Alternatives Integrating Omics Approaches for the Advancement of Human Skin Models: A Focus on Metagenomics, Metatranscriptomics, and Metaproteomics
Abstract
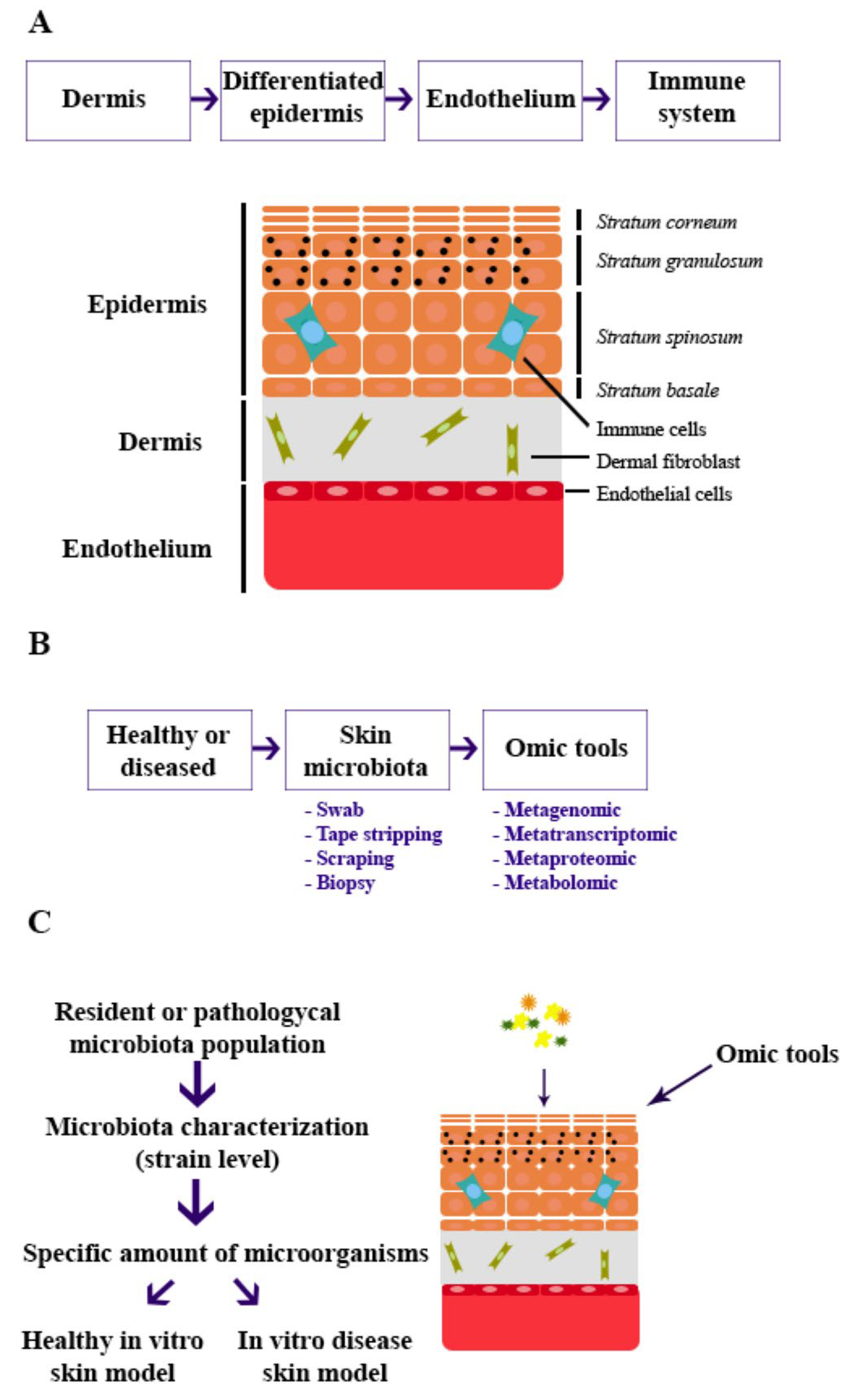
1. Introduction
2. Metagenomics
3. Metatranscriptomics
4. Metaproteomics
5. Metabolomics
6. Building In Vitro Skin Models
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Smythe, P.; Wilkinson, H.N. The Skin Microbiome: Current Landscape and Future Opportunities. Int. J. Mol. Sci. 2023, 24, 3950. [Google Scholar] [CrossRef]
- Nakatsuji, T.; Chiang, H.-I.; Jiang, S.B.; Nagarajan, H.; Zengler, K.; Gallo, R.L. The Microbiome Extends to Subepidermal Compartments of Normal Skin. Nat. Commun. 2013, 4, 1431. [Google Scholar] [CrossRef]
- Grice, E.A.; Segre, J.A. The Skin Microbiome. Nat. Rev. Microbiol. 2011, 9, 244–253. [Google Scholar] [CrossRef]
- Byrd, A.L.; Belkaid, Y.; Segre, J.A. The Human Skin Microbiome. Nat. Rev. Microbiol. 2018, 16, 143–155. [Google Scholar] [CrossRef]
- Dominguez-Bello, M.G.; Costello, E.K.; Contreras, M.; Magris, M.; Hidalgo, G.; Fierer, N.; Knight, R. Delivery Mode Shapes the Acquisition and Structure of the Initial Microbiota across Multiple Body Habitats in Newborns. Proc. Natl. Acad. Sci. USA 2010, 107, 11971–11975. [Google Scholar] [CrossRef]
- Chu, D.M.; Ma, J.; Prince, A.L.; Antony, K.M.; Seferovic, M.D.; Aagaard, K.M. Maturation of the Infant Microbiome Community Structure and Function across Multiple Body Sites and in Relation to Mode of Delivery. Nat. Med. 2017, 23, 314–326. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.; Conlan, S.; Polley, E.C.; Segre, J.A.; Kong, H.H. Shifts in Human Skin and Nares Microbiota of Healthy Children and Adults. Genome Med. 2012, 4, 77. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.; Byrd, A.L.; Park, M.; NISC Comparative Sequencing Program; Kong, H.H.; Segre, J.A. Temporal Stability of the Human Skin Microbiome. Cell 2016, 165, 854–866. [Google Scholar] [CrossRef]
- Grice, E.A.; Kong, H.H.; Conlan, S.; Deming, C.B.; Davis, J.; Young, A.C.; NISC Comparative Sequencing Program; Bouffard, G.G.; Blakesley, R.W.; Murray, P.R.; et al. Topographical and Temporal Diversity of the Human Skin Microbiome. Science 2009, 324, 1190–1192. [Google Scholar] [CrossRef] [PubMed]
- Celoria, V.; Rosset, F.; Pala, V.; Dapavo, P.; Ribero, S.; Quaglino, P.; Mastorino, L. The Skin Microbiome and Its Role in Psoriasis: A Review. Psoriasis 2023, 13, 71–78. [Google Scholar] [CrossRef]
- Martins, A.M.; Ascenso, A.; Ribeiro, H.M.; Marto, J. The Brain-Skin Connection and the Pathogenesis of Psoriasis: A Review with a Focus on the Serotonergic System. Cells 2020, 9, 796. [Google Scholar] [CrossRef] [PubMed]
