Genomic Insights into Edwardsiella ictaluri: Molecular Epidemiology and Antimicrobial Resistance in Striped Catfish (Pangasianodon hypophthalmus) Aquaculture in Vietnam
Abstract
1. Introduction
2. Materials and Methods
2.1. Edwardsiella ictaluri Strain Collection
2.2. Bacterial Species Confirmation by PCR
2.3. Antimicrobial Susceptibility Testing
2.4. Whole Genome Sequencing
2.5. Bioinformatic Analysis
3. Results
3.1. Species Identification and Whole Genome Sequencing
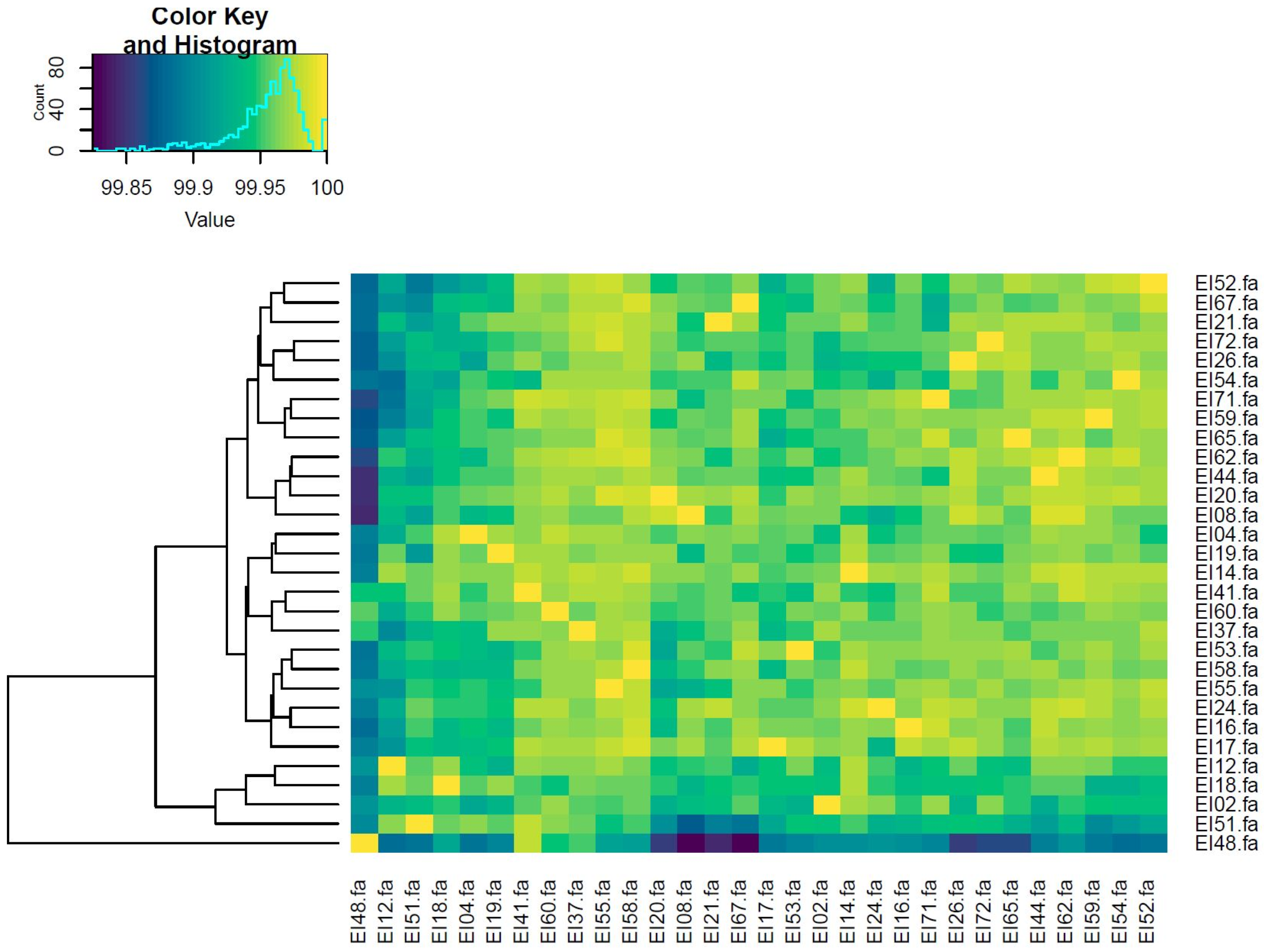
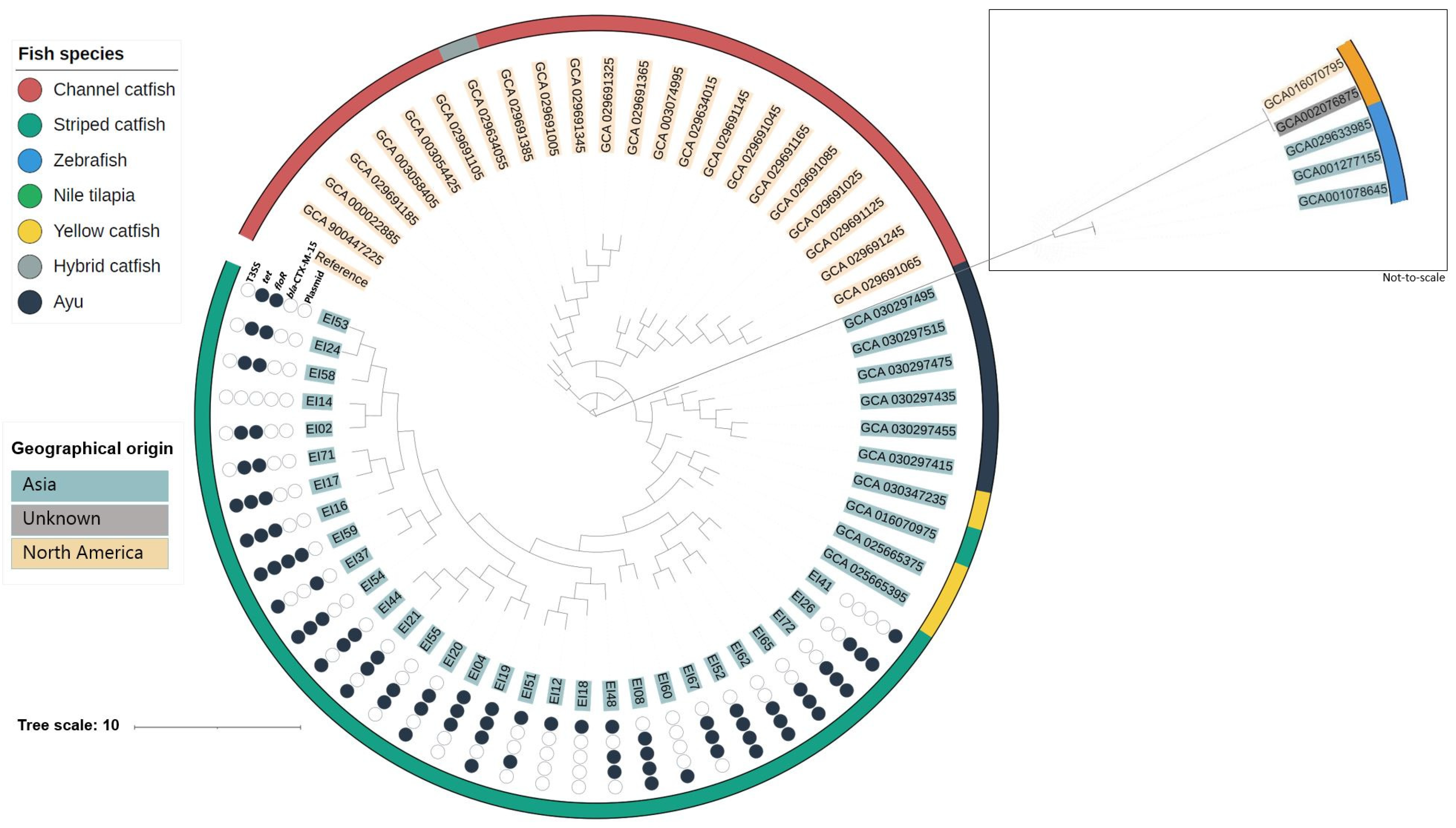
3.2. Phylogenetic Analysis
3.3. Pangenome Analysis
3.4. Antimicrobial Resistance Genes and Minimum Inhibitory Concentrations
3.5. Virulence Genes
3.6. Plasmids, Mobile Genetic Elements and Prophages
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Adeolu, M.; Alnajar, S.; Naushad, S.; Gupta, R.S. Genome-based phylogeny and taxonomy of the ‘Enterobacteriales’: Proposal for Enterobacterales ord. nov. divided into the families Enterobacteriaceae, Erwiniaceae fam. nov., Pectobacteriaceae fam. nov., Yersiniaceae fam. nov., Hafniaceae fam. nov., Morgane. Int. J. Syst. Evol. Microbiol. 2016, 66, 5575–5599. [Google Scholar] [CrossRef]
