Abstract
Cytochrome P450 monooxygenases (CYPs/P450s) are heme thiolate proteins present in species across the biological kingdoms. By virtue of their broad substrate promiscuity and regio- and stereo-selectivity, these enzymes enhance or attribute diversity to secondary metabolites. Actinomycetes species are well-known producers of secondary metabolites, especially Salinispora species. Despite the importance of P450s, a comprehensive comparative analysis of P450s and their role in secondary metabolism in Salinispora species is not reported. We therefore analyzed P450s in 126 strains from three different species Salinispora arenicola, S. pacifica, and S. tropica. The study revealed the presence of 2643 P450s that can be grouped into 45 families and 103 subfamilies. CYP107 and CYP125 families are conserved, and CYP105 and CYP107 families are bloomed (a P450 family with many members) across Salinispora species. Analysis of P450s that are part of secondary metabolite biosynthetic gene clusters (smBGCs) revealed Salinispora species have an unprecedented number of P450s (1236 P450s-47%) part of smBGCs compared to other bacterial species belonging to the genera Streptomyces (23%) and Mycobacterium (11%), phyla Cyanobacteria (8%) and Firmicutes (18%) and the classes Alphaproteobacteria (2%) and Gammaproteobacteria (18%). A peculiar characteristic of up to six P450s in smBGCs was observed in Salinispora species. Future characterization Salinispora species P450s and their smBGCs have the potential for discovering novel secondary metabolites.
1. Introduction
Cytochrome P450 monooxygenases (CYPs/P450s) comprise a superfamily of heme-thiolate proteins. P450s are present in all species of different biological kingdoms, including in viruses considered non-living entities [1,2]. This suggests that these enzymes play an important role in species’ primary and secondary metabolism. These enzymes were initially identified as monooxygenases due to their ability to introduce one oxygen atom into a substrate [3]. Subsequent research revealed that P450s are catalytically diverse enzymes performing some unusual enzymatic reactions [4,5,6,7,8]. The regio- and stereo-specific oxidation of many substrates by P450s caught the attention of researchers for biotechnological exploration of these enzymes [9,10,11,12]. P450s reactions are essential in designing drugs such that drug toxicity of prodrugs is primarily assessed against these enzymes [13]. Also, P450s play a vital role in xenobiotic compounds’ detoxification [14]. Microbial P450s, especially from lower eukaryotes such as fungal CYP51, have been used as an azole drug target [15,16]. The study also suggested that fungal CYP53 can act as a potential alternative drug target [17]. One of the best examples of P450s biotechnological applications includes the synthesis of antibiotics and anticancer drugs [18,19,20,21].
The utilization of P450s in the generation of secondary metabolites or natural products, organic compounds not directly involved in an organism’s normal growth, development, or reproduction, is gaining momentum as reactions catalyzed by these enzymes contribute to the secondary metabolite diversity [22,23]. Secondary metabolites, their structural diversity, bioactivity, and ecological functions, including their application in almost all areas of biology, have been thoroughly reviewed [24,25,26,27,28,29]. For example, secondary metabolites are widely used in human and veterinary medicine, agriculture, and manufacturing [30].
Secondary metabolites in organisms are produced by a set of genes usually located next to each other as a cluster known as secondary metabolite biosynthetic gene cluster (smBGCs) [30,31]. Earlier, researchers used to clone and sequence smBGCs to identify the genes/proteins involved in producing a particular secondary metabolite. The onset of genome sequencing and the advancement of science, especially in bioinformatics, led to the development of software programs that can automatically detect smBGCs [32]. Due to this advancement, many smBGCs were reported in species belonging to different biological kingdoms [30,31,33].
In the bacterial kingdom, species belonging to the phylum Actinobacteria are well-known for producing secondary metabolites [33,34,35,36], especially species of the genus Streptomyces [37]. It is a well-known fact that two-thirds of the clinically valuable antibiotics come from Streptomyces species [37]. Actinomycetes belonging to the genus Salinispora produce biotechnologically valuable secondary metabolites [38,39,40,41,42,43,44,45,46,47]. Salinosporamide A, a secondary metabolite, is one of the best examples, which is now under clinical trials as an anticancer drug [48].
Salinispora is the first genus of Actinobacteria identified for its requirement of seawater for growth [49]. This genus includes three distinct but closely related species Salinispora arenicola, S. pacifica, and S. tropica [36,50,51]. Salinispora species are widely distributed in tropical and subtropical marine environments with distinct geographical patterns [49,52]. The genome sequence of S. tropica revealed a large percentage of its genome (~9.9%) is dedicated to natural products biosynthesis, which was greater than any other natural product producing actinomycetes [47]. The genome sequencing analysis revealed that P450s were also part of smBGCs [47]. CYP107 from S. arenicola CNS-205 is involved in the biosynthesis of secondary metabolites, saliniketal, and rifampicin [53]. Apart from these notable mentions, no information is available on Salinispora species P450s.
Despite knowing that Salinispora species produce different types of human valuable secondary metabolites/natural products and the role of P450s in attributing diversity to these compounds, to date, comparative analysis of P450s and their role in secondary metabolism in Salinispora species is not reported. This study is aimed to address this research gap by performing genome-wide data mining, identification, annotation (assigning family and subfamily), and phylogenetic analysis of P450s in Salinispora species. The study also encompasses identification of P450s part of smBGCs, and comparative analysis of Salinispora P450 features with other bacterial species belonging to the genera, Streptomyces and Mycobacterium, phyla Firmicutes and Bacteroidetes, and the classes Alpha- and Gamma-proteobacteria.
2. Materials and Methods
2.1. Species and Database Information
A total of 126 Salinispora species genomes (permanent and finished draft genomes) are available for public use at the Joint Genome Institute Integrated Microbial Genomes and Microbiomes (JGI IMG/M) [54,55] were used in this study (last accessed on 2 February 2022). Information on the species and their genome IDs used in the study is provided in Table S1.
2.2. Genome Data Mining and Identification of P450s
Genome data mining and identification of P450s in Salinispora species were carried out following the protocol described elsewhere [56,57]. Each Salinispora species genome available at JGI IMG/M [54,55] was searched for P450s using the InterPro code “IPR001128”. The hit protein sequences were then searched for the presence of P450 characteristic motifs such as EXXR and CXG [58,59]. Proteins with one of these motifs or short amino acid length are considered P450-fragments. P450 fragments were not considered for the final P450 family and subfamily count.
2.3. Assigning Family and Subfamily to P450s
Above selected P450s were assigned to different families and subfamilies based on the International P450 Nomenclature Committee rule [60,61,62], proteins with a percentage identity greater than 40% were assigned to the same family as named homolog P450s, and those that had greater than 55% identity were assigned to the same subfamily as named homolog P450s. Proteins with a percentage identity of less than 40% were assigned to a new family. Salinispora species P450s, along with P450-fragments, are presented in Table S2.
2.4. Phylogenetic Analysis of P450s
Phylogenetic analysis of P450s was carried out following the procedure described elsewhere [63,64]. The phylogenetic tree of P450s was constructed using protein sequences. Firstly, the MAFFT v6.864 [65] was used to align the Trex web server’s protein sequences [66]. The alignments were then used to interpret the best tree by the Trex web server [66]. Finally, the best-inferred tree was visualized, colored, and generated by a web-based tool, VisuaLife [67].
2.5. Salinispora Species P450s Profile Heat-Maps
P450 profile heat-maps were generated following a method described elsewhere [64,68] to check the presence and absence of or co-presence of or conserved nature of P450 families in Salinispora species. Briefly, a tab-delimited file was imported into Multi-Experiment Viewer (Mev) [69], and hierarchical clustering using a Euclidean distance metric was used to cluster the data. 126 Salinispora species formed the vertical axis, and P450 families formed the horizontal axis. Data were presented as −3 for family absence (green) and 3 for family presence (red).
