of fic-like (A) and hipA (B) genes among 42 Cronobacter species was inferred using the Reltime method described by Nei et al. [61] and Tamura et al. [62,63]. Estimates of branch lengths were inferred using the neighbor-joining method [55]. The analysis involved 42 nucleotide sequences of fic-like and 35 nucleotide sequences of hipA genes. There was a total of 603 positions for fic-like (A) and 1,320 positions for hipA (B) in the final dataset. Evolutionary analyses were conducted in MEGA7 [54]. All positions containing gaps and missing data were eliminated. The accession numbers for nucleotide sequences can be found in Table S1.
3.4. TA Association with Cronobacter Plasmids
It is well-known that the maintenance and stabilization of bacterial plasmids is controlled through the interplay of TAs genes by coupling them with both host cell division and plasmid propagation [4]. To better understand this relationship, we aligned three C. sakazakii virulence plasmids, pESA3, pSP291-1, and pCSK29544_1p that were reported by Franco et al. [64], Kucerova et al. [65] Power et al. [11], Yan et al. [47], and Moine et al. [66] and reproduced the alignment in CGView Server from the Stothard Research Group (http://stothard.afns.ualberta.ca/cgview_server/; last accessed 4 October 2019). The results (shown in Figure S1) illustrated that all three C. sakazakii virulence plasmids possessed and shared the hipA toxin gene and its antitoxin gene xre (annotated as a helix-turn-helix XRE-family transcriptional regulator). Interestingly, these C. sakazakii virulence plasmids also shared a common backbone consisting of an origin of replication gene (repB), iron ABC transporter (eitCBAD), and the only Cronobacter siderophore (iucABCD/iutA/tonB family siderophore/update gene cluster) with pCTU1 possessed by C. turicensis strain z3032T (alias LMG 23827T), pCUNV1 possessed by C. universalis strain NCTC 9529T (alias 797T), and C. malonaticus strain LMG 23826T (alias CI825T) [64]. The presence of TA genes on Cronobacter plasmids needs further study using a larger sampling of isolates from different environmental and agricultural sources, to better understand their roles in persistence and pathogenicity.
3.5. TA Association with Cronobacter Phage
To understand the genomic relationship of TAs with that of prophage genomic regions that might be carried by Cronobacter strains, we first uploaded closed genome datasets for C. sakazakii strains BAA-894 (ST1, NCBI accession #: NC_009778) and ES15 (ST125, NCBI accession #: NC_017933) to the PHASTER web server and pipeline (https://phaster.ca/) for phage sequence identifications [67,68]. The PHASTER web server/pipeline identified two and three intact genomic regions in C. sakazakii strains BAA-894 and ES15, respectively. However, no TA genes were identified. Comparative sequence and phylogenetic analyses were then carried out using Geneious (http://www.geneious.com) and through annotation by BLAST it was observed that there was an intact Salmonella phage genomic region (identified as phage_118970_sal3, Refseq accession #:NC_031940) in the genomes of both BAA-894 and ES15 strains that contained a type II RelE/ParE family toxin-antitoxin system. In BAA-894, the Salmonella phage was only 40.2 kbp in size and in ES15, the phage was 42.8 kbp in size. Additionally, in BAA-894, the bicistronic TA operon was identified as a parE-relE/parD-relB type II TA system, where the antitoxin gene preceded the toxin gene and was located between base pair positions 98,5190–98,5426 (237 bp in size, encoding for a 79 amino acid protein) (originally annotated as a hypothetical protein). The toxin gene was located between 985,395–985,736 (342 bp in size, encoding for a 114 amino acid protein) and was identified as a RelE toxin. However, in ES15, the TA system was located from 1,346,920 to 1,369,940 bp and consisted of a bicistronic gene cluster where the antitoxin (location: 1,351,660–1,351,896, 237 bp in size, and encoding also for a 79 amino acid protein) was also originally annotated as a hypothetical protein, and was later annotated (through a wider BLAST analysis) as a parD/relB antitoxin that preceded the parE/relE toxin gene (location: 1,351,865–1,352,206, 342 bp in size, and encoding for a 114 amino acid protein). Other phages found in C. sakazakii strain ES15 included an intact Cronobacter phage (identified as phiES15, RefSeq accession #:NC_018454) that did not contain TAs within the phage genomic region. Although Salmonella phage_118970_sal3 was not as large as the P1 genome (93.6 kbp), the type II RelB/RelE-like toxin–antitoxins were found near the beginning of the phage genomic region (adjacent to a Holin encoding gene in both instances) where in P1 prophage, the type II Phd/Doc toxin–antitoxin gene cluster was found near the end of the genome region (adjacent to LP activation, packaging, and a repression of lytic function gene cluster) [7]. In contrast to P1, Salmonella phage118970_sal3 in the Cronobacter strains did not possess plasmid-like genes such as mobilization genes parA/parB, within its genome region, and the incompatibility class or origin of replication genes [7]. It might be speculated that the Cronobacter version of this Salmonella phage probably did not act as a plasmid like entity similar to that of prophage P1. Taken together, these results suggest that TAs associated with bacteriophage carried by C. sakazakii strains possess TAs that are both host-associated and phage-associated.
