Detection and Genomic Characteristics of NDM-19- and QnrS11-Producing O101:H5 Escherichia coli Strain Phylogroup A: ST167 from a Poultry Farm in Egypt
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation and Identification of E. coli M2-13-1
2.2. Minimum Inhibitory Concentration (MIC) of Various Antimicrobial Agents Against M2-13-1
2.3. Conjugation and Transformation Experiments
2.4. Complete Genome Sequencing (WGS) and Analysis
2.5. Nucleotide Sequence Accession Numbers
3. Results and Discussion
3.1. Minimum Inhibitory Concentration (MIC) of Various Antimicrobials Against M2-13-1
3.2. Genomic Characterization of E. coli M2-13-1
3.3. Plasmidome and Resistome of E. coli M2-13-1
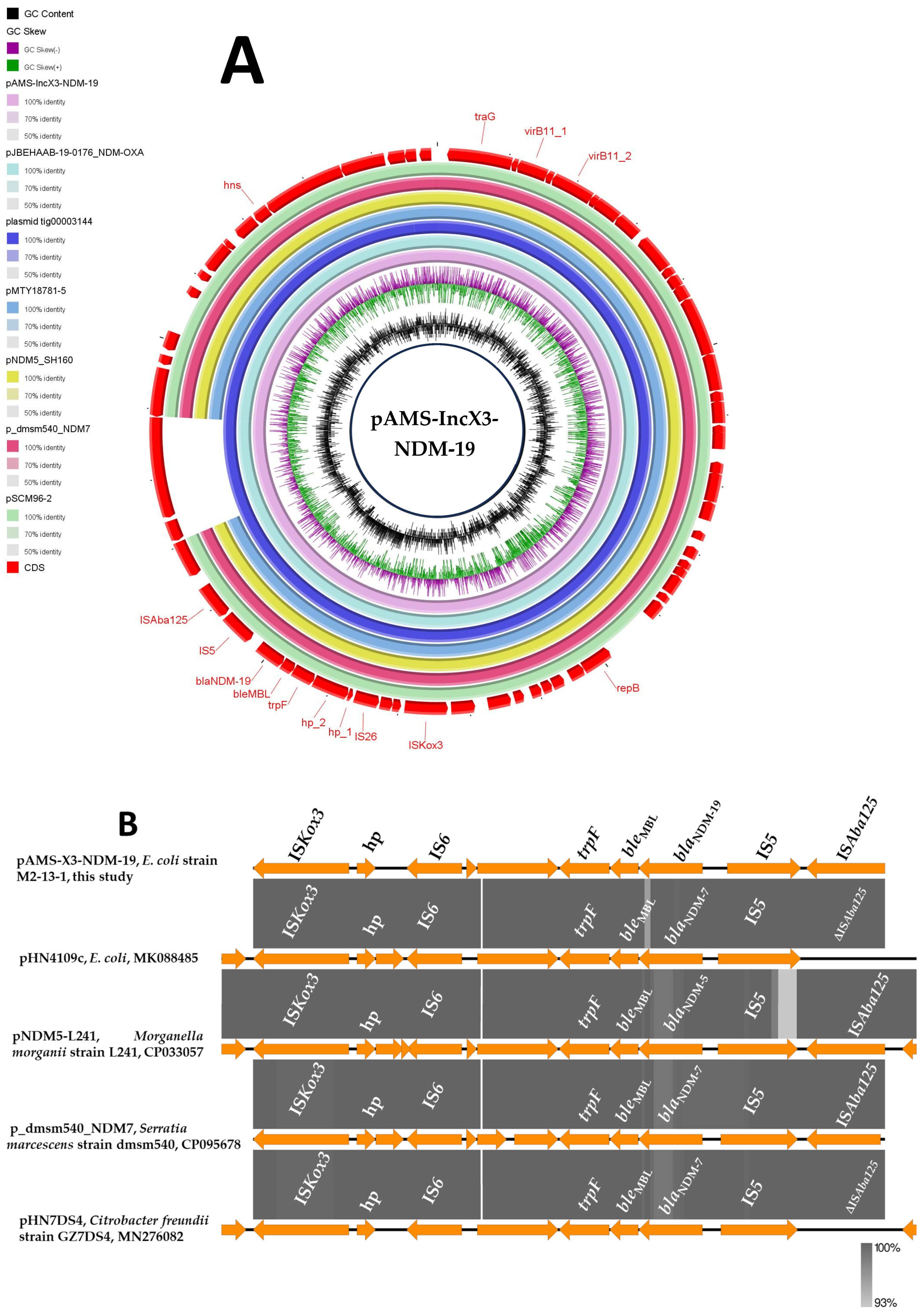
3.4. Identification of a blaNDM-19/IncX3 Plasmid in an Egyptian E. coli Poultry Strain
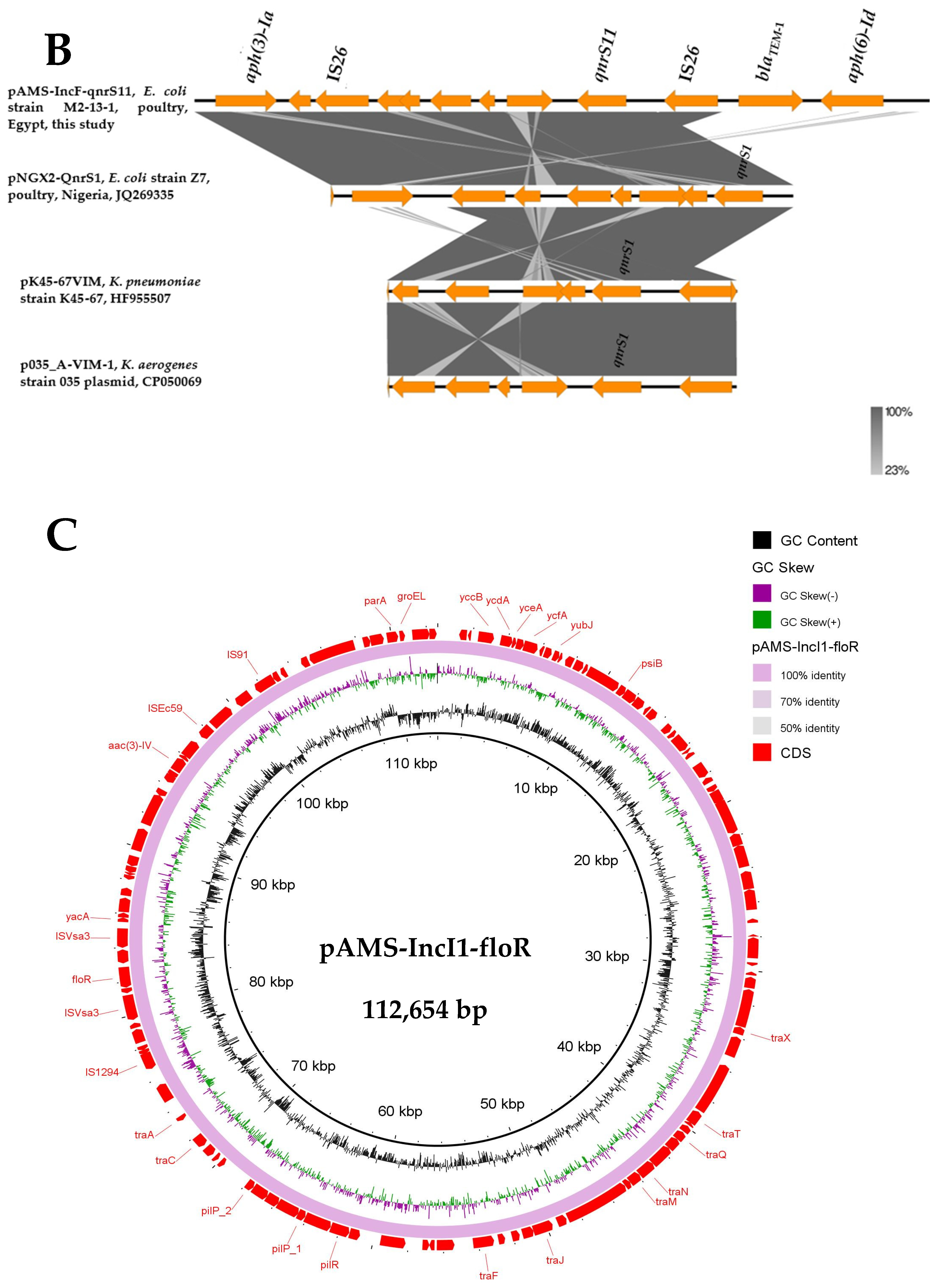
3.5. Detection of Other Multidrug-Resistant (MDR) Plasmids from E. coli M2-13-1
3.6. Analysis of the Virulome and Heavy Metal Resistance Genes
3.7. Clonal Relationship of ST167 E. coli Isolate
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- El-Gamal, M.I.; Brahim, I.; Hisham, N.; Aladdin, R.; Mohammed, H.; Bahaaeldin, A. Recent updates of carbapenem antibiotics. Eur. J. Med. Chem. 2017, 131, 185–195. [Google Scholar] [CrossRef]
- Gamal, D.; Egea, P.; Elías, C.; Fernández-Martínez, M.; Causse, M.; Pérez-Nadales, E.; Salem, D.; Fam, N.; Diab, M.; Aitta, A.A.; et al. High-risk clones and novel sequence type ST4497 of Klebsiella pneumoniae clinical isolates producing different alleles of NDM-type and other carbapenemases from a single tertiary-care centre in Egypt. Int. J. Antimicrob. Agents 2020, 56, 106164. [Google Scholar] [CrossRef]
- Soliman, A.M.; Zarad, H.O.; Nariya, H.; Shimamoto, T.; Shimamoto, T. Genetic analysis of carbapenemase-producing Gram-negative bacteria isolated from a university teaching hospital in Egypt. Infect. Genet. Evol. 2020, 77, 104065. [Google Scholar] [CrossRef]
- Soliman, A.M.; Khalifa, H.O.; Ahmed, A.M.; Shimamoto, T.; Shimamoto, T. Emergence of an NDM-5-producing clinical Escherichia coli isolate in Egypt. Int. J. Infect. Dis. 2016, 48, 46–48. [Google Scholar] [CrossRef] [PubMed]
- Khalifa, H.O.; Soliman, A.M.; Ahmed, A.M.; Shimamoto, T.; Shimamoto, T. NDM-4- and NDM-5-producing Klebsiella pneumoniae coinfection in a 6-month-old infant. Antimicrob. Agents Chemother. 2016, 60, 4416–4417. [Google Scholar] [CrossRef]