- Hawke, J.P. A Bacterium Associated with Disease of Pond Cultured Channel Catfish, Ictalurus punctatus. J. Fish. Res. Board Can. 1979, 36, 1508–1512. [Google Scholar] [CrossRef]
- Mitchell, A.J.; Goodwin, A.E. Evidence that Enteric Septicemia of Catfish (ESC) was Present in Arkansas by the Late 1960s: New Insights into the Epidemiology of ESC. J. Aquat. Anim. Health 1999, 11, 175–178. [Google Scholar] [CrossRef]
- Machimbirike, V.I.; Crumlish, M.; Dong, H.T.; Santander, J.; Khunrae, P.; Rattanarojpong, T. Edwardsiella ictaluri: A systemic review and future perspectives on disease management. Rev. Aquac. 2022, 14, 1613–1636. [Google Scholar] [CrossRef]
- Ferguson, H.W.; Turnbull, J.F.; Shinn, A.; Thompson, K.; Dung, T.T.; Crumlish, M. Bacillary necrosis in farmed Pangasius hypophthalmus (Sauvage) from the Mekong Delta, Vietnam. J. Fish Dis. 2001, 24, 509–513. [Google Scholar] [CrossRef]
- Crumlish, M.; Dung, T.T.; Turnbull, J.F.; Ngoc, N.T.N.; Ferguson, H.W. Identification of Edwardsiella ictaluri from diseased freshwater catfish, Pangasius hypophthalmus (Sauvage), cultured in the Mekong Delta, Vietnam. J. Fish Dis. 2002, 25, 733–736. [Google Scholar] [CrossRef]
- Nhinh, D.T.; Giang, N.T.H.; Van Van, K.; Dang, L.T.; Dong, H.T.; Hoai, T.D. Widespread presence of a highly virulent Edwardsiella ictaluri strain in farmed tilapia, Oreochromis spp. Transbound. Emerg. Dis. 2022, 69, e2276–e2290. [Google Scholar] [CrossRef]
- Hawke, J.P.; Kent, M.; Rogge, M.; Baumgartner, W.; Wiles, J.; Shelley, J.; Savolainen, L.C.; Wagner, R.; Murray, K.; Peterson, T.S. Edwardsiellosis Caused by Edwardsiella ictaluri in Laboratory Populations of Zebrafish Danio rerio. J. Aquat. Anim. Health 2013, 25, 171–183. [Google Scholar] [CrossRef]
- FAO. World Fisheries and Aquaculture; FAO: Rome, Italy, 2022; Available online: https://www.fao.org/3/ca9229en/online/ca9229en.html#chapter-1_1 (accessed on 20 February 2024).