4. Conclusions
On average, approximately 11 TAs such as relE-xre, COG5654-xre, fic-phd, fic-yhfG, and hipA loci with BLAST % nucleotide identity of >90% were found. A total of 109 TA homologs were confirmed to be present on the FDA Cronobacter microarray that represented the pan genome of the seven Cronobacter species. The TA allelic sequence divergence aligned along species taxa lines and within C. sakazakii aligned with ST lineages. WGS analysis of fic-like and a hipA orthologues of the seven species supported the phylogenetic relationship found by MA. Molecular clock analysis demonstrated that C. sakazakii strains possibly descended from an ancestral strain(s) that might have acquired TA genes at distinct evolutionary incidences that correlated with occurrence and evolution of ST lineages. We hypothesized that the possible acquisition of these genes could have occurred either as independent evolutionary events through horizontal gene transfer mechanisms (such as through the acquisition of plasmids or phage), or in certain situations TA genes could have been randomly acquired through a robust microevolutionary selective process, such as through transposition or plasmid acquisition, some of which might have provided functional advantages, e.g., the acquisition of plasmidborne virulence factor genes that provides the ability to survive in new econiches. Their identification is critical for a reliable prediction of gene function. Understanding the phylogenetic relatedness of TA genes among the Cronobacter spp. is essential for designing future studies to identify mechanisms used for survival and persistence in foods. These results demonstrate that in some species, TA homologs share a common phylogeny, while in other species, they follow species-specific phylogeny. Furthermore, these results add to the growing number of genomes of Cronobacter strains, many of which are from plant–host origins. Availability of genomic information from these strains would provide a better understanding of the genetic features linked to plant association and expand insights into the possible evolutionary history of this important foodborne pathogen.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2607/7/11/554/s1, Figure S1: Alignment of three C. sakazakii virulence plasmids, pESA3, pSP291-1, and pCSK29544_1p produced by using CGView Server from the Stothard Research Group (http://stothard.afns.ualberta.ca/cgview_server/; last accessed 25 June 2018) showing the alignment of hipA and its antitoxin xre among the three C. sakazakii virulence plasmids The software uses BLAST analysis to illustrate homology among the toxin homologs as well as presence of conserved or the absence of missing homologs. Two circular plasmids—pSP291-1 (NC_020263) and pCSK29544_1p (NZ_CP011048) – were compared against reference plasmid, pESA3 (NC_009780). hipA family toxin and a Helix-Turn-Helix (HTH-XRE) domain antitoxin (originally identified as XRE-family transcriptional regulator) are shown; Table S1: Cronobacter strains including the C. sakazakii strain names, sequence type (ST) assignment, source, country, and NCBI accession numbers used in the BLAST analysis experiment described in Supplemental Table S3; Table S2: One hundred and nine toxin-antitoxin probe, probeset, probe species ID that the probe sequence came from, % similarity to BAA-894 allele, NCBI protein accession #, NCBI annotations, loci, and genome region (GR) which are located on the FDA Cronobacter microarray as described by Tall et al. [53]. Fifty Cronobacter strains were evaluated using these homologs and the results were used to develop the phylogenetic tree shown in Figure 1, Table S3: Summary of local BLAST results using the 22 toxin-antitoxin loci against 234 C. sakazakii strains to identify which TA allele is present in each strain. Strains were identified as C. sakazakii by were identified using species-specific (rpoB and cgcA) PCR assays [47,48,49]. Multi-locus sequence typing (MLST) analysis of the strains was carried out following the protocol described by Joseph et al. [50].
