Abstract
Absidia is the most species-rich genus within the family Cunninghamellaceae, with its members commonly isolated from diverse substrates, particularly rhizosphere soil. In this study, four novel Absidia species, A. irregularis sp. nov., A. multiformis sp. nov., A. ovoidospora sp. nov., and A. verticilliformis sp. nov., were discovered from soil samples collected in southern and southwestern China, using integrated morphological and molecular analyses. Phylogenetic analyses based on concatenated ITS, SSU, LSU, Act, and TEF1α sequence data reconstructed trees that strongly supported the monophyly of each of these four new taxa. Key diagnostic features include A. irregularis (closely related to A. oblongispora) exhibiting irregular colony morphology, A. multiformis (sister to A. heterospora) demonstrating polymorphic sporangiospores, A. ovoidospora (forming a clade with A. panacisoli and A. abundans) producing distinctive ovoid sporangiospores, and A. verticilliformis (next to A. edaphica) displaying verticillately branched sporangiophores. Each novel species is formally described with comprehensive documentation, including morphological descriptions, illustrations, Fungal Names registration identifiers, designated type specimens, etymological explanations, maximum growth temperatures, and taxonomic comparisons. This work constitutes the sixth instalment in a series investigating early-diverging fungal diversity in China aiming to enhance our understanding of the diversity of fungi in tropical and subtropical ecosystems in Asia. In this paper, the known species of Absidia are expanded to 71.
1. Introduction
The genus Absidia Tiegh. (Mucoromycota Doweld, Mucoromycetes Doweld, Mucorales Dumort., Cunninghamellaceae Naumov ex R.K. Benj.) was established in 1878 and typified with A. reflexa [1,2]. It is currently the most numerous genus in the family Cunninghamellaceae, with 67 recognized species (https://www.catalogueoflife.org/data/taxon/LSM, accessed on 2 December 2024). Its taxonomic position has been controversial since its establishment. Initially, this genus was classified in Absidiaceae [3]. Then, Benny et al. transferred it to Mucoraceae in 2001 [4]. Finally, Ashton et al. accommodated it in Cunninghamellaceae in 2009 [5]. With the deepening of research, Absidia s.l. was divided into Lichtheimia Vuill. (thermotolerant, optimum growth temperature 37–45 °C), Absidia s.s. (mesophilic, optimum growth temperature 25–34 °C), and Lentamyces Kerst. Hoffm. & K. Voigt (parasitic on other mucoralean fungi, optimum growth temperature 14–25 °C) [6,7,8,9].
Strains of Absidia are distributed worldwide, ubiquitous in soil, dung, leaf litter, food, air, etc. [10]. In the GlobalFungi database (https://globalfungi.com/, accessed on 22 December 2024), Absidia members are recorded from soil (37,828 records, 44.52% of all records), root (15,865, 18.67%), shoot (11,740, 13.82%), topsoil (6923, 8.15%), rhizosphere soil (2530, 2.98%), deadwood (2253, 2.65%), air (2080, 2.45%), water (1549, 1.82%), litter (1406, 1.65%), mosses (265, 0.41%), lichen (217, 0.25%), coral (72, 0.08%), dust (53, 0.06%), fungal sporocarp (31, 0.04%), and glacial ice debris (3, 0%). In summary, soil and rhizosphere substrates account for approximately 74.32%.
Absidia coerulea, the most common species in this genus, plays an important role in bioengineering. It possesses the capability of transforming spirulina biomass into (-)-α-bisabolol [11]. It is able to catalyse the specific C-3 dehydrogenation for derivatives of ginsenoside-Rg1, as well as hydroxylation at the 7β and 15α positions, and some metabolites in this species exhibit moderate reversal activity towards multidrug-resistant tumour cells [12].
In order to enhance our understanding of the diversity of fungi in tropical and subtropical soil ecosystems in Asia, eight fungal strains were isolated from soil in the Hainan and Guizhou provinces, the south and southwest regions of China. According to ITS-SSU-LSU-Act-TEF1α molecular phylogenetic analyses and morphological comparisons, these strains were classified into four new species of Absidia and described herein as A. irregularis sp. nov., A. multiformis sp. nov., A. ovoidospora sp. nov., and A. verticilliformis sp. nov. This is the sixth report of a serial work on the diversity of Chinese early-diverging fungi [13,14,15,16,17].
2. Materials and Methods
2.1. Isolation and Morphology
Soil samples were collected in Hainan province in April 2023 and Guizhou province in August 2023, following the methods by Li et al. and Zou et al. [18,19]. Soil sample collection started with removal of surface contaminants using a sterilized stainless-steel shovel. About 100 g of homogenized soil was put into sample bags and labelled with collection date, administrative location, GPS coordinates, and altitude. Strains were obtained from the soil samples by serial dilution spread plate and single spore isolation.
About 1 g of soil samples was mixed with 10 mL of sterile water to prepare 10−1 soil suspension. One millilitre of the 10−1 suspension was transferred to 9 mL of sterile water to obtain a 10−2 soil suspension. In the same way, 10−3 and 10−4 soil suspensions were made. Approximately 0.2 mL of the final 10−4 soil suspension was dispersed evenly with sterilized coating rods on Rose-Bengal Chloramphenicol agar (RBC: peptone 5.00 g/L, glucose 10.00 g/L, KH2PO4 1.00 g/L, MgSO4·7H2O 0.50 g/L, rose red 0.05 g/L, chloramphenicol 0.10 g/L, agar 15.00 g/L) [20] and then cultured in the dark at 25 °C. Once visible, colonies were transferred and further cultured on Potato Dextrose Agar (PDA: glucose 20.00 g/L, potato 200.00 g/L, agar 20.00 g/L, pH 7) [14,21].
Appropriately, 0.1 g of soil samples was evenly sprinkled on the RBC medium with 0.06 mg/mL streptomycin and then incubated in darkness at 25 °C for 2–5 d. When sporangia formed, a sterilized inoculation needle was adopted to pick up a sporangium onto the PDA medium supplemented with 0.06 mg/mL streptomycin.
The maximum growth temperature was determined by a gradient method [13]. The strain was initially cultured at 25 °C for 2 d and then increased by 1 °C per day until there was no growth.