- Hoa, T.T.T.; Boerlage, A.S.; Duyen, T.T.M.; Thy, D.T.M.; Hang, N.T.T.; Humphry, R.W.; Phuong, N.T. Nursing stages of striped catfish (Pangasianodon hypophthalmus) in Vietnam: Pathogens, diseases and husbandry practices. Aquaculture 2021, 533, 736114. [Google Scholar] [CrossRef]
- Dung, T.T.; Haesebrouck, F.; Tuan, N.A.; Sorgeloos, P.; Baele, M.; Decostere, A. Antimicrobial susceptibility pattern of Edwardsiella ictaluri Isolates from natural outbreaks of bacillary necrosis of Pangasianodon hypophthalmus in Vietnam. Microb. Drug Resist. 2008, 14, 311–316. [Google Scholar] [CrossRef] [PubMed]
- Crumlish, M.; Thanh, P.C.; Koesling, J.; Tung, V.T.; Gravningen, K. Experimental challenge studies in Vietnamese catfish, Pangasianodon hypophthalmus (Sauvage), exposed to Edwardsiella ictaluri and Aeromonas hydrophila. J. Fish Dis. 2010, 33, 717–722. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.T.; Nguyen, V.V.; Phiwsaiya, K.; Gangnonngiw, W.; Withyachumnarnkul, B.; Rodkhum, C.; Senapin, S. Concurrent infections of Flavobacterium columnare and Edwardsiella ictaluri in striped catfish, Pangasianodon hypophthalmus in Thailand. Aquaculture 2015, 448, 142–150. [Google Scholar] [CrossRef]
- PHARMAQ. PHARMAQ Products—VACCINE PACKAGE INSERT [Internet]. 2018. Available online: https://d2fhlgkwzpo2gq.cloudfront.net/media/njgph0sn/pil-ajpanga2-vn-v3-english.pdf (accessed on 15 August 2023).
- Chambers, J.A.; Crumlish, M.; Comerford, D.A.; Phuoc, L.H.; Phuong, V.H.; O’carroll, R.E. Understanding Vaccine Hesitancy in Vietnamese Fish Farmers. Antibiotics 2022, 11, 878. [Google Scholar] [CrossRef] [PubMed]
- Sakai, T.; Yuasa, K.; Sano, M.; Iida, T. Identification of Edwardsiella ictaluri and E. tarda by species-specific polymerase chain reaction targeted to the upstream region of the fimbrial gene. J. Aquat. Anim. Health 2009, 21, 124–132. [Google Scholar] [CrossRef] [PubMed]
- Erickson, V.I.; Khoi, L.M.; Hounmanou, Y.M.G.; Dung, T.T.; Phu, T.M.; Dalsgaard, A. Comparative genomic analysis of Aeromonas dhakensis and Aeromonas hydrophila from diseased striped catfish fingerlings cultured in Vietnam. Front. Microbiol. 2023, 14, 1254781. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data [Online] [Internet]. 2010. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 10 September 2023).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Nurk, S.; Bankevich, A.; Antipov, D.; Gurevich, A.; Korobeynikov, A.; Lapidus, A.; Prjibelsky, A.; Pyshkin, A.; Sirotkin, A.; Sirotkin, Y.; et al. Assembling Genomes and Mini-Metagenomes from Highly Chimeric Reads BT—Research in Computational Molecular Biology; Deng, M., Jiang, R., Sun, F., Zhang, X., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 158–170. [Google Scholar]
- Mikheenko, A.; Prjibelski, A.; Saveliev, V.; Antipov, D.; Gurevich, A. Versatile genome assembly evaluation with QUAST-LG. Bioinformatics 2018, 34, i142–i150. [Google Scholar] [CrossRef] [PubMed]
- Wick, R.R.; Judd, L.M.; Holt, K.E. Performance of neural network basecalling tools for Oxford Nanopore sequencing. Genome Biol. 2019, 20, 129. [Google Scholar] [CrossRef] [PubMed]