Author Contributions
Conception of the work: B.D.T., S.F., and G.R.G.; Data acquisition: S.F., F.N., H.J., J.G., I.R.P., J.W., Y.L., H.R.C., C.Z.W., and L.W.; Interpretation: S.F., F.N., H.J., J.G., M.M., I.R.P., G.R.G., and B.D.T.; Writing and revising: S.F., F.N., H.J., J.G., M.M., I.R.P., H.R.C., J.W., Y.L., C.Z.W., L.W., B.D.T., and G.R.G.
Funding
This research was internally funded through U.S. FDA appropriations, and in part by the University of Maryland JIFSAN Program through a cooperative agreement with the FDA (no. FDU001418).
Acknowledgments
We thank the student internship programs sponsored by the Offices of International Affairs of Gachon University, Seongnam-si, Republic of Korea for supporting the student interns: Jungha Woo and Youyoung Lee. We thank the University of Maryland at College Park, Joint Institute for Food Safety and Applied Nutrition (JIFSAN) for supporting JIFSAN interns Samantha Finkelstein, Flavia Negrete, and Leah Weinstein. We also thank the Oak Ridge Institute for Science and Education of Oak Ridge, Tennessee for sponsoring research fellows Hannah R. Chase and Hyein Jang. We thank the FDA Pathways program for sponsoring Caroline Z. Wang. We thank summer interns, Deeksha Sesha and Sahithi Kondam for their help in preparing tables and in annotating figures.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Leplae, R.; Geeraerts, D.; Hallez, R.; Guglielmini, J.; Drèze, P.; Van Melderen, L. Diversity of bacterial type II toxin-antitoxin systems: A comprehensive search and functional analysis of novel families. Nucleic Acids Res. 2011, 39, 5513–5525. [Google Scholar] [CrossRef] [PubMed]
- Otsuka, Y. Prokaryotic toxin-antitoxin systems: Novel regulations of the toxins. Curr. Genet. 2016, 62, 379–382. [Google Scholar] [CrossRef] [PubMed]
- Rocker, A.; Meinhart, A. Type II toxin: Antitoxin systems. More than small selfish entities? Curr. Genet. 2016, 62, 287–290. [Google Scholar] [CrossRef] [PubMed]
- Ogura, T.; Hiraga, S. Mini-F plasmid genes that couple host cell division to plasmid proliferation. Proc. Natl. Acad. Sci. USA 1983, 80, 4784–4788. [Google Scholar] [CrossRef] [PubMed]
- Page, R.; Peti, W. Toxin-antitoxin systems in bacterial growth arrest and persistence. Nat. Chem. Biol. 2016, 12, 208–214. [Google Scholar] [CrossRef] [PubMed]
- Fernández-García, L.; Blasco, L.; Lopez, M.; Bou, G.; García-Contreras, R.; Wood, T.; Tomas, M. Toxin-antitoxin systems in clinical pathogens. Toxins 2016, 8, 227. [Google Scholar] [CrossRef] [PubMed]
- Lehnherr, H.; Maguin, E.; Jafri, S.; Yarmolinsky, M.B. Plasmid addiction genes of bacteriophage P1: Doc, which causes cell death on curing of prophage, and phd, which prevents host death when prophage is retained. J. Mol. Biol. 1993, 233, 414–428. [Google Scholar] [CrossRef] [PubMed]
- Goormaghtigh, F.; Fraikin, N.; Putrinš, M.; Hallaert, T.; Hauryliuk, V.; Garcia-Pino, A.; Sjödin, A.; Kasvandik, S.; Udekwu, K.; Tenson, T.; et al. Reassessing the role of type II toxin-antitoxin systems in formation of Escherichia coli type II persister cells. mBio 2018, 9, e00640-18. [Google Scholar] [CrossRef] [PubMed]