Pure cultivation was applied for observing anamorphs, and pairing experiments were carried out for observing zygospores by adding 0.1% lecithin to PDA and sealing Petri dishes to retain moisture. The morphological characteristics were observed with an optical microscope (Olympus BX53) and photographed with a high-definition colour digital camera (Olympus DP80). Each morphological character was statistically calculated against 30 measurements [22]. All strains were stored at 4 °C with 20% sterilized glycerine. Cultures were deposited in the China General Microbiological Culture Collection Center, Beijing, China (CGMCC) and Shandong Normal University, Jinan, Shandong, China (XG). Strains were deposited in the Herbarium Mycologicum Academiae Sinicae, Beijing, China (Fungarium, HMAS). Taxonomic information for the new taxa was registered in the Fungal Names repository (https://nmdc.cn/fungalnames/, accessed on 14 April 2025).
2.2. DNA Extraction and Amplification
Genomic DNA was extracted from mycelia using the CTAB method and GOMagTM Rapid Plant DNA Kit [21]. Information on the primers for PCR amplification is listed in Table 1. Amplification was performed in a final volume of 20 μL, containing 10 μL 2× Hieff Canace® Plus PCR Master Mix (Cat No.10154ES03; Yeasen Biotechnology, Shanghai, China), 0.5 μL of forward and reverse primers each (10 μM; TsingKe, Beijing, China), 1 μL of template genomic DNA (1 μM), and 8 μL of distilled deionised water. Molecular loci, PCR primers, and programmes used in this study are listed in Table 1. PCR products were electrophoresed with 1% agarose gel. DNA fragments were stained with TS-GelRed Nucleic Acid Gel Stain (10,000× in water; TSJ002; Beijing Tsingke Biotech Co., Ltd., Beijing, China) and observed under ultraviolet light. Then, a gel extraction kit (Cat# AE0101-C; Shandong Sparkiade Biotechnology Co., Ltd., Jinan, China) was used for gel recovery. Sanger sequencing was carried out by Biosune Co., Ltd. (Shanghai, China). Consensus sequences were assembled using MEGA v.7.0 [23]. Target sequences in some strains could not be obtained by conventional methods due to heterogeneous gene duplications; thus, they were extracted from genomic data [24]. The genomes were sequenced by Singke Biotech Co., Ltd. (Jinan, China). Genome sequencing was performed using Illumina, and the genomes were assembled using the St. Petersburg genome assembler (SPAdes). All sequences generated in this study were deposited at GenBank under the accession numbers in Table 2. The genome accession numbers of Absidia ovoidospora CGMCC 3.27811 and A. verticilliformis CGMCC 3.27810 are JBOEPW000000000 and JBOEPV000000000.

Table 1.
Molecular loci, PCR primers, and programmes used in this study.

Table 2.
Information on strains used in this study.
Relative sequences were obtained by BLAST search against the NCBI GenBank nucleotide database (https://blast.ncbi.nlm.nih.gov/, accessed on 13 October 2024). SSU, ITS, LSU, Act, and TEF1α sequences both generated herein and retrieved from GenBank (Table 2) were individually aligned using the MAFFT 7 online service. The aligned matrices were manually proofread and then jointly analysed. The optimal evolutionary model was determined for each partition and included in the analysis using MrModelTest v.2.3 [29]. Phylogenetic history was reconstructed using the maximum likelihood (ML) algorithm with RaxML-HPC2 on XSEDE (8.2.12) and the Bayesian inference (BI) algorithm with MrBayes [30,31,32,33]. Maximum likelihood analysis was performed using the best model with 1000 bootstrap replications. The BI analysis consisted of five million generations with four parallel runs under stopping rules and a sampling frequency of 100 generations. The burn-in score was set to 0.25, and the posterior probability (PP) was determined from the remaining trees. Initial adjustments to the phylogenetic tree were made using FigTree v.1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/, accessed on 23 October 2024), and the finalization was performed using Adobe Illustrator CC 2019 (https://adobe.com/products/illustrator, accessed on 23 October 2024).
3. Results
3.1. Phylogenetic Analyses
The sequence matrix included 87 strains in 64 species of Absidia, with Cunninghamella blakesleeana CBS 782.68 as an outgroup. A total of 4656 characters comprised ITS rDNA (1–975), SSU rDNA (976–2017), LSU rDNA (2018–3030), Act (3031–3689), and TEF1α (3690–4656). As many as 2737 characters were constant, while 706 and 1213 among the variable characters were parsimony-uninformative and informative, respectively (Supplementary File S1). MrModelTest suggested that the Dirichlet fundamental frequency and GTR-I-G evolution pattern for all partitions were adopted in Bayesian inference. The topology of the Bayesian tree, consistent with that of the ML tree, was used as a representative to summarise the evolutionary history within the genus Absidia (Figure 1), exhibiting the phylogenetic placement of the four new species. A. irregularis is related to A. oblongispora with full supports (MLBV = 100, BIPP = 1.00), and A. ovoidrospora is closely related to A. panacisoli and A. abundans with full supports (MLBV = 100, BIPP = 1.00). A. multiformis is most closely related to A. heterospora with high supports (BIPP = 0.99), and A. verticilliformis is closely related to A. edaphica with robust supports (MLBV = 94, BIPP = 1).
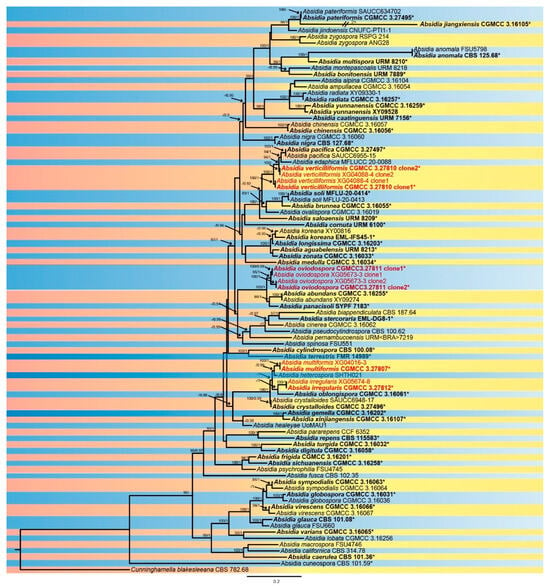
Figure 1.