2.6. Identification of P450s Part of smBGCs
P450s that are part of smBGCs were identified following the method described elsewhere [56,57]. Briefly, for each Salinispora species genome available at JGI IMG/M [54,55], the smBGCs were searched for the presence of P450s using the P450 gene ID. The cluster type is noted if a P450 is found as part of the cluster. Results were recorded on Excel spreadsheets and represented species-wise smBGCs, smBGC type, and P450s part of specific smBGCs. Among 126, only 103 Salinispora species smBGCs information is available at JGI IMG/M [54,55]. Thus the same 103 Salinispora species smBGCs were analyzed for the presence of P450s (Table S1).
2.7. Data Analysis
All calculations were carried out following the procedure reported previously by our laboratory [68]. The average number of P450s was calculated using the formula: Average number of P450s = Number of P450s/Number of species. The P450 diversity percentage was calculated using the formula: P450 diversity percentage = 100 × Total number of P450 families/Total number of P450s × Number of species with P450s. The percentage of P450s that formed part of BGCs was calculated using the formula: Percentage of P450s part of BGCs = 100 × Number of P450s part of BGCs/Total number of P450s present in species.
2.8. Comparative Analysis of P450s and smBGCs Data
For comparative analysis of P450s and smBGCs, information for bacterial species belonging to different groups such as classes, Alpha- and Gamma-proteobacteria [64,68], phyla, Firmicutes [70] and Cyanobacteria [71], and the genera, Streptomyces [56,72], Mycobacterium [72,73], was resourced from published articles.
3. Results and Discussion
3.1. Salinispora Species P450 Profiles
Genome-wide data mining and annotation of P450s in 126 Salinispora species revealed the presence of 2643 P450s in their genomes (Figure 1, Table 1 and Table 2). The P450 count in Salinispora species ranged from 10 to 35 P450s, with an average of 21 P450s (Table 1 and Table 2). Apart from the complete P450 sequences, 129 P450 fragments were also found in some Salinispora species (Table 2). P450 fragments in species are natural [58,70,74], and thus, these were excluded from further analysis. Among Salinispora species, S. arenicola CNY280 has the highest number of P450s (35 P450s), and S. pacifica CNS801 and S. pacifica CNT148 have the lowest number of P450s (10 P450s each) (Table 2). Comparative analysis revealed that Salinispora species have the highest average number of P450s than species belonging to Cyanobacteria, Firmicutes, Alphaproteobacteria, and Gammaproteobacteria (Table 1). However, Salinispora species had the lowest average number of P450s compared to species belonging to Streptomyces and Mycobacterium (Table 1). A point to be noted is that, among bacterial species, species belonging to the phylum Actinobacteria have the highest average number of P450s (Table 1). This indicates selective enrichment of P450s in these species due to their adaptation to ecological niches vis a vis P450s, helping them adapt to diverse ecological niches described elsewhere [58,74,75]. Salinispora species P450s, along with P450-fragments, are presented in Table S2.
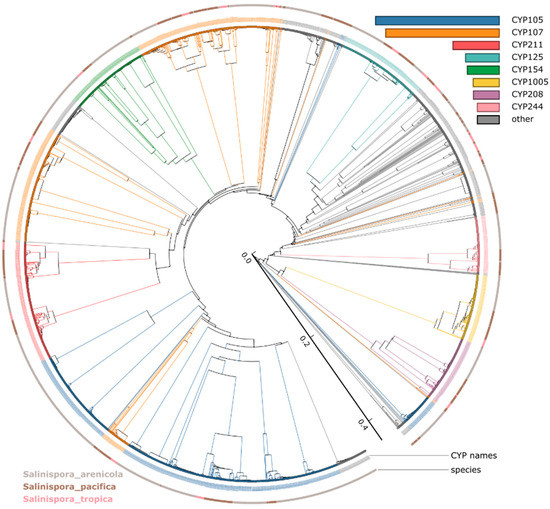
Figure 1.
Phylogenetic analysis of Salinispora species P450s. 2643 P450s were used to construct the tree, and the members of the eight most abundant P450 families are highlighted in different colors and indicated in the figure. P450 protein sequences used to build the tree are listed in Table S2. A high-resolution phylogenetic tree is provided in Figure S1.

Table 1.
Comparative analysis of key features of P450s and their association with secondary metabolism between Salinispora species and different bacterial species. Abbreviation: No., number of; BGCs: biosynthetic gene clusters.

Table 2.
Genome-wide data mining and annotation of P450s in 126 Salinispora species. Abbreviation, No. indicates the number in the table.
3.2. CYP105 and CYP107 Families Are Bloomed in Salinispora Species
Based on the International P450 Nomenclature Committee Rules [60,61,62], all 2643 P450s can be grouped into 45 families and 103 subfamilies (Table 1 and Table 3). Phylogenetic analysis revealed that large P450 families CYP105 and CYP107 were scattered across the evolutionary tree (Figure 1). Previously, this phenomenon was observed for these P450 families [56,72]. Authors suggested that phylogenetic-based annotation of P450s could detect similarity cues beyond a simple percentage identity cutoff [56,72]. Except for CYP105 and CYP107, the rest of the P450s are grouped as per their families (Figure 1). A point to be noted is that most of the P450s are orthologs considering the Salinispora species analyzed in this study are different strains of three species. Comparative analysis revealed that Salinispora species have the lowest number of P450 families and subfamilies compared to other actinomycetes such as Streptomyces and Mycobacterium (Table 1).

Table 3.
Comparative analysis of P450 families and subfamilies in Salinispora species.
Among Salinispora species, S. arenicola CNY280 had the highest number of P450 families (18) and P450 subfamilies (32) in its genome (Table 1). This is quite an interesting observation where a species with the highest number of P450s also had the highest number of P450 families and subfamilies. This phenomenon was not found in other actinomycetes such as Streptomyces [56] and Mycobacterium [72,73]. For example, in Streptomyces species, Streptomyces albulus ZPM had the highest number of P450s, but Streptomyces rimosus rimosus ATCC 10970, and Streptomyces clavuligerus had the highest number of P450 families and subfamilies, respectively [56]. Among mycobacterial species, Mycobacterium rhodesiae NBB3 had the highest P450s and P450 families, but M. marinum had the highest P450 subfamilies [72,73].
Analysis of P450 families and subfamilies suggested that P450s in Salinispora species bloomed (presence of more copies of the same P450 family in a species by duplication of an ancestral gene) (Table 3). Among P450 families, the CYP105 was dominant with 600 members, followed by CYP107 with 551 members, CYP211 with 225 members, CYP125 with 164 members, CYP154 with 155 members, CYP1005 with 127 members, and CYP208 with 126 members (Table 3). These P450 families contributed more than 70% to the total P450s (Table 3). This indicates that P450 families such as CYP105, CYP107, CYP211, CYP125, and CYP154 are bloomed, whereas CYP1005 and CYP208 families are expanded in these species. Comparing the dominant P450 families revealed that CYP105 is prevalent only in Salinispora species (Table 1), where this family was second most dominant in Streptomyces species (Table 1). Interestingly, the second most dominant P450 family of Salinispora species, CYP107, was dominant in species belonging to bacterial groups Streptomyces, Firmicutes and Gammaproteobacteria (Table 1). The blooming was also observed at the subfamily level, indicating these P450s are preferred by Salinispora species for a particular reason. For example, subfamily AB was dominant with 124 members in CYP105; Subfamily AY was dominant with 116 members in CYP107, subfamily A was dominant with 128 members in CYP125, Subfamily M was dominant with 150 members, subfamily A was dominant with 126 members in CYP208, and Subfamily B dominant with 124 members in CYP211 (Table 3). Due to the blooming of specific P450s at the family level, Salinispora species had the lowest P450 diversity percentage, the same as Firmicutes species (Table 1). The blooming or expansion of P450s is a common phenomenon in organisms and is observed in other bacterial species (Table 2). It has been hypothesized that species enrich specific P450s in their genomes that are beneficial to them, particularly to adapt to ecological niches [56,72].