- Aakre, C.D.; Phung, T.N.; Huang, D.; Laub, M.T. A bacterial toxin inhibits DNA replication elongation through a direct interaction with the β sliding clamp. Mol. Cell. 2013, 52, 617–628. [Google Scholar] [CrossRef] [PubMed]
- Grim, C.J.; Kotewicz, M.L.; Power, K.A.; Gopinath, G.; Franco, A.A.; Jarvis, K.G.; Yan, Q.Q.; Jackson, S.A.; Sathyamoorthy, V.; Hu, L.; et al. Genomic analysis of the pan genome of the emerging bacterial foodborne pathogen Cronobacter spp. suggests a species-level bidirectional divergence from a conserved genomic core. BMC Genom. 2013, 14, 366. [Google Scholar] [CrossRef] [PubMed]
- Power, K.A.; Yan, Q.; Fox, E.M.; Cooney, S.; Fanning, S. Genome sequence of Cronobacter sakazakii SP291, a persistent thermotolerant isolate derived from a factory producing powdered infant formula. Genome Announc. 2013, 1, e00082-13. [Google Scholar] [CrossRef] [PubMed]
- Iversen, C.; Mullane, N.; McCardell, B.; Tall, B.D.; Lehner, A.; Fanning, S.; Stephan, R.; Joosten, H. Cronobacter gen. nov., a new genus to accommodate the biogroups of Enterobacter sakazakii, and proposal of Cronobacter sakazakii gen. nov., comb. nov., Cronobacter malonaticus sp. nov., Cronobacter turicensis sp. nov., Cronobacter muytjensii sp. nov., Cronobacter dublinensis sp. nov., Cronobacter genomospecies 1, and of three subspecies, Cronobacter dublinensis subsp. dublinensis subsp. nov., Cronobacter dublinensis subsp. lausannensis subsp. nov. and Cronobacter dublinensis subsp. lactaridi subsp. nov. Int. J. Syst. Evol. Microbiol. 2008, 58, 1442–1447. [Google Scholar] [PubMed]
- Joseph, S.; Cetinkaya, E.; Drahovska, H.; Levican, A.; Figueras, M.J.; Forsythe, S.J. Cronobacter condimenti sp. nov., isolated from spiced meat, and Cronobacter universalis sp. nov., a species designation for Cronobacter sp. genomospecies 1, recovered from a leg infection, water and food ingredients. Int. J. Syst. Evol. Microbiol. 2011, 62, 1277–1283. [Google Scholar] [CrossRef] [PubMed]
- Stephan, R.; Lehner, A.; Tischler, P.; Rattei, T. Complete genome sequence of Cronobacter turicensis LMG 23827, a food-borne pathogen causing deaths in neonates. J. Bacteriol. 2011, 193, 309–310. [Google Scholar] [CrossRef] [PubMed]
- Bowen, A.B.; Braden, C.R. Invasive Enterobacter sakazakii disease in infants. Emerg. Infect. Dis. 2006, 12, 1185–1189. [Google Scholar] [CrossRef] [PubMed]
- Lai, K.K. Enterobacter sakazakii infections among neonates, infants, children, and adults. Case reports and a review of the literature. Medicine 2001, 80, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Jason, J. Prevention of invasive Cronobacter infections in young infants fed powdered infant formulas. Pediatrics 2012, 130, e1076–e1084. [Google Scholar] [CrossRef] [PubMed]
- Patrick, M.E.; Mahon, B.E.; Greene, S.A.; Rounds, J.; Cronquist, A.; Wymore, K.; Boothe, E.; Lathrop, S.; Palmer, A.; Bowen, A. Incidence of Cronobacter spp. infections, United States, 2003–2009. Emerg. Infect. Dis. 2014, 20, 1520–1523. [Google Scholar] [CrossRef] [PubMed]
- Holý, O.; Petrželová, J.; Hanulík, V.; Chromá, M.; Matoušková, I.; Forsythe, S.J. Epidemiology of Cronobacter spp. isolates from patients admitted to the Olomouc University Hospital (Czech Republic). Epidemiol. Mikrobiol. Imunol. 2014, 63, 69–72. [Google Scholar] [PubMed]
