Distribution of Interferon Lambda 4 Single Nucleotide Polymorphism rs11322783 Genotypes in Patients with COVID-19
Abstract
1. Introduction
2. Methods
2.1. Study Group
2.2. IFNL4 Genotyping
2.3. Data Analysis
3. Results
3.1. Clinical Features of SARS-CoV-2 Infected Patients
3.2. IFNL4 SNPs in Patients with COVID-19
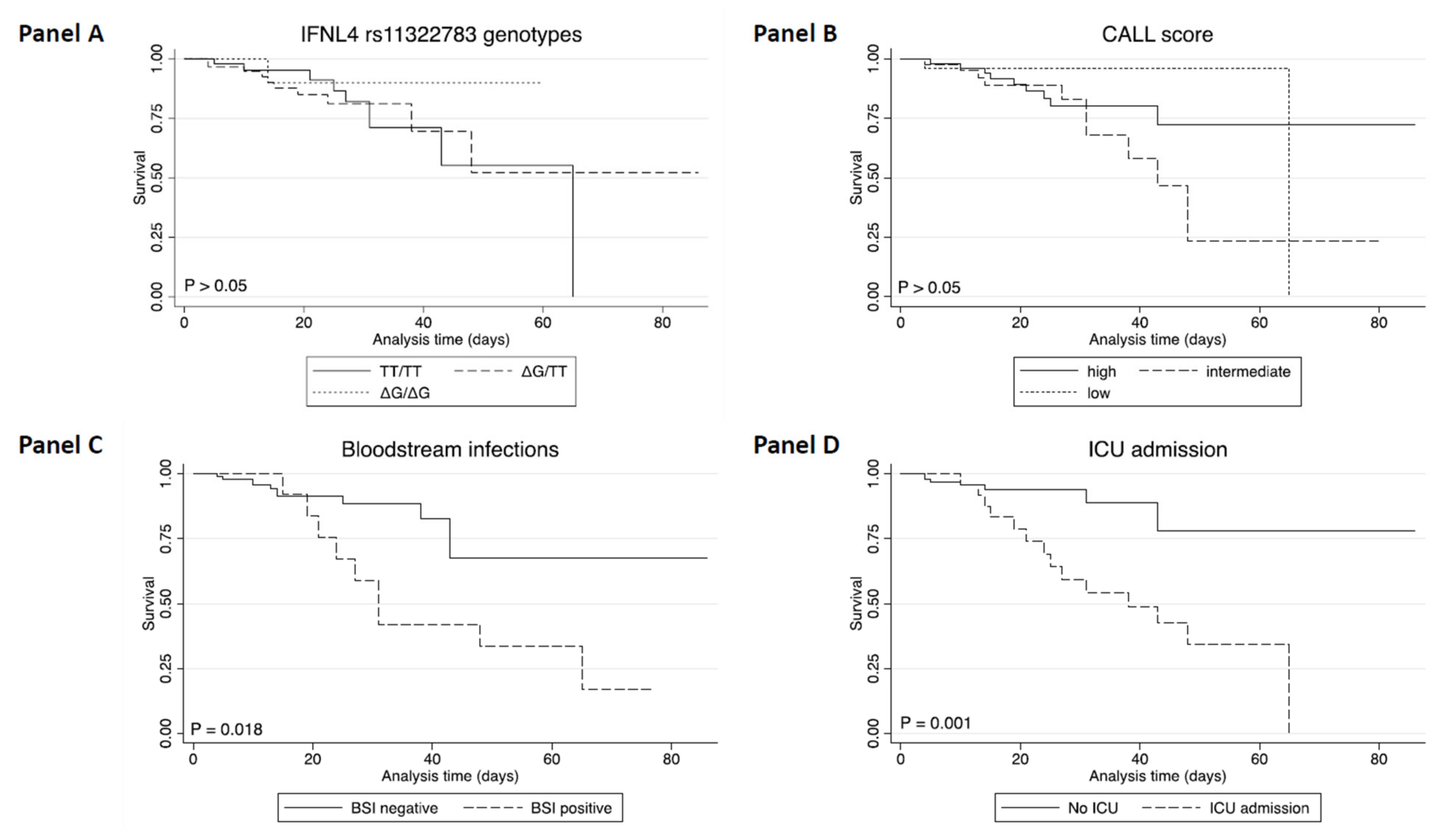
3.3. Survival Analysis in Patients with COVID-19
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kotenko, S.V.; Rivera, A.; Parker, D.; Durbin, J.E. Type III IFNs: Beyond antiviral protection. Sem. Immunol. 2019, 43, 101303. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, T.R.; Prokunina-Olsson, L.; Donnelly, R.P. IFN-λ4: The paradoxical new member of the interferon lambda family. J. Interferon Cytokine Res. 2014, 34, 829–838. [Google Scholar] [CrossRef] [PubMed]
- Lazear, H.M.; Schoggins, J.W.; Diamond, M.S. Shared and distinct functions of type i and type iii interferons. Immunity 2019, 50, 907–923. [Google Scholar] [CrossRef] [PubMed]
- Busnadiego, I.; Fernbach, S.; Pohl, M.O.; Karakus, U.; Huber, M.; Trkola, A.; Stertz, S.; Hale, B.G. Antiviral activity of type I, II, and III interferons counterbalances ace2 inducibility and restricts SARS-CoV-2. MBio 2020, 11, e01928-20. [Google Scholar] [CrossRef]
- Jeon, Y.J.; Gil, C.H.; Jo, A.; Won, J.; Kim, S.; Kim, H.J. The influence of interferon-lambda on restricting Middle East Respiratory Syndrome Coronavirus replication in the respiratory epithelium. Antiviral Res. 2020, 180, 104860. [Google Scholar] [CrossRef]
- Ge, D.; Fellay, J.; Thompson, A.J.; Simon, J.S.; Shianna, K.V.; Urban, T.J.; Heinzen, E.L.; Qiu, P.; Bertelsen, A.H.; Muir, A.J.; et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature 2009, 461, 399–401. [Google Scholar] [CrossRef]
- Prokunina-Olsson, L.; Muchmore, B.; Tang, W.; Pfeiffer, R.M.; Park, H.; Dickensheets, H.; Hergott, D.; Porter-Gill, P.; Mumy, A.; Kohaar, I.; et al. A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nat. Genet. 2013, 45, 164–171. [Google Scholar] [CrossRef]
- National Center for Biotechnology. 2021. Available online: https://www.ncbi.nlm.nih.gov/snp/rs368234815?vertical_tab=true (accessed on 30 December 2021).
- Noureddin, M.; Rotman, Y.; Zhang, F.; Park, H.; Rehermann, B.; Thomas, E.; Liang, T.J. Hepatic expression levels of interferons and interferon-stimulated genes in patients with chronic hepatitis C: A phenotype-genotype correlation study. Genes Immun. 2015, 16, 321–329. [Google Scholar] [CrossRef]