3.3. CYP107 and CYP125 Are Conserved in Salinispora Species
P450 family conservation analysis revealed that CYP107 and CYP125 families are conserved in 126 Salinispora species (Figure 2). Except for a few species, CYP208 (4 species), CYP105 (one species), CYP211 (one species), and CYP1005 (2 species), the rest of the Salinispora species have these families (Figure 2). In addition to this, P450 families such as CYP154, CYP244, CYP245, CYP166, CYP248, and CYP1056 are co-present in many species (Figure 2). This suggests a prominent role of these P450 families in these species, possibly in secondary metabolism as observed in other bacterial species [58,72,74]. Conservation or co-presence of specific P450s in other bacterial species was also reported. The CYP107 family is conserved in all 203 Streptomyces species, and P450 families such as CYP156, CYP105, CYP154, and CYP157 are also present in the majority of the Streptomyces species [56]. Ten P450 families, CYP51, CYP123, CYP125, CYP130, CYP135, CYP136, CYP138, CYP140, CYP144, and CYP1128, were conserved in mycobacterial species [73]. Analysis of conservation of P450 families in 229 Firmicutes species and 114 cyanobacterial species revealed no conservation of the P450 family [70,71]. Still, some of the P450 families were co-present in most of the species. The P450 families CYP152, CYP107, CYP012, and CYP109, were found to be a co-presence in most Firmicutes species [70], and the P450 families CYP110 and CYP120 were found to be a co-presence in most cyanobacterial species [71].
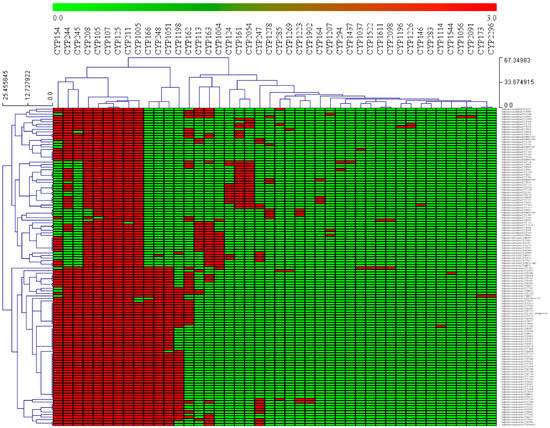
Figure 2.
Heat-map of P450 family conservation or co-presence analysis in Salinispora species. In the heat-map, the presence and absence of P450 families are indicated in red and green colors. The horizontal axis represents P450 families, and the vertical axis represents Salinispora species.
If a P450 family is conserved or few P450 families are co-presence, these families play an important role in a species’s primary- or secondary-metabolism. Previous studies showed that this type of P450s prominently plays a role in secondary metabolism, helping species adapt to diverse ecological niches [58,59,72,74,75]. The importance of P450 families that are conserved and co-presence in Salinispora species is discussed in detail in the next section.
3.4. Unprecedented Number of P450s Involved in smBGCs
Analysis of the P450s part of smBGCs revealed that many P450s (47%) are part of these clusters, indicating their involvement in producing different secondary metabolites in Salinispora species (Table 4 and Table S1). The percentage of P450s part of smBGCs in Salinispora species was found to be unprecedented compared to other bacterial species, including other actinomycetes Streptomyces species and mycobacterial species that had 30% and 27% of P450s as part of smBGCs (Table 1). This suggests that Salinispora species dedicated half of their P450s to the production of secondary metabolites.

Table 4.
Secondary metabolite biosynthetic gene cluster (smBGC) types and P450s are part of the cluster in Salinispora species. smBGC types were again classified into different varieties based on the P450s. The smBGCs type count and the total number of P450s in the cluster variety are also presented. The same smBGCs type names listed in the antibiotics and secondary metabolite analysis shell (anti-SMASH) database [74] were used in the table. Detailed information on secondary metabolite clusters, species, and P450s are shown in Table S1.
Among 2643 P450s, 1236 P450s belonging to the 35 P450 families were part of smBGCs (Figure 3 and Table 4 and Table S1). This means almost 78% of P450 families of Salinispora species are involved in secondary metabolism. Among the families that are part of smBGCs, CYP107 is dominant with 302 members (25%), followed by CYP105 with 220 members (18%), CYP208 with 87 members (7%), CYP244 with 79 members (7%), and CYP211 with 73 members (6%) (Figure 3 and Table S1). Analysis of the P450s part of smBGCs revealed a strong correlation between the dominant P450 families (Table 3) being dominant in smBGCs (Figure 3). This suggests that Salinispora species are enriched by blooming or expanding these P450 families (as discussed in the previous section) in their genome to produce secondary metabolites.
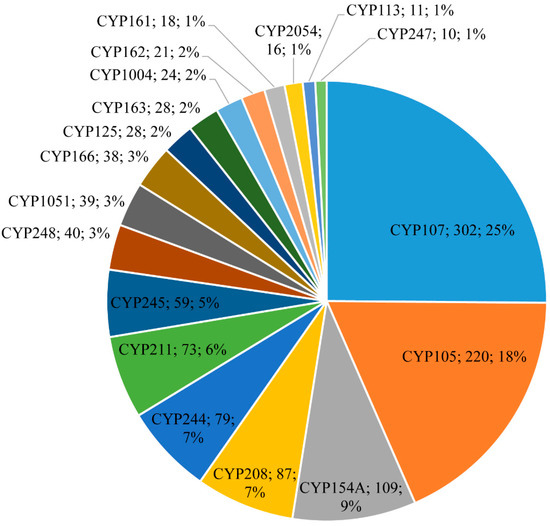
Figure 3.
Comparative analysis of P450s associated with secondary metabolism in Salinispora species. The P450 family name, number of P450s, and the percentage of the total number of P450s that are part of secondary metabolite biosynthetic gene clusters (smBGCs) are presented in the figure. Detailed information on secondary metabolite clusters, species, and P450s are shown in Table S1.
Analysis of P450 smBGCs revealed the presence of 18 types (Table 4 and Table S2). Among the types, Type I PKS (Polyketide synthase) (T1PKS) was dominant with 223 clusters, followed by nonribosomal peptides (NRPS) (205 clusters) and Type II PKS (T2PKS) (76 clusters) (Table 4 and Table S1). This suggests that most of the secondary metabolites produced by P450 smBGCs are T1PKS. When the P450 smBGCs were further analyzed for the number of P450s and P450 families, the dominant BGC type was not found to be dominant concerning the number of P450s being part of that smBGC type (Table 4 and Table S1). NRPS had the highest number of P450s (395 P450s), followed by T1PKS (275 P450s), oligosaccharide (121 P450s), and indole (105 P450s) (Table 4 and Table S1). The difference being not having more P450s despite being dominant smBGCs such as T1PKS is that the other smBGCs have more P450s per se more than one P450 being part of that type (Table 4 and Table S1). This phenomenon of more than one P450 being part of smBGCs has been reported earlier in other bacterial species [75]. However, having up to 6 P450s as part of smBGCs is unprecedented (Table 4), suggesting these clusters produce diverse secondary metabolites. The P450s co-present in different Salinispora species were part of the same cluster (Table 4). Based on the arrangement of P450s concerning their family/subfamily and the number of P450s in smBGCs, it is clear that these smBGCs are orthologs (Table 4). These smBGCs are passed into different Salinispora species from a single ancestor before diverging into S. arenicola, S. pacifica and S. tropica.