- Gosney, M.A.; Martin, M.V.; Wright, A.E.; Gallagher, M. Enterobacter sakazakii in the mouths of stroke patients and its association with aspiration pneumonia. Eur. J. Intern. Med. 2006, 17, 185–188. [Google Scholar] [CrossRef] [PubMed]
- Alsonosi, A.; Hariri, S.; Kajsík, M.; Oriešková, M.; Hanulík, V.; Röderová, M.; Petrželová, J.; Kollárová, H.; Drahovská, H.; Forsythe, S. The speciation and genotyping of Cronobacter isolates from hospitalized patients. Eur. J. Clin. Microbiol. Infect. Dis. 2015, 34, 1979–1988. [Google Scholar] [CrossRef] [PubMed]
- Yan, Q.Q.; Condell, O.; Power, K.; Butler, F.; Tall, B.D.; Fanning, S. Cronobacter species (formerly known as Enterobacter sakazakii) in powdered infant formula: A review of our current understanding of the biology of this bacterium. J. Appl. Microbiol. 2012, 113, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Himelright, I.; Harris, E.; Lorch, V.; Anderson, M.; Jones, T.; Craig, A.; Kuehnert, M.; Forster, T.; Arduino, M.; Jensen, B.; et al. Enterobacter sakazakii infections associated with the use of powdered infant formula—Tennessee, 2001. Morb. Mortal. Wkly. Rep. 2002, 51, 297–300. [Google Scholar]
- Tall, B.D.; Chen, Y.; Yan, Q.Q.; Gopinath, G.R.; Grim, C.J.; Jarvis, K.G.; Fanning, S.; Lampel, K.A. Cronobacter: An emergent pathogen using meningitis to neonates through their feeds. Sci. Prog. 2014, 97, 154–172. [Google Scholar] [PubMed]
- Noriega, F.R.; Kotloff, K.L.; Martin, M.A.; Schwalbe, R.S. Nosocomial bacteremia caused by Enterobacter sakazakii and Leuconostoc mesenteroides resulting from extrinsic contamination of infant formula. Pediatr. Infect. Dis. J. 1990, 9, 447–449. [Google Scholar] [PubMed]
- Friedemann, M. Enterobacter sakazakii in food and beverages (other than infant formula and milk powder). Int. J. Food Microbiol. 2007, 116, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Bowen, A.; Wiesenfeld, H.C.; Kloesz, J.L.; Pasculle, A.W.; Nowalk, A.J.; Brink, L.; Elliot, E.; Martin, H.; Tarr, C.L. Notes from the Field: Cronobacter sakazakii infection associated with feeding extrinsically contaminated expressed human milk to a premature infant—Pennsylvania. Morb. Mortal. Wkly. Rep. 2017, 66, 761–762. [Google Scholar] [CrossRef] [PubMed]
- McMullan, R.; Menon, V.; Beukers, A.G.; Jensen, S.O.; van Hal, S.J.; Davis, R. Cronobacter sakazakii infection from expressed breast milk, Australia. Emerg. Infect. Dis. 2018, 24, 393–394. [Google Scholar] [CrossRef] [PubMed]
- El-Sharoud, W.M.; El-Din, M.Z.; Ziada, D.M.; Ahmed, S.F.; Klena, J.D. Surveillance and genotyping of Enterobacter sakazakii suggest its potential transmission from milk powder into imitation recombined soft cheese. J. Appl. Microbiol. 2008, 105, 559–566. [Google Scholar] [CrossRef] [PubMed]
- Osaili, T.M.; Shaker, R.R.; Al-Haddaq, M.S.; Al-Nabulsi, A.A.; Holley, R.A. Heat resistance of Cronobacter species (Enterobacter sakazakii) in milk and special feeding formula. J. Appl. Microbiol. 2009, 107, 928–935. [Google Scholar] [CrossRef] [PubMed]
- Tall, B.D.; Gopinath, G.R.; Gangiredla, J.; Patel, I.R.; Fanning, S.; Lehner, A. Food Microbiology: Fundamentals and Frontiers, 5th ed.; Doyle, M.P., Diez-Gonzalez, F., Hill, C., Eds.; ASM Press: Washington, DC, USA, 2019; pp. 389–414. [Google Scholar]
