Proteomic Analysis of Spore Surface Proteins and Characteristics of a Novel Spore Wall Protein and Biomarker, EhSWP3, from the Shrimp Microsporidium Enterocytozoon hepatopenaei (EHP)
Abstract
:1. Introduction
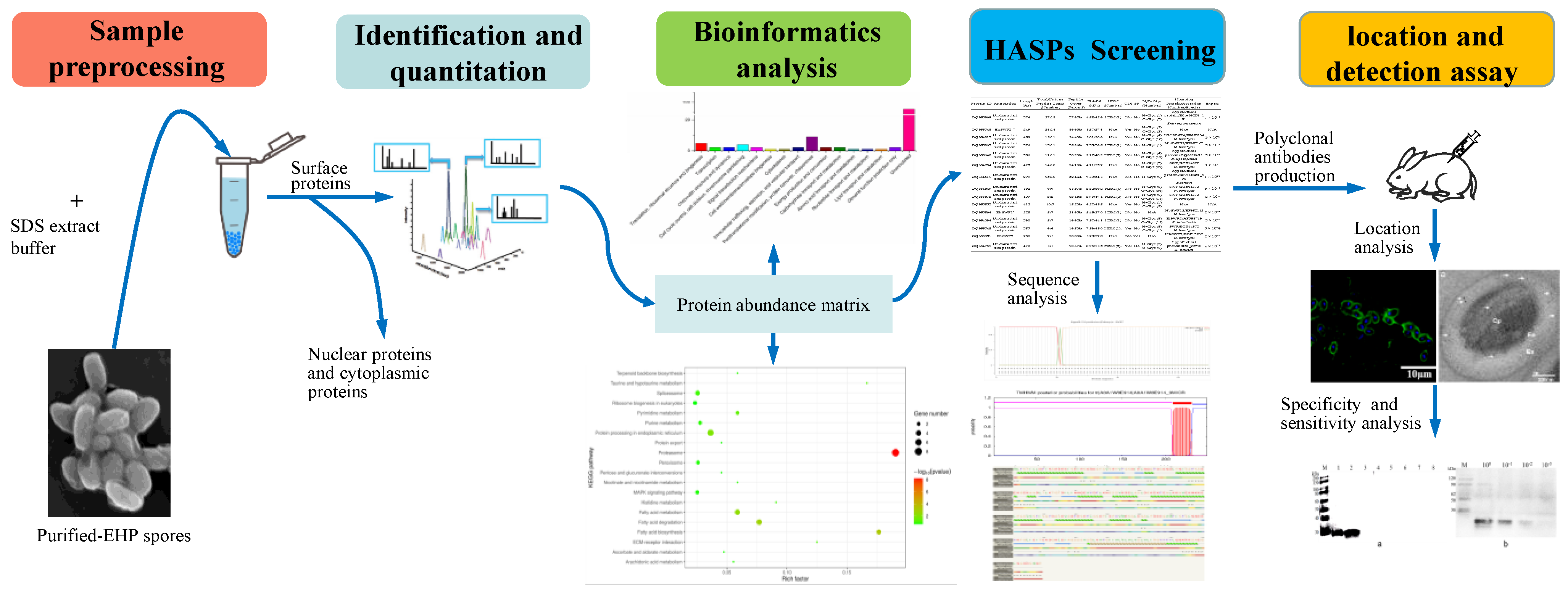
2. Materials and Methods
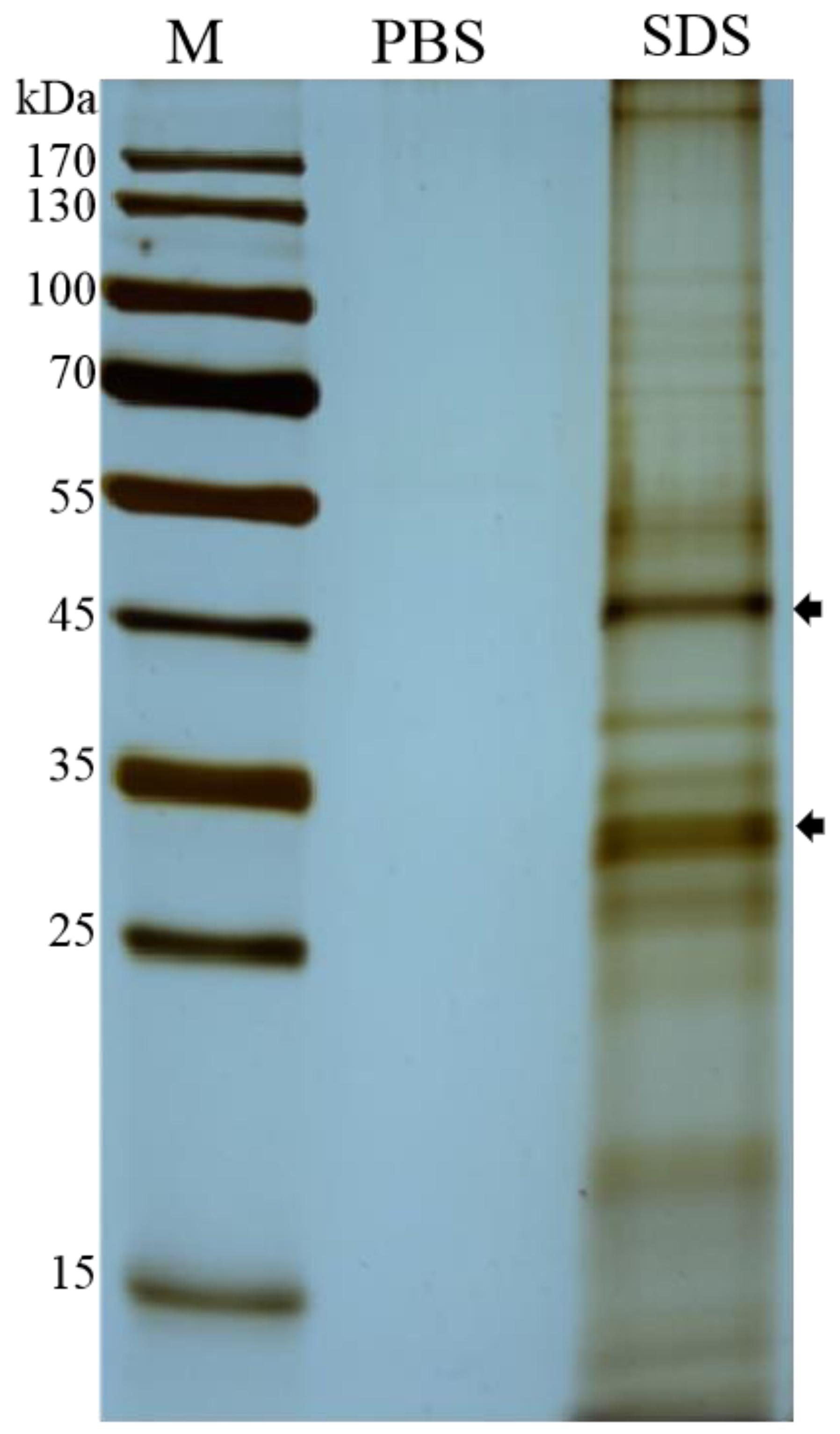
2.1. Purification of EHP Spores and Extraction of Spore Wall Proteins
2.2. LC-MS/MS Analysis and Protein Identification
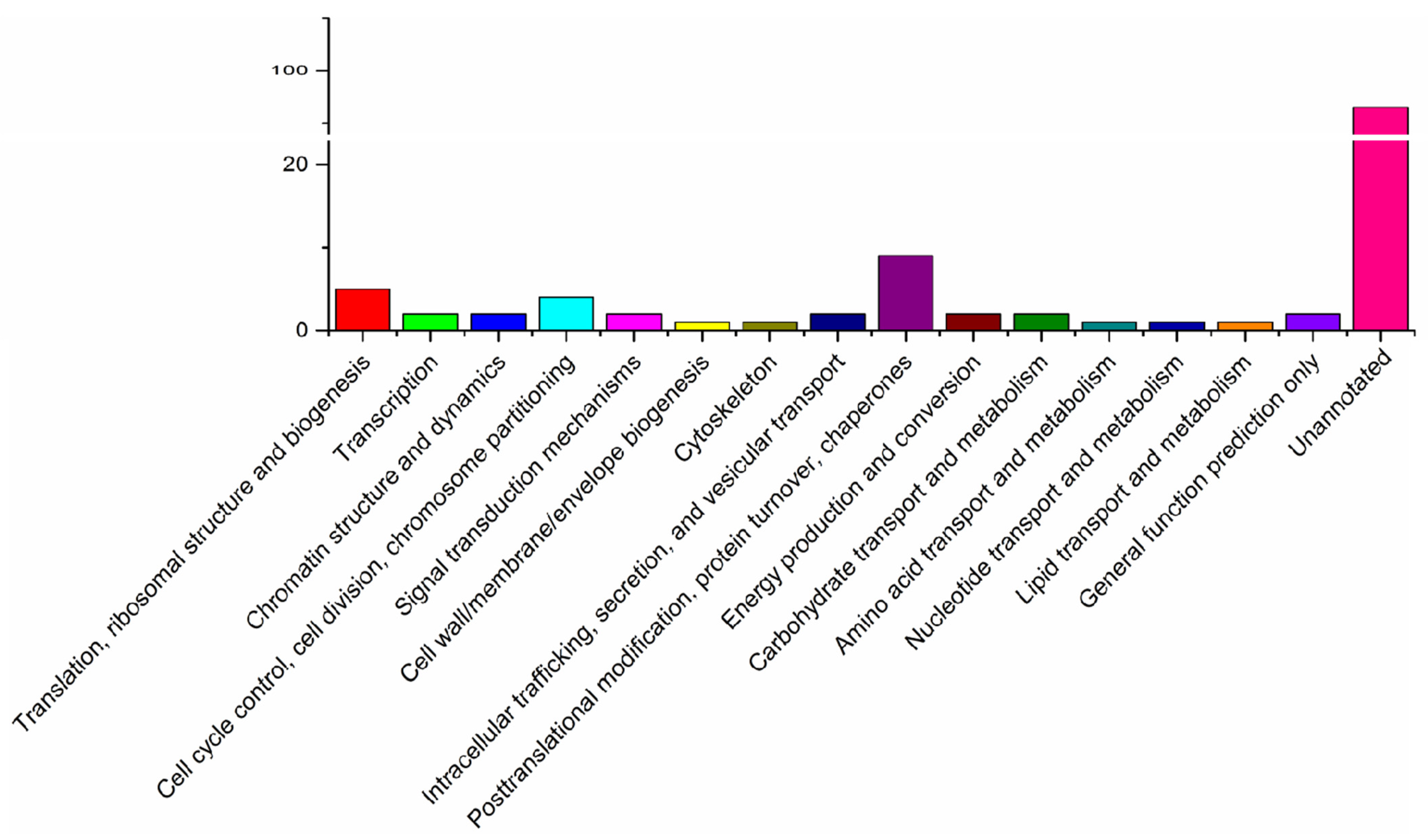
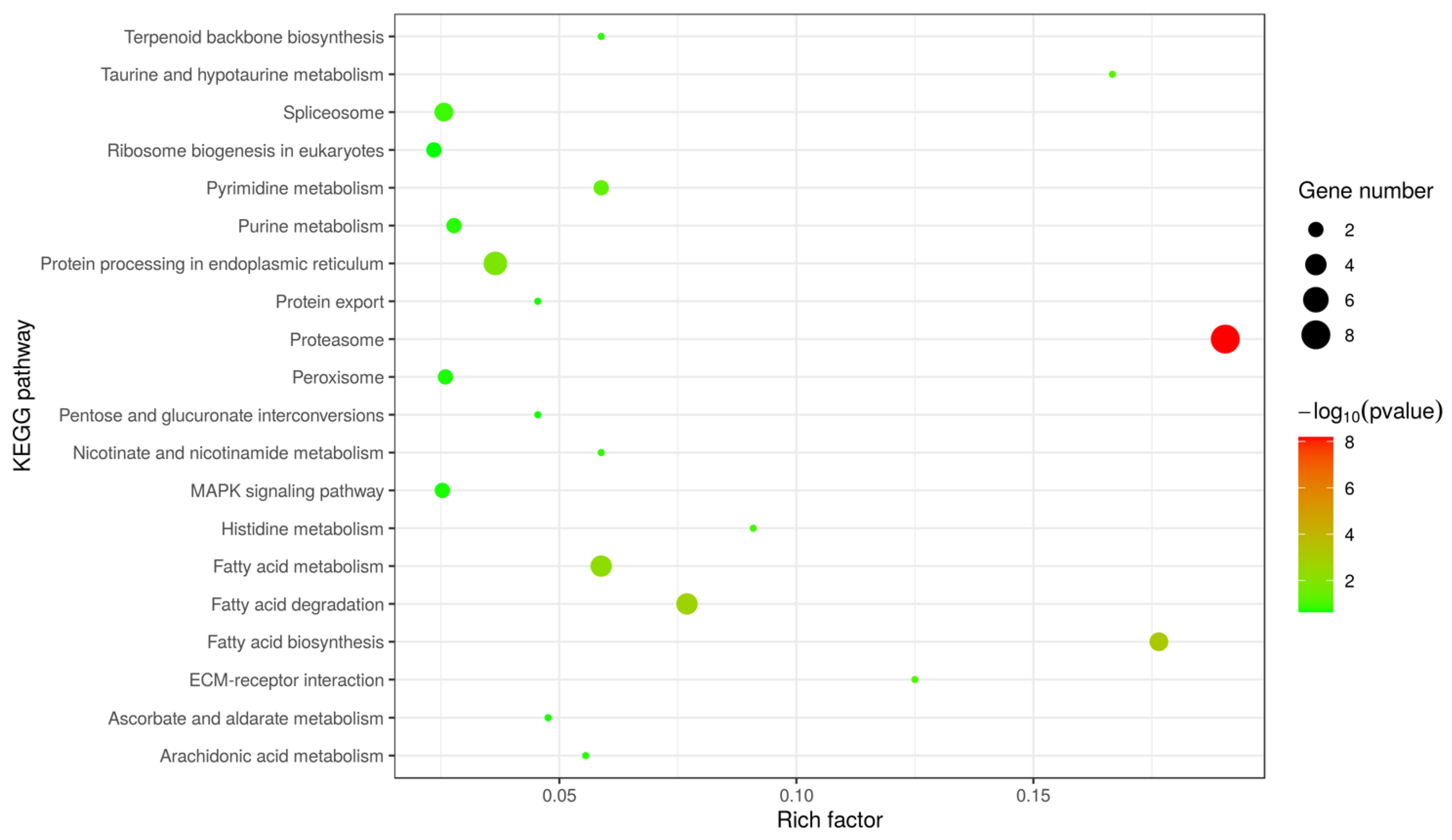
2.3. Bioinformatics Analysis
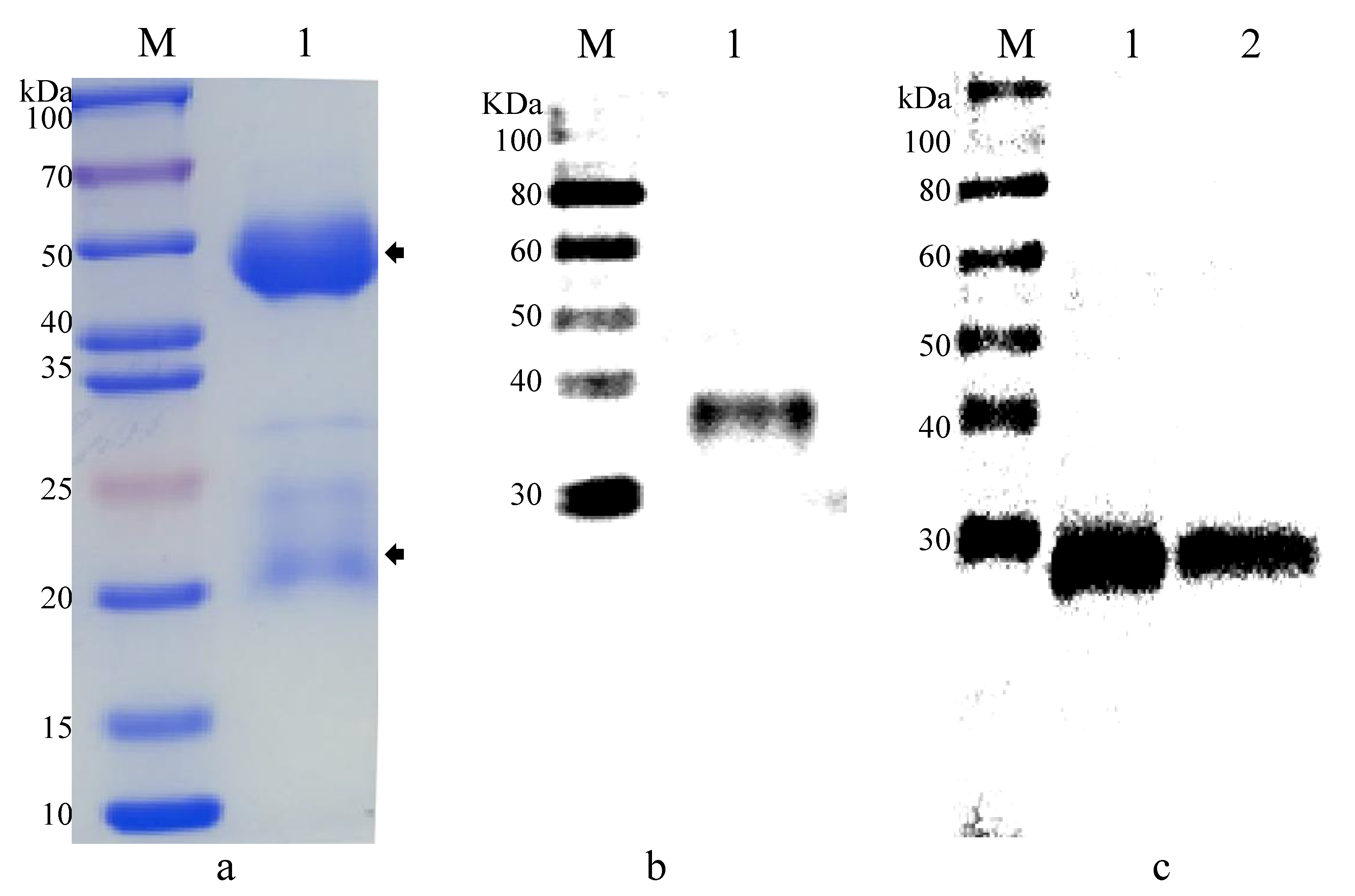
2.4. Gene Cloning and Recombinant Protein Expression of EhSWP3
2.5. Polyclonal Antibodies Preparation and Western Blotting Analysis
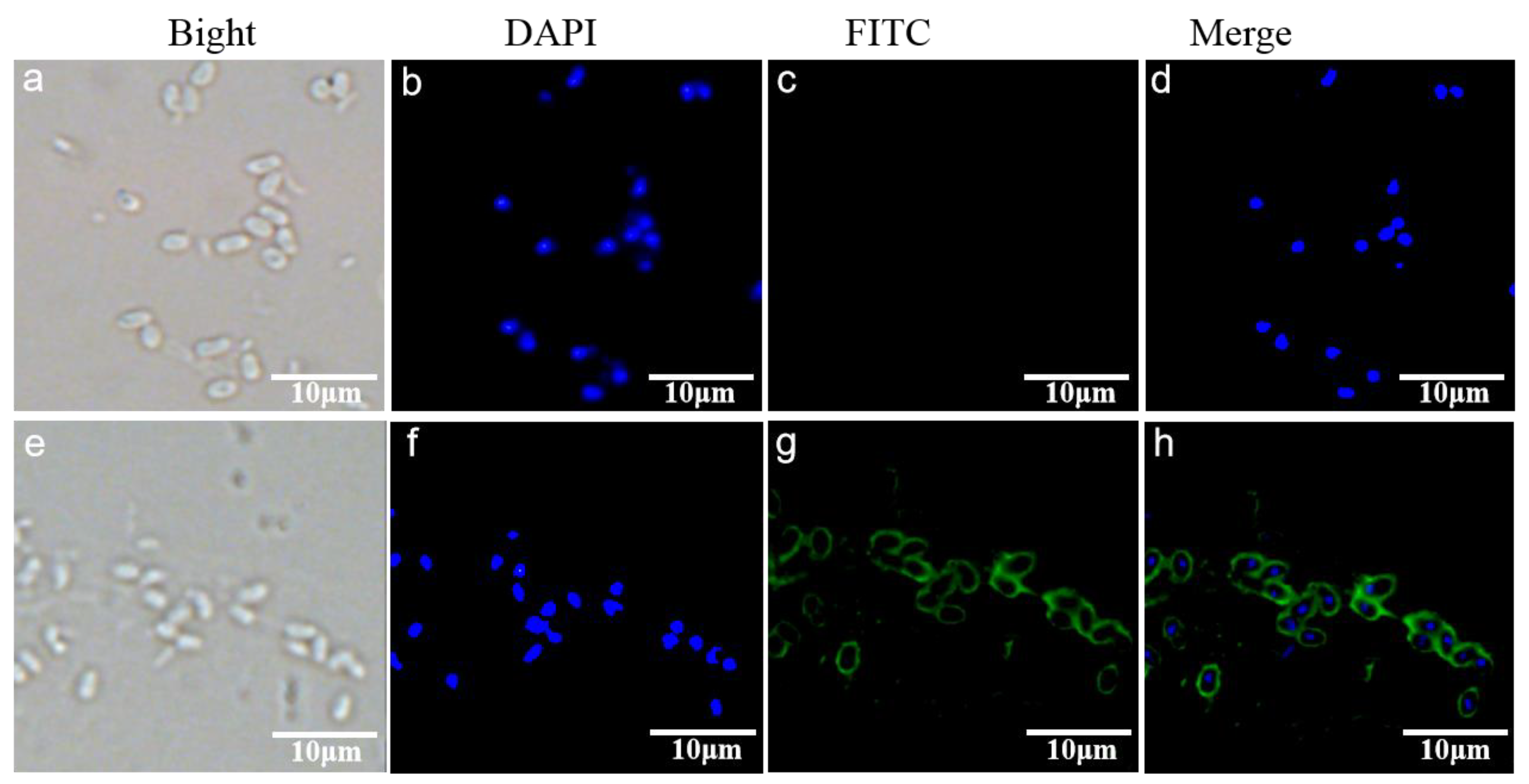
2.6. Indirect Immunofluorescence Assay (IFA)
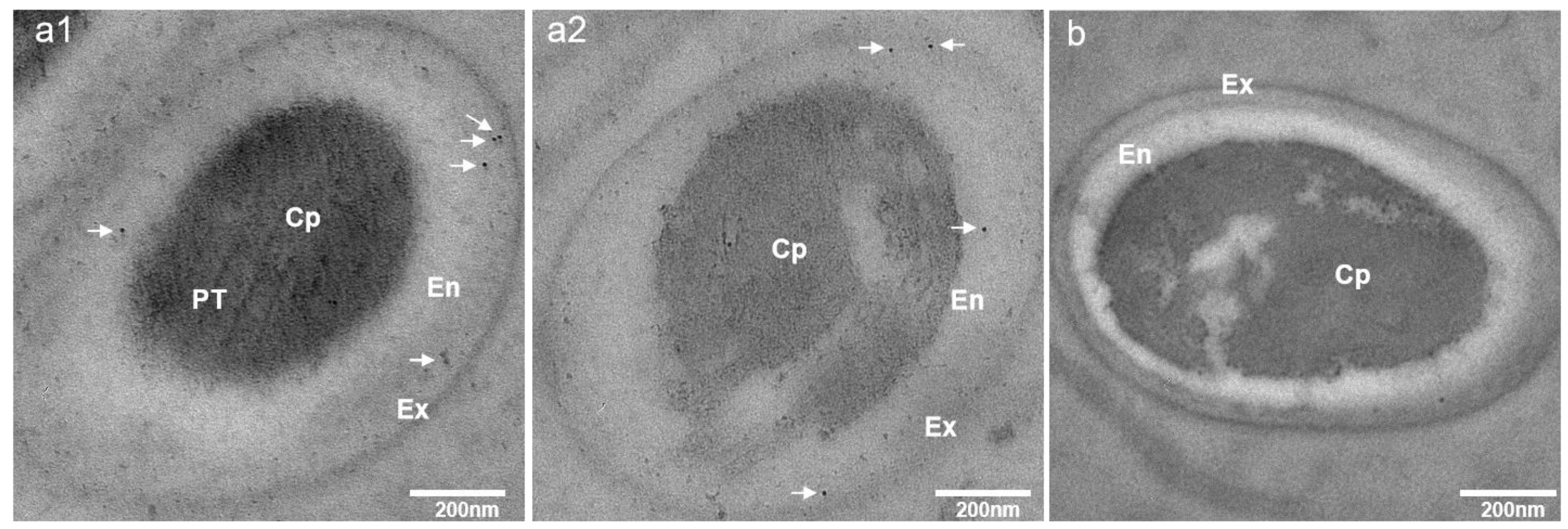
2.7. Immunoelectron Microscopy (IEM) Analysis
2.8. Specificity and Sensitivity of Ployclonal Antibodies
3. Results
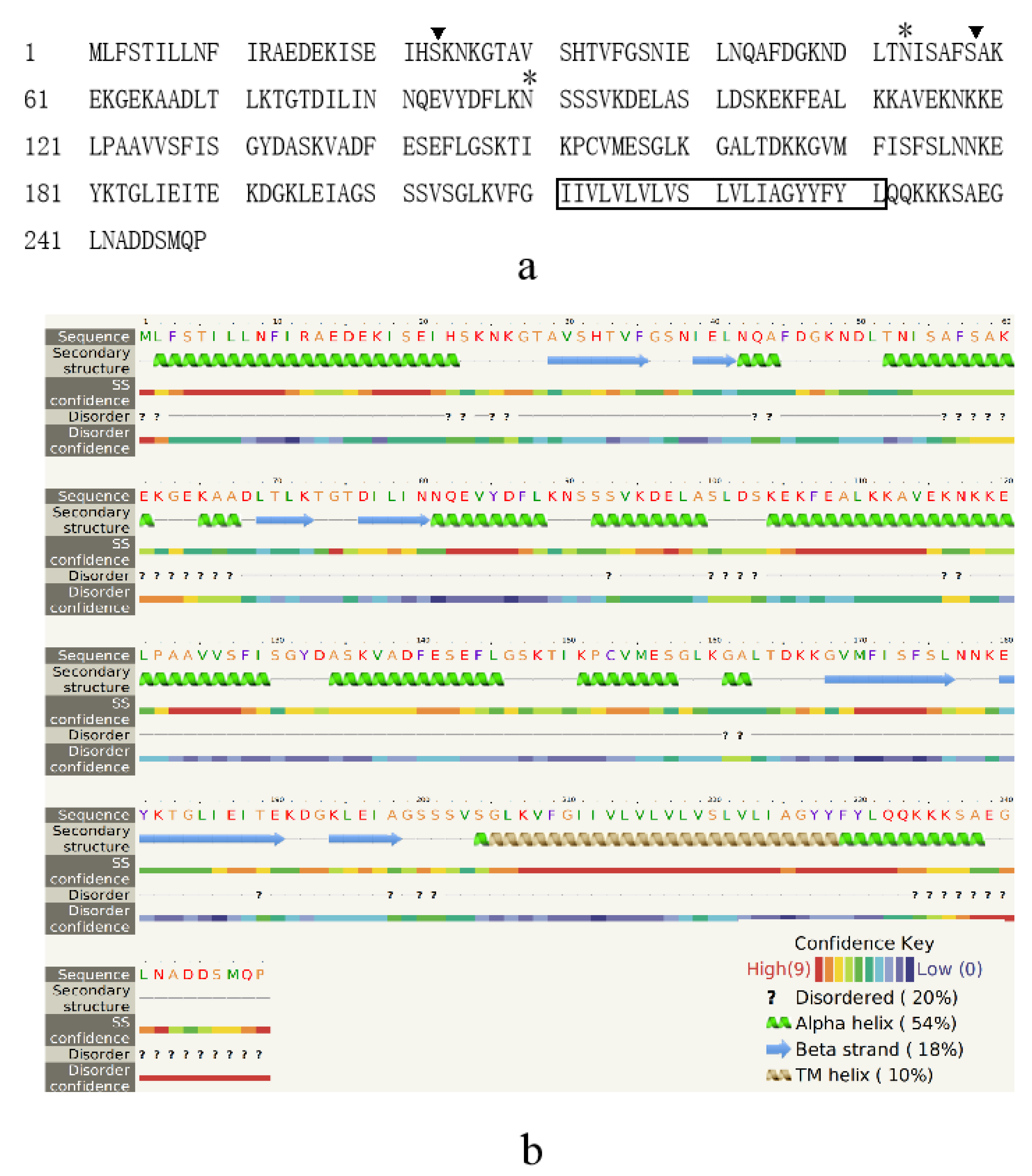
3.1. Character of the Surface Proteins of EHP Spores
3.2. Production of Polyclonal Antibodies against EhSWP3
3.3. Subcellular Localization of EhSWP3
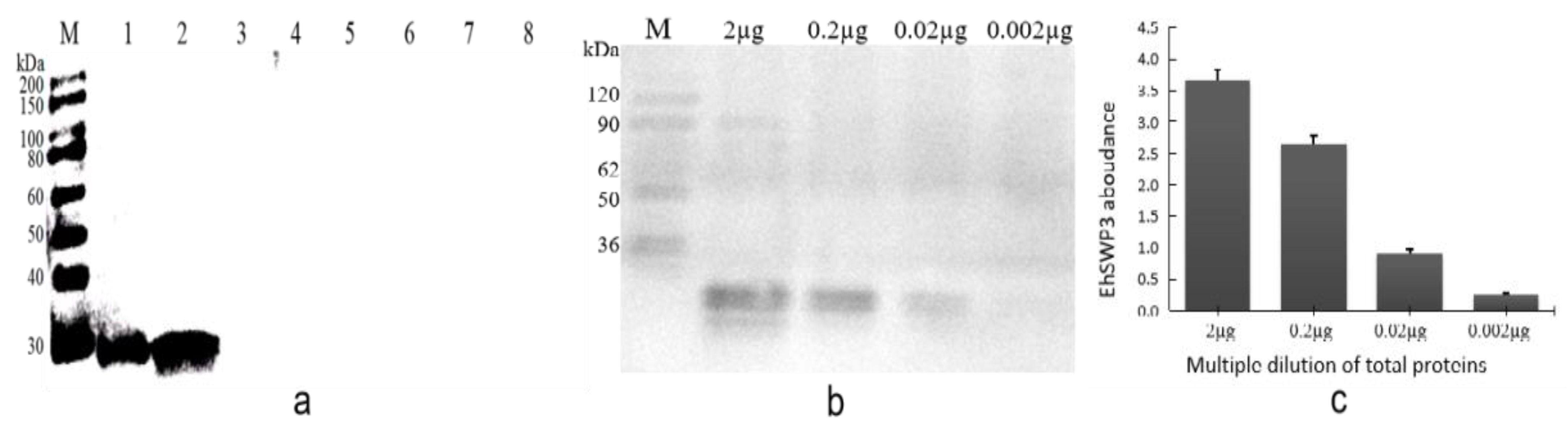
3.4. Specificity and Sensitivity of Polyclonal Antibodies Anti-EhSWP3
4. Discussion
4.1. High-Abundance Surface Proteins (HASPs) of EHP
4.2. Novel Species-Specific Protein Marker EhSWP3
4.3. Promising Potential of Anti-EhSWP3 for EHP Detection
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Capella-Gutiérrez, S.; Marcet-Houben, M.; Gabaldón, T. Phylogenomics supports microsporidia as the earliest diverging clade of sequenced fungi. BMC Biol. 2012, 10, 47. [Google Scholar] [CrossRef] [PubMed]
- Bass, D.; Czech, L.; Williams, B.A.P.; Berney, C.; Dunthorn, M.; Mahé, F.; Torruella, G.; Stentiford, G.D.; Williams, T.A. Clarifying the relationships between Microsporidia and Cryptomycota. J. Eukaryot. Microbiol. 2018, 65, 773–782. [Google Scholar] [CrossRef]
- Corsaro, D.; Wylezich, C.; Venditti, D.; Michel, R.; Walochnik, J.; Wegensteiner, R. Filling gaps in the microsporidian tree: rDNA phylogeny of Chytridiopsis typographi (Microsporidia: Chytridiopsida). Parasitol. Res. 2019, 118, 169–180. [Google Scholar] [CrossRef] [PubMed]
- Issi, I.V. Development of microsporidiology in Russia. Plant Prot. News. 2020, 103, 161–176. (In Russian) [Google Scholar] [CrossRef]
- Han, B.; Weiss, L.M. Microsporidia: Obligate intracellular pathogens within the fungal kingdom. Microbiol Spectr. 2017, 5, 2–5. [Google Scholar] [CrossRef]