3.5. Functional Prediction of Salinispora Species P450s
Most of the Salinispora species P450s are orphans without an assigned biological function. Based on the homolog P450s from other organisms and being part of smBGCs, some P450 functions can be predicted. CYP105 and CYP107 members are involved in the degradation/biotransformation of xenobiotics and biosynthesis of secondary metabolites [76,77,78,79,80]. CYP107 from S. arenicola CNS-205 is involved in secondary metabolite biosynthesis [53]. It catalyzes multiple oxidative rearrangement reactions in the biosynthesis of saliniketal and rifampin [53]. CYP105 and CYP107 members’ enzymatic functions could help Salinispora species utilize diverse compounds as carbon sources, detoxify toxic compounds, or kill other bacterial species to thrive in the environment. It is no doubt that due to these beneficial properties, Salinispora species enriched these family members in their genomes. CYP125 members conserved in Salinispora species are cholesterol and cholest-4-en-3-one hydroxylases [81,82]. One can assume that CYP125 members possibly help Salinispora species utilize cholesterol or cholesterol-like molecules as carbon sources. Growth of S. arenicola CNS-205 on cholesterol where complete degradation of cholesterol was observed [83] strongly supports this assumption considering these species do have CYP125 in their genome.
Interestingly, the presence of CYP125 members as part of smBGCs as observed in Salinispora species (Table 4) is also observed in mycobacterial species [75], indicating CYP125 members do have other functions apart from cholesterol oxidation. CYP146 members are involved in β-hydroxytyrosine formation, a precursor for the biosynthesis of vancomycin antibiotics [84]. Interestingly, only a single member was found in Salinispora species (Table 3) and is not part of smBGCs, complicating predicting its role in these species.
CYP154 members are involved in regio- and stereo-selective hydroxylation of different steroids [85,86]. CYP154 from Nocardia farcinica IFM10152 is a bifunctional enzyme with O-dealkylation and ortho-hydroxylation activities [87]. This P450 converts formononetin, an isoflavone compound, into ortho-dihydroxy-isoflavone [87]. In Salinispora species, CYP154 members are dominant, indicating they may attribute the above-said activities to these species. However, the role of CYP154 in the generation of secondary metabolites and these compounds’ properties concerning Salinispora species is of future interest (Figure 3 and Table 4).
CYP163A and CYP163B members produce novobiocin, aminocoumarin antibiotic [88], and skyllamycin, a potent inhibitor of the platelet-derived growth factor [89]. CYP162A members are involved in peptidyl nucleoside antibiotic nikkomycin synthesis [90,91]. CYP161A members are involved in the biosynthesis of antibiotics, pimaricin [92], and amphotericin [93]. CYP113 members are involved in the production a variety of antibiotics erythromycin [94,95], tylosin [96,97] and himastatin [98,99]. The presence of the CYP161-CYP163 and CYP113 members as part of smBGCs in Salinispora species (Figure 3 and Table 4) suggests that these members are certainly involved in the production of secondary metabolites in these species.
CYP244 and CYP245 members are involved in the biosynthesis of antibiotic rapamycin [100,101]. These two P450s together as part of smBGCs clusters in Salinispora species (Table 4) indicate they are working together in producing secondary metabolite. CYP248A members are involved in the production of antibiotic aureothin [102]. Salinispora species have 63 CYP248A members (Table 3), and 40 of them are part of smBGC (Figure 3 and Table 4), indicating their prominent role in secondary metabolites production. CYP124 members are known for their terminal hydroxylation of methyl branched-lipids in M. tuberculosis [103]. None of these members were found as part of smBGCs in Salinispora species (Table 4), indicating their limited role possibly in the oxidation of different methylated-aliphatic lipids in these species.
It is evident from the data presented in this article that close to half of Salinispora species P450s (1236 P450s) are part of smBGCs. Thus, we predict that these P450s play a role in producing different secondary metabolites characteristic of smBGC types (Table 4 and Table S2). The detailed information on species name, list of P450s part of smBGCs, their cluster information, and BGC type is presented in Table S1.
4. Conclusions
Salinispora species being marine organisms within the phylum Actinomycetes, are considered model organisms for studying bacterial diversity and secondary metabolite production. Compared to the genera Streptomyces and Mycobacterium, the genus Salinispora has an unprecedented number of P450s as part of secondary metabolite biosynthetic gene clusters (smBGCs), indicating a great diversity of secondary metabolites produced by these species. The presence of up to six P450s as part of smBGCs is unusual and not observed in other bacterial species. Future functional characterization of P450s sheds lighter on the untapped secondary metabolite biotechnological potentials from Salinispora species. Based on the data presented in this article and the literature published on P450s function, we predict that Salinispora species enriched or expanded specific P450s in their genome to utilize diverse compounds as carbon sources to detoxify toxic compounds or kill other bacterial species to thrive in the environment.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms10050871/s1. Figure S1: Phylogenetic analysis of Salinispora species P450s. 2643 P450s were used to construct the tree, and the members of the eight most abundant P450 families are highlighted in different colors and indicated in the figure. P450 protein sequences used to build the tree are listed in Table S2. Table S1: Identification of P450s that are part of secondary metabolite biosynthesis tic gene clusters (smBGCs) in Salinispora species. Cluster-ID and BGC type is retrieved from Integrated Microbial Genomes & Microbiomes (IMG/M) database [54,55]. BGC Type was indicated for consistency with the standard BGC Type name terminology available in the anti-SMASH database [74]. Table S2: P450 sequences identified and annotated in Salinispora species. Each P450 is presented with its assigned name followed by gene ID (in parenthesis) and species name.
Author Contributions
Conceptualization, K.S.; methodology, N.A.M., N.N., T.P., P.R.S., R.K., D.G., D.R.N. and K.S.; software, N.A.M., N.N., T.P., P.R.S., R.K., D.G., D.R.N. and K.S.; validation, N.A.M., N.N., T.P., P.R.S., R.K., D.G., D.R.N. and K.S.; formal analysis, N.A.M., N.N., T.P., P.R.S., R.K., D.G., D.R.N. and K.S.; investigation, N.A.M., N.N., T.P., P.R.S., R.K., D.G., D.R.N. and K.S.; resources, N.A.M., N.N., T.P., P.R.S., R.K., D.G., D.R.N. and K.S.; data curation, N.A.M., N.N., T.P., P.R.S., R.K., D.G., D.R.N. and K.S.; writing—original draft preparation, N.A.M., N.N., T.P., P.R.S., R.K., D.G., D.R.N. and K.S.; writing—review and editing, N.A.M., N.N., T.P., P.R.S., R.K., D.G., D.R.N. and K.S.; visualization, N.A.M., N.N., T.P., P.R.S., R.K., D.G., D.R.N. and K.S.; supervision, K.S.; project administration, K.S.; funding acquisition, K.S. All authors have read and agreed to the published version of the manuscript.
Funding
Khajamohiddin Syed expresses sincere gratitude to the University of Zululand (Grant number C686). Doctoral students Nomfundo Nzuza, Tiara Padayachee, and Puleng Rosinah Syed thank the National Research Foundation (NRF), South Africa, for postgraduate scholarships (Grant numbers MND210615611861, MND210504599108, and MND190606443406, respectively). Research in Dominik Gront’s laboratory is funded by the National Science Centre (Poland) 2018/29/B/ST6/01989.
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article and its Supplementary Materials.
Conflicts of Interest
The authors declare no conflict of interest, and the funders had no role in the study’s design, in the collection, analyses, or interpretation of data, in the writing of the manuscript, or in the decision to publish the results.