- Gopinath, G.R.; Chase, H.R.; Gangiredla, J.; Eshwar, A.; Jang, H.; Patel, I.; Negrete, F.; Finkelstein, S.; Park, E.; Chung, T.; et al. Genomic characterization of malonate positive Cronobacter sakazakii serotype O:2, sequence type 64 strains, isolated from clinical, food, and environment samples. Gut Pathog. 2018, 10, 11. [Google Scholar] [CrossRef] [PubMed]
- Berthold-Pluta, A.; Garbowska, M.; Stefanska, I.; Pluta, A. Microbiological quality of selected ready-to-eat leaf vegetables, sprouts and non-pasteurized fresh fruit-vegetable juices including the presence of Cronobacter spp. Food Microbiol. 2017, 65, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Vasconcellos, L.; Carvalho, C.T.; Tavares, R.O.; de Mello Medeiros, V.; de Oliveira Rosas, C.; Silva, J.N.; Dos Reis Lopes, S.M.; Forsythe, S.J.; Brandão, M.L.L. Isolation, molecular and phenotypic characterization of Cronobacter spp. in ready-to-eat salads and foods from Japanese cuisine commercialized in Brazil. Food Res. Int. 2018, 107, 353–359. [Google Scholar] [CrossRef] [PubMed]
- Pava-Ripoll, M.; Pearson, R.E.; Miller, A.K.; Ziobro, G.C. Prevalence and relative risk of Cronobacter spp., Salmonella spp., and Listeria monocytogenes associated with the body surfaces and guts of individual filth flies. Appl. Environ. Microbiol. 2012, 78, 7891–7902. [Google Scholar] [CrossRef] [PubMed]
- Lehner, A.; Tall, B.D.; Fanning, S. Cronobacter spp.-opportunistic foodborne pathogens: An update on evolution, osmotic adaptation and pathophysiology. Curr. Clin. Microbiol. Rep. 2017, 5, 97–105. [Google Scholar] [CrossRef]
- Cruz, A.; Xicohtencatl-Cortes, J.; Gonzalez-Pedrajo, B.; Bobadilla, M.; Eslava, C.; Rosas, I. Virulence traits in Cronobacter species isolated from different sources. Can. J. Microbiol. 2011, 57, 735–744. [Google Scholar] [CrossRef] [PubMed]
- Jang, H.; Chase, H.R.; Gangiredla, J.; Gopinath, G.R.; Grim, C.J.; Patel, I.R.; Kothary, M.H.; Jackson, S.A.; Mammel, M.K.; Carter, L.; et al. Analysis of the molecular diversity among Cronobacter species isolated from filth flies using a pan genomic DNA microarray and whole genome sequencing. Front. Microbiol. 2019. submitted. [Google Scholar]
- Umeda, N.S.; De Filippis, I.; Forsythe, S.J.; Brandão, M.L.L. Phenotypic characterization of Cronobacter spp. strains isolated from foods and clinical specimens in Brazil. Food Res. Int. 2017, 102, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Jang, H.; Addy, N.; Ewing, L.; Beaubrun, J.J.G.; Lee, Y.; Woo, J.; Negrete, F.; Finkelstein, S.; Tall, B.D.; Lehner, A.; et al. Whole Genome Sequences of Cronobacter sakazakii isolates obtained from plant-origin foods and dried food manufacturing environments. Genome Announc. 2018, 6, e00223-18. [Google Scholar] [CrossRef] [PubMed]
- Jang, H.; Gopinath, G.R.; Chase, H.R.; Gangiredla, J.; Eshwar, A.; Patel, I.; Addy, N.; Ewing, L.; Beaubrunm, J.J.G.; Negrete, F.; et al. Draft genomes of Cronobacter sakazakii strains isolated from dried spices bring unique insights into the epidemiology of plant-associated strains. Stand. Genom. Sci. 2018, 13, 35. [Google Scholar] [CrossRef] [PubMed]