- Adalsteinsson, J.A.; Kaushik, S.; Muzumdar, S.; Guttman-Yassky, E.; Ungar, J. An Update on the Microbiology, Immunology and Genetics of Seborrheic Dermatitis. Exp. Dermatol. 2020, 29, 481–489. [Google Scholar] [CrossRef] [PubMed]
- Sowell, J.; Pena, S.M.; Elewski, B.E. Seborrheic Dermatitis in Older Adults: Pathogenesis and Treatment Options. Drugs Aging 2022, 39, 315–321. [Google Scholar] [CrossRef] [PubMed]
- Ferretti, P.; Farina, S.; Cristofolini, M.; Girolomoni, G.; Tett, A.; Segata, N. Experimental Metagenomics and Ribosomal Profiling of the Human Skin Microbiome. Exp. Dermatol. 2017, 26, 211–219. [Google Scholar] [CrossRef]
- Sandhu, S.S.; Pourang, A.; Sivamani, R.K. A Review of next Generation Sequencing Technologies Used in the Evaluation of the Skin Microbiome: What a Time to Be Alive. Dermatol. Online J. 2019, 25. [Google Scholar] [CrossRef]
- Roux, P.-F.; Oddos, T.; Stamatas, G. Deciphering the Role of Skin Surface Microbiome in Skin Health: An Integrative Multiomics Approach Reveals Three Distinct Metabolite‒Microbe Clusters. JID 2022, 142, 469–479.e5. [Google Scholar] [CrossRef]
- Rusiñol, L.; Puig, L. Multi-Omics Approach to Improved Diagnosis and Treatment of Atopic Dermatitis and Psoriasis. Int. J. Mol. Sci. 2024, 25, 1042. [Google Scholar] [CrossRef]
- Guo, Y.; Luo, L.; Zhu, J.; Li, C. Multi-Omics Research Strategies for Psoriasis and Atopic Dermatitis. Int. J. Mol. Sci. 2023, 24, 8018. [Google Scholar] [CrossRef]
- Handelsman, J. Metagenomics: Application of Genomics to Uncultured Microorganisms. Microbiol. Mol. Biol. Rev. 2004, 68, 669–685. [Google Scholar] [CrossRef]
- Meisel, J.S.; Hannigan, G.D.; Tyldsley, A.S.; SanMiguel, A.J.; Hodkinson, B.P.; Zheng, Q.; Grice, E.A. Skin Microbiome Surveys Are Strongly Influenced by Experimental Design. JID 2016, 136, 947–956. [Google Scholar] [CrossRef]
- Kong, H.H.; Segre, J.A. The Molecular Revolution in Cutaneous Biology: Investigating the Skin Microbiome. JID 2017, 137, e119–e122. [Google Scholar] [CrossRef]
- De Filippis, F.; Laiola, M.; Blaiotta, G.; Ercolini, D. Different Amplicon Targets for Sequencing-Based Studies of Fungal Diversity. Appl. Environ. Microbiol. 2017, 83, e00905-17. [Google Scholar] [CrossRef] [PubMed]
- Jo, J.-H.; Kennedy, E.A.; Kong, H.H. Research Techniques Made Simple: Bacterial 16S Ribosomal RNA Gene Sequencing in Cutaneous Research. JID 2016, 136, e23–e27. [Google Scholar] [CrossRef]
- Jovel, J.; Patterson, J.; Wang, W.; Hotte, N.; O’Keefe, S.; Mitchel, T.; Perry, T.; Kao, D.; Mason, A.L.; Madsen, K.L.; et al. Characterization of the Gut Microbiome Using 16S or Shotgun Metagenomics. Front. Microbiol. 2016, 7, 459. [Google Scholar] [CrossRef] [PubMed]