References
- Nelson, D.R. Cytochrome P450 diversity in the tree of life. Biochim. Biophys. Acta Proteins Proteom. 2018, 1866, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Lamb, D.C.; Follmer, A.H.; Goldstone, J.V.; Nelson, D.R.; Warrilow, A.G.; Price, C.L.; True, M.Y.; Kelly, S.L.; Poulos, T.L.; Stegeman, J.J. On the occurrence of cytochrome P450 in viruses. Proc. Natl. Acad. Sci. USA 2019, 116, 12343–12352. [Google Scholar] [CrossRef] [PubMed]
- White, R.E.; Coon, M.J. Oxygen activation by cytochrome P-450. Annu. Rev. Biochem. 1980, 49, 315–356. [Google Scholar] [CrossRef] [PubMed]
- Sono, M.; Roach, M.P.; Coulter, E.D.; Dawson, J.H. Heme-containing oxygenases. Chem. Rev. 1996, 96, 2841–2888. [Google Scholar] [CrossRef]
- Bernhardt, R. Cytochromes P450 as versatile biocatalysts. J. Biotechnol. 2006, 124, 128–145. [Google Scholar] [CrossRef]
- Kelly, S.L.; Kelly, D.E. Microbial cytochromes P450: Biodiversity and biotechnology. Where do cytochromes P450 come from, what do they do and what can they do for us? Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2013, 368, 20120476. [Google Scholar] [CrossRef]
- Guengerich, F.P.; Munro, A.W. Unusual cytochrome p450 enzymes and reactions. J. Biol. Chem. 2013, 288, 17065–17073. [Google Scholar] [CrossRef]
- Lamb, D.C.; Waterman, M.R. Unusual properties of the cytochrome P450 superfamily. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2013, 368, 20120434. [Google Scholar] [CrossRef]
- Girvan, H.M.; Munro, A.W. Applications of microbial cytochrome P450 enzymes in biotechnology and synthetic biology. Curr. Opin. Chem. Biol. 2016, 31, 136–145. [Google Scholar] [CrossRef]
- Urlacher, V.B.; Eiben, S. Cytochrome P450 monooxygenases: Perspectives for synthetic application. Trends Biotechnol. 2006, 24, 324–330. [Google Scholar] [CrossRef]
- Bernhardt, R.; Urlacher, V.B. Cytochromes P450 as promising catalysts for biotechnological application: Chances and limitations. Appl. Microbiol. Biotechnol. 2014, 98, 6185–6203. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Jiang, Y.; Guengerich, F.P.; Ma, L.; Li, S.; Zhang, W. Engineering cytochrome P450 enzyme systems for biomedical and biotechnological applications. J. Biol. Chem. 2020, 295, 833–849. [Google Scholar] [CrossRef]
- Guengerich, F.P. A history of the roles of cytochrome P450 enzymes in the toxicity of drugs. Toxicol. Res. 2020, 37, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Esteves, F.; Rueff, J.; Kranendonk, M. The central role of cytochrome P450 in xenobiotic metabolism—A brief review on a fascinating enzyme family. J. Xenobiotics 2021, 11, 94–114. [Google Scholar] [CrossRef] [PubMed]
- Debnath, A.; Calvet, C.M.; Jennings, G.; Zhou, W.; Aksenov, A.; Luth, M.R.; Abagyan, R.; Nes, W.D.; McKerrow, J.H.; Podust, L.M. CYP51 is an essential drug target for the treatment of primary amoebic meningoencephalitis (PAM). PLoS Negl. Trop. Dis. 2017, 11, e0006104. [Google Scholar] [CrossRef]
- Lepesheva, G.I.; Friggeri, L.; Waterman, M.R. CYP51 as drug targets for fungi and protozoan parasites: Past, present and future. Parasitology 2018, 145, 1820–1836. [Google Scholar] [CrossRef]
- Jawallapersand, P.; Mashele, S.S.; Kovacic, L.; Stojan, J.; Komel, R.; Pakala, S.B.; Krasevec, N.; Syed, K. Cytochrome P450 monooxygenase CYP53 family in fungi: Comparative structural and evolutionary analysis and its role as a common alternative anti-fungal drug target. PLoS ONE 2014, 9, e107209. [Google Scholar] [CrossRef]
- Andersen, J.F.; Tatsuta, K.; Gunji, H.; Ishiyama, T.; Hutchinson, C.R. Substrate specificity of 6-deoxyerythronolide B hydroxylase, a bacterial cytochrome P450 of erythromycin A biosynthesis. Biochemistry 1993, 32, 1905–1913. [Google Scholar] [CrossRef]
- Bischoff, D.; Bister, B.; Bertazzo, M.; Pfeifer, V.; Stegmann, E.; Nicholson, G.J.; Keller, S.; Pelzer, S.; Wohlleben, W.; Süssmuth, R.D. The biosynthesis of vancomycin-type glycopeptide antibiotics—A model for oxidative side-chain cross-linking by oxygenases coupled to the action of peptide synthetases. Chembiochem 2005, 6, 267–272. [Google Scholar] [CrossRef]
- Jennewein, S.; Park, H.; DeJong, J.M.; Long, R.M.; Bollon, A.P.; Croteau, R.B. Coexpression in yeast of Taxus cytochrome P450 reductase with cytochrome P450 oxygenases involved in Taxol biosynthesis. Biotechnol. Bioeng. 2005, 89, 588–598. [Google Scholar] [CrossRef]
- van Beilen, J.B.; Holtackers, R.; Lüscher, D.; Bauer, U.; Witholt, B.; Duetz, W.A. Biocatalytic production of perillyl alcohol from limonene by using a novel Mycobacterium sp. cytochrome P450 alkane hydroxylase expressed in Pseudomonas putida. Appl. Environ. Microbiol. 2005, 71, 1737–1744. [Google Scholar] [CrossRef] [PubMed]
- Podust, L.M.; Sherman, D.H. Diversity of P450 enzymes in the biosynthesis of natural products. Nat. Prod. Rep. 2012, 29, 1251–1266. [Google Scholar] [CrossRef] [PubMed]
- Greule, A.; Stok, J.E.; De Voss, J.J.; Cryle, M.J. Unrivalled diversity: The many roles and reactions of bacterial cytochromes P450 in secondary metabolism. Nat. Prod. Rep. 2018, 35, 757–791. [Google Scholar] [CrossRef]
- Vaishnav, P.; Demain, A.L. Unexpected applications of secondary metabolites. Biotechnol. Adv. 2011, 29, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Demain, A.L.; Fang, A. The natural functions of secondary metabolites. In History of Modern Biotechnology I; Springer: Berlin/Heidelberg, Germany, 2000; pp. 1–39. [Google Scholar]
- Thirumurugan, D.; Cholarajan, A.; Raja, S.S.; Vijayakumar, R. An Introductory Chapter: Secondary Metabolites. In Secondary Metabolites-Sources and Applications; IntechOpen: London, UK, 2018. [Google Scholar]
- Sharma, A.; Kumari, N.; Menghani, E. Bioactive secondary metabolites: An overview. Int. J. Sci. Eng. Res. 2014, 5, 1395. [Google Scholar]
- Abegaz, B.M.; Kinfe, H.H. Secondary metabolites, their structural diversity, bioactivity, and ecological functions: An overview. Phys. Sci. Rev. 2019, 4. [Google Scholar] [CrossRef]
- Katz, L.; Baltz, R.H. Natural product discovery: Past, present, and future. J. Ind. Microbiol. Biotechnol. 2016, 43, 155–176. [Google Scholar] [CrossRef]