- Soliman, A.M.; Ramadan, H.; Sadek, M.; Nariya, H.; Shimamoto, T.; Hiott, L.M.; Frye, J.G.; Jackson, C.R.; Shimamoto, T. Draft genome sequence of a blaNDM-1- and blaOXA-244-carrying multidrug-resistant Escherichia coli D-ST69 clinical isolate from Egypt. J. Glob. Antimicrob. Resist. 2020, 22, 832–834. [Google Scholar] [CrossRef]
- Khalifa, H.O.; Soliman, A.M.; Ahmed, A.M.; Shimamoto, T.; Hara, T.; Ikeda, M.; Kuroo, Y.; Kayama, S.; Sugai, M.; Shimamoto, T. High carbapenem resistance in clinical Gram-negative pathogens isolated in Egypt. Microb. Drug Resist. 2017, 23, 838–844. [Google Scholar] [CrossRef] [PubMed]
- Mancini, S.; Keller, P.M.; Greiner, M.; Bruderer, V.; Imkamp, F. Detection of NDM-19, a novel variant of the New Delhi metallo-β-lactamase with increased carbapenemase activity under zinc-limited conditions, in Switzerland. Diagn. Microbiol. Infect. Dis. 2019, 95, 114851. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, H.; Zhang, X.; Jiang, N.; Zhang, Z.; Zhang, J.; Zhu, B.; Wang, G.; Zhao, K.; Zhou, Y. Characterization of an NDM-19-producing Klebsiella pneumoniae strain harboring 2 resistance plasmids from China. Diagn. Microbiol. Infect. Dis. 2019, 93, 355–361. [Google Scholar] [CrossRef]
- Rima, M.; Oueslati, S.; Dabos, L.; Daaboul, D.; Mallat, H.; Bou Raad, E.; Achkar, M.; Mawlawi, O.; Bernabeu, S.; Bonnin, R.A.; et al. Prevalence and molecular mechanisms of carbapenem resistance among Gram-negative Bacilli in three hospitals of northern Lebanon. Antibiotics 2022, 11, 1295. [Google Scholar] [CrossRef] [PubMed]
- El-Kholy, A.; El-Mahallawy, H.A.; Elsharnouby, N.; Abdel Aziz, M.; Helmy, A.M.; Kotb, R. Landscape of Multidrug-Resistant Gram-Negative Infections in Egypt: Survey and Literature Review. Infect. Drug Resist. 2021, 14, 1905–1920. [Google Scholar] [CrossRef]
- Thomsen, J.; Abdulrazzaq, N.M.; The UAE AMR Surveillance Consortium; Everett, D.B.; Menezes, G.A.; Senok, A.; Ayoub Moubareck, C. Carbapenem resistant Enterobacterales in the United Arab Emirates: A retrospective analysis from 2010 to 2021. Front. Public Health 2023, 11, 1244482. [Google Scholar] [CrossRef]