- Ranjan, R.; Rani, A.; Metwally, A.; McGee, H.S.; Perkins, D.L. Analysis of the Microbiome: Advantages of Whole Genome Shotgun versus 16S Amplicon Sequencing. Biochem. Biophys. Res. Commun. 2016, 469, 967–977. [Google Scholar] [CrossRef] [PubMed]
- Bashiardes, S.; Zilberman-Schapira, G.; Elinav, E. Use of Metatranscriptomics in Microbiome Research. Bioinform. Biol. Insights 2016, 10, BBI.S34610. [Google Scholar] [CrossRef]
- Mathieu, A.; Vogel, T.M.; Simonet, P. The Future of Skin Metagenomics. Res. Microbiol. 2014, 165, 69–76. [Google Scholar] [CrossRef]
- Gao, Z.; Tseng, C.; Strober, B.E.; Pei, Z.; Blaser, M.J. Substantial Alterations of the Cutaneous Bacterial Biota in Psoriatic Lesions. PLoS ONE 2008, 3, e2719. [Google Scholar] [CrossRef]
- Van Den Bossche, T.; Verschaffelt, P.; Moortele, T.V.; Dawyndt, P.; Martens, L.; Mesuere, B. Biodiversity Analysis of Metaproteomics Samples with Unipept: A Comprehensive Tutorial. In Protein Bioinformatics; Humana: New York, NY, USA, 2023. [Google Scholar]
- Kelly, D.; Yang, L.; Pei, Z. A Review of the Oesophageal Microbiome in Health and Disease. In Methods in Microbiology; Academic Press Inc.: Cambridge, MA, USA, 2017; pp. 19–35. [Google Scholar]
- Franzosa, E.A.; Hsu, T.; Sirota-Madi, A.; Shafquat, A.; Abu-Ali, G.; Morgan, X.C.; Huttenhower, C. Sequencing and beyond: Integrating Molecular “omics” for Microbial Community Profiling. Nat. Rev. Microbiol. 2015, 13, 360–372. [Google Scholar] [CrossRef]
- Benítez-Páez, A.; Belda-Ferre, P.; Simón-Soro, A.; Mira, A. Microbiota Diversity and Gene Expression Dynamics in Human Oral Biofilms. BMC Genom. 2014, 15, 311. [Google Scholar] [CrossRef]
- Wilmes, P.; Heintz-Buschart, A.; Bond, P.L. A Decade of Metaproteomics: Where We Stand and What the Future Holds. Proteomics 2015, 15, 3409–3417. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, W.; Ning, Z.; Mayne, J.; Mack, D.; Stintzi, A.; Tian, R.; Figeys, D. Deep Metaproteomics Approach for the Study of Human Microbiomes. Anal. Chem. 2017, 89, 9407–9415. [Google Scholar] [CrossRef]
- Heyer, R.; Schallert, K.; Zoun, R.; Becher, B.; Saake, G.; Benndorf, D. Challenges and Perspectives of Metaproteomic Data Analysis. J. Biotechnol. 2017, 261, 24–36. [Google Scholar] [CrossRef]
- Van Den Bossche, T.; Arntzen, M.Ø.; Becher, D.; Benndorf, D.; Eijsink, V.G.H.; Henry, C.; Jagtap, P.D.; Jehmlich, N.; Juste, C.; Kunath, B.J.; et al. The Metaproteomics Initiative: A Coordinated Approach for Propelling the Functional Characterization of Microbiomes. Microbiome 2021, 9, 243. [Google Scholar] [CrossRef]
- Armengaud, J. Metaproteomics to Understand How Microbiota Function: The Crystal Ball Predicts a Promising Future. Environ. Microbiol. 2023, 25, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Heintz-Buschart, A.; Wilmes, P. Human Gut Microbiome: Function Matters. Trends Microbiol. 2018, 26, 563–574. [Google Scholar] [CrossRef] [PubMed]
