Use of PCR Cycle Threshold and Clinical Interventions to Aid in the Management of Pediatric Clostridioides difficile Patients
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics
2.2. Use of PCR Ct Value
Specimen Collection and Study Design
2.3. Clinical Review
2.4. C. difficile Laboratory Diagnostic Tests
2.5. Statistical Analysis
2.6. Clinical Intervention
3. Results
3.1. Use of PCR Ct Value
3.2. Clinical Intervention
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alcalá, L.; Reigadas, E.; Marín, M.; Martín, A.; Catalán, P.; Bouza, E. Impact of Clinical Awareness and Diagnostic Tests on the Underdiagnosis of Clostridium Difficile Infection. Eur. J. Clin. Microbiol. Infect. Dis. 2015, 34, 1515–1525. [Google Scholar] [CrossRef] [PubMed]
- Antonara, S.; Leber, A.L. Diagnosis of Clostridium Difficile Infections in Children. J. Clin. Microbiol. 2016, 54, 1425–1433. [Google Scholar] [CrossRef] [PubMed]
- McDonald, L.C.; Gerding, D.N.; Johnson, S.; Bakken, J.S.; Carroll, K.C.; Coffin, S.E.; Dubberke, E.R.; Garey, K.W.; Gould, C.V.; Kelly, C.; et al. Clinical Practice Guidelines for Clostridium Difficile Infection in Adults and Children: 2017 Update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA). Clin. Infect. Dis. 2018, 66, e1–e48. [Google Scholar] [CrossRef] [PubMed]