- Sani, N.A.; Odeyemi, O.A. Occurrence and prevalence of Cronobacter spp. in plant and animal derived food sources: A systematic review and meta-analysis. SpringerPlus 2015, 4, 545. [Google Scholar] [CrossRef] [PubMed]
- Yong, W.; Guo, B.; Shi, X.; Cheng, T.; Chen, M.; Jiang, X.; Ye, Y.; Wang, J.; Xie, G.; Ding, J. An investigation of an acute gastroenteritis outbreak: Cronobacter sakazakii, a potential cause of food-borne illness. Front. Microbiol. 2018, 9, 2549. [Google Scholar] [CrossRef] [PubMed]
- Restaino, L.; Frampton, E.W.; Lionberg, W.C.; Becker, R.J. A chromogenic plating medium for the isolation and identification of Enterobacter sakazakii from foods, food ingredients, and environmental sources. J. Food Prot. 2006, 69, 315–322. [Google Scholar] [CrossRef] [PubMed]
- Jaradat, Z.W.; Ababneh, Q.O.; Saadoun, I.M.; Samara, N.A.; Rashdan, A.M. Isolation of Cronobacter spp. (formerly Enterobacter sakazakii) from infant food, herbs and environmental samples and the subsequent identification and confirmation of the isolates using biochemical, chromogenic assays, PCR and 16S rRNA sequencing. BMC Microbiol. 2009, 9, 225. [Google Scholar] [CrossRef] [PubMed]
- Chon, J.W.; Song, K.Y.; Kim, S.Y.; Hyeon, J.Y.; Seo, K.H. Isolation and characterization of Cronobacter from desiccated foods in Korea. J. Food Sci. 2012, 77, M354–M358. [Google Scholar] [CrossRef] [PubMed]
- Yan, Q.; Wang, J.; Gangiredla, J.; Cao, Y.; Martins, M.; Gopinath, G.R.; Stephan, R.; Lampel, K.; Tall, B.D.; Fanning, S. Comparative genotypic and phenotypic analysis of Cronobacter Species cultured from four powdered infant formula production facilities: Indication of pathoadaptation along the food chain. Appl. Environ. Microbiol. 2015, 81, 4388–4402. [Google Scholar] [CrossRef] [PubMed]
- Chase, H.R.; Gopinath, G.R.; Eshwar, A.K.; Stoller, A.; Fricker-Feer, C.; Gangiredla, J.; Patel, I.R.; Cinar, H.N.; Jeong, H.; Lee, C.; et al. Comparative genomic characterization of the highly persistent and potentially virulent Cronobacter sakazakii ST83, CC65 strain H322 and other ST83 strains. Front. Microbiol. 2017, 8, 1136. [Google Scholar] [CrossRef] [PubMed]
- Stoop, B.; Lehner, A.; Iversen, C.; Fanning, S.; Stephan, R. Development and evaluation of rpoB based PCR systems to differentiate the six proposed species within the genus Cronobacter. Int. J. Food Microbiol. 2009, 136, 165–168. [Google Scholar] [CrossRef] [PubMed]
- Lehner, A.; Fricker-Feer, C.; Stephan, R. Identification of the recently described Cronobacter condimenti by a rpoB based PCR system. J. Med. Microbiol. 2012, 61, 1034–1035. [Google Scholar] [CrossRef] [PubMed]
- Carter, L.; Lindsey, L.A.; Grim, C.J.; Sathyamoorthy, V.; Jarvis, K.G.; Gopinath, G.; Lee, C.; Sadowski, J.A.; Trach, L.; Pava-Ripoll, M.; et al. Multiplex PCR assay targeting a diguanylate cyclase-encoding gene, cgcA, to differentiate species within the genus Cronobacter. Appl. Environ. Microbiol. 2013, 79, 734–737. [Google Scholar] [CrossRef] [PubMed]
- Joseph, S.; Sonbol, H.; Hariri, S.; Desai, P.; McClelland, M.; Forsythe, S.J. Diversity of the Cronobacter genus as revealed by multilocus sequence typing. J. Clin. Microbiol. 2012, 50, 3031–3039. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, A.; Loughlin, M.; Caubilla-Barron, J.; Kucerova, E.; Manning, G.; Dowson, C.; Forsythe, S. Multilocus sequence typing of Cronobacter sakazakii and Cronobacter malonaticus reveals stable clonal structures with clinical significance which do not correlate with biotypes. BMC Microbiol. 2009, 9, 223. [Google Scholar] [CrossRef] [PubMed]