- Thitamadee, S.; Prachumwat, A.; Srisala, J.; Jaroenlak, P.; Salachan, P.V.; Sritunyalucksana, K.; Flegel, T.W.; Itsathitphaisarn, O. Review of current disease threats for cultivated penaeid shrimp in Asia. Aquaculture 2016, 452, 69–87. [Google Scholar] [CrossRef]
- Rodriguez-Tovar, L.E.; Speare, D.J.; Markham, R.J. Fish microsporidia: Immune response, immunomodulation and vaccination. Fish Shellfish Immunol. 2011, 30, 999–1006. [Google Scholar] [CrossRef]
- Tourtip, S.; Wongtripop, S.; Stentiford, G.D.; Bateman, K.S.; Sriurairatana, S.; Chavadej, J.; Sritunyalucksana, K.; Withyachumnarnkul, B. Enterocytozoon hepatopenaei sp. nov. (Microsporida: Enterocytozoonidae), a parasite of the black tiger shrimp Penaeus monodon (Decapoda: Penaeidae): Fine structure and phylogenetic relationships. J. Invertebr. Pathol. 2009, 102, 21–29. [Google Scholar] [CrossRef]
- Chayaburakul, K.; Nash, G.; Pratanpipat, P.; Sriurairatana, S.; Withyachumnarnkul, B. Multiple pathogens found in growth-retarded black tiger shrimp Penaeus monodon cultivated in Thailand. Dis. Aquat. Organ. 2004, 60, 89–96. [Google Scholar] [CrossRef]
- Tangprasittipap, A.; Srisala, J.; Chouwdee, S.; Somboon, M.; Chuchird, N.; Limsuwan, C.; Srisuvan, T.; Flegel, T.W.; Sritunyalucksana, K. The microsporidian Enterocytozoon hepatopenaei is not the cause of white feces syndrome in whiteleg shrimp Penaeus (Litopenaeus) vannamei. BMC Vet. Res. 2013, 9, 139. [Google Scholar] [CrossRef]
- Salachan, P.V.; Jaroenlak, P.; Thitamadee, S.; Itsathitphaisarn, O.; Sritunyalucksana, K. Laboratory cohabitation challenge model for shrimp hepatopancreatic microsporidiosis (HPM) caused by Enterocytozoon hepatopenaei (EHP). BMC Vet Res. 2017, 13, 9. [Google Scholar] [CrossRef] [PubMed]
- Chaijarasphong, T.; Munkongwongsiri, N.; Stentiford, G.D.; Aldama-Cano, D.J.; Thansa, K.; Flegel, T.W.; Sritunyalucksana, K.; Itsathitphaisarn, O. The shrimp microsporidian Enterocytozoon hepatopenaei (EHP): Biology, pathology, diagnostics and control. J. Invertebr. Pathol. 2020, 186, 107458. [Google Scholar] [CrossRef]
- Suebsing, R.; Prombun, P.; Srisala, J.; Kiatpathomchai, W. Loop-mediated isothermal amplification combined with colorimetric nanogold for detection of the microsporidian Enterocytozoon hepatopenaei in penaeid shrimp. J. Appl. Microbiol. 2013, 114, 1254–1263. [Google Scholar] [CrossRef] [PubMed]