- Sammons, J.S.; Localio, R.; Xiao, R.; Coffin, S.E.; Zaoutis, T. Clostridium difficile Infection Is Associated with Increased Risk of Death and Prolonged Hospitalization in Children. Clin. Infect. Dis. 2013, 57, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Mehrotra, P.; Jang, J.; Gidengil, C.; Sandora, T.J. Attributable Cost of Clostridium Difficile Infection in Pediatric Patients. Infect. Control Hosp. Epidemiol. 2017, 38, 1472–1477. [Google Scholar] [CrossRef] [PubMed]
- Marra, F.; Ng, K. Controversies Around Epidemiology, Diagnosis and Treatment of Clostridium difficile Infection. Drugs 2015, 75, 1095–1118. [Google Scholar] [CrossRef]
- Schwenk, H.T.; Pollock, N.R.; Vaughan-Malloy, A.M. Pediatric Clostridioides difficile Infection: Diagnosis and Diagnostic Stewardship. J. Pediatr. Infect. Dis. Soc. 2021, 10, S16–S21. [Google Scholar] [CrossRef]
- Gnocchi, M.; Gagliardi, M.; Gismondi, P.; Gaiani, F.; De’ Angelis, G.L.; Esposito, S. Updated Management Guidelines for Clostridioides difficile in Paediatrics. Pathogens 2020, 9, 291. [Google Scholar] [CrossRef]
- Schwenk, H.T.; Bio, L.L.; Kruger, J.F.; Banaei, N. Clinical Impact of Clostridium difficile PCR Cycle Threshold-Predicted Toxin Reporting in Pediatric Patients. J. Pediatr. Infect. Dis. Soc. 2019, 9, 44–50. [Google Scholar] [CrossRef]
- Polage, C.R.; Gyorke, C.E.; Kennedy, M.A.; Leslie, J.L.; Chin, D.L.; Wang, S.; Nguyen, H.H.; Huang, B.; Tang, Y.-W.; Lee, L.W.; et al. Overdiagnosis of Clostridium difficile Infection in the Molecular Test Era. JAMA Intern. Med. 2015, 175, 1792–1801. [Google Scholar] [CrossRef]