- Palmieri, O.; Ippolito, A.M.; Margaglione, M.; Valvano, M.R.; Gioffreda, D.; Fasano, M.; D’Andrea, G.; Corritore, G.; Milella, M.; Andriulli, N.; et al. Variation in genes encoding for interferon λ-3 and λ-4 in the prediction of HCV-1 treatment-induced viral clearance. Liver Int. 2014, 34, 1369–1377. [Google Scholar] [CrossRef]
- Melis, R.; Fauron, C.; McMillin, G.; Lyon, E.; Shirts, B.; Hubley, L.M.; Slev, P.R. Simultaneous genotyping of rs12979860 and rs8099917 variants near the IL28B locus associated with HCV clearance and treatment response. J. Mol. Diagn. 2011, 13, 446–451. [Google Scholar] [CrossRef]
- Sharafi, H.; Moayed Alavian, S.; Behnava, B.; Pouryasin, A.; Keshvari, M. The impact of IFNL4 rs12979860 polymorphism on spontaneous clearance of hepatitis c; a case-control study. Hepat Mon. 2014, 14, 10. [Google Scholar] [CrossRef]
- Lapiński, T.W.; Pogorzelska, J.; Kowalczuk, O.; Nikliński, J.; Flisiak, R. SNP RS12979860 related spontaneous clearance of hepatitis c virus infection in HCV/HIV-1 coinfected patients. Przegl Epidemiol. 2013, 67, 12–16. [Google Scholar]
- Franco, S.; Aparicio, E.; Parera, M.; Clotet, B.; Tural, C.; Martinez, M.A. IFNL4 ss469415590 variant is a better predictor than rs12979860 of pegylated interferon-alpha/ribavirin therapy failure in hepatitis C virus/HIV-1 coinfected patients. AIDS 2014, 28, 133–136. [Google Scholar] [CrossRef]
- Lin, C.Y.; Chen, J.Y.; Lin, T.N.; Jeng, W.J.; Huang, C.H.; Huang, C.W.; Chang, S.W.; Sheen, I.S. IL28B SNP rs12979860 is a critical predictor for on-treatment and sustained virologic response in patients with hepatitis C virus genotype-1 infection. PLoS ONE 2011, 6, e18322. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, S.J.; Dunnigan, C.M.; Russell, C.D.; Haas, J.G. The role of interferon-λ locus polymorphisms in Hepatitis C and other infectious diseases. J. Innate Immun. 2015, 7, 231–242. [Google Scholar] [CrossRef] [PubMed]
- Rugwizangoga, B.; Andersson, M.E.; Kabayiza, J.C.; Nilsson, M.S.; Ármannsdóttir, B.; Aurelius, J.; Nilsson, S.; Hellstrand, K.; Lindh, M.; Martner, A. IFNL4 genotypes predict clearance of RNA viruses in rwandan children with upper respiratory tract infections. Front. Cell Infect Microbiol. 2019, 9, 340. [Google Scholar] [CrossRef]
- Scagnolari, C.; Midulla, F.; Riva, E.; Monteleone, K.; Solimini, A.; Bonci, E.; Cangiano, G.; Papoff, P.; Moretti, C.; Pierangeli, A.; et al. Evaluation of interleukin 28B single nucleotide polymorphisms in infants suffering from bronchiolitis. Virus Res. 2012, 165, 236–240. [Google Scholar] [CrossRef]
- Agwa, S.H.A.; Kamel, M.M.; Elghazaly, H.; Abd Elsamee, A.M.; Hafez, H.; Girgis, S.A.; Elarab, H.E.; Ebeid, F.S.E.; Sayed, S.M.; Sherif, L.; et al. Association between interferon-lambda-3 rs12979860, tll1 rs17047200 and ddr1 rs4618569 variant polymorphisms with the course and outcome of SARS-CoV-2 patients. Genes 2021, 12, 830. [Google Scholar] [CrossRef]