The Bayesian phylogenetic tree of Absidia based on ITS-SSU-LSU-Act-TEF1α sequences, with Cunninghamella blakesleeana CBS 782.68 as outgroup. The maximum likelihood bootstrap value (MLBV) ≥ 75 and the Bayesian inference posterior probability (BIPP) ≥ 0.85 are shown at the first and second positions and separated by a slash “/” on relevant nodes. The ex-types or ex-holotypes are in bold and marked with an asterisk “*”, and strains involved in this study are in red. Branches shortened to fit the page are represented by double slashes “//” and folds “×”. The scale at the bottom centre indicates 0.2 substitutions per site.
3.2. Taxonomy
Absidia irregularis Yi Xin Wang & X.Y. Liu, sp. nov. Figure 2.
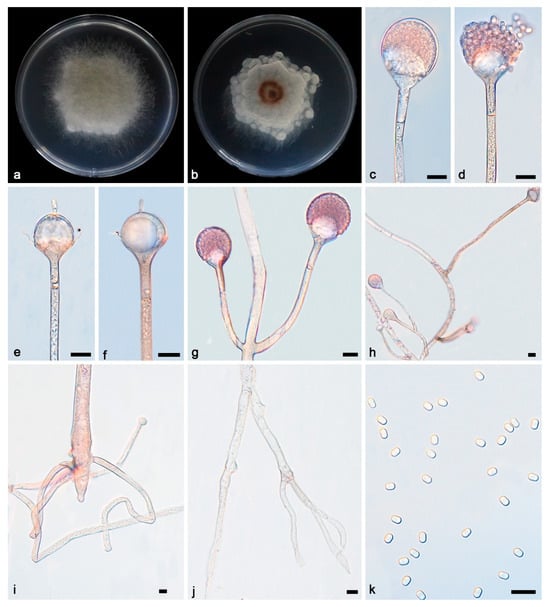
Figure 2.
Absidia irregularis ex-holotype CGMCC 3.27812. (a,b) Colonies on PDA ((a) obverse; (b) reverse); (c,d) an unbranched sporangiophore with a sporangium; (e,f) columellae, collars, sporangiospores, and apophyses; (g) branched sporangiophores with sporangia; (h) branched sporangiophores with columellae, collars, sporangia, and apophyses; (i,j) rhizoids; (k) sporangiospores; bars: (c–k) 10 μm.
Fungal Names—FN 572283.
Etymology—The epithet “irregularis” (Latin) pertains to irregular colonies.
Type—China, Hainan, Changjiang Li Autonomous County, Bawangling National Forest Park, 19.0859333° N, 109.122752° E, altitude 745.3 m, from soil sample, 9 August 2023, Yi-Xin Wang (Holotype HMAS 353186, ex-holotype strain CGMCC 3.27812 = XG05674-6).
Description—Colonies grow moderately on PDA in darkness at 25 °C for 7 d, reaching 43.2–54.8 mm in diameter, initially white, soon becoming grey to brown, irregular and scaly at the edge, cottony, and reversely white to grey. Hyphae hyaline at first, brownish when mature, branched, irregular, and 10.2–14.5 μm in diameter. Stolons are hyaline, branched, and smooth. Rhizoids are hyaline, branched, irregular, or root-like. Sporangiophores are located on aerial mycelia, hyaline, erect, or slightly curved, unbranched or slightly branched, swollen usually below sporangia, umbellately or sympodially branched, often with a septum below apophyses, 26.5–148.1 × 3.4–5.3 μm. Sporangia oval to subglobose, 27.2–32.4 × 26.5–28.9 μm, hyaline at first and then brown, deliquescent-walled, leaving a collar after releasing sporangiospores. Apophyses are hyaline, smooth, bowl-shaped, 2.9–9.6 × 8.6–19.0 μm. Collars are distinct. Columellae are hyaline or brown, hemispherical, 10.0–19.8 × 9.8–21.0 μm, and protruding (3.5–6.3 × 1.4–2.9 μm) at the top. Protrusions are always slightly contracted in the middle. Sporangiospores are hyaline, smooth, not uniform, mostly cylindrical, 3.6–4.6 × 2.3–2.9 μm. Chlamydospores are present. Zygospores were not observed.
Additional strain examined—China, Hainan, Changjiang Li Autonomous County, Bawangling National Forest Park, 19.0859333° N, 109.122752° E, altitude 745.3 m, from soil sample, 9 August 2023, Yi-Xin Wang (living culture XG05674-8).
GenBank accession numbers—CGMCC 3.27812 (ITS, PQ306325; LSU, PQ289020; Act, PQ807209; SSU, PQ799254; TEF1α, PV126019), XG05674-7 (ITS, PQ306326; LSU, PQ289021; Act, PQ807210; SSU, PQ799255; TEF1α, PV126020).
Maximum growth temperature—CGMCC 3.27812 (32 °C), XG05674-7 (32 °C).
Notes—Based on phylogenetic analyses of ITS-SSU-LSU-Act-TEF1α sequences, the two isolates of the new species Absidia irregularis formed an independent clade with full supports (MLBV = 100, BIPP = 1; Figure 1), which is closely related to A. oblongispora (BIPP = 1; Figure 1). This new species differs morphologically from A. oblongispora in apophysis and columella [34]. The new species is bigger than A. oblongispora in apophysis, 2.9–9.6 × 8.6–19.0 μm vs. 3.5–6.5 × 3.5–7.5 μm. The new species is also bigger than A. oblongispora in columellae, 10.0–19.8 × 9.8–21.0 μm vs. 7.0–15.0 × 8.5–16.5 μm. Combining the morphological and molecular phylogenetic analyses, the two isolates were classified as a new taxon, allied to A. oblongispora.
Absidia multiformis Yi Xin Wang & X.Y. Liu, sp. nov. Figure 3.
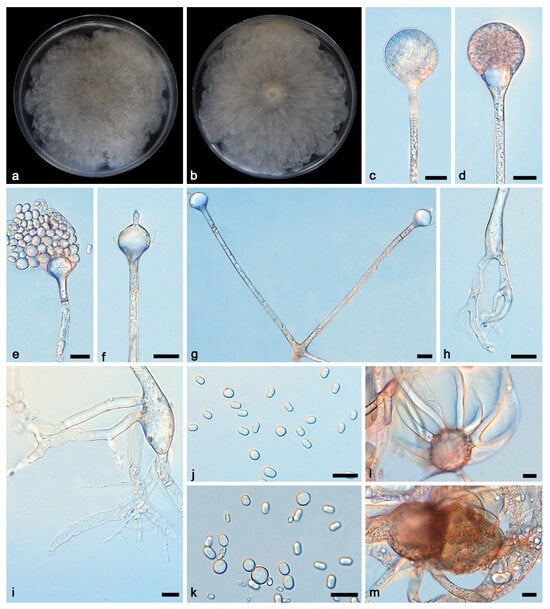
Figure 3.