- Longtin, Y.; Trottier, S.; Brochu, G.; Paquet-Bolduc, B.; Garenc, C.; Loungnarath, V.; Beaulieu, C.; Goulet, D.; Longtin, J. Impact of the Type of Diagnostic Assay on Clostridium difficile Infection and Complication Rates in a Mandatory Reporting Program. Clin. Infect. Dis. 2013, 56, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Kociolek, L.K.; Bovee, M.; Carter, D.; Ciolino, J.D.; Patel, R.; O’Donnell, A.; Rupp, A.H.; Zheng, X.; Shulman, S.T.; Patel, S.J. Impact of a Healthcare Provider Educational Intervention on Frequency of Clostridium difficile Polymerase Chain Reaction Testing in Children: A Segmented Regression Analysis. J. Pediatr. Infect. Dis. Soc. 2017, 6, 142–148. [Google Scholar] [CrossRef]
- Senchyna, F.; Gaur, R.L.; Gombar, S.; Truong, C.Y.; Schroeder, L.F.; Banaei, N. Clostridium difficile PCR Cycle Threshold Predicts Free Toxin. J. Clin. Microbiol. 2017, 55, 2651–2660. [Google Scholar] [CrossRef] [PubMed]
- Rao, K.; Micic, D.; Natarajan, M.; Winters, S.; Kiel, M.J.; Walk, S.T.; Santhosh, K.; Mogle, J.A.; Galecki, A.T.; LeBar, W.; et al. Clostridium difficile Ribotype 027: Relationship to Age, Detectability of Toxins A or B in Stool with Rapid Testing, Severe Infection, and Mortality. Clin. Infect. Dis. 2015, 61, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Sante, L.; Pedroso, Y.; Castro, B.; Lecuona, M. ¿Existe Relación Entre el Ciclo Umbral de la Reacción En Cadena de la Polimerasa y el Riesgo de Infección Grave por Clostridium difficile? Enfermedades Infecc. Microbiol. Clínica 2018, 36, 600–601. [Google Scholar] [CrossRef]