- Wood, D.E.; Salzberg, S.L. Kraken: Ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014, 15, R46. [Google Scholar] [CrossRef] [PubMed]
- Larsen, M.V.; Cosentino, S.; Lukjancenko, O.; Saputra, D.; Rasmussen, S.; Hasman, H.; Sicheritz-Pontén, T.; Aarestrup, F.M.; Ussery, D.W.; Lund, O. Benchmarking of methods for genomic taxonomy. J. Clin. Microbiol. 2014, 52, 1529–1539. [Google Scholar] [CrossRef]
- Hasman, H.; Saputra, D.; Sicheritz-Ponten, T.; Lund, O.; Svendsen, C.A.; Frimodt-Møller, N.; Aarestrup, F.M. Rapid whole-genome sequencing for detection and characterization of microorganisms directly from clinical samples. J. Clin. Microbiol. 2014, 52, 139–146. [Google Scholar] [CrossRef]
- Clausen, P.T.L.C.; Aarestrup, F.M.; Lund, O. Rapid and precise alignment of raw reads against redundant databases with KMA. BMC Bioinform. 2018, 19, 307. [Google Scholar] [CrossRef]
- Jolley, K.A.; Bray, J.E.; Maiden, M.C.J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018, 3, 124. [Google Scholar] [CrossRef]
- Bonin, N.; Doster, E.; Worley, H.; Pinnell, L.J.; Bravo, J.E.; Ferm, P.; Marini, S.; Prosperi, M.; Noyes, N.; Morley, P.S.; et al. MEGARes and AMR++, v3.0: An updated comprehensive database of antimicrobial resistance determinants and an improved software pipeline for classification using high-throughput sequencing. Nucleic Acids Res. 2023, 51, D744–D752. [Google Scholar] [CrossRef]
- Seemann, T. Abricate [Internet]. Available online: https://github.com/tseemann/abricate (accessed on 10 September 2023).
- Doster, E.; Lakin, S.M.; Dean, C.J.; Wolfe, C.; Young, J.G.; Boucher, C.; Belk, K.E.; Noyes, N.R.; Morley, P.S. MEGARes 2.0: A database for classification of antimicrobial drug, biocide and metal resistance determinants in metagenomic sequence data. Nucleic Acids Res. 2020, 48, D561–D569. [Google Scholar] [CrossRef]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Møller Aarestrup, F.; Hasman, H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Johansson, M.H.K.; Bortolaia, V.; Tansirichaiya, S.; Aarestrup, F.M.; Roberts, A.P.; Petersen, T.N. Detection of mobile genetic elements associated with antibiotic resistance in Salmonella enterica using a newly developed web tool: MobileElementFinder. J. Antimicrob. Chemother. 2021, 76, 101–109. [Google Scholar] [CrossRef]
- Zhou, Y.; Liang, Y.; Lynch, K.H.; Dennis, J.J.; Wishart, D.S. PHAST: A Fast Phage Search Tool. Nucleic Acids Res. 2011, 39, 347–352. [Google Scholar] [CrossRef]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.G.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef]
- Hadfield, J.; Croucher, N.J.; Goater, R.J.; Abudahab, K.; Aanensen, D.M.; Harris, S.R. Phandango: An interactive viewer for bacterial population genomics. Bioinformatics 2018, 34, 292–293. [Google Scholar] [CrossRef]
- Seemann, T. Snippy: Fast Bacterial Variant Calling from NGS Reads [Internet]. 2015. Available online: https://github.com/tseemann/snippy (accessed on 31 August 2023).