- Grenga, L.; Pible, O.; Miotello, G.; Culotta, K.; Ruat, S.; Roncato, M.; Gas, F.; Bellanger, L.; Claret, P.; Dunyach-Remy, C.; et al. Taxonomical and Functional Changes in COVID-19 Faecal Microbiome Could Be Related to SARS-CoV-2 Faecal Load. Environ. Microbiol. 2022, 24, 4299–4316. [Google Scholar] [CrossRef]
- Cheng, L.; Qi, C.; Yang, H.; Lu, M.; Cai, Y.; Fu, T.; Ren, J.; Jin, Q.; Zhang, X. GutMGene: A Comprehensive Database for Target Genes of Gut Microbes and Microbial Metabolites. Nucleic Acids Res. 2022, 50, D795–D800. [Google Scholar] [CrossRef]
- Xiong, W.; Abraham, P.E.; Li, Z.; Pan, C.; Hettich, R.L. Microbial Metaproteomics for Characterizing the Range of Metabolic Functions and Activities of Human Gut Microbiota. Proteomics 2015, 15, 3424–3438. [Google Scholar] [CrossRef]
- Xiao, M.; Yang, J.; Feng, Y.; Zhu, Y.; Chai, X.; Wang, Y. Metaproteomic Strategies and Applications for Gut Microbial Research. Appl. Microbiol. Biotechnol. 2017, 101, 3077–3088. [Google Scholar] [CrossRef] [PubMed]
- Aguiar-Pulido, V.; Huang, W.; Suarez-Ulloa, V.; Cickovski, T.; Mathee, K.; Narasimhan, G. Metagenomics, Metatranscriptomics, and Metabolomics Approaches for Microbiome Analysis. Evol. Bioinform. 2016, 12s1, EBO.S36436. [Google Scholar] [CrossRef]
- Kuehne, A.; Hildebrand, J.; Soehle, J.; Wenck, H.; Terstegen, L.; Gallinat, S.; Knott, A.; Winnefeld, M.; Zamboni, N. An Integrative Metabolomics and Transcriptomics Study to Identify Metabolic Alterations in Aged Skin of Humans in Vivo. BMC Genom. 2017, 18, 169. [Google Scholar] [CrossRef] [PubMed]
- Zeng, C.; Wen, B.; Hou, G.; Lei, L.; Mei, Z.; Jia, X.; Chen, X.; Zhu, W.; Li, J.; Kuang, Y.; et al. Lipidomics Profiling Reveals the Role of Glycerophospholipid Metabolism in Psoriasis. Gigascience 2017, 6, 1–11. [Google Scholar] [CrossRef]
- Xiong, Q.; Zhong, D.; Li, Q.; Yu, Y.; Zhang, S.; Liang, J.; Zhang, X. LC–MS Metabolomics Reveal Skin Metabolic Signature of Psoriasis Vulgaris. Exp. Dermatol. 2023, 32, 889–899. [Google Scholar] [CrossRef] [PubMed]
- Kalan, L.R.; Meisel, J.S.; Loesche, M.A.; Horwinski, J.; Soaita, I.; Chen, X.; Uberoi, A.; Gardner, S.E.; Grice, E.A. Strain- and Species-Level Variation in the Microbiome of Diabetic Wounds Is Associated with Clinical Outcomes and Therapeutic Efficacy. Cell Host Microbe 2019, 25, 641–655.e5. [Google Scholar] [CrossRef] [PubMed]
- Teertam, S.K.; Setaluri, V.; Ayuso, J.M. Advances in Microengineered Platforms for Skin Research. JID Innov. 2025, 5, 100315. [Google Scholar] [CrossRef]