- Jaroenlak, P.; Sanguanrut, P.; Williams, B.A.; Stentiford, G.D.; Flegel, T.W.; Sritunyalucksana, K.; Itsathitphaisarn, O. A nested PCR assay to avoid false positive detection of the microsporidian Enterocytozoon hepatopenaei (EHP) in environmental samples in shrimp farms. PLoS ONE. 2016, 11, e0166320. [Google Scholar] [CrossRef] [PubMed]
- Karthikeyan, K.; Sharma, A.; Mekata, T.; Itami, T.; Sudhakaran, R. Rapid and sensitive real-time loop meditated isothermal amplification for the detection of Enterocytozoon hepatopenaei of shrimp. Aquaculture 2017, 481, 119–123. [Google Scholar] [CrossRef]
- Cai, S.; Kong, F.; Xu, S.; Yao, C. Real-time loop-mediated isothermal amplification for rapid detection of Enterocytozoon hepatopenaei. PeerJ. 2018, 6, e5993. [Google Scholar] [CrossRef]
- Liu, Y.; Qiu, L.; Sheng, A.; Wan, X.; Cheng, D.; Huang, J. Quantitative detection method of Enterocytozoon hepatopenaei using TaqMan probe real-time PCR. J. Invertebr. Pathol. 2018, 151, 191–196. [Google Scholar] [CrossRef]
- Ma, B.; Yu, H.; Fang, J.; Sun, C.; Zhang, M. Employing DNA binding dye to improve detection of Enterocytozoon hepatopenaei in real-time LAMP. Sci. Rep. 2019, 9, 15860. [Google Scholar] [CrossRef]
- Kanitchinda, S.; Srisala, J.; Suebsing, R.; Prachumwat, A.; Chaijarasphong, T. CRISPR-Cas fluorescent cleavage assay coupled with recombinase polymerase amplification for sensitive and specific detection of Enterocytozoon hepatopenaei. Biotechnol. Rep. 2020, 27, e00485. [Google Scholar] [CrossRef]
- Wang, L.; Lv, Q.; He, Y.; Gu, R.; Zhou, B.; Chen, J.; Fan, X.; Pan, G.; Long, M.; Zhou, Z. Integrated qPCR and staining methods for detection and quantification of Enterocytozoon hepatopenaei in shrimp Litopenaeus vannamei. Microorganisms 2020, 8, 1366. [Google Scholar] [CrossRef]
- Zhao, R.; Gao, W.; Qiu, L.; Chen, X.; Dong, X.; Li, C.; Huang, J. A staining method for detection of Enterocytozoon hepatopenaei (EHP) spores with calcofluor white. J. Invertebr. Pathol. 2020, 172, 107347. [Google Scholar] [CrossRef]
- Arunrut, N.; Tondee, B.; Khumwan, P.; Kampeera, J.; Kiatpathomchai, W. Rapid and sensitive colorimetric detection of microsporidian Enterocytozoon hepatopenaei (EHP) based on spore wall protein (SWP) gene using loop-mediated isothermal amplification combined with DNA functionalized gold nanoparticles as probes. Aquaculture 2021, 533, 736206. [Google Scholar] [CrossRef]