- Saponi-Cortes, J.M.R.; Rivas, M.D.; Calle-Alonso, F.; Sanchez, J.F.; Costo, A.; Martin, C.; Zamorano, J. IFNL4 genetic variant can predispose to COVID-19. Sci. Rep. 2021, 11, 10–27. [Google Scholar] [CrossRef] [PubMed]
- Rahimi, P.; Tarharoudi, R.; Rahimpour, A.; Mosayebi Amroabadi, J.; Ahmadi, I.; Anvari, E.; Siadat, S.D.; Aghasadeghi, M.; Fateh, A. The association between interferon lambda 3 and 4 gene single-nucleotide polymorphisms and the recovery of COVID-19 patients. Virol. J. 2021, 18, 221. [Google Scholar] [CrossRef]
- Yosuke, F.; Homma, T.; Inoue, H.; Onitsuka, C.; Ikeda, H.; Goto, Y.; Sato, Y.; Kimura, T.; Hirai, K.; Ohta, S.; et al. Downregulation of type III interferons in patients with severe COVID-19. J. Med. Virol. 2021, 93, 4559–4563. [Google Scholar]
- Scagnolari, C.; Pierangeli, A.; Frasca, F.; Bitossi, C.; Viscido, A.; Oliveto, G.; Scordio, M.; Mazzuti, L.; di Carlo, D.; Gentile, M.; et al. Differential induction of type I and III interferon genes in the upper respiratory tract of patients with coronavirus disease 2019 (COVID-19). Virus Res. 2021, 295, 198283. [Google Scholar] [CrossRef] [PubMed]
- Sposito, B.; Broggi, A.; Pandolfi, L.; Crotta, S.; Clementi, N.; Ferrarese, R.; Sisti, S.; Criscuolo, E.; Spreafico, R.; Long, J.M.; et al. The interferon landscape along the respiratory tract impacts the severity of COVID-19. Cell 2021, 184, 4953–4968. [Google Scholar] [CrossRef] [PubMed]
- Raglow, Z.; Thoma-Perry, C.; Gilroy, R.; Wan, Y.J.Y. IL28B genotype and the expression of ISGs in normal liver. Liver Int. 2013, 33, 991–998. [Google Scholar] [CrossRef]
- Rosenberg, B.R.; Freije, C.A.; Imanaka, N.; Chen, S.T.; Eitson, J.L.; Caron, R.; Uhl, S.A.; Zeremski, M.; Talal, A.; Jacobson, I.M.; et al. Genetic variation at IFNL4 influences extrahepatic interferon-stimulated gene expression in chronic HCV patients. J. Infect. Dis. 2018, 217, 650–655. [Google Scholar] [CrossRef]
- Knapp, S.; Meghjee, N.; Cassidy, S.; Jamil, K.; Thursz, M. Detection of allele specific differences in IFNL3 (IL28B) mRNA expression. BMC Med. Genet. 2014, 15, 104. [Google Scholar] [CrossRef]
- Monteleone, K.; Scheri, G.C.; Statzu, M.; Selvaggi, C.; Falasca, F.; Giustini, N.; Mezzaroma, I.; Turriziani, O.; d’Ettorre, G.; Antonelli, G.; et al. IFN-stimulated gene expression is independent of the IFNL4 genotype in chronic HIV-1 infection. Arch. Virol. 2016, 161, 3263–3268. [Google Scholar] [CrossRef]