Absidia multiformis ex-holotype CGMCC 3.27807. (a,b) Colonies on PDA ((a) obverse; (b) reverse); (c,d) an unbranched sporangiophore with a sporangium; (e,f) columellae, collars, sporangiospores, and apophyses; (g) branched sporangiophores with columellae, collars, sporangium, and apophyses; (h,i) rhizoids; (j,k) sporangiospores; (l,m) zygospores; bars: (c–m) 10 μm.
Fungal Names—FN 572285.
Etymology—The epithet “multiformis” (Latin) pertains to polymorphic sporangiospores.
Type—China, Hainan, Lingshui Li Autonomous County, 18.6958768° N, 109.9407998° E, altitude 151.6 m, from soil sample, 23 April 2023, Yi-Xin Wang (Holotype HMAS 353194, ex-holotype strain CGMCC 3.27807 = XG04016-2).
Description—Colonies grow moderately on PDA in darkness at 25 °C for 7 d, reaching 82.2–90.0 mm in diameter, initially white, soon becoming grey to brown, irregular at the edge, cottony, and reversely white to grey. Hyphae are hyaline at first, brownish when mature, branched, irregular, and 4.2–12.3 μm in diameter. Stolons are hyaline, branched, and smooth. Rhizoids are hyaline, branched, irregular, or root-like. Sporangiophores on aerial mycelia are hyaline, erect, or slightly curved, unbranched, or slightly branched, swollen usually below sporangia, often with a septum below apophyses, 49.9–125.4 × 2.9–3.7 μm. Sporangia are oval to subglobose, and 20.1–36.1 × 23.1–36.3 μm, hyaline at first and then brown, and deliquescent-walled, leaving a collar after releasing sporangiospores. Apophyses are hyaline, smooth, shallow mouth bowl-shaped, and 2.5–7.9 × 8.0–19.8 μm. Collars are distinct. Columellae are hyaline or brown, hemispherical, 5.3–19.4 × 8.3–22.9 μm, with long or short cylindrical protrusions at top, and 1.7–6.4 × 1.1–4.0 μm. Sporangiospores are hyaline, smooth, not uniform, mainly cylindrical, and 4.5–6.1 × 2.5–4.4 μm, and some are ovoid, 4.4–5.8 × 3.9–6.1 μm, occasionally subglobose, and 4.9–7.2 μm. Chlamydospores are present. Zygospores are present, not uniform.
Additional strain examined—China, Hainan, Lingshui Li Autonomous County, 18.6958768° N, 109.9407998° E, altitude 151.6 m, from soil sample, 23 April 2023, Yi-Xin Wang (living culture XG04016-3).
GenBank accession numbers—CGMCC 3.27807 (ITS, PQ306319; LSU, PQ803168; Act, PQ807203; SSU, PQ799260; TEF1α, PV126021), XG0 4016-3 (ITS, PQ306320; LSU, PQ803169; Act, PQ807204; SSU, PQ799261; TEF1α, PV126022).
Maximum growth temperature—CGMCC 3.27807 (32 °C), XG0 4016-3 (32 °C).
Notes—Based on phylogenetic analyses of ITS-SSU-LSU-Act-TEF1α sequences, the two isolates of the new species Absidia multiformis formed an independent clade with full supports (MLBV = 100, BIPP = 1; Figure 1), which is closely related to A. heterospora (BIPP = 0.99; Figure 1). This new species differs morphologically from A. heterospora in sporangiophore, sporangium, apophysis, and columella [35]. It differs from A. heterospora by narrower and shorter sporangiophores, 49.9–125.4 × 2.9–3.7 μm vs. 230–1700 × 4.3–10.0 µm. It is smaller than A. heterospora in sporangia, 20.1–36.3 μm vs. 15.0–55.0 µm. In columellae, it differs from A. heterospora by more shapes and smaller size. In detail, it is hemispherical and 5.3–19.4 × 8.3–22.9 μm, while A. heterospora is regularly dorsiventrally flattened and 10.5–34 µm in diameter. Combining morphological and molecular phylogenetic analyses, the two isolates were classified as a new taxon, allied to A. heterospora.
Absidia ovoidospora Yi Xin Wang & X.Y. Liu, sp. nov. Figure 4.
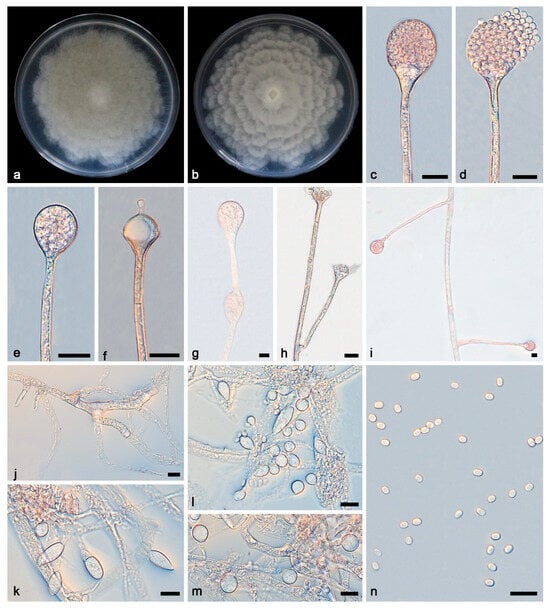
Figure 4.
Absidia ovoidospora ex-holotype CGMCC 3.27811. (a,b) Colonies on PDA ((a) obverse; (b) reverse); (c–e) an unbranched sporangiophore with a sporangium; (f) columellae, collars, sporangiospores, and apophyses; (g) unbranched sporangiophores with swelling and sporangium; (h) branched sporangiophores with columellae, collars, and apophyses; (i) branched sporangiophores with sporangia; (j) rhizoids; (k–m) giant cells; (n) sporangiospores; bars: (c–n) 10 μm.
Fungal Names—FN 572284.
Etymology—The epithet “ovoidospora” (Latin) pertains to ovoid sporangiospores.