- Tuhamize, B.; Bazira, J. Carbapenem-resistant Enterobacteriaceae in the livestock, humans and environmental samples around the globe: A systematic review and meta-analysis. Sci. Rep. 2024, 14, 16333. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Castillo, F.Y.; Guerrero-Barrera, A.L.; Avelar-González, F.J. An overview of carbapenem-resistant organisms from food-producing animals, seafood, aquaculture, companion animals, and wildlife. Front. Vet. Sci. 2023, 10, 1158588. [Google Scholar] [CrossRef]
- Poirel, L.; Walsh, T.R.; Cuvillier, V.; Nordmann, P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn. Microbiol. Infect. Dis. 2011, 70, 119–123. [Google Scholar] [CrossRef] [PubMed]
- Soliman, A.M.; Maruyama, F.; Zarad, H.O.; Ota, A.; Nariya, H.; Shimamoto, T.; Shimamoto, T. Emergence of a multidrug-resistant Enterobacter hormaechei clinical isolate from Egypt co-harboring mcr-9 and blaVIM-4. Microorganisms 2020, 8, 595. [Google Scholar] [CrossRef] [PubMed]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Tanizawa, Y.; Fujisawa, T.; Nakamura, Y. DFAST: A flexible prokaryotic genome annotation pipeline for faster genome publication. Bioinformatics 2018, 34, 1037–1039. [Google Scholar] [CrossRef]
- Alikhan, N.F.; Petty, N.K.; Zakour, N.L.; Beatson, S.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genom. 2011, 12, 402. [Google Scholar] [CrossRef]
- Zhou, Z.; Alikhan, N.F.; Mohamed, K.; Fan, Y.; Achtman, M.; Agama Study Group. The EnteroBase user’s guide, with case studies on Salmonella transmissions, Yersinia pestis phylogeny, and Escherichia core genomic diversity. Genome Res. 2020, 30, 138–152. [Google Scholar] [CrossRef]
- Peirano, G.; Chen, L.; Nobrega, D.; Finn, T.J.; Kreiswirth, B.N.; DeVinney, R.; Pitout, J.D. Genomic epidemiology of global carbapenemase-producing Escherichia coli, 2015–2017. Emerg. Infect. Dis. 2022, 28, 924–931. [Google Scholar] [CrossRef]
- Shen, P.; Yi, M.; Fu, Y.; Ruan, Z.; Du, X.; Yu, Y.; Xie, X. Detection of an Escherichia coli sequence type 167 strain with two tandem copies of blaNDM-1 in the chromosome. J. Clin. Microbiol. 2016, 55, 199–205. [Google Scholar] [CrossRef]
- Xia, S.; Wang, W.; Cheng, J.; Zhang, T.; Xia, Z.; Zhao, X.; Han, Y.; Li, Y.; Shi, X.; Qin, S. Emergence of a novel hybrid mcr-1-bearing plasmid in an NDM-7-producing ST167 Escherichia coli strain of clinical origin. Front. Microbiol. 2020, 13, 950087. [Google Scholar] [CrossRef]
- Johnson, T.J.; Wannemuehler, Y.; Doetkott, C.; Johnson, S.J.; Rosenberger, S.C.; Nolan, L.K. Identification of minimal predictors of avian pathogenic Escherichia coli virulence for use as a rapid diagnostic tool. J. Clin. Microbiol. 2008, 46, 3987–3996. [Google Scholar] [CrossRef]