- Cimermancic, P.; Medema, M.H.; Claesen, J.; Kurita, K.; Wieland Brown, L.C.; Mavrommatis, K.; Pati, A.; Godfrey, P.A.; Koehrsen, M.; Clardy, J.; et al. Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell 2014, 158, 412–421. [Google Scholar] [CrossRef]
- Medema, M.H.; Kottmann, R.; Yilmaz, P.; Cummings, M.; Biggins, J.B.; Blin, K.; de Bruijn, I.; Chooi, Y.H.; Claesen, J.; Coates, R.C.; et al. Minimum Information about a Biosynthetic Gene cluster. Nat. Chem. Biol. 2015, 11, 625–631. [Google Scholar] [CrossRef]
- Weber, T.; Kim, H.U. The secondary metabolite bioinformatics portal: Computational tools to facilitate synthetic biology of secondary metabolite production. Synth. Syst. Biotechnol. 2016, 1, 69–79. [Google Scholar] [CrossRef]
- Nair, S.; Abraham, J. Natural products from actinobacteria for drug discovery. In Advances in Pharmaceutical Biotechnology; Springer: Berlin/Heidelberg, Germany, 2020; pp. 333–363. [Google Scholar]
- Jose, P.A.; Maharshi, A.; Jha, B. Actinobacteria in natural products research: Progress and prospects. Microbiol. Res. 2021, 246, 126708. [Google Scholar] [CrossRef] [PubMed]
- Berdy, J. Bioactive microbial metabolites. J. Antibiot. 2005, 58, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Barka, E.A.; Vatsa, P.; Sanchez, L.; Gaveau-Vaillant, N.; Jacquard, C.; Klenk, H.-P.; Clément, C.; Ouhdouch, Y.; van Wezel, G.P. Taxonomy, physiology, and natural products of Actinobacteria. Microbiol. Mol. Biol. Rev. 2016, 80, 1–43. [Google Scholar] [CrossRef] [PubMed]
- de Lima Procópio, R.E.; da Silva, I.R.; Martins, M.K.; de Azevedo, J.L.; de Araújo, J.M. Antibiotics produced by Streptomyces. Braz. J. Infect. Dis. 2012, 16, 466–471. [Google Scholar] [CrossRef]
- Bonet, B.; Teufel, R.; Crusemann, M.; Ziemert, N.; Moore, B.S. Direct capture and heterologous expression of Salinispora natural product genes for the biosynthesis of enterocin. J. Nat. Prod. 2015, 78, 539–542. [Google Scholar] [CrossRef]
- Tyc, O.; Song, C.; Dickschat, J.S.; Vos, M.; Garbeva, P. The Ecological Role of Volatile and Soluble Secondary Metabolites Produced by Soil Bacteria. Trends Microbiol. 2017, 25, 280–292. [Google Scholar] [CrossRef]
- Penn, K.; Jenkins, C.; Nett, M.; Udwary, D.W.; Gontang, E.A.; McGlinchey, R.P.; Foster, B.; Lapidus, A.; Podell, S.; Allen, E.E.; et al. Genomic islands link secondary metabolism to functional adaptation in marine Actinobacteria. ISME J. 2009, 3, 1193–1203. [Google Scholar] [CrossRef]
- Asolkar, R.N.; Kirkland, T.N.; Jensen, P.R.; Fenical, W. Arenimycin, an antibiotic effective against rifampin- and methicillin-resistant Staphylococcus aureus from the marine actinomycete Salinispora arenicola. J. Antibiot. 2010, 63, 37–39. [Google Scholar] [CrossRef]
- Eustaquio, A.S.; Nam, S.J.; Penn, K.; Lechner, A.; Wilson, M.C.; Fenical, W.; Jensen, P.R.; Moore, B.S. The discovery of salinosporamide K from the marine bacterium “Salinispora” pacifica by genome mining gives insight into pathway evolution. Chembiochem 2011, 12, 61–64. [Google Scholar] [CrossRef]
- Jensen, P.R.; Moore, B.S.; Fenical, W. The marine actinomycete genus Salinispora: A model organism for secondary metabolite discovery. Nat. Prod. Rep. 2015, 32, 738–751. [Google Scholar] [CrossRef]
- Jensen, P.R.; Williams, P.G.; Oh, D.C.; Zeigler, L.; Fenical, W. Species-specific secondary metabolite production in marine actinomycetes of the genus Salinispora. Appl. Environ. Microbiol. 2007, 73, 1146–1152. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, S.; Adachi, K.; Matsuo, Y.; Nukina, M.; Shizuri, Y. Salinisporamycin, a novel metabolite from Salinispora arenicola. [corrected]. J. Antibiot. 2009, 62, 519–526. [Google Scholar] [CrossRef] [PubMed]
- Ziemert, N.; Lechner, A.; Wietz, M.; Millán-Aguiñaga, N.; Chavarria, K.L.; Jensen, P.R. Diversity and evolution of secondary metabolism in the marine actinomycete genus Salinispora. Proc. Natl. Acad. Sci. USA 2014, 111, E1130–E1139. [Google Scholar] [CrossRef] [PubMed]
- Udwary, D.W.; Zeigler, L.; Asolkar, R.N.; Singan, V.; Lapidus, A.; Fenical, W.; Jensen, P.R.; Moore, B.S. Genome sequencing reveals complex secondary metabolome in the marine actinomycete Salinispora tropica. Proc. Natl. Acad. Sci. USA 2007, 104, 10376–10381. [Google Scholar] [CrossRef]
- Fenical, W.; Jensen, P.R.; Palladino, M.A.; Lam, K.S.; Lloyd, G.K.; Potts, B.C. Discovery and development of the anticancer agent salinosporamide A (NPI-0052). Bioorganic Med. Chem. 2009, 17, 2175–2180. [Google Scholar] [CrossRef]
- Jensen, P.R.; Mafnas, C. Biogeography of the marine actinomycete Salinispora. Environ. Microbiol. 2006, 8, 1881–1888. [Google Scholar] [CrossRef]
- Maldonado, L.A.; Fenical, W.; Jensen, P.R.; Kauffman, C.A.; Mincer, T.J.; Ward, A.C.; Bull, A.T.; Goodfellow, M. Salinispora arenicola gen. nov., sp. nov. and Salinispora tropica sp. nov., obligate marine actinomycetes belonging to the family Micromonosporaceae. Int. J. Syst. Evol. Microbiol. 2005, 55, 1759–1766. [Google Scholar] [CrossRef]
- Ahmed, L.; Jensen, P.R.; Freel, K.C.; Brown, R.; Jones, A.L.; Kim, B.-Y.; Goodfellow, M. Salinispora pacifica sp. nov., an actinomycete from marine sediments. Antonie Van Leeuwenhoek 2013, 103, 1069–1078. [Google Scholar] [CrossRef]
- Contador, C.A.; Rodríguez, V.; Andrews, B.A.; Asenjo, J.A. Use of genome-scale models to get new insights into the marine actinomycete genus Salinispora. BMC Syst. Biol. 2019, 13, 11. [Google Scholar] [CrossRef]
- Wilson, M.C.; Gulder, T.A.; Mahmud, T.; Moore, B.S. Shared biosynthesis of the saliniketals and rifamycins in Salinispora arenicola is controlled by the sare1259-encoded cytochrome P450. J. Am. Chem. Soc. 2010, 132, 12757–12765. [Google Scholar] [CrossRef]
- Chen, I.-M.A.; Chu, K.; Palaniappan, K.; Ratner, A.; Huang, J.; Huntemann, M.; Hajek, P.; Ritter, S.; Varghese, N.; Seshadri, R. The IMG/M data management and analysis system v. 6.0: New tools and advanced capabilities. Nucleic Acids Res. 2021, 49, D751–D763. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Stamatis, D.; Bertsch, J.; Ovchinnikova, G.; Sundaramurthi, J.C.; Lee, J.; Kandimalla, M.; Chen, I.-M.A.; Kyrpides, N.C.; Reddy, T. Genomes OnLine Database (GOLD) v. 8: Overview and updates. Nucleic Acids Res. 2021, 49, D723–D733. [Google Scholar] [CrossRef] [PubMed]