- Ansari, M.A.; Marchi, E.; Ramamurthy, N.; Klenerman, P. In vivo negative regulation of SARS-CoV-2 receptor, ACE2, by interferons and its genetic control [version 1; peer review: 1 approved with reservations]. Wellcome Open Res. 2021, 6, 47. [Google Scholar] [CrossRef]
- Scagnolari, C.; Bitossi, C.; Viscido, A.; Frasca, F.; Oliveto, G.; Scordio, M.; Petrarca, L.; Mancino, E.; Nenna, R.; Riva, E.; et al. ACE2 expression is related to the interferon response in airway epithelial cells but is that functional for SARS-CoV-2 entry? Cytokine 2021, 140, 155430. [Google Scholar] [CrossRef] [PubMed]
- National Center for Biotechnology. 2021. Available online: https://www.ncbi.nlm.nih.gov/snp/rs11322783?vertical_tab=true#frequency_tab (accessed on 30 December 2021).
- D’Ettorre, G.; Recchia, G.; Ridolfi, M.; Siccardi, G.; Pinacchio, C.; Innocenti, G.P.; Santinelli, L.; Frasca, F.; Bitossi, C.; Ceccarelli, G.; et al. Analysis of type I IFN response and T cell activation in severe COVID-19/HIV-1 coinfection: A case report. Medicine 2020, 99, e21803. [Google Scholar] [CrossRef]
- Ji, D.; Zhang, D.; Xu, J.; Chen, Z.; Yang, T.; Zhao, P.; Chen, G.; Cheng, G.; Wang, Y.; Bi, J.; et al. Prediction for progression risk in patients with COVID-19 pneumonia: The CALL score. Clin. Infect. Dis. 2020, 71, 1393–1399. [Google Scholar] [CrossRef] [PubMed]
- Kalinowski, A.; Galen, B.T.; Ueki, I.F.; Sun, Y.; Mulenos, A.; Osafo-Addo, A.; Clark, B.; Joerns, J.; Liu, W.; Nadel, J.A.; et al. Respiratory syncytial virus activates epidermal growth factor receptor to suppress interferon regulatory factor 1-dependent interferon-lambda and antiviral defense in airway epithelium. Mucosal Immunol. 2018, 11, 958–967. [Google Scholar] [CrossRef] [PubMed]
- Davidson, S.; McCabe, T.M.; Crotta, S.; Gad, H.H.; Hessel, E.M.; Beinke, S.; Hartmann, R.; Wack, A. IFN λ is a potent anti-influenza therapeutic without the inflammatory side effects of IFN α treatment. EMBO Mol. Med. 2016, 8, 1099–1112. [Google Scholar] [CrossRef] [PubMed]
- Vanderheiden, A.; Ralfs, P.; Chirkova, T.; Upadhyay, A.A.; Zimmerman, M.G.; Bedoya, S.; Aoued, H.; Tharp, G.M.; Pellegrini, K.L.; Manfredi, C.; et al. Type I and type iii interferons restrict SARS-CoV-2 infection of human airway epithelial cultures. J. Virol. 2020, 94, e00985-20. [Google Scholar] [CrossRef] [PubMed]
- Amodio, E.; Pipitone, R.M.; Grimaudo, S.; Immordino, P.; Maida, C.M.; Prestileo, T.; Restivo, V.; Tramuto, F.; Vitale, F.; Craxì, A.; et al. SARS-CoV-2 viral load, ifnλ polymorphisms and the course of COVID-19: An observational study. J. Clin. Med. 2020, 9, 3315. [Google Scholar] [CrossRef]
- Schönrich, G.; Raftery, M.J. Neutrophil extracellular traps go viral. Front. Immunol. 2016, 7, 366. [Google Scholar] [CrossRef]