- Pan, S.; Liu, S.; Tai, S.; Yu, J.; Yuan, E.; Duan, Y. Genomic analysis of an Escherichia coli sequence type 167 isolate harboring a multidrug-resistant conjugative plasmid, suggesting the potential transmission of the type strains from animals to humans. Infect. Drug Resist. 2023, 16, 5077–5084. [Google Scholar] [CrossRef] [PubMed]
- Boyd, S.E.; Livermore, D.M.; Hooper, D.C.; Hope, W.W. Metallo-β-lactamases: Structure, function, epidemiology, treatment options, and the development pipeline. Antimicrob. Agents Chemother. 2020, 64, e00397-20. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Chen, R.; Wang, Q.; Li, C.; Ge, H.; Qiao, J.; Li, Y. Global prevalence, characteristics, and future prospects of IncX3 plasmids: A review. Front. Microbiol. 2022, 13, 979558. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Lu, Y.; Yu, J.; Cai, X.; Wang, C.; Lv, L.; Moran, R.A.; Zhao, X.; Hu, Z.; Deng, M.; et al. Comprehensive analysis of Enterobacteriaceae IncX plasmids reveals robust conjugation regulators PrfaH, H-NS, and conjugation-fitness tradeoff. Commun. Biol. 2025, 8, 363. [Google Scholar] [CrossRef]
- Carattoli, A. Plasmids and the spread of resistance. Int. J. Med. Microbiol. 2013, 303, 298–304. [Google Scholar] [CrossRef]
- Aworh, M.K.; Kwaga, J.K.P.; Hendriksen, R.S.; Okolocha, E.C.; Harrell, E.; Thakur, S. Quinolone-resistant Escherichia coli at the interface between humans, poultry and their shared environment—A potential public health risk. One Health Outlook 2023, 5, 2. [Google Scholar] [CrossRef]
- Rendón, M.A.; Saldaña, Z.; Erdem, A.L.; Monteiro-Neto, V.; Vázquez, A.; Kaper, J.B.; Puente, J.L.; Girón, J.A. Commensal and pathogenic Escherichia coli use a common pilus adherence factor for epithelial cell colonization. Proc. Natl. Acad. Sci. USA 2007, 104, 10637–10642. [Google Scholar] [CrossRef] [PubMed]
- Wells, T.J.; McNeilly, T.N.; Totsika, M.; Mahajan, A.; Gally, D.L.; Schembri, M.A. The Escherichia coli O157:H7 EhaB autotransporter protein binds to laminin and collagen I and induces a serum IgA response in O157:H7 challenged cattle. Environ. Microbiol. 2009, 11, 1803–1814. [Google Scholar] [CrossRef]
- Clarke, K.R.; Hor, L.; Pilapitiya, A.; Luirink, J.; Paxman, J.J.; Heras, B. Phylogenetic classification and functional review of autotransporters. Front. Immunol. 2022, 13, 921272. [Google Scholar] [CrossRef]