- Barros, N.R.; Kang, R.; Kim, J.; Ermis, M.; Kim, H.-J.; Dokmeci, M.R.; Lee, J. A Human Skin-on-a-Chip Platform for Microneedling-Driven Skin Cancer Treatment. Mater. Today Bio 2025, 30, 101399. [Google Scholar] [CrossRef]
- Ko, B.; Son, J.; In Won, J.; Kang, B.M.; Choi, C.W.; Kim, R.; Sung, J.H. Gut Microbe–Skin Axis on a Chip for Reproducing the Inflammatory Crosstalk. Lab Chip 2025, 25, 2609–2619. [Google Scholar] [CrossRef]
- Holland, D.B.; Bojar, R.A.; Jeremy, A.H.T.; Ingham, E.; Holland, K.T. Microbial Colonization of an in Vitro Model of a Tissue Engineered Human Skin Equivalent--a Novel Approach. FEMS Microbiol. Lett. 2008, 279, 110–115. [Google Scholar] [CrossRef]
- Laclaverie, M.; Rouaud-Tinguely, P.; Grimaldi, C.; Jugé, R.; Marchand, L.; Aymard, E.; Closs, B. Development and Characterization of a 3D in Vitro Model Mimicking Acneic Skin. Exp. Dermatol. 2021, 30, 347–357. [Google Scholar] [CrossRef] [PubMed]
- Kanwar, I.L.; Haider, T.; Kumari, A.; Dubey, S.; Jain, P.; Soni, V. Models for Acne: A Comprehensive Study. Drug Discov. Ther. 2018, 12, 329–340. [Google Scholar] [CrossRef]
- Fitz-Gibbon, S.; Tomida, S.; Chiu, B.-H.; Nguyen, L.; Du, C.; Liu, M.; Elashoff, D.; Erfe, M.C.; Loncaric, A.; Kim, J.; et al. Propionibacterium Acnes Strain Populations in the Human Skin Microbiome Associated with Acne. J. Investig. Dermatol. 2013, 133, 2152–2160. [Google Scholar] [CrossRef] [PubMed]
- Zouboulis, C.C.; Eady, A.; Philpott, M.; Goldsmith, L.A.; Orfanos, C.; Cunliffe, W.C.; Rosenfield, R. What Is the Pathogenesis of Acne? Exp. Dermatol. 2005, 14, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Byrd, A.L.; Deming, C.; Cassidy, S.K.B.; Harrison, O.J.; Ng, W.-I.; Conlan, S.; NISC Comparative Sequencing Program; Belkaid, Y.; Segre, J.A.; Kong, H.H. Staphylococcus Aureus and Staphylococcus Epidermidis Strain Diversity Underlying Pediatric Atopic Dermatitis. Sci. Transl. Med. 2017, 9, eaal4651. [Google Scholar] [CrossRef]
- Ahn, K.; Kim, B.E.; Kim, J.; Leung, D.Y. Recent Advances in Atopic Dermatitis. Curr. Opin. Immunol. 2020, 66, 14–21. [Google Scholar] [CrossRef]
- Wu, X.; Zhao, Y.; Gu, Y.; Li, K.; Wang, X.; Zhang, J. Interferon-Lambda 1 Inhibits Staphylococcus Aureus Colonization in Human Primary Keratinocytes. Front. Pharmacol. 2021, 12, 652302. [Google Scholar] [CrossRef]
- Jung, S.-Y.; You, H.J.; Kim, M.-J.; Ko, G.; Lee, S.; Kang, K.-S. Wnt-Activating Human Skin Organoid Model of Atopic Dermatitis Induced by Staphylococcus Aureus and Its Protective Effects by Cutibacterium Acnes. iScience 2022, 25, 105150. [Google Scholar] [CrossRef]