- Bruijnesteijn van Coppenraet, L.E.S.; Dullaert-de Boer, M.; Ruijs, G.J.H.M.; van der Reijden, W.A.; van der Zanden, A.G.M.; Weel, J.F.L.; Schuurs, T.A. Case-Control Comparison of Bacterial and Protozoan Microorganisms Associated with Gastroenteritis: Application of Molecular Detection. Clin. Microbiol. Infect. 2015, 21, 592.e9–592.e19. [Google Scholar] [CrossRef] [PubMed]
- Davies, K.A.; Planche, T.; Wilcox, M.H. The Predictive Value of Quantitative Nucleic Acid Amplification Detection of Clostridium difficile Toxin Gene for Faecal Sample Toxin Status and Patient Outcome. PLoS ONE 2018, 13, e0205941. [Google Scholar] [CrossRef]
- Kamboj, M.; Brite, J.; McMillen, T.; Robilotti, E.; Herrera, A.; Sepkowitz, K.; Babady, N.E. Potential of Real-Time PCR Threshold Cycle (CT) to Predict Presence of Free Toxin and Clinically Relevant C. difficile Infection (CDI) in Patients with Cancer. J. Infect. 2018, 76, 369–375. [Google Scholar] [CrossRef]
- De Francesco, M.A.; Lorenzin, G.; Piccinelli, G.; Corbellini, S.; Bonfanti, C.; Caruso, A. Correlation between tcdB Gene PCR Cycle Threshold and Severe Clostridium difficile Disease. Anaerobe 2019, 59, 141–144. [Google Scholar] [CrossRef]
- Reigadas, E.; Alcalá, L.; Valerio, M.; Marín, M.; Martin, A.; Bouza, E. Toxin B PCR Cycle Threshold as a Predictor of Poor Outcome of Clostridium difficile Infection: A Derivation and Validation Cohort Study. J. Antimicrob. Chemother. 2016, 71, 1380–1385. [Google Scholar] [CrossRef]
- Jazmati, N.; Hellmich, M.; Ličanin, B.; Plum, G.; Kaasch, A.J. PCR Cycle Threshold Value Predicts the Course of Clostridium difficile Infection. Clin. Microbiol. Infect. 2016, 22, e7–e8. [Google Scholar] [CrossRef] [PubMed]