- Tall, B.D.; Gangiredla, J.; Gopinath, G.R.; Yan, Q.Q.; Chase, H.R.; Lee, B.; Hwang, S.; Trach, L.; Park, E.; Yoo, Y.J.; et al. Development of a custom-designed, pan genomic DNA microarray to characterize strain-level diversity among Cronobacter spp. Front. Pediatr. 2015, 3, 36. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Wei, Y.; Shen, Y.; Li, X.; Zhou, H.; Tai, C.; Deng, Z.; Ou, H.Y. TADB 2.0: An updated database of bacterial type II toxin-antitoxin loci. Nucleic Acids Res. 2018, 46, D749–D753. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Kamruzzaman, M.; Iredell, J. A ParDE-family toxin antitoxin system in major resistance plasmids of Enterobacteriaceae confers antibiotic and heat tolerance. Sci. Rep. 2019, 9, 9872. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Jolley, K.A.; Maiden, M.C.J. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinform. 2010, 11, 595. [Google Scholar] [CrossRef] [PubMed]
- Negrete, F.; Gangiredla, J.; Jang, H.; Chase, H.R.; Woo, J.; Lee, Y.; Patel, I.; Finkelstein, S.B.; Tall, B.D.; Gopinath, G.R. Prevalence and distribution of efflux pump complexes genes in Cronobacter sakazakii using whole genome and pan-genomic datasets. Curr. Opin. Food Sci. 2019, 30, 32–42. [Google Scholar] [CrossRef]
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics; Thomas, R.H., Ed.; Oxford University Press: Oxford, UK, 2000; p. 333. [Google Scholar]
- Tamura, K.; Nei, M.; Kumar, S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. USA 2004, 101, 11030–11035. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Battistuzzi, F.U.; Billing-Ross, P.; Murillo, O.; Filipski, A.; Kumar, S. Estimating divergence times in large molecular phylogenies. Proc. Natl. Acad. Sci. USA 2012, 109, 19333–19338. [Google Scholar] [CrossRef] [PubMed]
- Franco, A.A.; Hu, L.; Grim, C.J.; Gopinath, G.; Sathyamoorthy, V.; Jarvis, K.G.; Lee, C.; Sadowski, J.; Kim, J.; Kothary, M.H.; et al. Characterization of putative virulence genes on the related RepFIB plasmids harbored by Cronobacter spp. Appl. Environ. Microbiol. 2011, 77, 3255–3267. [Google Scholar] [CrossRef] [PubMed]
- Kucerova, E.; Clifton, S.W.; Xia, X.Q.; Long, F.; Porwollik, S.; Fulton, L.; Fronick, C.; Minx, P.; Kyung, K.; Warren, W.; et al. Genome sequence of Cronobacter sakazakii BAA-894 and comparative genomic hybridization analysis with other Cronobacter species. PLoS ONE 2010, 5, e9556. [Google Scholar] [CrossRef] [PubMed]
- Moine, D.; Kassam, M.; Baert, L.; Tang, Y.; Barretto, C.; Ngom Bru, C.; Klijn, A.; Descombes, P. Fully closed genome sequences of five type strains of the genus Cronobacter and one Cronobacter sakazakii Strain. Genome Announc. 2016, 4, e00142-16. [Google Scholar] [CrossRef] [PubMed]
- Arndt, D.; Grant, J.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Liang, Y.; Lynch, K.H.; Dennis, J.J.; Wishart, D.S. PHAST: A fast phage search tool. Nucleic Acids Res. 2011, 39, W347–W352. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).