- Schwartz, J.R.; Messenger, A.G.; Tosti, A.; Todd, G.; Hordinsky, M.; Hay, R.J.; Wang, X.; Zachariae, C.; Kerr, K.M.; Henry, J.P.; et al. A Comprehensive Pathophysiology of Dandruff and Seborrheic Dermatitis—Towards a More Precise Definition of Scalp Health. Acta Derm. Venereol. 2013, 93, 131–137. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, R.A.; Janusz, C.A.; Janniger, C.K. Seborrheic Dermatitis: An Overview. Am. Fam. Physician 2006, 74, 125–130. [Google Scholar]
- Dawson, T.L. Malassezia Globosa and Restricta: Breakthrough Understanding of the Etiology and Treatment of Dandruff and Seborrheic Dermatitis through Whole-Genome Analysis. J. Investig. Dermatol. Symp. Proc. 2007, 12, 15–19. [Google Scholar] [CrossRef]
- Donnarumma, G.; Perfetto, B.; Paoletti, I.; Oliviero, G.; Clavaud, C.; Del Bufalo, A.; Guéniche, A.; Jourdain, R.; Tufano, M.A.; Breton, L. Analysis of the Response of Human Keratinocytes to Malassezia Globosa and Restricta Strains. Arch. Dermatol. Res. 2014, 306, 763–768. [Google Scholar] [CrossRef] [PubMed]
- Meloni, M.; Balzaretti, S.; Collard, N.; Desaint, S.; Laperdrix, C. Reproducing the Scalp Microbiota Community: Co-Colonization of a 3D Reconstructed Human Epidermis with C. Acnes and M. Restricta. Int. J. Cosmet. Sci. 2021, 43, 235–245. [Google Scholar] [CrossRef]
- Cui, M.; Wiraja, C.; Zheng, M.; Singh, G.; Yong, K.; Xu, C. Recent Progress in Skin-on-a-Chip Platforms. Adv. Ther. 2022, 5, 2100138. [Google Scholar] [CrossRef]
- Fernandez-Carro, E.; Remacha, A.R.; Orera, I.; Lattanzio, G.; Garcia-Barrios, A.; del Barrio, J.; Alcaine, C.; Ciriza, J. Human Dermal Decellularized ECM Hydrogels as Scaffolds for 3D In Vitro Skin Aging Models. Int. J. Mol. Sci. 2024, 25, 4020. [Google Scholar] [CrossRef]
- Risueño, I.; Valencia, L.; Jorcano, J.L.; Velasco, D. Skin-on-a-Chip Models: General Overview and Future Perspectives. APL Bioeng. 2021, 5, 030901. [Google Scholar] [CrossRef]
- Pontiggia, L.; Klar, A.S.; Michalak-Micka, K.; Moehrlen, U.; Biedermann, T. Isolation, Characterization, and Utilization of Human Skin Basal and Suprabasal Epidermal Stem Cells. In Skin Stem Cells; Humana: New York, NY, USA, 2024; pp. 1–15. [Google Scholar] [CrossRef]
- Kwak, B.S.; Jin, S.; Kim, S.J.; Kim, E.J.; Chung, J.H.; Sung, J.H. Microfluidic Skin Chip with Vasculature for Recapitulating the Immune Response of the Skin Tissue. Biotechnol. Bioeng. 2020, 117, 1853–1863. [Google Scholar] [CrossRef]
- Ogai, K.; Nagase, S.; Mukai, K.; Iuchi, T.; Mori, Y.; Matsue, M.; Sugitani, K.; Sugama, J.; Okamoto, S. A Comparison of Techniques for Collecting Skin Microbiome Samples: Swabbing Versus Tape-Stripping. Front. Microbiol. 2018, 9, 2362. [Google Scholar] [CrossRef] [PubMed]
- Gotschlich, E.C.; Colbert, R.A.; Gill, T. Methods in Microbiome Research: Past, Present, and Future. Best. Pract. Res. Clin. Rheumatol. 2019, 33, 101498. [Google Scholar] [CrossRef]