- Hou, Z.; Yu, J.; Wang, J.; Li, T.; Chang, L.; Fang, Y.; Yan, D. Development of a PCR assay for the effective detection of Enterocytozoon hepatopenaei (EHP) and investigation of EHP prevalence in Shandong Province, China. J. Invertebr. Pathol. 2021, 184, 107653. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Fan, S.; Wang, Y.; Yang, H.; Qiao, Y.; Jiang, G.; Lyu, M.; Dong, J.; Shen, H.; Gao, S. Rapid detection of Enterocytozoon hepatopenaei infection in shrimp with a real-time isothermal recombinase polymerase amplification assay. Front. Cell. Infect. Microbiol. 2021, 11, 631960. [Google Scholar] [CrossRef] [PubMed]
- Sathish Kumar, T.; Radhika, K.; Joseph Sahaya Rajan, J.; Makesh, M.; Alavandi, S.V.; Vijayan, K.K. Closed-tube field-deployable loop-mediated isothermal amplification (LAMP) assay based on spore wall protein (SWP) for the visual detection of Enterocytozoon hepatopenaei (EHP). J. Invertebr. Pathol. 2021, 183, 107624. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Pan, L.; Chen, Z.; Du, H.; Luo, B.; Luo, J.; Pan, G. The roles of microsporidia spore wall proteins in the spore wall formation and polar tube anchorage to spore wall during development and infection processes. Exp. Parasitol. 2018, 187, 93–100. [Google Scholar] [CrossRef]
- Han, B.; Takvorian, P.M.; Weiss, L.M. Invasion of host cells by microsporidia. Front. Microbiol. 2020, 11, 172. [Google Scholar] [CrossRef]
- Franzen, C. Microsporidia: How can they invade other cells? Trends Parasitol. 2004, 20, 275–279. [Google Scholar] [CrossRef]
- Hayman, J.R.; Southern, T.R.; Nash, T.E. Role of sulfated glycans in adherence of the microsporidian Encephalitozoon intestinalis to host cells in vitro. Infect. Immun. 2005, 73, 841–848. [Google Scholar] [CrossRef]
- Southern, T.R.; Jolly, C.E.; Lester, M.E.; Hayman, J.R. EnP1, a microsporidian spore wall protein that enables spores to adhere to and infect host cells in vitro. Eukaryot. Cell 2007, 6, 1354–1362. [Google Scholar] [CrossRef]
- Bohne, W.; Ferguson, D.J.; Kohler, K.; Gross, U. Developmental expression of a tandemly repeated, glycine- and serine-rich spore wall protein in the microsporidian pathogen Encephalitozoon cuniculi. Infect. Immun. 2000, 68, 2268–2275. [Google Scholar] [CrossRef]
- Hayman, J.R.; Hayes, S.F.; Amon, J.; Nash, T.E. Developmental expression of two spore wall proteins during maturation of the microsporidian Encephalitozoon intestinalis. Infect. Immun. 2001, 69, 7057–7066. [Google Scholar] [CrossRef]
- Brosson, D.; Kuhn, L.; Prensier, G.; Vivares, C.P.; Texier, C. The putative chitin deacetylase of Encephalitozoon cuniculi: A surface protein implicated in microsporidian spore-wall formation. FEMS Microbiol. Lett. 2005, 247, 81–90. [Google Scholar] [CrossRef]