- Croucher, N.J.; Page, A.J.; Connor, T.R.; Delaney, A.J.; Keane, J.A.; Bentley, S.D.; Parkhill, J.; Harris, S.R. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015, 43, e15. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Jain, C.; Rodriguez-R, L.M.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 2018, 9, 5114. [Google Scholar] [CrossRef]
- Edrees, A.; Abdelhamed, H.; Nho, S.W.; Park, S.B.; Karsi, A.; Austin, F.W.; Essa, M.; Pechan, T.; Lawrence, M.L. Construction and evaluation of type III secretion system mutants of the catfish pathogen Edwardsiella piscicida. J. Fish Dis. 2018, 41, 805–816. [Google Scholar] [CrossRef]
- Qian, Z.; Tian, H.T.; You, L.D.; Yi, L.L.; Pin, N.; Xia, X.H. The Edwardsiella piscicida Type III Effector EseJ Suppresses Expression of Type 1 Fimbriae, Leading to Decreased Bacterial Adherence to Host Cells. Infect. Immun. 2019, 87, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Gai, C.; Ou, R.; Lu, L.; Yang, X.; Cao, H. Providencia Rettgeri: An emerging pathogen for freshwater cultured whiteleg shrimp (Penaeus vannamei). Isr. J. Aquac.-Bamidgeh 2017, 69, 1–6. [Google Scholar]
- Payne, C.J.; Grace, K.; Phuong, V.H.; Phuoc, N.N.; Dung, T.T.; Phuoc, L.H.; Crumlish, M. Exploring the genetic diversity of Edwardsiella ictaluri in Vietnamese striped catfish (Pangasianodon hypophthalmus) farms over a 20-year period. Front. Mar. Sci. 2023, 10, 1270968. [Google Scholar] [CrossRef]
- Cai, W.; Arias, C.R. Biofilm Formation on Aquaculture Substrates by Selected Bacterial Fish Pathogens. J. Aquat. Anim. Health 2017, 29, 95–104. [Google Scholar] [CrossRef]
- Karunasagar, I.; Otta, S.K.; Karunasagar, I. Biofilm formation by Vibrio harveyi on surfaces. Aquaculture 1996, 140, 241–245. [Google Scholar] [CrossRef]
- Tam, J.P. 12—Synthesis and Applications of Branched Peptides in Immunological Methods and Vaccines. In Peptides; Academic Press: San Diego, CA, USA, 1995; pp. 455–500. Available online: https://www.sciencedirect.com/science/article/pii/B9780123109200500139 (accessed on 15 March 2024).
- Wang, Q.; Yang, M.; Xiao, J.; Wu, H.; Wang, X.; Lv, Y.; Xu, L.; Zheng, H.; Wang, S.; Zhao, G.; et al. Genome sequence of the versatile fish pathogen Edwardsiella tarda provides insights into its adaptation to broad host ranges and intracellular niches. PLoS ONE 2009, 4, e7646. [Google Scholar] [CrossRef]
- Hoa, T.T.T.; Nakayama, T.; Huyen, H.M.; Harada, K.; Hinenoya, A.; Phuong, N.T.; Yamamoto, Y. Extended-spectrum beta-lactamase-producing Escherichia coli harbouring sul and mcr-1 genes isolates from fish gut contents in the Mekong Delta, Vietnam. Lett. Appl. Microbiol. 2020, 71, 78–85. [Google Scholar] [CrossRef]
- Hoa, T.T.T.; Huyen, H.M.; Nakayama, T.; Minh, D.T.N.; Hoang, O.N.; Le Thi, H.; Thanh, P.N.; Hoai, P.H.; Yamaguchi, T.; Jinnai, M.; et al. Frequent contamination of edible freshwater fish with colistin-resistant Escherichia coli harbouring the plasmid-mediated mcr-1 gene. Mar. Pollut. Bull. 2022, 184, 114108. [Google Scholar] [CrossRef]
- Rogge, M.L.; Dubytska, L.; Jung, T.S.; Wiles, J.; Elkamel, A.A.; Rennhoff, A.; Oanh, D.T.H.; Thune, R.L. Comparison of Vietnamese and US isolates of Edwardsiella ictaluri. Dis. Aquat. Organ. 2013, 106, 17–29. [Google Scholar] [CrossRef]
- Fernandez, D.H.; Pittman-Cooley, L.; Thune, R.L. Sequencing and Analysis of the Edwardsiella ictaluri Plasmids. Plasmid 2001, 45, 52–56. [Google Scholar] [CrossRef]
- Pham-duc, P.; Meghan, C.A.; Cong-hong, H.; Nguyen-thuy, H.; Padungtod, P.; Nguyen-Thi, H.; Dang-Xuan, S. Knowledge, attitudes and practices of livestock and aquaculture producers regarding antimicrobial use and resistance in Vietnam. PLoS ONE 2019, 10, e0223115. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute. VET04 Performance Standards For Antimicrobial Susceptibility Testing of Bacteria Isolated from Aquatic Animals, 3rd ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2020. [Google Scholar]
- European Committee on Antimicrobial Susceptibility Testing. MIC Distributions and Epidemiological Cut-Off Value (ECOFF) Setting. Eucast Sop 102 [Internet]. 2021. Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/EUCAST_SOPs/2021/EUCAST_SOP_10.2_MIC_distributions_and_epidemiological_cut-off_value__ECOFF__setting_20211202.pdf (accessed on 15 March 2024).