- Ederveen, T.H.A.; Smits, J.P.H.; Boekhorst, J.; Schalkwijk, J.; van den Bogaard, E.H.; Zeeuwen, P.L.J.M. Skin Microbiota in Health and Disease: From Sequencing to Biology. J. Dermatol. 2020, 47, 1110–1118. [Google Scholar] [CrossRef] [PubMed]
- Dariush Gholami, Z.E.A.R.N.S.A. Advances in Bacterial Identification and Characterization: Methods and Applications. Microbiol. Metab. Biotechnol. 2019, 2, 119–136. [Google Scholar]
- van den Bogaard, E.; Ilic, D.; Dubrac, S.; Tomic-Canic, M.; Bouwstra, J.; Celli, A.; Mauro, T. Perspective and Consensus Opinion: Good Practices for Using Organotypic Skin and Epidermal Equivalents in Experimental Dermatology Research. JID 2021, 141, 203–205. [Google Scholar] [CrossRef] [PubMed]
| Collecting Area | Advantages | Disadvantages | |
|---|---|---|---|
| Swab | Surface and epidermal layers | Easy No invasive Commercially available Standardized | Often contain small bacterial yields |
| Tape stripping | Deep epidermal layers | Easy Commercially available More cultivable bacteria collected | Different adhesives Less efficient in oily, wet, or undulating skin |
| Scraping | Deep epidermal layers | Easy | Invasive Mechanical scraping damages the skin. DNA contamination (host DNA) |
| Biopsy | Epidermis and dermis | Most representative skin-microbiota detection | Invasive DNA contamination (host DNA) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fernández-Carro, E.; Letsiou, S.; Tsironi, S.; Chaniotis, D.; Ciriza, J.; Beloukas, A. Alternatives Integrating Omics Approaches for the Advancement of Human Skin Models: A Focus on Metagenomics, Metatranscriptomics, and Metaproteomics. Microorganisms 2025, 13, 1771. https://doi.org/10.3390/microorganisms13081771
Fernández-Carro E, Letsiou S, Tsironi S, Chaniotis D, Ciriza J, Beloukas A. Alternatives Integrating Omics Approaches for the Advancement of Human Skin Models: A Focus on Metagenomics, Metatranscriptomics, and Metaproteomics. Microorganisms. 2025; 13(8):1771. https://doi.org/10.3390/microorganisms13081771
Chicago/Turabian StyleFernández-Carro, Estibaliz, Sophia Letsiou, Stella Tsironi, Dimitrios Chaniotis, Jesús Ciriza, and Apostolos Beloukas. 2025. "Alternatives Integrating Omics Approaches for the Advancement of Human Skin Models: A Focus on Metagenomics, Metatranscriptomics, and Metaproteomics" Microorganisms 13, no. 8: 1771. https://doi.org/10.3390/microorganisms13081771
APA StyleFernández-Carro, E., Letsiou, S., Tsironi, S., Chaniotis, D., Ciriza, J., & Beloukas, A. (2025). Alternatives Integrating Omics Approaches for the Advancement of Human Skin Models: A Focus on Metagenomics, Metatranscriptomics, and Metaproteomics. Microorganisms, 13(8), 1771. https://doi.org/10.3390/microorganisms13081771