- Zhu, N.; Liu, W.; Prakash, A.; Zhang, C.; Kim, K.S. Targeting E. coli invasion of the blood-brain barrier for investigating the pathogenesis and therapeutic development of E. coli meningitis. Cell. Microbiol. 2020, 22, e13231. [Google Scholar] [CrossRef]
- Zhang, Y.; Tan, P.; Zhao, Y.; Ma, X. Enterotoxigenic Escherichia coli: Intestinal pathogenesis mechanisms and colonization resistance by gut microbiota. Gut Microbes 2022, 14, 2055943. [Google Scholar] [CrossRef]
- de Pace, F.; Nakazato, G.; Pacheco, A.; de Paiva, J.B.; Sperandio, V.; da Silveira, W.D. The type VI secretion system plays a role in type 1 fimbria expression and pathogenesis of an avian pathogenic Escherichia coli strain. Infect. Immun. 2010, 78, 4990–4998. [Google Scholar] [CrossRef] [PubMed]
- Quainoo, S.; Coolen, J.P.M.; van Hijum, S.; Huynen, M.A.; Melchers, W.J.G.; van Schaik, W.; Wertheim, H.F. Whole-genome sequencing of bacterial pathogens: The future of nosocomial outbreak analysis. Clin. Microbiol. Rev. 2017, 30, 1015–1063. [Google Scholar] [CrossRef] [PubMed]


| Antibiotic | MIC (mg/L) a for Strain: | |||
|---|---|---|---|---|
| M2-13-1 (Wild type) b | M2-13-1 TS1 (E. coli DH5α + pAMS-X3-NDM-19) | M2-13-1 TC (E. coli J53 + pAMS-IncF-qnrs11) | E. coli ATCC 25922 | |
| Aztreonam | <2 | <2 | <2 | <2 |
| Ceftriaxone | >256 | 64 | <2 | <2 |
| Meropenem | 64 | 4 | <0.5 | <2 |
| Doripenem | 16 | 4 | ND | <2 |
| Ertapenem | 256 | 8 | <0.5 | <2 |
| Tetracycline | 64 | <2 | 64 | <2 |
| Ciprofloxacin | 128 | <2 | 0.5 | <2 |
| Kanamycin | >256 | <2 | >256 | <2 |
| Gentamicin | 32 | <2 | ND | <2 |
| Chloramphenicol | 256 | <2 | 4 | 4 |
| Fosfomycin | 64 | <4 | 4 | 64 |
| Colistin | <0.5 | <0.5 | <0.5 | <0.5 |
| Tigecycline | 1 | 1 | <0.5 | 1 |
| Sample | Size (bp) | GC% | No. of CDSs | MLST or pMLST | Incompatibility Group | Antimicrobial Resistance Genes | QRDR Point Mutations | Virulence Genes |
|---|---|---|---|---|---|---|---|---|
| Chromosome | 4,738,278 | 50.8 | 4557 | ST167 | NA | ND | parC: S80I, parE: S458A, gyrA: S83L, and D87N | gad, terC, hra, capU, iss, csgA, hha, hlyE, hra, nlpI, yehB, yehC, yehD |
| pAMS-F-qnrs11 | 113,285 | 51.5 | 130 | F34: A-: B53 | IncFII: IncFIB | aph(6)-Id, aph(3′)-Ia, aph(3″)-Ib, qnrS11, dfrA14, sul2, tet(A), blaTEM-1B | NA | anr, traJ, traT |
| pAMS-IncI1-floR | 112,645 | 51.1 | 119 | ST3 | IncI1-Iγ | aac(3)-IV, aph(4)-Ia, sul2, floR | NA | cib |