- Peuvel-Fanget, I.; Polonais, V.; Brosson, D.; Texier, C.; Kuhn, L.; Peyret, P.; Vivarès, C.; Delbac, F. EnP1 and EnP2, two proteins associated with the Encephalitozoon cuniculi endospore, the chitin-rich inner layer of the microsporidian spore wall. Int. J. Parasitol. 2006, 36, 309–318. [Google Scholar] [CrossRef]
- Xu, Y.; Takvorian, P.; Cali, A.; Wang, F.; Zhang, H.; Orr, G.; Weiss, L.M. Identification of a new spore wall protein from Encephalitozoon cuniculi. Infect. Immun. 2006, 74, 239–247. [Google Scholar] [CrossRef]
- Wu, Z.; Li, Y.; Pan, G.; Tan, X.; Hu, J.; Zhou, Z.; Xiang, Z. Proteomic analysis of spore wall proteins and identification of two spore wall proteins from Nosema bombycis (Microsporidia). Proteomics 2008, 8, 2447–2461. [Google Scholar] [CrossRef]
- Li, Z.; Pan, G.; Li, T.; Huang, W.; Chen, J.; Geng, L.; Yang, D.; Wang, L.; Zhou, Z. SWP5, a spore wall protein, interacts with polar tube proteins in the parasitic microsporidian Nosema bombycis. Eukaryot. Cell 2012, 11, 229–237. [Google Scholar] [CrossRef]
- Chen, J.; Geng, L.; Long, M.; Li, T.; Li, Z.; Yang, D.; Ma, C.; Wu, H.; Ma, Z.; Li, C.; et al. Identification of a novel chitin-binding spore wall protein (NbSWP12) with a BAR-2 domain from Nosema bombycis (microsporidia). Parasitology 2013, 140, 1394–1402. [Google Scholar] [CrossRef]
- Yang, D.; Dang, X.; Peng, P.; Long, M.; Ma, C.; Jia, J.; Qin, G.; Wu, H.; Liu, T.; Zhou, X.; et al. NbHSWP11, a microsporidia Nosema bombycis protein, localizing in the spore wall and membranes, reduces spore adherence to host cell BME. J. Parasitol. 2014, 100, 623–632. [Google Scholar] [CrossRef]
- Yang, D.; Pan, G.; Dang, X.; Shi, Y.; Li, C.; Peng, P.; Luo, B.; Bian, M.; Song, Y.; Ma, C.; et al. Interaction and assembly of two novel proteins in the spore wall of the microsporidian species Nosema bombycis and their roles in adherence to and infection of host cells. Infect. Immun. 2015, 83, 1715–1731. [Google Scholar] [CrossRef]
- Yang, D.; Pan, L.; Peng, P.; Dang, X.; Li, C.; Li, T.; Long, M.; Chen, J.; Wu, Y.; Du, H.; et al. Interaction between SWP9 and polar tube proteins of the microsporidian Nosema bombycis and function of SWP9 as a scaffolding protein contribute to polar tube tethering to the spore wall. Infect. Immun. 2017, 85, e00872-16. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wu, Z.; Pan, G.; He, W.; Zhang, R.; Hu, J.; Zhou, Z. Identification of a novel spore wall protein (SWP26) from microsporidia Nosema bombycis. Int. J. Parasitol. 