- Fuchs, T.A.; Abed, U.; Goosmann, C.; Hurwitz, R.; Schulze, I.; Wahn, V.; Weinrauch, Y.; Brinkmann, V.; Zychlinsky, A. Novel cell death program leads to neutrophil extracellular traps. J. Cell Biol. 2007, 176, 231–241. [Google Scholar] [CrossRef]
- Thierry, A.R.; Roch, B. Neutrophil extracellular traps and by-products play a key role in COVID-19: Pathogenesis, Risk factors, and therapy. J. Clin. Med. 2020, 9, 2942. [Google Scholar] [CrossRef]
- Barnes, B.J.; Adrover, J.M.; Baxter-Stoltzfus, A.; Borczuk, A.; Cools-Lartigue, J.; Crawford, J.M.; Daßler-Plenker, J.; Guerci, P.; Huynh, C.; Knight, J.S.; et al. Targeting potential drivers of COVID-19: Neutrophil extracellular traps. J. Exp. Med. 2020, 217, e20200652. [Google Scholar] [CrossRef]
- Middleton, E.A.; He, X.Y.; Denorme, F.; Campbell, R.A.; Ng, D.; Salvatore, S.P.; Mostyka, M.; Baxter-Stoltzfus, A.; Borczuk, A.C.; Loda, M.; et al. Neutrophil extracellular traps contribute to immunothrombosis in COVID-19 acute respiratory distress syndrome. Blood 2020, 136, 1169–1179. [Google Scholar] [CrossRef]
- Masso-Silva, J.A.; Moshensky, A.; Lam, M.; Odish, M.; Patel, A.; Xu, L.; Hansen, E.; Trescott, S.; Nguyen, C.; Kim, R.; et al. Increased peripheral blood neutrophil activation phenotypes and NETosis in critically ill COVID-19 patients: A case series and review of the literature. Clin. Infect. Dis. 2021, 437. online ahead of print. [Google Scholar]
- Arcanjo, A.; Logullo, J.; Menezes, C.; de Souza Carvalho Giangiarulo, T.C.; Dos Reis, M.C.; de Castro, G.; da Silva Fontes, Y.; Todeschini, A.R.; Freire-de-Lima, L.; Decoté-Ricardo, D.; et al. The emerging role of neutrophil extracellular traps in severe acute respiratory syndrome coronavirus 2 (COVID-19). Sci. Rep. 2020, 10, 19630. [Google Scholar] [CrossRef]
- Palermo, E.; di Carlo, D.; Sgarbanti, M.; Hiscott, J. Type i interferons in COVID-19 pathogenesis. Biology 2021, 10, 829. [Google Scholar] [CrossRef] [PubMed]
- Jafarzadeh, A.; Nemati, M.; Saha, B.; Bansode, Y.D.; Jafarzadeh, S. Protective potentials of type III interferons in COVID-19 patients: Lessons from differential properties of type I- and III interferons. Viral Immunol. 2021, 34, 307–320. [Google Scholar] [CrossRef] [PubMed]
- Chua, R.L.; Lukassen, S.; Trump, S.; Hennig, B.P.; Wendisch, D.; Pott, F.; Debnath, O.; Thürmann, L.; Kurth, F.; Völker, M.T.; et al. COVID-19 severity correlates with airway epithelium–immune cell interactions identified by single-cell analysis. Nat. Biotechnol. 2020, 38, 970–979. [Google Scholar] [CrossRef] [PubMed]
- Liao, M.; Liu, Y.; Yuan, J.; Wen, Y.; Xu, G.; Zhao, J.; Cheng, L.; Li, J.; Wang, X.; Wang, F.; et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020, 26, 842–844. [Google Scholar] [CrossRef] [PubMed]