- Hitchcock, M.M.; Holubar, M.; Hogan, C.A.; Tompkins, L.S.; Banaei, N. Dual Reporting of Clostridioides difficile PCR and Predicted Toxin Result Based on PCR Cycle Threshold Reduces Treatment of Toxin-Negative Patients without Increases in Adverse Outcomes. J. Clin. Microbiol. 2019, 57, e01288-19. [Google Scholar] [CrossRef] [PubMed]
- Pahud, B.A.; Hassan, F.; Harrison, C.J.; Halasa, N.B.; Chappell, J.D.; Englund, J.A.; Klein, E.J.; Szilagyi, P.G.; Weinberg, G.A.; Sherman, A.K.; et al. Detection of Clostridioides difficile by Real-Time Pcr in Young Children Does Not Predict Disease. Hosp. Pediatr. 2020, 10, 555–562. [Google Scholar] [CrossRef] [PubMed]
- Sandora, T.J.; Williams, D.N.; Daugherty, K.; Geer, C.; Cuddemi, C.; Kociolek, L.K.; Chen, X.; Xu, H.; Savage, T.J.; Banz, A.; et al. Stool Toxin Concentration Does Not Distinguish Clostridioides Difficile Infection from Colonization in Children Less Than 3 Years of Age. J. Pediatr. Infect. Dis. Soc. 2022, 11, 454–458. [Google Scholar] [CrossRef]
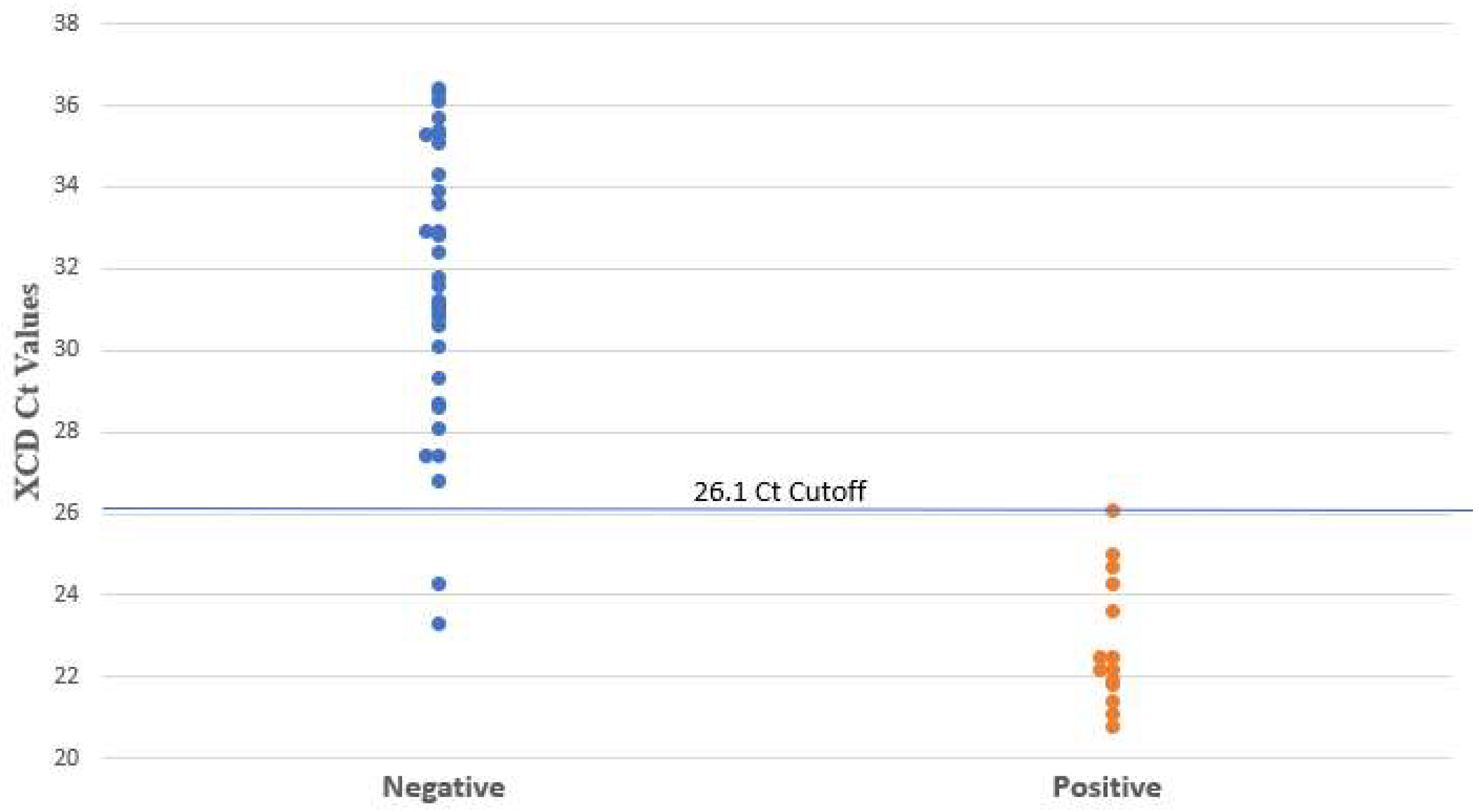
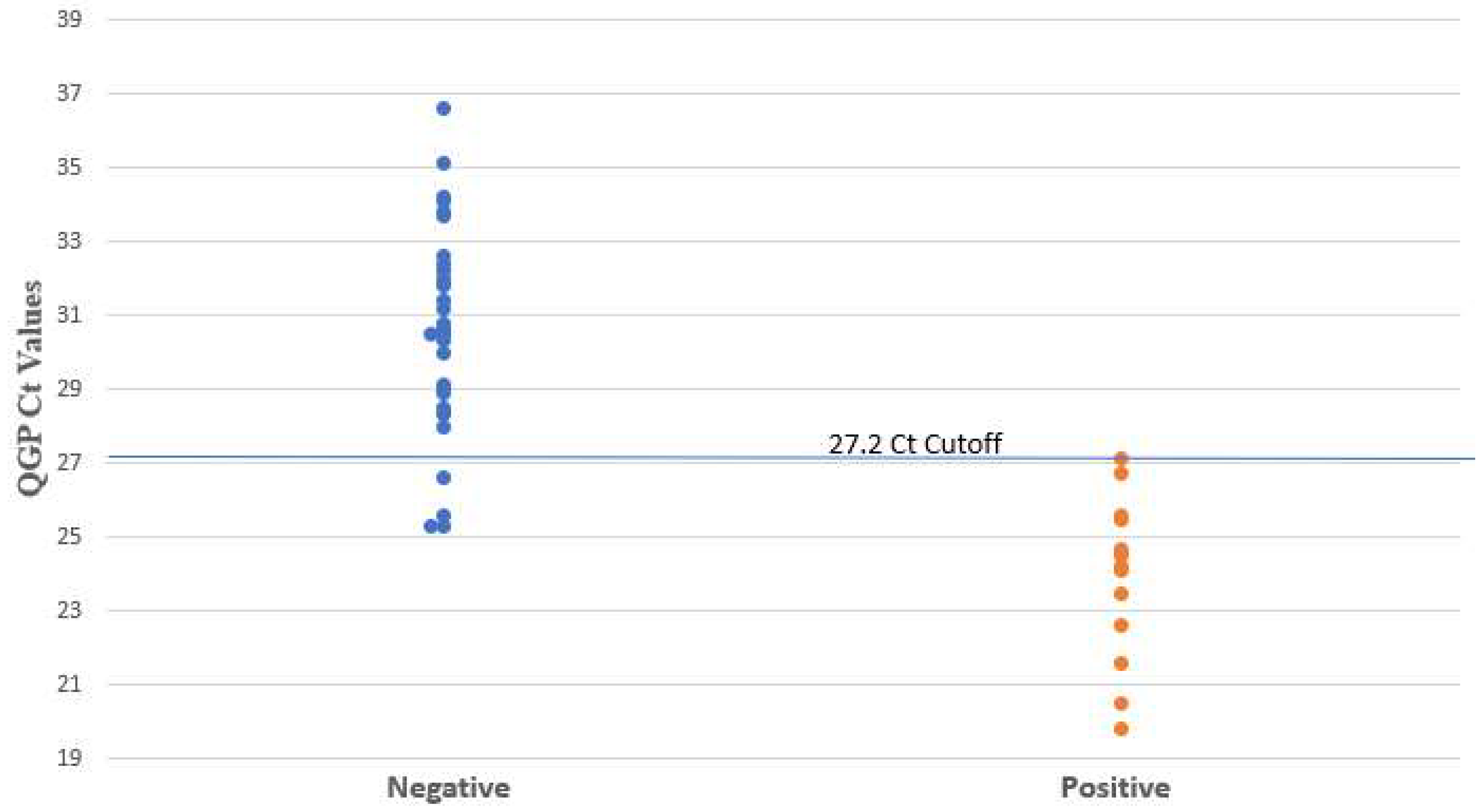
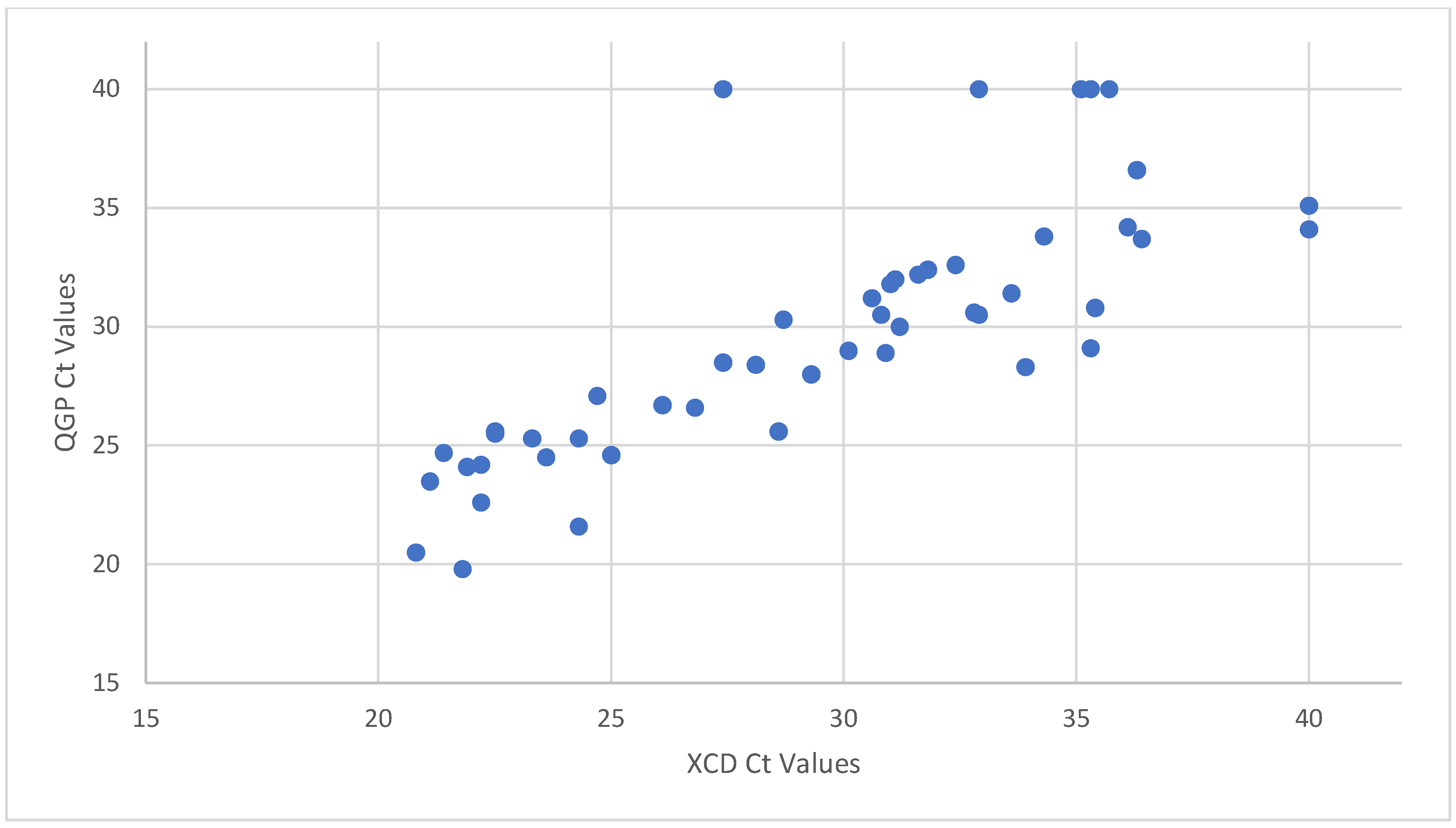
| Method | Ct Cutoff | Reference | TP | TN | FP | FN | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) |
|---|---|---|---|---|---|---|---|---|---|---|
| XCD | 26.1 | Clinical review | 14 | 33 | 2 | 0 | 100 (78–100) | 94 (81–98) | 88 (64–97) | 100 (90–100) |