- Leung, K.Y.; Siame, B.A.; Tenkink, B.J.; Noort, R.J.; Mok, Y.-K. Edwardsiella tarda—Virulence mechanisms of an emerging gastroenteritis pathogen. Microbes Infect. 2012, 14, 26–34. [Google Scholar] [CrossRef]



| Isolate | PCR | Species in KmerFinder | MLST | GC% | Contigs | Size (Mbp) | Weight of Fish (g) | Year | Province |
|---|---|---|---|---|---|---|---|---|---|
| EI02 | E. ictaluri | E. ictaluri | 26 | 57.4 | 209 | 3.62 | 20 | 2017 | Dong Thap |
| EI04 | E. ictaluri | E. ictaluri | 26 | 57.1 | 233 | 3.75 | 20 | 2017 | Dong Thap |
| EI08 | E. ictaluri | E. ictaluri | 26 | 57.3 | 219 | 3.74 | 5 | 2017 | An Giang |
| EI12 | E. ictaluri | E. ictaluri | 26 | 57.1 | 231 | 3.70 | 100 | 2017 | Can Tho |
| EI14 | E. ictaluri | E. ictaluri | 26 | 57.4 | 190 | 3.61 | 100 | 2017 | Can Tho |
| EI16 | E. ictaluri | E. ictaluri | 26 | 57.4 | 196 | 3.67 | 20 | 2018 | Tien Giang |
| EI17 | E. ictaluri | E. ictaluri | 26 | 57.4 | 200 | 3.67 | 30 | 2018 | Tien Giang |
| EI18 | E. ictaluri | E. ictaluri | 26 | 57.0 | 219 | 3.74 | 40 | 2018 | Dong Thap |
| EI19 | E. ictaluri | E. ictaluri | 26 | 57.1 | 226 | 3.82 | 40 | 2018 | Dong Thap |
| EI20 | E. ictaluri | E. ictaluri | 26 | 57.3 | 230 | 3.74 | 100 | 2019 | Can Tho |
| EI21 | E. ictaluri | E. ictaluri | 26 | 57.3 | 214 | 3.75 | 75 | 2019 | Tien Giang |
| EI24 | E. ictaluri | E. ictaluri | 26 | 57.4 | 202 | 3.55 | 75 | 2019 | Can Tho |
| EI26 | E. ictaluri | E. ictaluri | 26 | 57.4 | 201 | 3.66 | 55 | 2019 | Vinh Long |
| EI37 | E. ictaluri | E. ictaluri | 26 | 57.6 | 183 | 3.63 | 50 | 2019 | Can Tho |
| EI41 | E. ictaluri | E. ictaluri | 26 | 57.7 | 195 | 3.56 | 40 | 2019 | An Giang |
| EI44 | E. ictaluri | E. ictaluri | 26 | 57.3 | 208 | 3.74 | 35 | 2019 | Ben Tre |
| EI48 | E. ictaluri | E. ictaluri | 26 | 57.1 | 190 | 3.75 | 30 | 2019 | Dong Thap |
| EI51 | E. ictaluri | E. ictaluri | 26 | 57.1 | 214 | 3.75 | 15 | 2020 | An Giang |
| EI52 | E. ictaluri | E. ictaluri | 26 | 57.3 | 195 | 3.74 | 100 | 2020 | Dong Thap |
| EI53 | E. ictaluri | E. ictaluri | 26 | 57.4 | 222 | 3.63 | 750 | 2020 | An Giang |
| EI54 | E. ictaluri | E. ictaluri | 26 | 57.4 | 201 | 3.65 | 25 | 2020 | Can Tho |
| EI55 | E. ictaluri | E. ictaluri | 26 | 57.4 | 183 | 3.63 | 520 | 2020 | Dong Thap |
| EI58 | E. ictaluri | E. ictaluri | 26 | 57.4 | 185 | 3.62 | 250 | 2020 | Dong Thap |
| EI59 | E. ictaluri | E. ictaluri | 26 | 57.3 | 193 | 3.74 | 50 | 2021 | Dong Thap |