- Lachowicz-Scroggins, M.E.; Dunican, E.M.; Charbit, A.R.; Raymond, W.; Looney, M.R.; Peters, M.C.; Gordon, E.D.; Woodruff, P.G.; Lefrançais, E.; Phillips, B.R.; et al. Extracellular DNA, neutrophil extracellular traps, and inflammasome activation in severe asthma. Am. J. Resp. Crit. Care Med. 2019, 199, 1076–1085. [Google Scholar] [CrossRef]
- Blazek, K.; Eames, H.L.; Weiss, M.; Byrne, A.J.; Perocheau, D.; Pease, J.E.; Doyle, S.; McCann, F.; Williams, R.O.; Udalova, I.A. IFN-λ resolves inflammation via suppression of neutrophil infiltration and IL-1β production. J. Exp. Med. 2015, 212, 845–853. [Google Scholar] [CrossRef]
- Nick, J.A.; Caceres, S.M.; Kret, J.E.; Poch, K.R.; Strand, M.; Faino, A.V.; Nichols, D.P.; Saavedra, M.T.; Taylor-Cousar, J.L.; Geraci, M.W.; et al. Extremes of Interferon-stimulated gene expression associate with worse outcomes in the acute respiratory distress syndrome. PLoS ONE 2016, 11, e0162490. [Google Scholar] [CrossRef]
- Malcolm, K.C.; Kret, J.E.; Young, R.L.; Poch, K.R.; Caceres, S.M.; Douglas, I.S.; Coldren, C.D.; Burnham, E.L.; Moss, M.; Nick, J.A. Bacteria-specific neutrophil dysfunction associated with interferon-stimulated gene expression in the acute respiratory distress syndrome. PLoS ONE 2011, 6, e21958. [Google Scholar] [CrossRef]
- Broggi, A.; Tan, Y.; Granucci, F.; Zanoni, I. IFN-λ suppresses intestinal inflammation by non-translational regulation of neutrophil function. Nat. Immunol. 2017, 18, 1084–1093. [Google Scholar] [CrossRef]
- Severe COVID-19 GWAS Group; Ellinghaus, D.; Degenhardt, F.; Bujanda, L.; Buti, M.; Albillos, A.; Invernizzi, P.; Fernández, J.; Prati, D.; Baselli, G.; et al. Genomewide association study of severe COVID-19 with respiratory failure. N. Engl. J. Med. 2020, 383, 1522–1534. [Google Scholar] [CrossRef] [PubMed]
- Mousa, M.; Vurivi, H.; Kannout, H.; Uddin, M.; Alkaabi, N.; Mahboub, B.; Tay, G.K.; Alsafar, H.S.; UAE COVID-19 Collaborative Partnership. Genome-wide association study of hospitalized COVID-19 patients in the United Arab Emirates. EBioMedicine 2021, 74, 103695. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ke, Y.; Xia, X.; Wang, Y.; Cheng, F.; Liu, X.; Jin, X.; Li, B.; Xie, C.; Liu, S.; et al. Genome-wide association study of COVID-19 severity among the Chinese population. Cell Discov. 2021, 7, 76. [Google Scholar] [CrossRef] [PubMed]
| Features | COVID-19 Patients (n = 128) |
|---|---|
| Age at diagnosis (years) (mean (range)) | 63.9 (25–95) |
| Gender (N (percentage)) | |
| Male | 79 (61.7) |
| Female | 49 (38.3) |
| CALL score (N (percentage)) | |
| Low severity (4–6) | 29 (22.6) |