2009, 39, 391–398. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Shen, Z.; Hou, J.; Zhang, J.; Geng, T.; Tang, X.; Xu, L.; Guo, X. Identification of a protein interacting with the spore wall protein SWP26 of Nosema bombycis in a cultured BmN cell line of silkworm. Infect. Genet. and Evol. 2013, 17, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Tang, X.; Xiao, S.; Wang, H.; Zhang, Y.; Shao, Y.; Tang, F.; Chen, S.; Bai, X. The role of Bombyx mori Bmtutl-519 protein in the infection of BmN cells by Nosema bombycis. Dev. Comp. Immunol. 2019, 92, 283–290. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Li, R.; You, Y.; Zhang, K.; Zhang, L. A novel spore wall protein from Antonospora locustae (Microsporidia: Nosematidae) contributes to sporulation. J. Eukaryot. Microbiol. 2017, 64, 779–791. [Google Scholar] [CrossRef]
- Jaroenlak, P.; Boakye, D.W.; Vanichviriyakit, R.; Williams, B.A.P.; Sritunyalucksana, K.; Itsathitphaisarn, O. Identification, characterization and heparin binding capacity of a spore-wall, virulence protein from the shrimp microsporidian, Enterocytozoon hepatopenaei (EHP). Parasite. Vector. 2018, 11, 177. [Google Scholar] [CrossRef]
- Li, Z.; Wang, Y.; Fang, W.; Zhou, J.; Li, X. Identification, sequence characteristics and expression analyses of four spore wall protien genes of Enterocytozoon hepatopenaei (EHP) in Litopenaeus vannamei. Mar. Fish. 2021, 43, 81–92. [Google Scholar] [CrossRef]
- Shen, W.; Guo, Q.; Liu, L.; Niu, B.; Weng, H.; Lou, B. Cloning, expression and application in detection of SWP2 gene in Enterocytozoon hepatopenaei. Acta Agric. Zhejiangensis 2021, 33, 993–1000. [Google Scholar]
- Boakye, D.W.; Jaroenlak, P.; Prachumwat, A.; Williams, T.A.; Bateman, K.S.; Itsathitphaisarn, O.; Sritunyalucksana, K.; Paszkiewicz, K.H.; Moore, K.A.; Stentiford, G.D.; et al. Decay of the glycolytic pathway and adaptation to intranuclear parasitism within Enterocytozoonidae microsporidia. Environ. Microbiol. 2017, 19, 2077–2089. [Google Scholar] [CrossRef]
- Chen, J.; Liao, G.; Wu, Y.; Zhang, Q.; Yang, X.; Fan, X.; Long, M.; Pan, G.; Zhou, Z. Three methods for light microscopic detection of Enterocytozoon hepatopenaei. J. Southwest Univ. Nat. Sci. 2021, 43, 17–23. [Google Scholar]