| pAMS-IncY | 106,741 | 47.6 | 118 | ND | IncY | ND | NA | ND |
| pAMS-X3-NDM-19 | 49,816 | 47.3 | 64 | ND | IncX3 | blaNDM-19 | NA | ND |
| Chromosome | pAMS-IncI1-floR | |
|---|---|---|
| Adherence | E. coli common pilus (ecpABCDE) E. coli laminin-binding fimbriae (elfACD) eaeH Hemorrhagic E. coli pilus (hcpABC) Type I fimbriae (fimDF) | Type IV pili (pilQRSVW) |
| Autotransporter | EhaB autotransporter protein (ehaB) | NA |
| Invasion | Invasion of brain endothelial cells (ibeBC) Epithelial cell adherence (tia) | NA |
| Non-LEE encoded TTSS effectors | espL1, espL4, espR1, espX1, espX4, espX5, espY1 | NA |
| Secretion systems | ACE T6SS (aec15, aec16, aec17, aec18, aec19, aec22, aec24, aec25, aec26, aec27/clpV, aec28, aec29, aec30, aec31, aec32) | NA |
| Toxins | Hemolysin/cytolysin A (hlyE/clyA) | NA |
| Antiphagocytosis | capsule | NA |
| Fimbrial adherence determinants | stjC | NA |
| Immune evasion | Exopolysaccharide (galE) | NA |
| Serum resistance | LPS rfb locus (rmlD) | NA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soliman, A.M.; Ramadan, H.; Shimamoto, T.; Komatsu, T.; Maruyama, F.; Shimamoto, T. Detection and Genomic Characteristics of NDM-19- and QnrS11-Producing O101:H5 Escherichia coli Strain Phylogroup A: ST167 from a Poultry Farm in Egypt. Microorganisms 2025, 13, 1769. https://doi.org/10.3390/microorganisms13081769
Soliman AM, Ramadan H, Shimamoto T, Komatsu T, Maruyama F, Shimamoto T. Detection and Genomic Characteristics of NDM-19- and QnrS11-Producing O101:H5 Escherichia coli Strain Phylogroup A: ST167 from a Poultry Farm in Egypt. Microorganisms. 2025; 13(8):1769. https://doi.org/10.3390/microorganisms13081769
Chicago/Turabian StyleSoliman, Ahmed M., Hazem Ramadan, Toshi Shimamoto, Tetsuya Komatsu, Fumito Maruyama, and Tadashi Shimamoto. 2025. "Detection and Genomic Characteristics of NDM-19- and QnrS11-Producing O101:H5 Escherichia coli Strain Phylogroup A: ST167 from a Poultry Farm in Egypt" Microorganisms 13, no. 8: 1769. https://doi.org/10.3390/microorganisms13081769
APA StyleSoliman, A. M., Ramadan, H., Shimamoto, T., Komatsu, T., Maruyama, F., & Shimamoto, T. (2025). Detection and Genomic Characteristics of NDM-19- and QnrS11-Producing O101:H5 Escherichia coli Strain Phylogroup A: ST167 from a Poultry Farm in Egypt. Microorganisms, 13(8), 1769. https://doi.org/10.3390/microorganisms13081769