- Mnguni, F.C.; Padayachee, T.; Chen, W.; Gront, D.; Yu, J.-H.; Nelson, D.R.; Syed, K. More P450s are involved in secondary metabolite biosynthesis in Streptomyces compared to Bacillus, Cyanobacteria and Mycobacterium. Int. J. Mol. Sci. 2020, 21, 4814. [Google Scholar] [CrossRef] [PubMed]
- Syed, P.R.; Chen, W.; Nelson, D.R.; Kappo, A.P.; Yu, J.H.; Karpoormath, R.; Syed, K. Cytochrome P450 Monooxygenase CYP139 Family Involved in the Synthesis of Secondary Metabolites in 824 Mycobacterial Species. Int. J. Mol. Sci. 2019, 20, 2690. [Google Scholar] [CrossRef]
- Syed, K.; Mashele, S.S. Comparative analysis of P450 signature motifs EXXR and CXG in the large and diverse kingdom of fungi: Identification of evolutionarily conserved amino acid patterns characteristic of P450 family. PLoS ONE 2014, 9, e95616. [Google Scholar] [CrossRef]
- Gotoh, O. Substrate recognition sites in cytochrome P450 family 2 (CYP2) proteins inferred from comparative analyses of amino acid and coding nucleotide sequences. J. Biol. Chem. 1992, 267, 83–90. [Google Scholar] [CrossRef]
- Nelson, D.R.; Kamataki, T.; Waxman, D.J.; Guengerich, F.P.; Estabrook, R.W.; Feyereisen, R.; Gonzalez, F.J.; Coon, M.J.; Gunsalus, I.C.; Gotoh, O.; et al. The P450 superfamily: Update on new sequences, gene mapping, accession numbers, early trivial names of enzymes, and nomenclature. DNA Cell Biol. 1993, 12, 1–51. [Google Scholar] [CrossRef]
- Nelson, D.R. Cytochrome P450 nomenclature, 2004. Methods Mol. Biol. 2006, 320, 1–10. [Google Scholar] [CrossRef]
- Nelson, D.R. Cytochrome P450 nomenclature. Methods Mol. Biol. 1998, 107, 15–24. [Google Scholar] [CrossRef]
- Nzuza, N.; Padayachee, T.; Chen, W.; Gront, D.; Nelson, D.R.; Syed, K. Diversification of Ferredoxins across Living Organisms. Curr. Issues Mol. Biol. 2021, 43, 1374–1390. [Google Scholar] [CrossRef]
- Nzuza, N.; Padayachee, T.; Syed, P.R.; Kryś, J.D.; Chen, W.; Gront, D.; Nelson, D.R.; Syed, K. Ancient Bacterial Class Alphaproteobacteria Cytochrome P450 Monooxygenases Can Be Found in Other Bacterial Species. Int. J. Mol. Sci. 2021, 22, 5542. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Kuma, K.; Toh, H.; Miyata, T. MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005, 33, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Boc, A.; Diallo, A.B.; Makarenkov, V. T-REX: A web server for inferring, validating and visualizing phylogenetic trees and networks. Nucleic Acids Res. 2012, 40, W573–W579. [Google Scholar] [CrossRef] [PubMed]
- Kryś, J.D.; Gront, D. VisuaLife: Library for interactive visualization in rich web applications. Bioinformatics 2021, 37, 3662–3663. [Google Scholar] [CrossRef] [PubMed]
- Msomi, N.N.; Padayachee, T.; Nzuza, N.; Syed, P.R.; Kryś, J.D.; Chen, W.; Gront, D.; Nelson, D.R.; Syed, K. In silico analysis of P450s and their role in secondary metabolism in the bacterial class Gammaproteobacteria. Molecules 2021, 26, 1538. [Google Scholar] [CrossRef]
- Howe, E.A.; Sinha, R.; Schlauch, D.; Quackenbush, J. RNA-Seq analysis in MeV. Bioinformatics 2011, 27, 3209–3210. [Google Scholar] [CrossRef]
- Padayachee, T.; Nzuza, N.; Chen, W.; Nelson, D.R.; Syed, K. Impact of lifestyle on cytochrome P450 monooxygenase repertoire is clearly evident in the bacterial phylum Firmicutes. Sci. Rep. 2020, 10, 13982. [Google Scholar] [CrossRef]
- Khumalo, M.J.; Nzuza, N.; Padayachee, T.; Chen, W.; Yu, J.-H.; Nelson, D.; Syed, K. Comprehensive analyses of cytochrome P450 monoxygenases and secondary metabolite biosynthetic gene clusters in Cyanobacteria. Int. J. Mol. Sci. 2020, 21, 656. [Google Scholar] [CrossRef]
- Senate, L.M.; Tjatji, M.P.; Pillay, K.; Chen, W.; Zondo, N.M.; Syed, P.R.; Mnguni, F.C.; Chiliza, Z.E.; Bamal, H.D.; Karpoormath, R.; et al. Similarities, variations, and evolution of cytochrome P450s in Streptomyces versus Mycobacterium. Sci. Rep. 2019, 9, 3962. [Google Scholar] [CrossRef]
- Parvez, M.; Qhanya, L.B.; Mthakathi, N.T.; Kgosiemang, I.K.; Bamal, H.D.; Pagadala, N.S.; Xie, T.; Yang, H.; Chen, H.; Theron, C.W.; et al. Molecular evolutionary dynamics of cytochrome P450 monooxygenases across kingdoms: Special focus on mycobacterial P450s. Sci. Rep. 2016, 6, 33099. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; Van Wezel, G.P.; Medema, M.H.; Weber, T. antiSMASH 6.0: Improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef] [PubMed]
- Ngcobo, N.S.; Chiliza, Z.E.; Chen, W.; Yu, J.-H.; Nelson, D.R.; Tuszynski, J.A.; Preto, J.; Syed, K. Comparative Analysis, Structural Insights, and Substrate/Drug Interaction of CYP128A1 in Mycobacterium Tuberculosis. Int. J. Mol. Sci. 2020, 21, 4816. [Google Scholar] [CrossRef] [PubMed]
- Weber, J.; Leung, J.; Swanson, S.; Idler, K.; McAlpine, J. An erythromycin derivative produced by targeted gene disruption in Saccharopolyspora erythraea. Science 1991, 252, 114–117. [Google Scholar] [CrossRef]
- Trefzer, A.; Jungmann, V.; Molnár, I.; Botejue, A.; Buckel, D.; Frey, G.; Hill, D.S.; Jörg, M.; Ligon, J.M.; Mason, D. Biocatalytic conversion of avermectin to 4″-oxo-avermectin: Improvement of cytochrome p450 monooxygenase specificity by directed evolution. Appl. Environ. Microbiol. 2007, 73, 4317–4325. [Google Scholar] [CrossRef] [PubMed]
- Fujii, Y.; Kabumoto, H.; Nishimura, K.; Fujii, T.; Yanai, S.; Takeda, K.; Tamura, N.; Arisawa, A.; Tamura, T. Purification, characterization, and directed evolution study of a vitamin D3 hydroxylase from Pseudonocardia autotrophica. Biochem. Biophys. Res. Commun. 2009, 385, 170–175. [Google Scholar] [CrossRef] [PubMed]
- Prior, J.E.; Shokati, T.; Christians, U.; Gill, R.T. Identification and characterization of a bacterial cytochrome P450 for the metabolism of diclofenac. Appl. Microbiol. Biotechnol. 2010, 85, 625–633. [Google Scholar] [CrossRef]