| Intermediate severity (7–9) | 45 (35.2) |
| High severity (10–13) | 54 (42.2) |
| Clinical features (N (percentage)) | |
| ICU | 24 (18.7) |
| Thrombotic events | 15 (11.7) |
| Death | 21 (16.4) |
| BSI | 13 (10.2) |
| Bacterial pulmonary superinfection | 12 (9.4) |
| Blood parameters (mean (range)) | |
| WBC cell/mm3 | 6293.6 (2110–19150) |
| Neutrophils cell/mm3 | 4691.2 (1120–18000) |
| Lymphocytes cell/mm3 | 1067.7 (110–4760) |
| Monocytes cell/mm3 | 361.7 (150–1040) |
| CRP µg/L | 98380 (300–540000) |
| D-dimer µg/L | 1690 (176–4610) |
| Albumin g/L | 36.9 (19–46) |
| LDH U/L | 335 (111–1249) |
| Platelets cell/mm3 | 221 × 103 (65–516) |
| Features | Ranges * | IFNL4 SNP TT/TT | IFNL4 SNP ΔG/TT | IFNL4 SNP ΔG/ΔG | p-Value |
|---|---|---|---|---|---|
| SARS-CoV-2 patients | 53 (41.4) | 61 (47.7) | 14 (10.9) | ||
| WBC cell/mm3 | 4.5–11.0 × 103 | 40 (75.5) | 52 (86.7) | 7 (50.0) | 0.036 |
| <4.5 × 103 | 7 (13.2) | 5 (8.3) | 5 (35.7) | ||
| >11.0 × 103 | 6 (11.3) | 3 (5.0) | 2 (14.3) | ||
| Neutrophils cell/mm3 | 1.5–8.0 × 103 | 43 (81.1) | 52 (86.6) | 8 (57.1) | 0.042 |
| <1.5 × 103 | 3 (5.7) | 4 (6.7) | 4 (28.6) | ||
| >8.0 × 103 | 7 (13.2) | 4 (6.7) | 2 (14.3) | ||
| Lymphocytes cell/mm3 | 1.0–4.0 × 103 | 43 (81.1) | 50 (83.3) | 10 (71.4) | 0.59 |
| <1.0 × 103 | 10 (18.9) | 10 (16.7) | 4 (28.6) | ||
| >4.0 × 103 | - | - | - | ||
| Monocytes cell/mm3 | 0.1–0.7 × 103 | 49 (92.4) | 60 (98.4) | 14 (100) | 0.47 |
| <0.1 × 103 | 3 (5.7) | 1 (1.6) | 0 (0.0) | ||
| >0.7 × 103 | 1 (1.9) | 0 (0.0) | 0 (0.0) | ||
| CRP µg/L | <8.0 × 103 | 8 (15.1) | 6 (9.8) | 3 (21.4) | 0.45 |
| >8.0 × 103 | 45 (84.9) | 55 (90.2) | 11 (78.6) | ||
| D-dimer µg/L | <500 | 5 (11.4) | 9 (16.4) | 2 (14.3) | 0.78 |
| >500 | 39 (88.6) | 46 (83.6) | 12 (85.7) | ||
| Albumin g/L | 35–55 | 30 (66.7) | 30 (53.6) | 8 (61.5) | 0.41 |
| <35 | 15 (33.3) | 26 (46.4) | 5 (38.5) | ||
| >55 | - | - | - | ||
| LDH U/L | 80–300 × 103 | 16 (38.8) | 21 (35.0) | 4 (28.6) | 0.47 |
| <80 × 103 | 4 (7.7) | 1 (1.7) | 0 (0.0) | ||
| >300 × 103 | 32 (61.5) | 38 (63.3) | 10 (71.4) | ||
| Platelets cell/mm3 | 150–450 × 103 | 44 (83.0) | 45 (76.3) | 12 (85.7) | 0.8 |
| <150 × 103 | 7 (13.2) | 12 (20.3) | 2 (14.3) | ||
| >450 × 103 | 2 (3.8) | 2 (3.4) | 0 (0.0) | ||
| Call | Low severity (4–6) | 13 (24.6) | 13 (21.3) | 3 (21.4) | 0.94 |
| Intermediate severity (7–9) | 20 (37.7) | 20 (32.8) | 5 (35.7) | ||
| High severity (10–13) | 20 (37.7) | 28 (45.9) | 6 (42.9) | ||
| ICU admission rate | yes | 11 (20.7) | 11 (18.0) | 2 (14.3) | 0.84 |
| no | 42 (79.3) | 50 (82.0) | 12 (85.7) | ||
| Thrombotic events | Positive | 5 (9.4) | 8 (13.3) | 2 (14.3) | 0.78 |