- Cardin, A.D.; Weintraub, H.J. Molecular modeling of protein-glycosaminoglycan interactions. Arteriosclerosis 1989, 9, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Brosson, D.; Kuhn, L.; Delbac, F.; Garin, J.; Vivarès, C.P.; Texier, C. Proteomic analysis of the eukaryotic parasite Encephalitozoon cuniculi (microsporidia): A reference map for proteins expressed in late sporogonial stages. Proteomics 2006, 6, 3625–3635. [Google Scholar] [CrossRef] [PubMed]
- Han, B.; Ma, Y.; Tu, V.; Tomita, T.; Mayoral, J.; Williams, T.; Horta, A.; Huang, H.; Weiss, L.M. Microsporidia interact with host cell mitochondria via voltage-dependent anion channels using sporoplasm surface protein 1. mBio 2019, 10, e01944-19. [Google Scholar] [CrossRef] [PubMed]

| Category | Protein Number |
|---|---|
| Peptides | 427 |
| Proteins | 130 |
| Uncharacterized proteins | 71 (54.62%) |
| GO categories of biological process | 67 (51.54%) |
| GO categories of cellular component | 62 (47.69%) |
| GO categories of molecular function | 71 (54.62%) |
| KEGG pathway | 32 (24.62%) |
| Protein ID | Annotation | Length (Aa) | Total/Unique Peptide Count (Number) | Peptide Cover (Percent) | PI/MW (kDa) | HBM (Number) | TM | SP | N/O-Glyc (Number) | Homolog Protein/Accession Number/Species | Expect |
|---|---|---|---|---|---|---|---|---|---|---|---|
| OQS53969 | Uncharacterized protein | 374 | 27/15 | 37.97% | 4.88/42.6 | HBM (1) | No | No | N-Glyc (1) O-Glyc (3) | hypothetical protein/ECANGB1_151 Enterospora canceri | 7 × 10−18 |
| OQS55745 | EhSWP3 ※ | 249 | 21/14 | 56.63% | 5.87/27.1 | N/A | Yes | No | N-Glyc (2) O-Glyc (2) | N/A | N/A |
| OQS54917 | Uncharacterized protein | 459 | 15/11 | 24.40% | 5.01/50.6 | N/A | Yes | No | N-Glyc (4) O-Glyc (10) | NbHSWP4/EF683104 N. bombycis | 3 × 10−5 |
| OQS53967 | Uncharacterized protein | 326 | 13/11 | 38.96% | 7.33/36.8 | HBM (1) | No | No | N-Glyc (1) | NbSWP32/EF683103 N. bombycis | 3 × 10−5 |
| OQS55648 | Uncharacterized protein | 356 | 11/11 | 30.90% | 9.12/40.9 | HBM (3), | Yes | No | N-Glyc (4) O-Glyc (12) | hypothetical protein/OQS55748.1 E.hepatopenaei | 3 × 10−5 |
| OQS54254 | Uncharacterized protein | 473 | 14/10 | 24.10% | 4.11/53.7 | N/A | No | No | N-Glyc (3) O-Glyc (29) | SWP/EOB14572 N. bombycis | 1 × 10−7 |
| OQS54511 | Uncharacterized protein | 299 | 13/10 | 32.44% | 7.50/34.5 | N/A | No | No | N-Glyc (1) | hypothetical protein/ECANGB1_595 E.canceri | 1 × 10−21 |
| OQS54349 | Uncharacterized protein | 592 | 9/9 | 15.37% | 8.62/69.2 | HBM (4) | No | No | N-Glyc (6) O-Glyc (36) | SWP/EOB14572 N. bombycis | 3 × 10−13 |