Type—China, Hainan, Changjiang Li Autonomous County, Bawangling National Forest Park, 25.905722° N, 107.279063° E, altitude 745.3 m, from soil sample, 9 August 2023, Yi-Xin Wang (Holotype HMAS 353185, ex-holotype strain CGMCC 3.27811 = XG05673-2).
Description—Colonies grow fast on PDA in darkness at 25 °C for 7 d, reaching 90 mm in diameter, initially white, soon becoming grey to brown, irregular at the edge, cottony, and reversely white to grey. Hyphae are hyaline at first, brownish when mature, branched, irregular, and 5.8–13.7 μm in diameter. Stolons are hyaline, branched, and smooth. Rhizoids are hyaline, branched, irregular, or root-like. Sporangiophores are on aerial mycelia, hyaline, erect or slightly curved, unbranched or slightly branched, swollen usually below sporangia, umbellately or sympodially branched, often with a septum below apophyses, and 45.3–355.3 × 2.5–4.0 μm. Sporangia are oval to subglobose, 13.6–29.0 × 13.3–28.5 μm, hyaline at first and then brown, deliquescent-walled, leaving a collar after releasing sporangiospores. Apophyses are hyaline, smooth, bowl-shaped and long funnel-shaped, and 4.1–6.9 × 8.6–10.9 μm. Columellae are hyaline or brown, hemispherical with a short or long protruding at the top, and 7.9–12.9 × 9.0–11.6 μm. Protrusions are always slightly contracted in the middle and 1.9–4.6 × 1.7–2.6 μm. Sporangiospores are hyaline, smooth, not uniform, mostly ovoid, and 3.2–3.8 × 2.4–3.1 μm, and some are cylindrical and 3.4–4.6 × 2.2–3.0 μm μm. Chlamydospores are present. Zygospores were not observed.
Additional strains examined—China, Hainan, Changjiang Li Autonomous County, Bawangling National Forest Park, 25.905722° N, 107.279063° E, altitude 745.3 m, from soil sample, 9 August 2023, Yi-Xin Wang (living culture XG05673-3).
Genome accession numbers—JBOEPW000000000.
GenBank accession numbers—CGMCC 3.27811 clone1 (ITS, PQ306327; LSU, PQ803164; Act, PQ807207; SSU, PQ799256; TEF1α, PV126015), CGMCC 3.27811 clone2 (ITS, PV069753; LSU, PQ803165; Act, PV126023; SSU, PQ799257; TEF1α, PV126017), XG05673-3 clone 1 (ITS, PQ306328; LSU, PQ803166; Act, PQ807208; SSU, PQ799258; TEF1α, PV126016), XG05673-3 clone 2 (ITS, PV069754; LSU, PQ803167; Act, PV126024; SSU, PQ799259; TEF1α, PV126018).
Maximum growth temperature—CGMCC 3.27811 (30 °C), XG05673-3 (30 °C).
Notes—Based on phylogenetic analyses of ITS-SSU-LSU-Act-TEF1α sequences, the new species Absidia ovoidospora formed two sister clades with high supports (MLBV = 96, BIPP = 1; Figure 1), which are closely related to A. panacisoli and A. abundans with full supports (MLBV = 100, BIPP = 1; Figure 1). These two clades resulted from two clones. This new species differs morphologically from A. oblongispora in sporangiophore and sporangium [36]. The new species differs from A. panacisoli by a wider sporangiophore, 2.5–4.0 μm vs. 1.7–2.8 μm. In sporangia, the new species is bigger than A. panacisoli, 13.6–29.0 × 13.3–28.5 μm vs. 13.0–21.5 × 9.4–15.5 μm. This new species differs morphologically from A. abundans in sporangiophore, sporangium, and columella [9]. The new species differs from A. abundans by bigger sporangiophores, 45.3–355.3 × 2.5–4.0 μm vs. 35.0–170.0 × 2.0–3.5 μm. The new species is bigger than A. abundans in sporangia, 13.6–29.0 × 13.3–28.5 μm vs. 8.0–16.5 × 8.5–16.0 μm. In columellae, the new species is bigger than A. abundans, 7.9–12.9 × 9.0–11.6 μm vs. 4.5–10.0 × 3.5–8.0 μm. Combining morphological and molecular phylogenetic analyses, the two were classified isolates as a new taxon, allied with A. panacisoli and A. abundans.
Absidia verticilliformis, Yi Xin Wang & X.Y. Liu sp. nov. Figure 5.
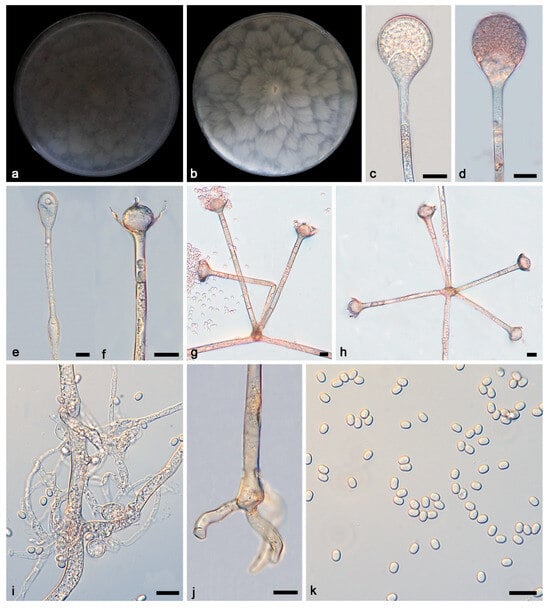
Figure 5.
Absidia verticilliformis ex-holotype CGMCC 3.27810. (a,b) Colonies on PDA ((a) obverse; (b) reverse); (c–e) an unbranched sporangiophore with a sporangium; (f) columellae, collars, sporangiospores, and apophyses; (g,h) branched sporangiophores with columellae, collars, sporangiospores, and apophyses; (i,j) rhizoids; (k) sporangiospores; bars: (c–k) 10 μm.
Fungal Names—FN 572287.
Etymology—The epithet “verticilliformis” (Latin) pertains to verticillate branches of sporangiophores.
Type—China, Hainan, Sanya City, Jiyang District, G224 Haiyu Middle Line, 18.391817° N, 109.641068° E, altitude 193.0 m, from soil sample, 24 April 2023, Yi-Xin Wang (Holotype HMAS 353183, ex-holotype strain CGMCC 3.27810 = XG04088-3).