| Negative | 48 (90.6) | 52 (86.7) | 12 (85.7) | ||
| Bloodstream infections (BSI) | Positive | 6 (11.8) | 6 (10.7) | 1 (7.7) | 0.91 |
| Negative | 45 (88.2) | 50 (89.3) | 13 (92.3) | ||
| Bacterial pulmonary superinfections | Positive | 6 (12.2) | 4 (7.3) | 2 (15.4) | 0.57 |
| Negative | 43 (87.8) | 51 (92.7) | 11 (84.6) |
| Allele Frequencies Comparison | Heterozygous and Homozygous Comparison | Homozygous and Homozygous Comparison | Allele Positivity Comparison | Armitage’s Trend Test | |||
|---|---|---|---|---|---|---|---|
| WBC | Normal levels vs. low levels | allele T | 0.22 | 0.33 | 0.04 | 0.95 | 0.20 |
| allele ∆G | 0.22 | 0.003 | 0.04 | 0.005 | 0.20 | ||
| Normal levels vs. high levels | allele T | 0.89 | 0.18 | 0.48 | 0.37 | 0.88 | |
| allele ∆G | 0.89 | 0.08 | 0.48 | 0.20 | 0.88 | ||
| Neutrophils | Normal levels vs. low levels | allele T | 0.04 | 0.90 | 0.01 | 0.35 | 0.04 |
| allele ∆G | 0.04 | 0.01 | 0.01 | 0.003 | 0.04 | ||
| Normal levels vs. high levels | allele T | 0.82 | 0.25 | 0.63 | 0.41 | 0.81 | |
| allele ∆G | 0.82 | 0.19 | 0.63 | 0.36 | 0.81 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sorrentino, L.; Silvestri, V.; Oliveto, G.; Scordio, M.; Frasca, F.; Fracella, M.; Bitossi, C.; D’Auria, A.; Santinelli, L.; Gabriele, L.; et al. Distribution of Interferon Lambda 4 Single Nucleotide Polymorphism rs11322783 Genotypes in Patients with COVID-19. Microorganisms 2022, 10, 363. https://doi.org/10.3390/microorganisms10020363
Sorrentino L, Silvestri V, Oliveto G, Scordio M, Frasca F, Fracella M, Bitossi C, D’Auria A, Santinelli L, Gabriele L, et al. Distribution of Interferon Lambda 4 Single Nucleotide Polymorphism rs11322783 Genotypes in Patients with COVID-19. Microorganisms. 2022; 10(2):363. https://doi.org/10.3390/microorganisms10020363
Chicago/Turabian StyleSorrentino, Leonardo, Valentina Silvestri, Giuseppe Oliveto, Mirko Scordio, Federica Frasca, Matteo Fracella, Camilla Bitossi, Alessandra D’Auria, Letizia Santinelli, Lucia Gabriele, and et al. 2022. "Distribution of Interferon Lambda 4 Single Nucleotide Polymorphism rs11322783 Genotypes in Patients with COVID-19" Microorganisms 10, no. 2: 363. https://doi.org/10.3390/microorganisms10020363
APA StyleSorrentino, L., Silvestri, V., Oliveto, G., Scordio, M., Frasca, F., Fracella, M., Bitossi, C., D’Auria, A., Santinelli, L., Gabriele, L., Pierangeli, A., Mastroianni, C. M., d’Ettorre, G., Antonelli, G., Caruz, A., Ottini, L., & Scagnolari, C. (2022). Distribution of Interferon Lambda 4 Single Nucleotide Polymorphism rs11322783 Genotypes in Patients with COVID-19. Microorganisms, 10(2), 363. https://doi.org/10.3390/microorganisms10020363