| OQS55378 | Uncharacterized protein | 407 | 8/8 | 18.43% | 8.78/47.4 | HBM (2) | No | No | N-Glyc (1) O-Glyc (15) | SWP/EOB14572 N. bombycis | 2 × 10−6 |
| OQS53833 | Uncharacterized protein | 412 | 10/7 | 18.20% | 9.27/45.8 | N/A | Yes | No | N-Glyc (1) O-Glyc (5) | N/A | N/A |
| OQS53864 | EhSWP1 | 228 | 8/7 | 21.93% | 8.45/27.0 | HBM (1) | No | No | N/A | NbSWP12/EF683112 N. bombycis | 2 × 10−29 |
| OQS54094 | Uncharacterized protein | 390 | 8/7 | 16.92% | 7.57/44.1 | HBM (1) | No | No | N-Glyc (5) O-Glyc (12) | EiSWP2/AF355749 E. intestinalis | 3 × 10−5 |
| OQS55748 | Uncharacterized protein | 387 | 6/6 | 16.80% | 7.59/45.0 | HBM (1), | Yes | No | N-Glyc (5) O-Glyc (1) | SWP/EOB14572 N. bombycis | 3 × 10−6 |
| OQS55031 | EhSWP7 | 250 | 7/5 | 20.00% | 5.28/27.8 | N/A | No | Yes | N/A | NbSWP7/EOB13707 N. bombycis | 2 × 10−25 |
| OQS54755 | Uncharacterized protein | 478 | 5/5 | 10.67% | 8.95/55.3 | HBM (3), | Yes | No | N-Glyc (2) O-Glyc (5) | hypothetical protein/EBI_22790 E. bieneusi | 4 × 10−73 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, X.; Wei, C.; Yang, X.; Xiao, A.; Tan, N.; Chen, J.; Long, M.; Pan, G.; Wan, Y.; Zhou, Z. Proteomic Analysis of Spore Surface Proteins and Characteristics of a Novel Spore Wall Protein and Biomarker, EhSWP3, from the Shrimp Microsporidium Enterocytozoon hepatopenaei (EHP). Microorganisms 2022, 10, 367. https://doi.org/10.3390/microorganisms10020367
Fan X, Wei C, Yang X, Xiao A, Tan N, Chen J, Long M, Pan G, Wan Y, Zhou Z. Proteomic Analysis of Spore Surface Proteins and Characteristics of a Novel Spore Wall Protein and Biomarker, EhSWP3, from the Shrimp Microsporidium Enterocytozoon hepatopenaei (EHP). Microorganisms. 2022; 10(2):367. https://doi.org/10.3390/microorganisms10020367
Chicago/Turabian StyleFan, Xiaodong, Chunmei Wei, Xiaojuan Yang, Ai Xiao, Nianqiu Tan, Jie Chen, Mengxian Long, Guoqing Pan, Yongji Wan, and Zeyang Zhou. 2022. "Proteomic Analysis of Spore Surface Proteins and Characteristics of a Novel Spore Wall Protein and Biomarker, EhSWP3, from the Shrimp Microsporidium Enterocytozoon hepatopenaei (EHP)" Microorganisms 10, no. 2: 367. https://doi.org/10.3390/microorganisms10020367
APA StyleFan, X., Wei, C., Yang, X., Xiao, A., Tan, N., Chen, J., Long, M., Pan, G., Wan, Y., & Zhou, Z. (2022). Proteomic Analysis of Spore Surface Proteins and Characteristics of a Novel Spore Wall Protein and Biomarker, EhSWP3, from the Shrimp Microsporidium Enterocytozoon hepatopenaei (EHP). Microorganisms, 10(2), 367. https://doi.org/10.3390/microorganisms10020367