- Moody, S.C.; Loveridge, E.J. CYP105-diverse structures, functions and roles in an intriguing family of enzymes in Streptomyces. J. Appl. Microbiol. 2014, 117, 1549–1563. [Google Scholar] [CrossRef]
- McLean, K.J.; Lafite, P.; Levy, C.; Cheesman, M.R.; Mast, N.; Pikuleva, I.A.; Leys, D.; Munro, A.W. The Structure of Mycobacterium tuberculosis CYP125: Molecular basis for cholesterol binding in a P450 needed for host infection. J. Biol. Chem. 2009, 284, 35524–35533. [Google Scholar] [CrossRef]
- Ouellet, H.; Guan, S.; Johnston, J.B.; Chow, E.D.; Kells, P.M.; Burlingame, A.L.; Cox, J.S.; Podust, L.M.; de Montellano, P.R.O. Mycobacterium tuberculosis CYP125A1, a steroid C27 monooxygenase that detoxifies intracellularly generated cholest-4-en-3-one. Mol. Microbiol. 2010, 77, 730–742. [Google Scholar] [CrossRef]
- Bergstrand, L.H.; Cardenas, E.; Holert, J.; Van Hamme, J.D.; Mohn, W.W. Delineation of steroid-degrading microorganisms through comparative genomic analysis. mBio 2016, 7, e00166-16. [Google Scholar] [CrossRef]
- Cryle, M.J.; Schlichting, I. Structural insights from a P450 Carrier Protein complex reveal how specificity is achieved in the P450BioI ACP complex. Proc. Natl. Acad. Sci. USA 2008, 105, 15696–15701. [Google Scholar] [CrossRef] [PubMed]
- Bracco, P.; Janssen, D.B.; Schallmey, A. Selective steroid oxyfunctionalisation by CYP154C5, a bacterial cytochrome P450. Microb. Cell Factories 2013, 12, 95. [Google Scholar] [CrossRef] [PubMed]
- Subedi, P.; Kim, K.-H.; Hong, Y.-S.; Lee, J.-H.; Oh, T.-J. Enzymatic characterization and comparison of two steroid hydroxylases CYP154C3-1 and CYP154C3-2 from Streptomyces species. J. Microbiol. Biotechnol. 2021, 31, 464–474. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.-Y.; Park, H.-Y.; Kim, B.-G. Characterization of bi-functional CYP154 from Nocardia farcinica IFM10152 in the O-dealkylation and ortho-hydroxylation of formononetin. Enzym. Microb. Technol. 2010, 47, 327–334. [Google Scholar] [CrossRef]
- Chen, H.; Walsh, C.T. Coumarin formation in novobiocin biosynthesis: Beta-hydroxylation of the aminoacyl enzyme tyrosyl-S-NovH by a cytochrome P450 NovI. Chem. Biol. 2001, 8, 301–312. [Google Scholar] [CrossRef][Green Version]
- Uhlmann, S.; Sussmuth, R.D.; Cryle, M.J. Cytochrome p450sky interacts directly with the nonribosomal peptide synthetase to generate three amino acid precursors in skyllamycin biosynthesis. ACS Chem. Biol. 2013, 8, 2586–2596. [Google Scholar] [CrossRef]
- Lauer, B.; Russwurm, R.; Bormann, C. Molecular characterization of two genes from Streptomyces tendae Tu901 required for the formation of the 4-formyl-4-imidazolin-2-one-containing nucleoside moiety of the peptidyl nucleoside antibiotic nikkomycin. Eur. J. Biochem. 2000, 267, 1698–1706. [Google Scholar] [CrossRef][Green Version]
- Xie, Z.; Niu, G.; Li, R.; Liu, G.; Tan, H. Identification and characterization of sanH and sanI involved in the hydroxylation of pyridyl residue during nikkomycin biosynthesis in Streptomyces ansochromogenes. Curr. Microbiol. 2007, 55, 537–542. [Google Scholar] [CrossRef]
- Mendes, M.V.; Anton, N.; Martin, J.F.; Aparicio, J.F. Characterization of the polyene macrolide P450 epoxidase from Streptomyces natalensis that converts de-epoxypimaricin into pimaricin. Biochem. J. 2005, 386, 57–62. [Google Scholar] [CrossRef][Green Version]
- Caffrey, P.; Lynch, S.; Flood, E.; Finnan, S.; Oliynyk, M. Amphotericin biosynthesis in Streptomyces nodosus: Deductions from analysis of polyketide synthase and late genes. Chem. Biol. 2001, 8, 713–723. [Google Scholar] [CrossRef]
- Shafiee, A.; Hutchinson, C.R. Macrolide antibiotic biosynthesis: Isolation and properties of two forms of 6-deoxyerythronolide B hydroxylase from Saccharopolyspora erythraea (Streptomyces erythreus). Biochemistry 1987, 26, 6204–6210. [Google Scholar] [CrossRef] [PubMed]
- Stassi, D.; Donadio, S.; Staver, M.J.; Katz, L. Identification of a Saccharopolyspora erythraea gene required for the final hydroxylation step in erythromycin biosynthesis. J. Bacteriol. 1993, 175, 182–189. [Google Scholar] [CrossRef] [PubMed]
- Merson-Davies, L.A.; Cundliffe, E. Analysis of five tylosin biosynthetic genes from the tyllBA region of the Streptomyces fradiae genome. Mol. Microbiol. 1994, 13, 349–355. [Google Scholar] [CrossRef] [PubMed]
- Fouces, R.; Mellado, E.; Diez, B.; Barredo, J.L. The tylosin biosynthetic cluster from Streptomyces fradiae: Genetic organization of the left region. Microbiology 1999, 145 Pt 4, 855–868. [Google Scholar] [CrossRef]
- Zhang, H.; Chen, J.; Wang, H.; Xie, Y.; Ju, J.; Yan, Y.; Zhang, H. Structural analysis of HmtT and HmtN involved in the tailoring steps of himastatin biosynthesis. FEBS Lett. 2013, 587, 1675–1680. [Google Scholar] [CrossRef]
- Ma, J.; Wang, Z.; Huang, H.; Luo, M.; Zuo, D.; Wang, B.; Sun, A.; Cheng, Y.Q.; Zhang, C.; Ju, J. Biosynthesis of himastatin: Assembly line and characterization of three cytochrome P450 enzymes involved in the post-tailoring oxidative steps. Angew. Chem. 2011, 50, 7797–7802. [Google Scholar] [CrossRef]
- Aparicio, J.F.; Molnar, I.; Schwecke, T.; Konig, A.; Haydock, S.F.; Khaw, L.E.; Staunton, J.; Leadlay, P.F. Organization of the biosynthetic gene cluster for rapamycin in Streptomyces hygroscopicus: Analysis of the enzymatic domains in the modular polyketide synthase. Gene 1996, 169, 9–16. [Google Scholar] [CrossRef]
- Molnar, I.; Aparicio, J.F.; Haydock, S.F.; Khaw, L.E.; Schwecke, T.; Konig, A.; Staunton, J.; Leadlay, P.F. Organisation of the biosynthetic gene cluster for rapamycin in Streptomyces hygroscopicus: Analysis of genes flanking the polyketide synthase. Gene 1996, 169, 1–7. [Google Scholar] [CrossRef]
- Zocher, G.; Richter, M.E.; Mueller, U.; Hertweck, C. Structural fine-tuning of a multifunctional cytochrome P450 monooxygenase. J. Am. Chem. Soc. 2011, 133, 2292–2302. [Google Scholar] [CrossRef]
- Johnston, J.B.; Kells, P.M.; Podust, L.M.; Ortiz de Montellano, P.R. Biochemical and structural characterization of CYP124: A methyl-branched lipid omega-hydroxylase from Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 2009, 106, 20687–20692. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).