Description—Colonies grow fast on PDA in darkness at 25 °C for 7 days, reaching 90 mm in diameter, initially white, soon becoming grey to brown, irregular at the edge, cottony, and reversely white to grey, growing outward in a petal shape. Hyphae are hyaline at first, brownish when mature, branched, and 4.4–8.9 μm in diameter. Stolons are hyaline, branched, and smooth. Rhizoids are hyaline, branched, irregular, or root-like. Sporangiophores are located on aerial mycelia, hyaline, erect or slightly curved, unbranched or slightly branched, and mostly umbellately branched, and swelling is usually present below the sporangia, often with a septum below apophyses, 22.3–368.7 × 3.3–4.6 μm. Sporangia are oval to subglobose, 15.7–29.0 × 17.5–26.0 μm, hyaline at first and then brown, and deliquescent-walled, mostly leaving a collar after releasing sporangiospores. Apophyses are hyaline, smooth, shallow mouth bowl-shaped, and 4.5–14.4 × 11.1–20.4 μm. Collars are distinct. Columellae are hyaline or brown, hemispherical, with tips with short or long cylindrical protrusions, and 12.6–25.6 × 14.7–26.2 μm. Protrusions are 3.1–6.3 × 1.8–2.2 μm. Sporangiospores are hyaline, smooth, not uniform, mostly ovoid, and 3.4–4.3 × 2.3–3.1 μm, and some are cylindrical, 3.9–4.4 × 2.1–3.0 μm. Chlamydospores are present. Zygospores were not observed.
Additional strain examined—China, Hainan, Sanya City, Jiyang District, G224 Haiyu Middle Line, 18.391817° N, 109.641068° E, altitude 193.0 m, from soil sample, 24 April 2023, Yi-Xin Wang (living culture XG04088-4).
Genome accession number—JBOEPV000000000.
GenBank accession numbers—CGMCC 3.27810 clone 1 (ITS, PQ306315; LSU, PQ803170; Act, PQ807205; SSU, PQ799262; TEF1α, PV126011), CGMCC 3.27810 clone 2 (ITS, PV069755; LSU, PQ803171; Act, PV126025; SSU, PQ799263; TEF1α, PV126013), XG04088-4 clone1 (ITS, PQ306316; LSU, PQ803172; Act, PQ807206; SSU, PQ799264; TEF1α, PV126012), XG04088-4 clone2 (ITS, PV069756; LSU, PQ803173; Act, PV126026; SSU, PQ799265; TEF1α, PV126014).
Maximum growth temperature—CGMCC 3.27810 (34 °C), XG04088-4 (34 °C).
Notes—Based on phylogenetic analyses of ITS-SSU-LSU-Act-TEF1α sequences, the two isolates of the new species Absidia verticilliformis formed two sister clades with full supports (MLBV = 100, BIPP = 1; Figure 1), which is closely related to A. edaphica (MLBV = 94, BIPP = 1; Figure 1). This new species differs morphologically from A. edaphica in its sporangia, columellae, and sporangiospores [37]. The new species is smaller than A. edaphica in sporangia, 15.7–29.0 × 17.5–26.0 μm vs. 30.5–35.5 × 24–27 μm. In columellae, the new species is bigger than A. edaphica, 12.6–25.6 × 14.7–26.2 μm vs. 5–9.5 × 6.5–20 μm. The new species is smaller than A. edaphica in sporangiospores, 3.4–4.4 × 2.1–3.1 μm vs. 3.5–5.5 × 2–3.5 μm. Combining morphological and molecular phylogenetic analyses, the two isolates were classified as a new taxon, allied to A. edaphica.
4. Discussion
The genus Absidia was established nearly 150 years ago. Between 1878 and 2010, the taxonomic status of the genus was changed several times, and the species of the genus were divided according to the optimum growth temperature, phylogenetic relationships, and microscopic morphology. Finally, Absidia s. l. has been divided into three genera, namely Absidia s. s., Lichtheimia, and Lentamyces [7,8]. Nowadays, Absidia is the most species-rich genus in the family Cunninghamellaceae. Together with the four new species proposed in this study, the world diversity of Absidia s.s. reaches 71 recognized species.
According to Hoffmann’s study [6], Absidia s.s. is not thermotolerant, with an optimal growth temperature above 30 °C, and does not grow above 37 °C, rapidly growing. The sporangiophore has a subsporangial septum, and the zygospore has sterile hair-like, mycelial appendages on the suspensors. It does not parasitize on other Mucorales fungi; Lichtheimia is thermotolerant, with an optimal temperature above 34 °C and growth above 37 °C, rapidly growing. The sporangiophore has a subsporangial septum, and zygospore has no mycelial appendages on the suspensors. It does not parasitize on other Mucorales fungi; Lentamyces is not thermotolerant, and its temperature maximum is below 30 °C, slowly growing, sporangiophores with subsporangial septum, potentially parasitic on other Mucorales, homothallic, and warty zygospores without appendaged suspensors.
Phylogenetically, based on the ITS sequence, a maximum likelihood method (ML) tree was reconstructed for main species of the genus Absidia s.s., Lichtheimia, and Lentamyces, as well as the four new species proposed herein. Three groups with relatively high supports emerged, which was consistent with the division mentioned above, and the four new species in this article were clustered in the Absidia s.s. group (Supplementary File S3). Physiologically, thermal tolerance thresholds of fungal strains were determined using a temperature gradient cultivation technique. Growth characterization revealed distinct maximum growth temperatures for the four Absidia species: A. ovoidospora (30 °C), A. irregularis (32 °C), A. multiformis (34 °C), and A. verticilliformis (34 °C). These thermal parameters align with the established physiological profile of Absidia s.s. (maximum growth temperature at 30–37 °C) [7]. Morphologically, the sporangiophore has a subsporangial septum of the four species, and Absidia multiformis captured the presence of sterile hair-like, mycelial appendages on the suspensors of zygospore.
Based on the above phylogenetic, morphological, and optimal growth temperature evidence, the four species in this paper belong to Absidia s.s.