| QGP | 27.2 | 14 | 31 | 4 | 0 | 100 (78–100) | 89 (74–95) | 78 (54–91) | 100 (89–100) | |
| Toxin A/B EIA | N/A | 9 | 35 | 0 | 5 | 64 (39–84) | 100 (90–100) | 100 (70–100) | 88 (74–96) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Suleiman, M.; Tang, P.; Imam, O.; Morales, P.; Altrmanini, D.; Barr, K.L.; Roberts, J.C.; Pérez-López, A. Use of PCR Cycle Threshold and Clinical Interventions to Aid in the Management of Pediatric Clostridioides difficile Patients. Microorganisms 2024, 12, 1181. https://doi.org/10.3390/microorganisms12061181
Suleiman M, Tang P, Imam O, Morales P, Altrmanini D, Barr KL, Roberts JC, Pérez-López A. Use of PCR Cycle Threshold and Clinical Interventions to Aid in the Management of Pediatric Clostridioides difficile Patients. Microorganisms. 2024; 12(6):1181. https://doi.org/10.3390/microorganisms12061181
Chicago/Turabian StyleSuleiman, Mohammed, Patrick Tang, Omar Imam, Princess Morales, Diyna Altrmanini, Kelli L. Barr, Jill C. Roberts, and Andrés Pérez-López. 2024. "Use of PCR Cycle Threshold and Clinical Interventions to Aid in the Management of Pediatric Clostridioides difficile Patients" Microorganisms 12, no. 6: 1181. https://doi.org/10.3390/microorganisms12061181
APA StyleSuleiman, M., Tang, P., Imam, O., Morales, P., Altrmanini, D., Barr, K. L., Roberts, J. C., & Pérez-López, A. (2024). Use of PCR Cycle Threshold and Clinical Interventions to Aid in the Management of Pediatric Clostridioides difficile Patients. Microorganisms, 12(6), 1181. https://doi.org/10.3390/microorganisms12061181