| EI60 | E. ictaluri | E. ictaluri | 26 | 57.7 | 186 | 3.56 | 800 | 2021 | Dong Thap |
| EI62 | E. ictaluri | E. ictaluri | 26 | 57.4 | 188 | 3.67 | 400 | 2021 | Dong Thap |
| EI65 | E. ictaluri | E. ictaluri | 26 | 57.4 | 183 | 3.67 | 180 | 2021 | Ben Tre |
| EI67 | E. ictaluri | E. ictaluri | 26 | 57.3 | 192 | 3.70 | 45 | 2021 | Ben Tre |
| EI71 | E. ictaluri | E. ictaluri | 26 | 57.4 | 183 | 3.55 | 60 | 2021 | Can Tho |
| EI72 | E. ictaluri | E. ictaluri | 26 | 57.4 | 203 | 3.66 | 25 | 2021 | Long An |
| Isolate | Plasmid | Coverage (%) | Identity (%) | Plasmid Accession |
|---|---|---|---|---|
| EI04 | IncA/C2 | 100 | 92.1 | JN157804 |
| EI12 | IncA/C2 | 100 | 92.1 | JN157804 |
| p0111 | 100 | 98.5 | AP010962 | |
| EI18 | IncA/C2 | 100 | 92.1 | JN157804 |
| p0111 | 100 | 98.5 | AP010962 | |
| EI19 | IncA/C2 | 100 | 92.1 | JN157804 |
| EI48 | p0111 | 100 | 98.5 | AP010962 |
| IncA/C2 | 100 | 92.1 | JN157804 | |
| IncQ1 | 93.9 | 89.9 | M28829.1 | |
| EI51 | IncA/C2 | 100 | 92.1 | JN157804 |
| p0111 | 100 | 98.5 | AP010962 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Erickson, V.I.; Dung, T.T.; Khoi, L.M.; Hounmanou, Y.M.G.; Phu, T.M.; Dalsgaard, A. Genomic Insights into Edwardsiella ictaluri: Molecular Epidemiology and Antimicrobial Resistance in Striped Catfish (Pangasianodon hypophthalmus) Aquaculture in Vietnam. Microorganisms 2024, 12, 1182. https://doi.org/10.3390/microorganisms12061182
Erickson VI, Dung TT, Khoi LM, Hounmanou YMG, Phu TM, Dalsgaard A. Genomic Insights into Edwardsiella ictaluri: Molecular Epidemiology and Antimicrobial Resistance in Striped Catfish (Pangasianodon hypophthalmus) Aquaculture in Vietnam. Microorganisms. 2024; 12(6):1182. https://doi.org/10.3390/microorganisms12061182
Chicago/Turabian StyleErickson, Vera Irene, Tu Thanh Dung, Le Minh Khoi, Yaovi Mahuton Gildas Hounmanou, Tran Minh Phu, and Anders Dalsgaard. 2024. "Genomic Insights into Edwardsiella ictaluri: Molecular Epidemiology and Antimicrobial Resistance in Striped Catfish (Pangasianodon hypophthalmus) Aquaculture in Vietnam" Microorganisms 12, no. 6: 1182. https://doi.org/10.3390/microorganisms12061182
APA StyleErickson, V. I., Dung, T. T., Khoi, L. M., Hounmanou, Y. M. G., Phu, T. M., & Dalsgaard, A. (2024). Genomic Insights into Edwardsiella ictaluri: Molecular Epidemiology and Antimicrobial Resistance in Striped Catfish (Pangasianodon hypophthalmus) Aquaculture in Vietnam. Microorganisms, 12(6), 1182. https://doi.org/10.3390/microorganisms12061182