Prior to 2020, the phylogenetic tree of this genus was predominantly reconstructed utilizing the combination of ITS and LSU sequences [36]. Subsequently, the SSU sequence and the protein sequences Act and TEF1α were gradually added, yielding results that were largely in line with previous findings [13,17,37,38]. Aiming at the problem of overlapping peaks in the PCR amplification of Act protein sequences of some species, genomic sequencing was performed on them in this paper, and the corresponding sequences were extracted from the assembled genomes. It is speculated that the chromatogram overlapping problem is due to the existence of duplicate copies in this section of the gene sequence, and this problem has also been confirmed through genomic extraction sequences. Extracting sequences by assembling genomes might become a solution to the problem of overlapping in PCR sequencing of some species.
In this study, the phylogenetic history was inferred using ITS-SSU-LSU-Act-TEF1α, and newly isolated strains were grouped into four individual clades with high supports, namely Absidia irregularis sp. nov., A. multiformis sp. nov., A. ovoidospora sp. nov., and A. verticilliformis sp. nov.
Absidia irregularis is related to A. oblongispora with high supports (MLBV = 100, BIPP = 1; Figure 1), while distinguished by bigger apophyses and columellae. Absidia multiformis is most closely related to A. heterospora with high support (BIPP = 0.99; Figure 1). Morphologically, A. multiformis has narrower and shorter sporangiophores, smaller sporangia size, and different columellae shape and size. Absidia ovoidospora is closely related to A. panacisoli and A. abundans. However, compared with A. panacisoli, A. ovoidospora had wider sporangiophore width and a larger sporangium. At the same time, compared with A. abundans, the microscopical measurements of the sporangiophore, sporangium, and columella of A. ovoidospora were larger. A. verticilliformis is closely related to A. edaphica. A. verticilliformis differs from A. edaphica by smaller sporangia, bigger columellae, and smaller sporangiospores.
In summary, the molecular phylogenetic and morphological results both support the identification of the four new species. These findings further enhance our understanding of mucoralean biodiversity in Asian tropical and subtropical ecosystems.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms13061315/s1; File S1: ITS-SSU-LSU-Act-TEF1a; File S2: ITS.fas; File S3: Phylogenetic tree based on ITS; and File S4: ITS GenBank accession numbers.
Author Contributions
Y.-X.W. took charge of the microscopy, DNA sequencing, data analyses, and manuscript draft; Z.-Y.D. and Y.-X.W. collected samples and isolated living cultures; X.-Y.J. and Z.-Y.D. made dry cultures; Z.M. revised the manuscript; X.-Y.L. contributed to new species proposal, manuscript revision, financial support, and funding acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the Key Technological Innovation Program of Shandong Province, China (2022CXGC020710), the National Natural Science Foundation of China (Nos. 32170012, 32470004, 32300011), Ji’nan City’s “New University 20 Policies” Initiative for Innovative Research Teams Project (202228028), and the Innovative Agricultural Application Technology Project of Jinan City (CX202210).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The sequences were deposited in the GenBank database.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- van Tieghem, P. Troisieme memoire sur les Mucorinees. Ann. Sci. Nat. Bot. Ser. 1876, 4, 312–398. [Google Scholar]
- Tedersoo, L.; Sánchez-Ramírez, S.; Kõljalg, U.; Bahram, M.; Döring, M.; Schigel, D.; May, T.; Ryberg, M.; Abarenkov, K. High-level classification of the Fungi and a tool for evolutionary ecological analyses. Fungal Divers. 2018, 90, 135–159. [Google Scholar] [CrossRef]
- Von Arx, J.A. On Mucoraceae s. str. and other families of the Mucorales. Sydowia 1982, 35, 10–26. [Google Scholar]
- Benny, G.; Humber, R.; Morton, J. Zygomycota: Zygomycetes. In The Mycota. VII. Systematics and Evolution: Part A; McLaughlin, D.J., McLaughlin, E.G., Lemke, P.A., Eds.; Springer: Berlin/Heidelberg, Germany, 2001; pp. 113–146. [Google Scholar]
- Kirk, P.; Cannon, P.; Minter, D.; Stalpers, J. Ainsworth and Bisby’s Dictionary of the Fungi (10th edition). Ref. Rev. 2009, 23, 42. [Google Scholar]
- Hoffmann, K. Identification of the genus Absidia (Mucorales, Zygomycetes): A comprehensive taxonomic revision. In Molecular Identification of Fungi; Gherbawy, Y., Voigt, K., Eds.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 439–460. [Google Scholar]
- Hoffmann, K.; Discher, S.; Voigt, K. Revision of the genus Absidia (Mucorales, Zygomycetes) based on physiological, phylogenetic, and morphological characters; thermotolerant Absidia spp. form a coherent group, Mycocladiaceae fam. nov. Mycol. Res. 2007, 111, 1169–1183. [Google Scholar] [CrossRef]
- Hoffmann, K.; Voigt, K. Absidia parricida plays a dominant role in biotrophic fusion parasitism among mucoralean fungi (Zygomycetes): Lentamyces, a new genus for A. parricida and A. zychae. Plant Biol. 2009, 11, 537–554. [Google Scholar] [CrossRef]
- Zhao, H.; Nie, Y.; Tongkai, Z.; Wang, Y.-J.; Wang, M.; Dai, Y.-C.; Liu, X.-Y. Species diversity and ecological habitat of Absidia (Cunninghamellaceae, Mucorales) with emphasis on five new species from forest and grassland soil in China. J. Fungi 2022, 8, 471. [Google Scholar] [CrossRef]
- Zhao, H.; Nie, Y.; Zong, T.; Dai, Y.; Liu, X. Three new species of Absidia (Mucoromycota) from China based on phylogeny, morphology and physiology. Diversity 2022, 14, 132. [Google Scholar] [CrossRef]
- Braga, A.R.; Nunes, M.C.; Raymundo, A. The experimental development of emulsions enriched and stabilized by recovering matter from spirulina biomass: Valorization of residue into a sustainable protein source. Molecules 2023, 28, 6179. [Google Scholar] [CrossRef]
- Liu, X.; Qiao, L.; Xie, D.; Zhang, Y.; Zou, J.; Chen, X.; Dai, J. Microbial transformation of ginsenoside-Rg1 by Absidia coerulea and the reversal activity of the metabolites towards multi-drug resistant tumor cells. Fitoterapia 2011, 82, 1313–1317. [Google Scholar] [CrossRef]
- Tao, M.-F.; Ding, Z.-Y.; Wang, Y.-X.; Zhang, Z.-X.; Zhao, H.; Meng, Z.; Liu, X.-Y. Unveiling species diversity within early-diverging fungi from China II: Three new species of Absidia (Cunninghamellaceae, Mucoromycota) from Hainan Province. MycoKeys 2024, 110, 255–272. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-X.; Zhao, H.; Jiang, Y.; Liu, X.-Y.; Tao, M.-F.; Liu, X.-Y. Unveiling species diversity within early-diverging fungi from China III: Six new species and a new record of Gongronella (Cunninghamellaceae, Mucoromycota). MycoKeys 2024, 110, 287–317. [Google Scholar] [CrossRef]
- Zhao, H.; Nie, Y.; Huang, B.; Liu, X.-Y. Unveiling species diversity within early-diverging fungi from China I: Three new species of Backusella (Backusellaceae, Mucoromycota). MycoKeys 2024, 109, 285–304. [Google Scholar] [CrossRef]
- Ding, Z.-Y.; Ji, X.-Y.; Tao, M.-F.; Jiang, Y.; Liu, W.-X.; Wang, Y.-X.; Meng, Z.; Liu, X.-Y. Unveiling species diversity within early-diverging fungi from China IV: Four new species of Absidia (Cunninghamellaceae, Mucoromycota). MycoKeys 2025, 117, 267. [Google Scholar]
- Ji, X.-Y.; Ding, Z.-Y.; Nie, Y.; Zhao, H.; Wang, S.; Huang, B.; Liu, X.-Y. Unveiling species diversity within early-diverging fungi from China V: Five new species of Absidia (Cunninghamellaceae, Mucoromycota). MycoKeys 2025, 117, 267–288. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wei, Y.; Zou, Y.; Liu, P.; Li, Z.; Gontcharov, A.; Stephenson, S.; Wang, Q.; Zhang, S.; Li, Y. Dictyostelid cellular slime molds from the russian far east. Protist 2020, 171, 125756. [Google Scholar] [CrossRef]
- Zou, Y.; Hou, J.; Guo, S.; Li, C.; Li, Z.; Stephenson, S.L.; Pavlov, I.N.; Liu, P.; Li, Y. Diversity of dictyostelid cellular slime molds, including two species new to science, in forest soils of Changbai Mountain, China. Microbiol. Spectr. 2022, 10, e0240222. [Google Scholar] [CrossRef]
- Corry, J.E.L.; Curtis, G.D.W.; Baird, R.M. Rose Bengal Chloramphenicol (RBC) agar. Prog. Ind. Microbiol. 1995, 34, 431–433. [Google Scholar]
- Wang, Y.; Zhao, H.; Ding, Z.; Ji, X.; Zhang, Z.; Wang, S.; Zhang, X.; Liu, X. Three new species of Gongronella (Cunninghamellaceae, Mucorales) from soil in Hainan, China based on morphology and molecular phylogeny. J. Fungi 2023, 9, 1182. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, R.; Liu, S.; Mu, T.; Zhang, X.; Xia, J. Morphological and phylogenetic analyses reveal two new species of Sporocadaceae from Hainan, China. MycoKeys 2022, 88, 171–192. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Voigt, K.; Wostemeyer, J. Reliable amplification of actin genes facilitates deep-level phylogeny. Microbiol. Res. 2000, 155, 179–195. [Google Scholar] [CrossRef]
- White, T.; Bruns, T.; Lee, S.; Taylor, J.; Innis, M.; Gelfand, D.; Sninsky, J. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics; PCR Protocols; Academic Press, Inc.: Cambridge, MA, USA, 1990; Volume 31, pp. 315–322. [Google Scholar]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef]
- Carbone, I.; Kohn, L.M. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 1999, 91, 553–556. [Google Scholar] [CrossRef]
- O’Donnell, K.; Lutzoni, F.; Ward, T.; Benny, G. Evolutionary relationships among mucoralean fungi (Zygomycota): Evidence for family polyphyly on a large scale. Mycologia 2001, 93, 286–296. [Google Scholar] [CrossRef]
- Nylander, J. MrModeltest V2. Program distributed by the author. Bioinformatics 2004, 24, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Mark, P.; Ayres, D.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.; Huelsenbeck, J. MrBayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Zhao, H.; Nie, Y.; Zong, T.-K.; Wang, K.; Lv, M.-L.; Cui, Y.-J.; Tohtirjap, A.; Chen, J.-J.; Zhao, C.-L.; Wu, F.; et al. Species diversity, updated classification and divergence times of the phylum Mucoromycota. Fungal Divers. 2023, 123, 49–157. [Google Scholar] [CrossRef]
- Zhang, Z.; Shang, Y.; Liu, Q.; Li, D.; Yin, C.; Liu, X.; Tao, M.; Jiang, Y.; Wang, Y.; Zhang, M.; et al. Deciphering the evolutionary and taxonomic complexity of Diaporthales (Sordariomycetes, Ascomycota) through integrated phylogenomic and divergence time estimation. Fungal Divers. 2025. [Google Scholar] [CrossRef]
- Hesseltine, C.W.; Ellis, J.J. The genus Absidia: Gongronella and cylindrical-spored species of Absidia. Mycologia 1964, 4, 568–601. [Google Scholar] [CrossRef]
- Zhang, T.-Y.; Yu, Y.; Zhu, H.; Yang, S.-Z.; Yang, T.-M.; Zhang, M.-y.; Zhang, Y.-X. Absidia panacisoli sp. nov., isolated from rhizosphere of Panax notoginseng. Int. J. Syst. Evol. Microbiol. 2018, 68, 2468–2472. [Google Scholar] [CrossRef] [PubMed]
- Hurdeal, V.G.; Gentekaki, E.; Lee, H.B.; Jeewon, R.; Hyde, K.D.; Tibpromma, S.; Mortimer, P.E.; Xu, J. Mucoralean fungi in Thailand: Novel species of Absidia from tropical forest soil. Cryptogam. Mycol. 2021, 42, 39–61. [Google Scholar] [CrossRef]
- Nguyen, T.T.T.; Santiago, A.; Hallsworth, J.E.; Cordeiro, T.; Voigt, K.; Kirk, P.; Crous, P.W.; Júnior, M.A.M.; Elsztein, C.; Lee, H. New Mucorales from opposite ends of the world. Stud. Mycol. 2024, 109, 273–321. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).